Abstract
Background
Influenza pandemics are usually caused by the re-assortment of several influenza viruses, results in the emergence of new influenza virus strains that can infect the entire population. These pandemic strains, as well as seasonal influenza viruses, are subjected to extensive antigenic change that has, so far, prevented the generation of a universal vaccine.
Methods
Samples of patients hospitalized due to infection with the pandemic H1N1 influenza virus (A(H1N1)pdm09) from 2009, when the virus first appeared, until 2013 were analyzed.
Results
While many patients were hospitalized in 2009 due to infection with the pandemic H1N1 influenza virus, only small percentages of patients were hospitalized later in 2010–2012. Surprisingly, however in 2012–2013, we noticed that the percentages of patients hospitalized due to the pandemic H1N1 influenza infection increased significantly. Moreover, the ages of hospitalized patients differed throughout this entire period (2009–2013) and pregnant women were especially vulnerable to the infection.
Conclusions
High percentages of patients (especially pregnant women) were hospitalized in 2013 due to the A(H1N1)pdm09 infection, which may have been enabled by an antigenic drift from those which circulated at the onset of the pandemic.
Electronic supplementary material
The online version of this article (doi:10.1186/s12879-014-0710-1) contains supplementary material, which is available to authorized users.
Background
Influenza viruses are responsible for most respiratory infections, affecting all age groups, particularly the elderly population [1],[2]. Annual epidemics of seasonal influenza are estimated to result in approximately three to five million cases of severe illness and about 250,000 to 500,000 deaths worldwide (WHO, 2009). The virus contains a single strand RNA molecule which encodes at least 12 proteins; of these proteins, M2, an ion channel protein, neuraminidase (NA) and hemagglutinin (HA) are expressed on the cell surface of infected cells [3].
In influenza virus-infected cells, the assembly and the budding of progeny viruses is the final and critical step in the life cycle of the virus. This step, which significantly affects disease progression [4], requires the coordinated localization of the viral HA and NA proteins to lipid rafts domains on the apical cell membrane [5]-[7]. Membrane localization of HA and NA allows the virus to acquire these glycoproteins by simply budding through the host cell membrane [8]. To complete viral particle budding, the viral NA enzyme cleaves sialic acid residues that link the progeny virus to the infected cells [5].
Because the “foreign” HA and the NA proteins are found on the virus and on the infected cells, they elicit innate, humoral and cellular immune responses, directed against the virus. To avoid this immune attack, both the HA and the NA proteins undergo extensive antigenic changes, complicating design of a universal vaccine that would be effective against all influenza strains. The WHO vaccine that is administered each year is based on the antigenic properties of emerging and circulating strains and contains three influenza virus strains. Nevertheless, every year, millions are infected with influenza.
In addition, influenza pandemics, that occur every few decades, pose a major global threat. Three influenza pandemic outbreaks occurred in the 20th century and one has already been recorded in the current century. The most devastating, was the 1918 Spanish flu pandemic (A/H1N1), which lasted only two years (1918–1919), but took the lives of approximately 50 million people [9]. Two later influenza pandemics, that occurred in 1957 (Asian flu, H2N2) and in 1968 (Hong Kong flu, H3N2), resulted in approximately 70,000 and 34,000 deaths, respectively, in the US alone [1]. The most recent influenza (A(H1N1)pdm09) pandemic emerged in 2009. Although the mortality rate was relatively low, many people around the world were infected by the virus [9]. The emergence of new pandemic in recent years provides us with a unique opportunity to study the virus spread and evolution through out the years from the time it first appeared until today.
Here, we analyzed data obtained from patients hospitalized in the period between May 2009 and May 2013 due to A(H1N1)pdm09 infection. We observed that in 2013 a significant number of patients (including increasing percentages of pregnant women) were hospitalized due the A(H1N1)pdm09 infection and we suggest that the virus re-appeared because of antigenic changes it acquired over the last 4 years.
Methods
Ethics
This is a retrospective study performed on anonymous patient samples that were analyzed for the presence of various viruses, as part of the routine tests performed in the Sheba Medical Center. No extra samples were obtained for this research. Therefore, informed consent was not required. The institutional review board (IRB) of the Sheba Medical Center approved this research (Helsinki Number 0402-13-SMC).
