Summary
Detection of substances tasting bitter to humans occurs in diverse organisms including the social amoeba Dictyostelium discoideum. To establish a molecular mechanism for bitter tastant detection in Dictyostelium, we screened a mutant library for resistance to a commonly used bitter standard, phenylthiourea. This approach identified a G-protein-coupled receptor mutant, grlJ−, which showed a significantly increased tolerance to phenylthiourea in growth, survival and movement. This mutant was not resistant to a structurally dissimilar potent bitter tastant, denatonium benzoate, suggesting it is not a target for at least one other bitter tastant. Analysis of the cell-signalling pathway involved in the detection of phenylthiourea showed dependence upon heterotrimeric G protein and phosphatidylinositol 3-kinase activity, suggesting that this signalling pathway is responsible for the cellular effects of phenylthiourea. This is further supported by a phenylthiourea-dependent block in the transient cAMP-induced production of phosphatidylinositol (3,4,5)-trisphosphate (PIP3) in wild-type but not grlJ− cells. Finally, we have identified an uncharacterized human protein γ-aminobutyric acid (GABA) type B receptor subunit 1 isoform with weak homology to GrlJ that restored grlJ− sensitivity to phenylthiourea in cell movement and PIP3 regulation. Our results thus identify a novel pathway for the detection of the standard bitter tastant phenylthiourea in Dictyostelium and implicate a poorly characterized human protein in phenylthiourea-dependent cell responses.
Key words: Dictyostelium, Bitter taste, Denatonium, GABABR1, grlJ, Phenylthiourea, PI3K, Q8NHA5, Taste perception
Introduction
Ingestion of bitter tastants can lead to aversive behaviour, reduced gastric emptying, nausea and vomiting in mammals (Stern et al., 2011), and as such, bitter tastants are thought to provide a potentially vital warning sign of toxicity (Chaudhari and Roper, 2010). The taste-type-2 receptor (T2R) family of G-protein-coupled receptors provides the accepted mechanism for the detection of bitter tastants in mammalian systems (Chandrashekar et al., 2000). This family of receptors currently includes 25 members in humans, which show varying affinities for different bitter tastants (Chandrashekar et al., 2000; Pronin et al., 2004). Activation of these receptors leads to dissociation of the G proteins Gα-gustucin and Gβγ from the cell membrane, which subsequently regulates downstream signalling (Chaudhari and Roper, 2010; Kinnamon, 2012). In addition to T2R receptors, other mammalian receptors have also been shown to be involved in bitter tastant detection, including the family III G-protein-coupled receptors and the calcium-sensing receptor (CASR) (Rogachevskaja et al., 2011). More receptors and downstream kinases that do not belong to the T2R family have also been shown to play a role in detection of bitter tastants (Zubare-Samuelov et al., 2003; Zubare-Samuelov et al., 2005). A comprehensive examination of the molecular mechanisms of tastant function is therefore still needed.
Bitter tastant research, originally focused on vertebrates, has more recently extended to invertebrates including Drosophila melanogaster (McBride et al., 2007) and Caenorhabditis elegans (Hilliard et al., 2004), which are both capable of bitter tastant detection. Our recent research has also reported a simpler organism, the social amoeba Dictyostelium discoideum, to be capable of detecting bitter-tasting compounds (Robery et al., 2011). This organism represents one of the earliest branches from the common ancestor of all eukaryotes (Eichinger et al., 2005). The conserved ability to detect bitter tastants suggests that Dictyostelium might provide a useful model for analysing human tastants, and suggests common evolutionary mechanisms of tastant detection.
Phenylthiourea is an alkaloid that is used as a standard bitter tastant in research and was initially thought to provide an example of a compound with a single protein controlling its perception in humans, a T2R receptor encoded by the TAS2R38 gene (Bufe et al., 2005; Tepper et al., 2009). Single amino acid polymorphisms in this receptor give rise to differences in perception of phenylthiourea bitterness, where combinations of mutations create ‘supertaster’, ‘taster’ and ‘non-taster’ phenotypes to compounds containing an N-C = S group (Bufe et al., 2005). However, although the various TAS2R38 genotypes determine a threshold of phenylthiourea-tasting ability, differences in tasting among threshold groups cannot explain the variation in taste perception within each genotype (Hayes et al., 2008). These results suggest that other mechanisms are involved in phenylthiourea tastant detection.
We have previously reported a potent and rapid onset block in Dictyostelium cell behaviour (shape and movement) during chemotaxis for several bitter tastants, including phenylthiourea (Robery et al., 2011). This response was unexpected because Dictyostelium does not contain genes encoding homologues to T2R proteins associated with phenylthiourea detection. Here, we explore the molecular mechanisms responsible for phenylthiourea detection in Dictyostelium. By screening for mutants resistant to the effect of phenylthiourea on growth (Kuspa, 2006), we identified a putative G-protein-coupled receptor mutant, grlJ−, which showed resistance to phenylthiourea in both growth and cell movement, suggesting that phenylthiourea acts through this receptor in Dictyostelium. We further show that G protein and phosphatidylinositol 3-kinase (PI3K) signalling are probably downstream effectors of phenylthiourea in Dictyostelium. Translation of this discovery to a human context identified an uncharacterised human γ-aminobutyric acid (GABA) type-B isoform that is weakly homologous to GrlJ, which restored grlJ− sensitivity to phenylthiourea, implicating this human protein as a novel receptor for phenylthiourea detection.
Results
Identification of a phenylthiourea receptor in Dictyostelium
Our previous study identified a strong and rapid effect of phenylthiourea on Dictyostelium cell movement (Robery et al., 2011), but a molecular mechanism for this effect was unclear. In order to identify this mechanism, we first established conditions necessary for a mutant screen, based upon resistance to a block in growth caused by phenylthiourea in shaking suspension. Phenylthiourea caused an increasing inhibitory effect on Dictyostelium cell growth compared with control conditions (in the absence of phenylthiourea) to 0.5, 1, 2 and 5 mM phenylthiourea over a 7 day period (Fig. 1A). Under these conditions, 1 and 2 mM phenylthiourea caused a significant (P<0.05) reduction in Dictyostelium cell growth and a 5 mM concentration blocked growth (Fig. 1A). This indicated an approximate IC50 of 1.95 mM (Fig. 1B). On the basis of these results, we then carried out a growth screen using a library of insertional mutants (Kuspa, 2006) in shaking suspension with 1 mM phenylthiourea over a 21 day period and characterised the resulting mutants. This approach identified a range of putative loci that controlled the effect of phenylthiourea on growth (supplementary material Table S1). Among the proteins identified in this screen was a seven-transmembrane G-protein-coupled receptor, GrlJ (Prabhu et al., 2007). This receptor plays a role in Dictyostelium development, but a ligand for the protein has not been identified.
Fig. 1.
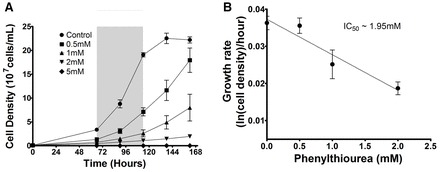
Dictyostelium proliferation in varying concentrations of phenylthiourea. Dictyostelium cells were grown in axenic medium over 168 hours in shaking suspension in the presence of phenylthiourea. (A) Phenylthiourea concentrations from 0.5–5 mM provided a range of inhibitory effects on cell growth; 5 mM was found to be toxic to cells. Growth rate determined between 66 and 114 hours revealed a significant (P<0.05) reduction between control conditions and both 1 mM and 2 mM phenylthiourea. (B) Secondary plot during log growth phase (66–114 hours; grey box in A) for 0.5–2 mM phenylthiourea showing the change in growth rate at each concentration to estimate a potency of inhibition (IC50) of 1.95 mM. Data from A are presented as means ± s.e.m. of triplicate experiments.
To investigate a role for GrlJ in phenylthiourea detection in Dictyostelium, we first knocked out the encoding gene (grlJ) (supplementary material Fig. S1). As previously reported, this null mutant (grlJ−) showed no gross morphological changes in development (fruiting body formation) but produced abnormally shaped spores (Prabhu et al., 2007) (supplementary material Fig. S2). The enhanced growth rate previously published for grlJ− was not recapitulated here (Fig. 2A), which could be due to differences in background (Ax2) laboratory strains or might be caused by our deletion of the central region of the open reading frame and excision of the selection cassette through Cre-lox technology (Faix et al., 2004) rather than through insertional inactivation. We then measured the effect of phenylthiourea on grlJ− cells in shaking suspension (Fig. 2B). In these experiments, wild-type (WT) and grlJ− cells mid-way through log-phase growth (Fig. 2A, 96 hours and ∼1.8×106 cells/ml), were transferred to different concentrations phenylthiourea and growth was monitored for a further 48 hours (Fig. 2B,C). WT cells showed a significant reduction in cell growth during the first 24 hours of phenylthiourea exposure at concentrations over 1 mM (Fig. 2B). By contrast, grlJ− cells were sensitive to phenylthiourea at 3 mM and above (Fig. 2C). This resistant phenotype is shown in a significant shift in cell density at increasing phenylthiourea concentrations (F = 26.12; P<0.001) upon comparison of WT and grlJ− cells (Fig. 2D). Using an independent approach, we also assessed the effect of phenylthiourea on Dictyostelium survival (colony number) and growth (colony size) on a bacterial lawn. In these experiments, grlJ− cells showed a significantly improved resistance to 3 mM phenylthiourea in both colony number and colony size compared with WT cells (Fig. 2E,F). grlJ− cells also showed no change in colony size in the presence of 1 mM phenylthiourea, whereas WT cells showed a significant reduction (Fig. 2F). These data suggest that ablation of grlJ increases resistance to phenylthiourea in cell survival and growth.
Fig. 2.