Patients and samples
Respiratory clinical samples (nasopharyngeal swabs or aspirates) were collected from 29,956 patients hospitalized at Chaim Sheba Medical Center, Israel, due to respiratory illnesses, during the winter seasons between May 2009 and May 2013. 4,674 patients were positive for A(H1N1)pdm09 virus infection. All patients were from the same geographical area.
Detection of A(H1N1)pdm09 virus infection
Viral genomic RNA was extracted from patient samples by using the NucliSENS easyMAG (BioMerieux, France). A(H1N1)pdm09 infection was detected by using a panel of real-time reverse transcription-PCR (rRT-PCR), as previously described [10],[11].
Phylogenetic analysis
For the phylogenetic analysis, influenza hemagglutinin protein-specific primers were used by WHO Swine genome set [12] (Table 1). The Sequencher® 5.0 program (Gencodes Corporation, Ann Arbor, MI) was used to compare the nucleotide sequences. Phylogenetic trees were prepared by nearest neighbor joining analysis using Clustal X with 1000 bootstraps and trees were visualized using TreeView or NJ plot software. The sequences were submitted to the NCBI.
Table 1.
Primer sequences
| Fragment | HA forward | Primers 5'-3' | HA reverse | Primers 5'-3' |
|---|---|---|---|---|
| Fragment 1 | 1 | TGT AAA ACG ACG AGT ATA CGA CTA GCA AAA GCA GGG G | 461 | CAG GAA ACA GCT ATG ACC TCA TGA TTG GGC CAY GA |
| Fragment 2 | 351 | TGT AAA ACG ACG GCC AGT ACR TGT TAC CCW GGR GAT TTC A | 943 | CAG GAA ACA GCT ATG ACC GAA AKG GGA GRC TGG TGT TTA |
| Fragment 3 | 379 | TGT AAA ACG ACG GCC AGT ACR TGT TAC CCA GGR GAT TTC | 1204 | CAG GAA ACA GCT ATG ACC TCT TTA CCY RCT GTG AA |
| Fragment 4 | 736 | TGT AAA ACG ACG GCC AGT AGR ATG RAC TAT TAC TGG AC | 1340 | CAG GAA ACA GCT ATG ACC TTC TKC ATT RTA WGT CCA AA |
| Fragment 5 | 1124 | TGT AAA ACG ACG GCC AGT TGG ATG GTA YGG TTA YCA YCA | 1541 | CAG GAA ACA GCT ATG ACC TCA TAA GTY CCA TTT YTGA |
| Fragment 6 | 1204 | TGT AAA ACG ACG GCC AGT AAG ATG AAY ACR CAR TTC ACAG | 1778 | CAG GAA ACA GCT TCA GTA GAA ACA ACA AGG GTG TTT |
Statistical analysis
The positivity percent was calculated by dividing the number of positive H1N1pdm samples by the total number of tests performed in the laboratory. A Binomial 95% confidence interval was calculated for this rate. The chi-square test was applied to evaluate the differences in positivity percent between the compared years and between the different age groups within the same year. A p value <0.05 was considered to be statistically significant.
All analyses were performed using SPSS (version 21.0.0. SPSS Inc., Chicago, IL, USA), SAS (SAS 9.1, SAS Institute Inc, Cary, NC, USA) and Excel softwares.
Results
Rebound of pandemic H1N1 influenza
We demonstrate in this paper that the pandemic H1N1 influenza virus reappeared in Israel in 2012–2013. We noticed that the percentages of patients hospitalized due to the pandemic H1N1 influenza infection increased significantly in this year. Interestingly, we also observed that pregnant women were especially vulnerable to the infection. We have started this research by analyzing the percentages of patients infected with the A(H1N1)pdm09 virus from the time it first appeared in the country until 2013. The new influenza A(H1N1)pdm09 virus was observed in Israel throughout the entire year of 2009 following its initial identification in May 2009. A(H1N1)pdm09-infected patients were hospitalized from May 2009 until the virus ceased to be prevalent, at the end of the first quarter of 2010 (Figure 1). From the end of 2010 until the beginning of 2011, relatively fewer patients were hospitalized due to the A(H1N1)pdm09 infection. Only sporadic cases of A(H1N1)pdm09 infection were reported in the winter season of 2011–12. However, surprisingly, at the beginning of 2013, the percentages of patients hospitalized due to A(H1N1)pdm09 infection increased dramatically (Figure 1).
Figure 1.