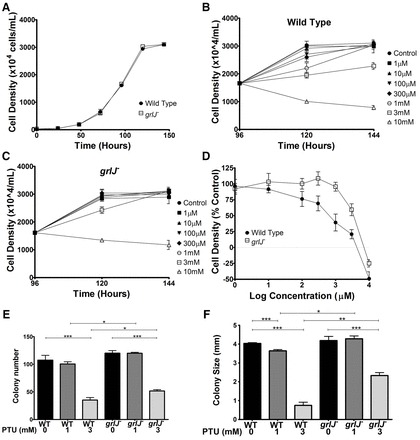
WT and grlJ− growth in the presence of phenylthiourea. (A) WT and grlJ− Dictyostelium cells were grown under control conditions until cells reached stationary phase. (B) WT cells were then grown for 96 hours under control conditions until cells reached ∼50% of stationary phase, before exposure to 1 µM, 100 µM, 300 µM, 1 mM, 3 mM and 10 mM concentrations of phenylthiourea. A significant decrease in cell density was reached at 1 mM phenylthiourea (P<0.05) in comparison to control conditions after 24 hours exposure. (C) Under the same conditions, grlJ− cells showed resistance to phenylthiourea, where a significant decrease in cell density was reached at 3 mM (P<0.05). (D) Cell density 24 hours after exposure to phenylthiourea was then plotted as a percentage of control growth, where a two-way ANOVA identified a significant difference in cell density as the concentration increased between WT and grlJ− cells (F = 26.12; P<0.001). (E) Growth on bacterial lawns indicated significant reduction in WT (P<0.001) and grlJ− (P<0.001) colony formation in the presence of 3 mM phenylthiourea but not 1 mM. In addition, significantly (P<0.05) more grlJ− colonies formed in the presence of 1 mM and 3 mM phenylthiourea. (F) In analyzing colony size, a significant (P<0.001) decrease in size was identified at 3 mM phenylthiourea for both WT and grlJ− cells, and at 1 mM (P<0.05) for WT cells. In addition, significantly larger colonies were formed at 1 mM (P<0.05) and 3 mM (P<0.001) in grlJ− cells. Error bars represent + s.e.m., *P<0.05, **P<0.01, ***P<0.001.
A mechanism for the effect of phenylthiourea on cell movement
To investigate the effect of phenylthiourea on cell movement in grlJ− cells, we then developed a series of cell behaviour measurements based upon random cell movement (Fig. 3). In these assays, three independent experiments were used to assess ∼10 Dictyostelium cells per experiment for changes in cell behaviour following exposure to solvent (control) or phenylthiourea (to give a final concentration of 3 mM). Cell behaviour was monitored over a 288 second period prior to phenylthiourea addition, and recorded for a further 612 seconds. Cell behaviour was defined by displacement travelled for each cell, cell shape (circularity), number of protrusions (pseudopodia) and average cell motility (magnitude of all membrane protrusions and retractions summed up over time). No significant difference was found in any behavioural measurements before or after solvent addition for WT and grlJ− strains.
Fig. 3.
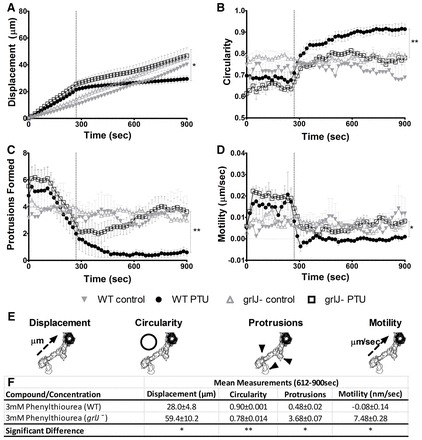
Random cell movement of WT and grlJ− Dictyostelium. After 288 seconds, 3 mM phenylthiourea was added and cells were monitored for the remainder of the assay. (A) A significant (P = 0.0228) difference in displacement at the final cumulative time point was observed between WT and grlJ− cells. (B) WT cells were found to show significantly (P<0.0085) increased circularity compared with grlJ− cells during the final 612–900 seconds. (C) Number of protrusions within a 10 frame window. Significantly (P = 0.0101) fewer protrusions were formed in WT cells than grlJ− cells during the final 612–900 seconds. (D) WT cells were found to be significantly (P<0.0146) less active than grlJ− cells during the final 612–900 seconds. (E) Image overlays of WT cells showing quantification of measurements taken over the course of the assay. (F) Data quantification showing mean measurements taken during the final 612–900 seconds of the assay for displacement, circularity, number of protrusions and motility. In all measurements, no significant difference was observed between WT and grlJ− cells under control conditions (0–288 seconds). Measurements were derived from an average of 40 cells measured from quadruplicate experiments. Error bars represent ± s.e.m.; *P<0.05, **P<0.01, ***P<0.001.