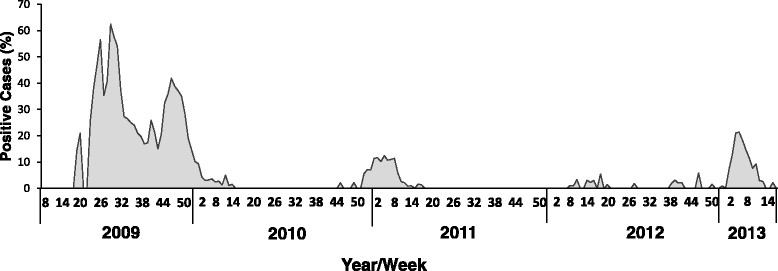
Distribution of H1N1 pandemic virus infection in hospitalized patients. The percentage of patients hospitalized (on a weekly basis, X axis) due to influenza-like syndrome and infected with the pandemic 2009 influenza virus.
Figure 2 summarizes the infection peaks and the annual percentages of patients infected with A(H1N1)pdm09 virus. As can be seen, in 2009–10, when the virus first appeared, approximately 60% of the patients admitted with respiratory disease at week 29 of the year were infected with the virus strain. In the following years, infection rates dropped significantly and in 2011–12 (week 16), at the peak of infection for that year, only 5% of the patients were infected with A(H1N1)pdm09 (Figure 2a) and less than 1% of patients were hospitalized due to A(H1N1)pdm09 infection throughout the entire year (Figure 2b). In week 6 of 2012–13, a dramatic increase in the percentages of hospitalized patients was observed, with around 20% of the patients presenting infection with the A(H1N1)pdm09 virus (Figure 2a). Similarly, the annual percentages of patients hospitalized due to the A(H1N1)pdm09 infection was higher as compared to 2011–2012 (Figure 2b).
Figure 2.
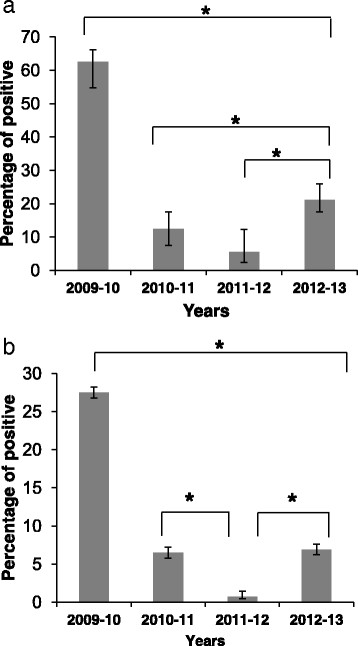
Infection peaks and annual infection percentages. The figure shows the percentages of patients infected with the pandemic 2009 influenza virus between 2009 and 2013. The percentages of patients hospitalized due to influenza-like syndrome are presented either at the peak of the infection in each year (a) or as an annual average of the infection (b). *P < 0 · 05 using the chi-square test.
To investigate the reasons accounting for the return of the pandemic H1N1 influenza virus in 2013, we analyzed the ages of the infected patients over three periods: when the virus first appeared (2009–10), when only a low percentage of patients were infected with the virus (2010–12; the age of the patients in each year was not analyzed separately due to the small number of patients hospitalized in these years) and at 2013, when a relatively high percentage of patients was hospitalized due to the A(H1N1)pdm09 infection. In 2009–10, most of the infected patients were 21–60 years of age (Figure 3 and Table 2). In 2010–12, equal infection rates were observed throughout all age groups, with the exception of adolescent patients, aged 11–20 (Figure 3 and Table 2). Similarly, in 2012–13, patients 11–20 years of age were less affected by the A(H1N1)pdm09 virus when compared to other age groups. However, in 2012–13, most of the A(H1N1)pdm09-infected patients were 21–40 or 61–100 years of age group.
Figure 3.
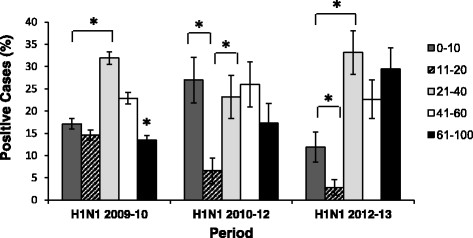
Age distribution of the infected patients. The age percentages of patients hospitalized due to influenza-like syndrome and infected with the pandemic 2009 influenza virus. Three periods of infection are presented (X axis). *P < 0 · 05 using the chi-square test.
Table 2.
Male and female patient distribution
| Age | H1N1pdm 2009–10 (1y) | H1N1pdm 2009–12 (1y) | H1N1pdm 2009–13 (1y) | P value | |
|---|---|---|---|---|---|
| 0-10 | Patients n | 5112 | 3001 | 1367 | |
| Positive n (%) | 879 (17.2) | 80.(2.7) | 51 (3.7) | <.0001 | |
| Male n (%) | 2908 (56.9) | 1784 (59.4) | 780 (57.1) | 0.0682 | |
| Male Positive n (%) | 499 (17.2) | 39 (.2) | 24 (3.1) | <.0001 | |
| Female n (%) | 2204 (43.1) | 1217 (40.6) | 587 (42.9) | 0.0682 | |
| Female positive n (%) | 380 (17.2) | 41 (3.4) | 27 (4.6) | <.0001 | |
| 11-20 | Patients n | 1621 | 390 | 205 | |
| Positive n (%) | 733 (45.2) | 20 (5.1) | 10 (4.9) | <.0001 | |
| Male n (%) | 927 (57.2) | 197 (50.5) | 113 (55.1) | 0.0573 | |
| Male Positive n (%) | 412 (44.4) | 11 (5.6) | 6 (5.3) | <.0001 | |
| Female n (%) | 694 (42.8) | 193 (49.5) | 92 (44.9) | 0.0573 | |
| Female positive n (%) | 321 (46.3) | 9 (4.7) | 4 (4.3) | <.0001 | |
| 21-40 | Patients n | 3888 | 990 | 800 | |
| Positive n (%) | 1606 (41.3) | 74 (7.5) | 121 (15.1) | <.0001 | |
| Male n (%) | 1978 (50.9) | 481 (48.6) | 401 (50.1) | 0.4317 | |
| Male Positive n (%) | 828 (41.8) | 31 (6.44) | 30 (7.48) | <.0001 | |
| Female n (%) | 1910 (49.1) | 509 (51.4) | 399 (49.9) | 0.4317 | |
| Female positive n (%) | 778 (40.7) | 43 (8.44) | 91 (22.8) | <.0001 | |
| Pregnant positive n (%) | 158 (20.3) | 17 (39.5) | 59 (64.8) | <.0001 | |
| 41-60 | Patients n | 3033 | 928 | 864 | |
| Positive n (%) | 904 (29.8) | 76 (8.2) | 80 (9.3) | <.0001 | |
| Male n (%) | 1563 (51.5) | 503 (54.2) | 507 (58.7) | 0.008 | |
| Male Positive n (%) | 464 (29.7) | 43 (8.5) | 38 (7.5) | <.0001 | |
| Female n (%) | 1470 (48.5) | 425 (45.8) | 357 (41.3) | 0.008 | |
| Female positive n (%) | 440 (29.9) | 33 (7.8) | 42 (11.8) | <.0001 | |
| 61-100 | Patients n | 4497 | 1975 | 1872 | |
| Positive n (%) | 378 (8.4) | 50 (2.5) | 105 (5.6) | <.0001 | |
| Male n (%) | 2550 (56.7) | 1063 (53.8) | 976 (52.1) | 0.0015 | |
| Male Positive n (%) | 189 (7.4) | 28 (2.6) | 58 (5.9) | <.0001 | |
| Female n (%) | 1942 (43.2) | 912 (46.2) | 896 (47.9) | 0.0015 | |
| Female positive n (%) | 189 (9.7) | 22 (2.4) | 47 (5.2) | <.0001 |
We included in this table only patients that we were able to define by gender. In bold are the pregnant women.
Pregnant women are particularly sensitive to A(H1N1)pdm09 infection
We next investigated the percentages of males and females that were hospitalized due to A(H1N1)pdm09 virus infection. Particular focus was placed upon the 21–40 age group, as in 2013, this group was particularly affected by the A(H1N1)pdm09 virus. The highest number of patients (between the ages of 21–40) hospitalized due to influenza symptoms was observed in 2009–10, when the A(H1N1)pdm09 first appeared (Table 2), with similar prevalence among males and females (Table 2). In 2010–12, fewer patients were hospitalized with influenza-like illness, which equally affected both males and females (Table 2). In contrast, in 2013, when a new wave of A(H1N1)pdm09 virus infection arrived, the number and the percentages of women hospitalized due to A(H1N1)pdm09 infection were much higher than those of the men (Table 2). More specifically, the percentages of pregnant women hospitalized due to the pandemic H1N1 influenza virus infection gradually increased from 2009 until 2013, accounting for <60% of the women (ages 21–40) infected in 2013 (Figure 4). A simple logistic regression analysis (Table 3) showed that within the group of pregnant women, no differences in the ages of the infected women were noticed throughout the year, however, as we demonstrate here, differences in the increased percentages of infected women were observed in 2012–13.