Exposure of WT cells to 3 mM phenylthiourea caused an acute block in Dictyostelium cell behaviour, where cells stopped moving, lost shape (to become round), showed a large reduction in the number of cellular protrusions and reduced cell motility (Fig. 3A–E; supplementary material Movie 1). Exposure of grlJ− cells to 3 mM phenylthiourea caused a less severe change in cell behaviour, in which cells reduced movement, and showed less change in shape, number of protrusions and motility (Fig. 3A–F; supplementary material Movie 2). With regards to displacement, analysis of the rate of movement (defined as the slopes of regression lines fitted to data in Fig. 3A) during the first 288 seconds (prior to phenylthiourea addition) showed no significant difference between grlJ− and WT cells. Furthermore, the rate of movement comparing grlJ− cells during treatment with phenylthiourea and solvent only was not different. By contrast, a significant difference in rate of movement was shown when comparing WT and grlJ− cells following addition of phenylthiourea (P<0.001). Comparison of the rate of movement before and after addition of phenylthiourea (defined as the ratio of slopes before and after addition) between WT (9.62±1.26) and grlJ− (2.4±0.21) cells was also significantly different (P<0.05) suggesting that grlJ− cells show resistance to phenylthiourea. grlJ− cells thus showed resistance to reduced displacement, in addition to reduced loss in cell shape, number of protrusions, and drop in cell motility compared with the WT background strain following phenylthiourea treatment (Fig. 3). This partial resistance phenotype is also seen in the chemotaxis assays used previously (Robery et al., 2011) (supplementary material Fig. S3). These experiments suggest that GrlJ provides at least one mechanism by which phenylthiourea modulates Dictyostelium cell behaviour.
To confirm that GrlJ controlled partial resistance to the effect of phenylthiourea on Dictyostelium cell behaviour, grlJ was expressed with a C-terminal GFP tag (GrlJ-GFP) in the null mutant (grlJ−/+). This cell line showed enhanced membrane fluorescence (Fig. 4A,B) and elevated expression (compared with endogenous grlJ expression levels; Fig. 4C). Behavioural analysis of this overexpression mutant upon exposure to phenylthiourea (supplementary material Movie 3), showed significantly reduced cell displacement compared with grlJ− cells (Fig. 4D), increased (although not significant) cell rounding (Fig. 4E), and complete loss of protrusions (Fig. 4F) and cell motility (Fig. 4G) in a similar manner to that shown for WT cells.
Fig. 4.
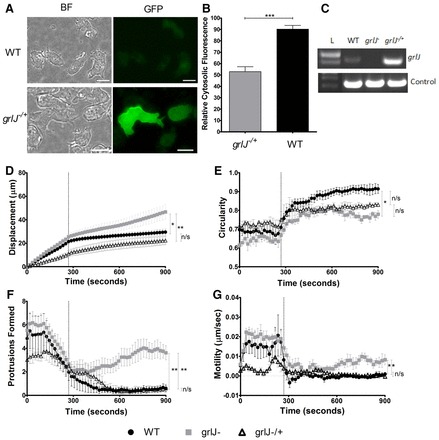
GrlJ overexpression in Dictyostelium. (A) grlJ−/+ cells showed membrane-bound GFP fluorescence when compared with WT cells containing an empty GFP vector. BF, brightfield. Scale bars: 10 µM. (B) Quantification of membrane fluorescence showed significantly less cytosolic fluorescence observed in grlJ−/+ cells. (C) RT-PCR showing grlJ gene expression in both WT and grlJ−/+ but not grlJ− cell lines. Control bands represent constitutively active gene, IG7. (D–G) During random cell movement, 3 mM phenylthiourea was added to cells after 288 seconds. Changes in cell behaviour were monitored and a one-way ANOVA with subsequent post-hoc Tukey's test was performed. As shown in D, a significant difference in the means (P = 0.0067) was observed at the final cumulative time point of the displacement assay between WT and grlJ− (P<0.05) as well as grlJ− and grlJ−/+ (P<0.01) cell lines. As shown in E, a significant difference in the means (P = 0.0060) was observed and WT cells were found to show a significant (P<0.05) increase in circularity compared with grlJ− cells during the final 612–900 seconds. As shown in F, number of protrusions detected within a running window of 10 time points (subsequently, affects of phenylthiourea are apparent before 288 sec). Significant (P = 0.0026) variance between cell lines was shown with both WT and grlJ−/+ cell lines producing significantly fewer protrusions compared with grlJ− cells during the final 612–900 seconds. As shown in G, a significant (P = 0.0083) variance was found between cell lines, with post-hoc analysis showing WT and grlJ−/+ cells to be significantly (P<0.05) less active than grlJ− cells during the final 612–900 seconds. In all measurements, no significant difference was observed between WT, grlJ− and grlJ−/+ cells under control conditions (0–288 seconds). Measurements were derived from 30 cells measured from a minimum of triplicate experiments. Error bars represent ± s.e.m.; *P<0.05, **P<0.01, ***P<0.001.
To test specificity of GrlJ in bitter tastant perception, we then measured cell behaviour in the presence of other bitter tastants. We found that both WT and grlJ− cells showed sensitivity when subjected to 1 mM of a structurally unrelated bitter tastant, denatonium benzoate, or 5 mM of propylthiouracil (supplementary material Table S2), indicating that grlJ− cells were not resistant to at least two other bitter tastants. Furthermore, we also assessed phenylthiourea effects on a non-related G-protein-coupled receptor mutant in Dictyostelium grlE− (Anjard and Loomis, 2006) to test the specificity of other receptors for this tastant. grlE− cells were sensitive to 3 mM phenylthiourea, which was comparable to that of WT cells (Table 1). These data suggest that GrlJ partially controls phenylthiourea detection but not detection of all bitter tastants, and this partial resistance is not evident for other related G-protein-coupled receptors.
Table 1. Summary of WT and grlE− measurements taken during the final 288 seconds of random cell movement assays in the presence of 3 mM phenylthiourea.