Figure 4.
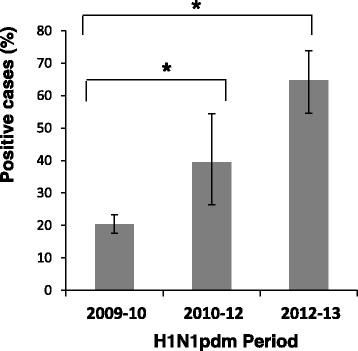
The percentage of positive cases for pregnant women. The percentages (out of the 21–40 age group) of pregnant women hospitalized due to influenza-like syndrome and infected with the pandemic 2009 influenza virus from 2009-today (2013). Three periods of infection are presented (x axis). *P < 0 · 05 using the chi-square test.
Table 3.
Logistic regression
| Variable | Odds ratio | p-value |
|---|---|---|
| Period 2009–10 (Reference) | 1 | |
| Period 2010-12 | (1.33-4.77) 2.52 | 0.0045 |
| Period 2010-13 | (4.53-11.5) 7.21 | <.0001 |
| Age (as continuous) | 1.01 (0.99-1.04) | 0.3486 |
Circulation of genetic variants of the pandemic H1N1 influenza virus strains
The differences in the ages of the hospitalized patients observed throughout the years and the rebound of the A(H1N1)pdm09 virus infection observed in 2012–13, led us to hypothesize that the A(H1N1)pdm09 virus had undergone changes during this short period. Several influenza viruses were isolated from the samples collected during these years, to amplify their hemagglutinin gene using the primers described in Table 1. The obtained sequences were used to build a phylogenetic tree. Viruses isolated in 2009 and in 2013 diverged differently in the phylogenetic tree (Figure 5). While the influenza strains obtained in 2009 resemble the strain used for the influenza vaccine of that year, those who were isolated in 2011–13 resembled other strains, in particular, group 6 viruses.
Figure 5.
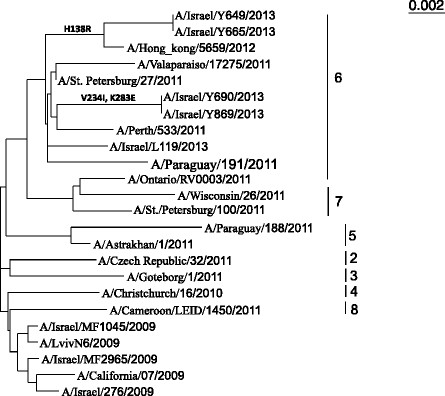
Phylogenetic tree. Phylogenetic tree was generated using the primers listed in Table 1. The HA genes of H1N1 viruses cluster into eight genetic groups defined by amino acid substitution in HA1. The numbers indicate the different subgroups of the pandemic influenza viruses. Mutations are indicated.
Discussion and conclusions
Influenza pandemics that occur every few decades put the entire population at risk [13], and present a huge economic burden. The last influenza pandemic, which occurred in 2009 and was caused by the A(H1N1)pdm09 virus, is the second documented pandemic involving H1N1 influenza viruses. The first one was the 1918 influenza pandemic which killed tens of millions of people [9]. The 2009 influenza virus resulted from triple reassortment of bird, swine and human influenza viruses further combined with a Eurasian pig flu virus [14].
As shown here and by others [15], at the beginning of 2009, unlike most strains of influenza viruses [16], adults older than 60 years of age were less affected by the A(H1N1)pdm09 virus [17], while the most affected group of patients were within the 21–40 year-old age. In contrast, we show here, as far as we know, for the first time, that when the virus returned in 2013, that in addition to the 21–40 year-old adults, adults over 60 years of age were particularly sensitive to infection. The reasons accounting for the age-dependent sensitivity of the different age groups are unknown. Moreover, more women were hospitalized in 2013 due to A(H1N1)pdm09 virus infection, which we attribute to the increased hospitalization of pregnant women observed in 2013. Already in 2009, when the virus first appeared, pregnant women and obese individuals were shown to be more susceptible to infection [18],[19]. In 2009–2010 when the A(H1N1)pdm09virus first appeared other influenza virus strains were hardly detected [20],[21]. In later years infections with the following H3N2 viruses A/Perth/16/2009-like and A/Victoria/361/2011-like were detected. Furthermore, the pattern of respiratory virus infections (other than influenza) changed during this time period, especially around 2009 [22]-[24]. We don’t think however that this is the reason of why more pregnant women were hospitalized in 2013 due to the A(H1N1)pdm09 infection. Thus, it is currently unknown why pregnant women are more sensitive to infection with the pandemic virus.
The 2009 pandemic influenza began in Mexico [9]. The Mexican government closed most of Mexico City's public and private facilities in an attempt to contain the spread of the virus; however, it continued to spread globally. In June 2009, the World Health Organization (WHO) and the U.S. CDC stopped counting cases of people infected with the virus and WHO declared the outbreak of a pandemic.
As reported here and by others [25],[26], the pandemic declined around the world at the beginning of 2010. In August 2010, the Director-General of the WHO announced the end of the H1N1 pandemic [27], further informing that the pandemic influenza virus had moved into the post-pandemic period and could be considered a seasonal influenza virus. Today, experts, including at the WHO, have agreed that an estimated 284,500 people were killed by the disease, much higher than the initial estimates of the death toll. We demonstrated here, however, that the virus did not disappear, as reported by others as well [28].
However, we submit that the risk of infection with the H1N1pdm09 virus should still be taken seriously and that its ability to cause severe disease has not abated: while in 2010–2012 the virus was detected at low percentages in hospitalized patients, in 2013, infections with the pandemic influenza virus increased dramatically and affected a variety of age groups. We suggest that genetic alterations, which resulted in reduced viral recognition by antibodies generated against the 2009 pandemic influenza virus, underlie the return of the A(H1N1)pdm09 virus in 2013. This is despite observations, we made previously in which we show that around half of the Israeli population had antibodies against A(H1N1)pdm09 [29]. Nevertheless, the fact that the virus, in 2013, primarily infected elderly individuals and people at the ages between 21–40, support the claim that it transformed from a pandemic virus to a seasonal influenza virus. Still, the dramatic increase in the percentages of hospitalized patients, especially pregnant women, in 2013, suggests that the threat from the pandemic virus still exists.
Acknowledgments
This work was supported by internal sources.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
HS, JM, TM, MH reviewed and collected the data. MM conceived the study, and participated in its design, analyzed and interpreted the data. MM drafted the manuscript. EM reviewed the manuscript. All authors approved the final manuscript.
Contributor Information
Hilda Sherbany, Email: hilda.sherbany@sheba.health.gov.il.
John McCauley, Email: jmccaul@nimr.mrc.ac.uk.
Tal Meningher, Email: talsegman@gmail.com.
Musa Hindiyeh, Email: musa.hindiyeh@sheba.health.gov.il.
Rita Dichtiar, Email: Rita.Dichtiar@MOH.HEALTH.GOV.IL.
Michal Perry Markovich, Email: Michal.Perry@MOH.HEALTH.GOV.IL.
Ella Mendelson, Email: ella.mendelson@sheba.health.gov.il.
Michal Mandelboim, Email: michal.mandelboim@sheba.health.gov.il.