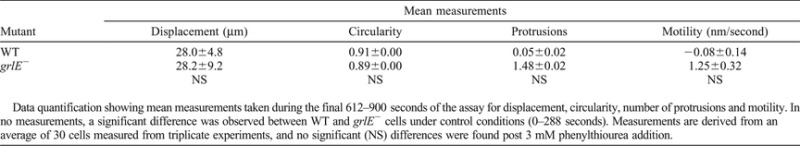
Data quantification showing mean measurements taken during the final 612–900 seconds of the assay for displacement, circularity, number of protrusions and motility. In no measurements, a significant difference was observed between WT and grlE− cells under control conditions (0–288 seconds). Measurements are derived from an average of 30 cells measured from triplicate experiments, and no significant (NS) differences were found post 3 mM phenylthiourea addition.
Establishing a molecular pathway for phenylthiourea detection
To investigate the signal transduction pathway regulated by phenylthiourea in Dictyostelium, we initially measured the effect of phenylthiourea on cell behaviour in a range of Dictyostelium mutants lacking specific pathways, using gene knockout mutants. We first examined a mutant lacking the single copy of the hetrotrimeric Gβ protein (Lilly et al., 1993), thus lacking any heterotrimeric G-protein activity. Although significant differences in random cell movement in the Gβ− cells and the WT background cell line (JH10) were observed, this mutant was resistant to the effect of phenylthiourea on cell behaviour (Fig. 5), showing no loss of cell displacement, circularity, protrusions or cell motility following addition of phenylthiourea. This result is consistent with phenylthiourea activating a GrlJ as a G-protein-coupled receptor in bitter tastant detection.
Fig. 5.
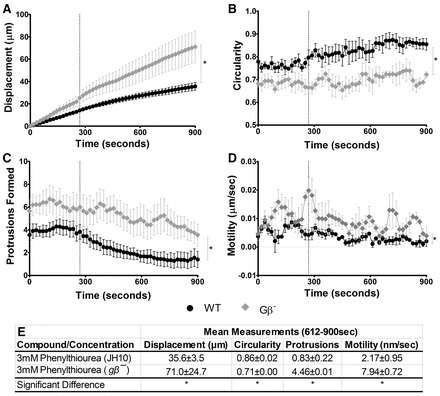
Dictyostelium WT and Gβ− random cell movement. To examine the effect of phenylthiourea on cell behaviour, 3 mM phenylthiourea was added to WT and Gβ− cells after 288 seconds, and cells were monitored for the remainder of the assay. (A) A significant (P = 0.0380) difference in displacement at the final cumulative time point was observed between WT and Gβ− cells. (B) WT cells were found to show significantly (P = 0.0203) increased circularity compared with Gβ− cells during the final 612–900 seconds. (C) Number of protrusions within a 10-frame window. Significantly (P = 0.0324) fewer protrusions were formed in WT cells than Gβ− cells during the final 612–900 seconds. (D) Motility. WT cells were found to be significantly (P = 0.0176) less active than Gβ− cells during the final 612–900 seconds. (E) Data quantification showing mean measurements taken during the final 612–900 seconds of the assay for displacement, circularity, number of protrusions and motility (*P<0.05). In all measurements, no significant difference was observed between WT and Gβ−cells under control conditions (0–288 seconds). Measurements were derived from an average of 30 cells measured from triplicate experiments. Error bars represent ± s.e.m. Dotted vertical line indicates time of addition of phenylthiourea.
We continued to investigate the effect of phenylthiourea on signal transduction by analysing one pathway targeted by Gβ proteins, phosphatidylinositol 3-kinase (PI3K) that function in the production of phosphatidylinositol (3,4,5)-trisphosphate (PIP3). In Dictyostelium, ablation of multiple genes is possible, enabling the analysis of a single cell line lacking all five type-1 PI3K enzymes and the single phosphatase and tensin (pTEN) homolog (PI31–5K−/PTEN−) (Hoeller and Kay, 2007). Ablation of these genes also caused partial resistance to the effects of phenylthiourea on cell behaviour (Fig. 6A), suggesting the involvement of PIP3 production in the phenylthiourea detection pathway. To analyse any PIP3 dependence of phenylthiourea action, we then overexpressed a PHcrac-GFP construct in WT and grlJ− cells, which allowed us to monitor transient PIP3 production following cAMP stimulation by fluorescent microscopy (Xu et al., 2007). Stimulating WT cells containing this fluorescent protein with 1 µM cAMP showed a transient increase in membrane fluorescence 10 seconds after treatment, indicating transient production of PIP3 at the cell membrane under control conditions (Fig. 6C,E). Pre-treatment of these cells with phenylthiourea (3 mM 10 minutes), blocked this transient PIP3 production, in agreement with a mechanism of phenylthiourea acting through PIP3. Repeating these experiments with grlJ− cells showed both a WT response to cAMP in PIP3 production under control conditions and a similar response following phenylthiourea treatment (3 mM 10 minutes incubation), again suggesting that grlJ− is resistant to phenylthiourea (Fig. 6C). We also examined the transient production of PIP3 by direct mass assay, using an ELISA-based method (Fig. 6F,G). Stimulating cells with 1 µM cAMP and extracting PIP3 at three time points, before (control) and 5 and 20 seconds after global stimulation, and quantifying PIP3 for WT and grlJ− cells. In WT cells under control conditions, PIP3 levels showed a two- to threefold increase after 5 seconds and then returned to basal level 20 seconds after cAMP stimulation (Fig. 6F). Incubation of cells for 10 minutes with phenylthiourea (3 mM) blocked this PIP3 increase at 5 seconds in WT cells. As shown in the previous experimental approaches, grlJ− showed WT production of PIP3 under control conditions (Fig. 6F), but was resistant to the effect of phenylthiourea at 5 seconds (Fig. 6E,G). These data indicate that phenylthiourea functions through a GrlJ and subsequent heterotrimeric G-protein signalling to block PI3K-dependent production of PIP3.