References
- 1.Taubenberger JK, Kash JC. Influenza virus evolution, host adaptation, and pandemic formation. Cell Host Microbe. 2010;7(6):440–451. doi: 10.1016/j.chom.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thompson WW, Shay DK, Weintraub E, Brammer L, Bridges CB, Cox NJ, Fukuda K. Influenza-associated hospitalizations in the United States. JAMA. 2004;292(11):1333–1340. doi: 10.1001/jama.292.11.1333. [DOI] [PubMed] [Google Scholar]
- 3.Jagger BW, Wise HM, Kash JC, Walters KA, Wills NM, Xiao YL, Dunfee RL, Schwartzman LM, Ozinsky A, Bell GL, Dalton RM, Lo A, Efstathiou S, Atkins JF, Firth AE, Taubenberger JK, Digard P. An overlapping protein-coding region in influenza A virus segment 3 modulates the host response. Science. 2012;337(6091):199–204. doi: 10.1126/science.1222213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rossman JS, Lamb RA. Influenza virus assembly and budding. Virology. 2011;411(2):229–236. doi: 10.1016/j.virol.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nayak DP, Hui EK, Barman S. Assembly and budding of influenza virus. Virus Res. 2004;106(2):147–165. doi: 10.1016/j.virusres.2004.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang X, Hinson ER, Cresswell P. The interferon-inducible protein viperin inhibits influenza virus release by perturbing lipid rafts. Cell Host Microbe. 2007;2(2):96–105. doi: 10.1016/j.chom.2007.06.009. [DOI] [PubMed] [Google Scholar]
- 7.Schmitt AP, Lamb RA. Influenza virus assembly and budding at the viral budozone. Adv Virus Res. 2005;64:383–416. doi: 10.1016/S0065-3527(05)64012-2. [DOI] [PubMed] [Google Scholar]
- 8.Rifkin DB, Quigley JP. Virus-induced modification of cellular membranes related to viral structure. Annu Rev Microbiol. 1974;28:325–351. doi: 10.1146/annurev.mi.28.100174.001545. [DOI] [PubMed] [Google Scholar]
- 9.Neumann G, Noda T, Kawaoka Y. Emergence and pandemic potential of swine-origin H1N1 influenza virus. Nature. 2009;459(7249):931–939. doi: 10.1038/nature08157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hindiyeh M, Ram D, Mandelboim M, Meningher T, Hirsh S, Robinov J, Levy V, Orzitzer S, Azar R, Grossman Z, Mendelson E. Rapid detection of influenza A pandemic (H1N1) 2009 virus neuraminidase resistance mutation H275Y by real-time reverse transcriptase PCR. J Clin Microbiol. 2010;48(5):1884–1887. doi: 10.1128/JCM.02540-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Panning M, Eickmann M, Landt O, Monazahian M, Olschlager S, Baumgarte S, Reischl U, Wenzel JJ, Niller HH, Gunther S Hollmann B, Huzly D, Drexler JF, Helmer A, Becker S, Matz B, Eis-Hübinger A, Drosten C: Detection of influenza A(H1N1)v virus by real-time RT-PCR. Euro Surveill 2009, 14(36): pii: 19329 [PubMed]
- 12.WHO: website.World Health Organization. Sequencing primers and protocol. Available: http://www.who.int/csr/resources/publications/swineflu/GenomePrimers_20090512.pdf. Accessed: 29 April 2009.
- 13.Miller MA, Viboud C, Olson DR, Grais RF, Rabaa MA, Simonsen L. Prioritization of influenza pandemic vaccination to minimize years of life lost. J Infect Dis. 2008;198(3):305–311. doi: 10.1086/589716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qu Y, Zhang R, Cui P, Song G, Duan Z, Lei F. Evolutionary genomics of the pandemic 2009 H1N1 influenza viruses (pH1N 1v) Virol J. 2011;8:250. doi: 10.1186/1743-422X-8-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kelly HA, Grant KA, Williams S, Fielding J, Smith D. Epidemiological characteristics of pandemic influenza H1N1 2009 and seasonal influenza infection. Med J Aust. 2009;191(3):146–149. doi: 10.5694/j.1326-5377.2009.tb02723.x. [DOI] [PubMed] [Google Scholar]
- 16.Belongia EA, Irving SA, Waring SC, Coleman LA, Meece JK, Vandermause M, Lindstrom S, Kempf D, Shay DK. Clinical characteristics and 30-day outcomes for influenza A 2009 (H1N1), 2008–2009 (H1N1), and 2007–2008 (H3N2) infections. Jama. 2010;304(10):1091–1098. doi: 10.1001/jama.2010.1277. [DOI] [PubMed] [Google Scholar]
- 17.Hancock K, Veguilla V, Lu X, Zhong W, Butler EN, Sun H, Liu F, Dong L, DeVos JR, Gargiullo PM, Brammer TL, Tumpey TM, Katz JMox NJ. Cross-reactive antibody responses to the 2009 pandemic H1N1 influenza virus. N Engl J Med. 2009;361(20):1945–1952. doi: 10.1056/NEJMoa0906453. [DOI] [PubMed] [Google Scholar]
- 18.Louie JK, Acosta M, Jamieson DJ, Honein MA. Severe 2009 H1N1 influenza in pregnant and postpartum women in California. N Engl J Med. 2010;362(1):27–35. doi: 10.1056/NEJMoa0910444. [DOI] [PubMed] [Google Scholar]
- 19.Siston AM, Rasmussen SA, Honein MA, Fry AM, Seib K, Callaghan WM, Louie J, Doyle TJ, Crockett M, Lynfield R, Moore Z, Wiedeman C, Anand M, Tabony L, Nielsen CF, Waller K, Page S, Thompson JM, Avery C, Springs CB, Jones T, Williams JL, Newsome K, Finelli L, Jamieson DJ, Pandemic H1N1 Influenza in Pregnancy Working Group Pandemic 2009 influenza A(H1N1) virus illness among pregnant women in the United States. JAMA. 2010;303(15):1517–1525. doi: 10.1001/jama.2010.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dapat IC, Dapat C, Baranovich T, Suzuki Y, Kondo H, Shobugawa Y, Saito R, Suzuki H. Genetic characterization of human influenza viruses in the pandemic (2009–2010) and post-pandemic (2010–2011) periods in Japan. PLoS One. 2012;7(6):e36455. doi: 10.1371/journal.pone.0036455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li T, Fu C, Di B, Wu J, Yang Z, Wang Y, Li M, Lu J, Chen Y, Lu E, Geng J, Hu W, Dong Z, Li MF, Zheng BJ, Cao KY, Wang M. A two-year surveillance of 2009 pandemic influenza A (H1N1) in Guangzhou, China: from pandemic to seasonal influenza? PLoS One. 2011;6(11):e28027. doi: 10.1371/journal.pone.0028027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meningher T, Hindiyeh M, Regev L, Sherbany H, Mendelson E, Mandelboim M. Relationships between A(H1N1)pdm09 influenza infection and infections with other respiratory viruses. Influenza Other Respir Viruses. 2014;8(4):422–430. doi: 10.1111/irv.12249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nisii C, Meschi S, Selleri M, Bordi L, Castilletti C, Valli MB, Lalle E, Lauria FN, Piselli P, Lanini S, Ippolito G, Di Caro A, Capobianchi MR. Frequency of detection of upper respiratory tract viruses in patients tested for pandemic H1N1/09 viral infection. J Clin Microbiol. 2010;48(9):3383–3385. doi: 10.1128/JCM.01179-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pascalis H, Temmam S, Turpin M, Rollot O, Flahault A, Carrat F, de Lamballerie X, Gerardin P, Dellagi K. Intense co-circulation of non-influenza respiratory viruses during the first wave of pandemic influenza pH1N1/2009: a cohort study in Reunion Island. PLoS One. 2012;7(9):e44755. doi: 10.1371/journal.pone.0044755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.De Florentiis D, Parodi V, Orsi A, Rossi A, Altomonte F, Canepa P, Ceravolo A, Valle L, Zancolli M, Piccotti E, Renna S, Macrina G, Martini M, Durando P, Padrone D, Moscatelli P, Orengo G, Icardi G, Ansaldi F. Impact of influenza during the post-pandemic season: epidemiological picture from syndromic and virological surveillance. J Prev Med Hyg. 2011;52(3):134–136. [PubMed] [Google Scholar]
- 26.Viasus D, Anton A, Pumarola T, Carratala J. Influenza A(H1N1)pdm09: beyond the pandemic. Enferm Infecc Microbiol Clin. 2012;30(Suppl 4):54–57. doi: 10.1016/S0213-005X(12)70106-4. [DOI] [PubMed] [Google Scholar]
- 27.Martirosyan L, Paget WJ, Jorgensen P, Brown CS, Meerhoff TJ, Pereyaslov D, Mott JA. The community impact of the 2009 influenza pandemic in the WHO European region: a comparison with historical seasonal data from 28 countries. BMC Infect Dis. 2012;12:36. doi: 10.1186/1471-2334-12-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lehners N, Geis S, Eisenbach C, Neben K, Schnitzler P. Changes in severity of influenza A(H1N1)pdm09 infection from pandemic to first postpandemic season. Germany. Emerg Infect Dis. 2013;19(5):748–755. doi: 10.3201/eid1905.130034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weil M, Shohat T, Bromberg M, Bassal R, Dichtiar R, Mandelboim M, Sofer D, Cohen D, Mendelson E: The dynamics of infection and the persistence of immunity to A(H1N1)pdm09 virus in Israel. Influenza Other Respi Viruses 2012, 7(5):838–46., [DOI] [PMC free article] [PubMed]