Fig. 6.
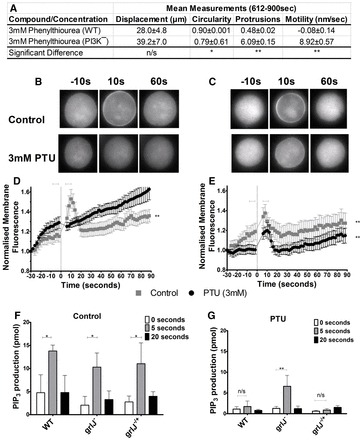
PI3K protein function in phenylthiourea detection. (A) Data quantification showing mean measurements taken during the final 612–900 seconds during random cell movement assays following exposure to phenylthiourea (3 mM) for displacement, circularity, number of protrusions and motility. A significant difference between WT and PI3K− cells in circularity (P = 0.0374), protrusion formation (P = 0.009) and motility (P = 0.004) was observed during the final 612 seconds of the assay. In all measurements, no significant difference was observed between WT and PI3K− cells under control conditions (0–288 seconds). Measurements were derived from an average of 30 cells measured from triplicate experiments. Error bars represent ± s.e.m. (B,D) Relative PHcrac-GFP fluorescence of cells during control conditions and after incubation for 10 minutes with 3 mM phenylthiourea. Cells were stimulated with 1 µM cAMP after 30 seconds. WT cells showed a significant (P = 0.0017) increase in membrane fluorescence upon cAMP stimulation under control conditions. Upon pre-treatment with 3 mM phenylthiourea, cells showed no significant changes in membrane fluorescence upon cAMP stimulation. (C,E) grlJ− cells showed significant (P = 0.0016 and P = 0.0012) increases in membrane fluorescence upon cAMP stimulation for both control and phenylthiourea conditions, respectively. Figures are representative of five independent experiments using two-tailed Student's t-tests for statistical comparisons. (F) PIP3 mass ELISA 1 µM cAMP, showing a significant increase in PIP3 production 5 seconds after global stimulation in WT, grlJ− and grlJ−/+ cells. (G) Incubation with 3 mM phenylthiourea inhibited PIP3 production in WT and grlJ−/+ but not grlJ− cell lines. Figures are representative of triplicate experiments using a two-way ANOVA for statistical comparisons. Error bars represent + s.e.m.
A candidate human gene involved in phenylthiourea detection
Because the molecular mechanism of phenylthiourea in humans has not been fully explained, we searched the human genome for a protein related to GrlJ. BLAST analysis revealed a uncharacterised human protein, encoded by Q8HNA5, showing 26% identity and 48% similarity (with a BLASTP probability of 2×10−20; supplementary material Table S3A). Hydrophobicity analysis of the encoded protein suggested a seven-transmembrane receptor (see schematic Fig. 7A), representing a poorly characterised variant of the GABABR1 protein. We then examined the functionality of this protein in Dictyostelium by overexpression of Q8NHA5 using an open reading frame corrected for Dictyostelium codon bias and expressed in grlJ− cells (grlJ−/ Q8HNA5+). Expression of the transgene was confirmed by reverse transcriptase PCR (Fig. 7B). Expression of this transgene in grlJ−/ cells restored WT sensitivity to phenylthiourea in PIP3 production (Fig. 7C), and in cell movement (Fig. 7D–G; supplementary material Movie 4), suggesting that this human receptor provides a novel candidate for detection of phenylthiourea, suggesting that this human receptor provides a novel candidate for detection of phenylthiourea.
Fig. 7.
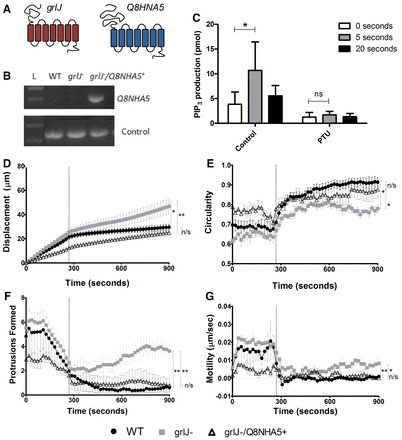
Overexpression of Q8NHA5 in grlJ− Dictyostelium. (A) Schematic of GrlJ and Q8NHA5 protein structure. (B) RT-PCR showing Q8NHA5 gene expression in grlJ− Q8NHA5+ but not WT and grlJ− cell lines. Control bands represent the constitutively expressed gene IG7. (C) PIP3 mass ELISA with 1 µM cAMP, showing a significant increase in PIP3 production 5 seconds after global stimulation in grlJ− Q8NHA5+ cells. Incubation with 3 mM phenylthiourea inhibited PIP3 production. Figures are representative of triplicate experiments using a two-way ANOVA for statistical comparisons. During random cell movement, 3 mM phenylthiourea was added to cells after 288 seconds and changes in cell behaviour monitored. (D) A significant difference in total distance at the final time point was observed between WT and grlJ− cells as well as grlJ− Q8NHA5+ and grlJ−. (E) WT cells were found to show significantly (P<0.05) increased circularity compared with grlJ− cells during the final 612–900 seconds. (F) Number of Protrusions within a 10 frame window. Significantly (P<0.05) fewer protrusions were formed in WT and grlJ− Q8NHA5+ cells than grlJ− cells during the final 612–900 seconds. (G) Motility. WT and grlJ−/+ cells were found to be significantly (P<0.005) less active motile than grlJ− cells during the final 612–900 seconds. In all measurements, no significant difference was observed between WT and grlJ− cells under control conditions (0–288 seconds). Measurements were derived from an average of 40 cells measured from quadruplicate experiments. Dotted vertical lines indicate addition of phenylthiourea. Error bars represent ± s.e.m., *P<0.05, **P<0.01, ***P<0.001; n/s, not significant.
Discussion
This paper explores the underlying mechanism of a standard bitter tastant, phenylthiourea, using the simple model system Dictyostelium. We describe the identification of a seven-transmembrane G-protein-coupled receptor mutant, grlJ− (Prabhu et al., 2007), in a screen for resistance to the inhibitory effect of phenylthiourea on Dictyostelium growth in shaking suspension. This mutant shows a reduction in the inhibitory effect of phenylthiourea in growth (in shaking suspension and on a bacteria lawn), in survival, and in quantitative measures of cell behaviour during random cell movement. This evidence supports a role for GrlJ in phenylthiourea detection in Dictyostelium. However, because both growth and cell behaviour are still affected by phenylthiourea, our data suggest that other proteins are also involved in the detection of this bitter tastant.
Analysis of the signal transduction mechanism involved in phenylthiourea detection showed that the effect of the tastant on cell behaviour was dependent upon heterotrimeric G-protein function. In these studies, ablation of the single gene encoding the Gβ protein in Dictyostelium (hence blocking all heterotrimeric signalling events) protected cells against the effect of phenylthiourea on cell behaviour. This result also suggests that the other (non-GrlJ) mechanism(s) for phenylthiourea detection are also controlled by heterotrimeric G-protein signalling in Dictyostelium. These results support a role of other G-protein-coupled receptor proteins, distinct from the mammalian T2R receptors and Drosophila gustatory receptors (Chaudhari and Roper, 2010; Yarmolinsky et al., 2009), in the detection of phenylthiourea.
A phenylthiourea-dependent effect on cell behaviour was also regulated by PI3K signalling. This was shown through ablation of all PI3K and PTEN activity leading to a partial resistance to the effect of phenylthiourea, and in a phenylthiourea-dependent block in transient PIP3 production. The PI3K pathway in Dictyostelium controls a range of cell functions involved in movement (Manahan et al., 2004). In developing cells, directional movement involves cell polarisation, where cAMP binds a specific receptor cAR1 at the leading edge of the cell (Jin et al., 2009), to activate heterotrimeric G-protein function, leading to Ras activation followed by subsequent activation of PI3K (Gerisch et al., 2012). This triggers the phosphorylation of PIP2 to PIP3 and the activation of a range of downstream cell-signalling processes (Chung and Firtel, 2002; Takeda et al., 2007). Our data suggest that the overactivation of GrlJ induced by phenylthiourea leads to dysregulation of the cAR1–G-protein–Ras–PI3K pathway because ablation of grlJ does not block cAMP-induced PIP3 production (meaning the cellular mechanism of cAMP-induced PIP3 production is intact in this mutant), and phenylthiourea exposure blocks PIP3 production following cAMP stimulation (Fig. 8). This mechanism is supported by the resistance of grlJ− to the block in PIP3 production caused by phenylthiourea.
Fig. 8.
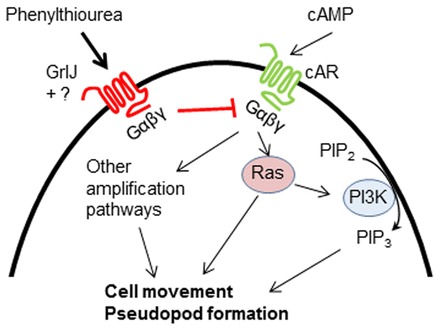
A novel mechanism for the effect of phenylthiourea through GrlJ in Dictyostelium cell behaviour. Under control conditions, during chemotactic and random cell movement, cAMP binds to the cAR receptor, resulting in G-protein dissociation and Ras activation. PI3K activity is subsequently increased, leading to PIP3 synthesis from PIP2. Heterotrimeric G-protein- and Ras-mediated activation of PI3K along with other amplification pathways results in normal cell behaviour. GrlJ activation by phenylthiourea through a heterotrimeric G-protein-dependent mechanism subsequently inhibits PI3K activity and regulates cell behaviour. Partial resistance of grlJ− to phenylthiourea suggests other heterotrimeric G-protein dependent pathway(s) might exist in Dictyostelium.
Studies into the molecular mechanism of phenylthiourea as a tastant have implicated TAS2R38 as the target receptor (Bufe et al., 2005; Kim et al., 2003). However, it has also been suggested that detection of phenylthiourea through TAS2R38 cannot fully explain variation in tastant strength in human populations (Hayes et al., 2008). The identification of an uncharacterised human GABA type B receptor subunit 1 isoform, GABABR1 (Q8NHA5), might provide this alternative mechanism. Overexpression of this receptor in Dictyostelium restored sensitivity to phenylthiourea in cell behaviour studies, supporting a function for the receptor in the detection of this tastant. The receptor also restored sensitivity to cAMP-induced PIP3 production, further supporting its role in phenylthiourea detection in Dictyostelium. Further studies in mammalian models will be necessary to examine the role of this protein as a mechanism for bitter tastant action.
In mammalian systems, a heteromeric assembly of GABABR1 and GABABR2BR2 provides GABAB function as a metabotropic receptor, exerting a slow and prolonged response to binding the main inhibitory neurotransmitter GABA (Tao et al., 2013). The GABAB receptor subunit 1 has been associated with Ashkenazi Jewish Crohn's disease (Kenny et al., 2012), temporal lobe epilepsy (Xi et al., 2011), schizophrenia and mood disorders (Fatemi et al., 2011) and other conditions. Interestingly phenylthiourea tastant status (non-tasters) has also been linked to epilepsy occurrence in patient populations (Pal et al., 2004). Although no information is currently available on the cellular function of Q8NHA5, it shares the highest identity (93%) with GABAB-1B, closely followed by GABAB-1C (91%), and contains extended regions of identity with both proteins (supplementary material Table S3B). Interestingly, analysis of N-glycosylation site potential (using NetNGlyc 1.0 Server), suggests five putative sites in GrlJ (supplementary material Fig. S4A) with similar sites predicted in both Q8NHA5 and GABAB-1B (supplementary material Fig. S4B,C), where they have been associated with bitter taste receptor function in previous studies (Bufe et al., 2002; Reichling et al., 2008). Because our data support a role for the overactivation of Q8NHA5 in response to phenylthiourea, we conclude that this receptor could be activated by phenylthiourea, with the potential of regulating neuropsychiatric function. It remains to be determined whether the other GABAB isoforms also respond to phenylthiourea.
It remains unclear whether such a role for Q8NHA5 in the regulation of PIP3 production is found only in Dictyostelium. Inhibition of PI3K has not been shown to occur in response to activation of taste receptors; however, this response has been shown in olfactory neurons upon exposure to specific odorants such as the bittersweet compound citral (Brunert et al., 2010; Ukhanov et al., 2011). Further studies will be necessary to see whether either GrlJ or Q8NHA5 function in a similar manner to mammalian T2R proteins through phospholipase Cβ2 cleavage of PIP2 to produce inositol trisphosphate with a subsequent release of calcium (Kinnamon, 2012; Yarmolinsky et al., 2009).
The sensation of bitterness is an important factor in the development of novel pharmaceutical drugs because for orally ingested treatments, strong bitter taste of medications can lead to a reduction in compliance. Bitter taste sensation has also been associated with nausea (Peyrot des Gachons et al., 2011) and emesis (Sibert and Frude, 1991) in humans. It is, therefore, important to fully understand bitter taste detection and the molecular mechanisms associated with bitter compounds. Identifying molecular targets for these compounds might also enable in vitro screening of compounds in drug development (Holmes et al., 2009) to reduce the number of animal experiments used in pharmaceutical research. Medium- to high-throughput in vitro screens could also be used to screen for bitter taste perception in early drug development, thereby eliminating compounds early in the development pipeline, providing a major cost-saving advantage in the production of new pharmaceutical treatments.
Materials and Methods
Materials
Axenic medium and SM agar was purchased from ForMedium Ltd (Norfolk, UK). All restriction enzymes were purchased from Fermentas. Cyclic adenosine-monophosphate, DMSO, phenylthiourea, propylthiouracil, denatonium benzoate and trichloroacetic acid were purchased from Sigma-Aldrich (Dorset, UK).
Cell culture, strains and plasmids
All Dictyostelium strains were grown in Axenic medium (Formedium, Norfolk, UK) containing 100 µg/ml penicillin and 100 µg/ml streptomycin while maintained at 22°C. WT (Ax2) strain was used to generate grlJ− mutants as well as the overexpressors for grlJ and Q8NHA5. grlE−, gβ−, pI31-5K−/PTEN− mutants as well as PHcracGFP plasmid vectors were obtained from Dictybase (www.dictybase.org (Fey et al 2013)).
Knockout constructs were created as previously described (Pakes et al., 2012; Terbach et al., 2011). Briefly, 5′ and 3′ fragments flanking the gene of interest were amplified by PCR (peqSTAR 96 Universal Gradient, Erlangen, Germany) from Ax2 genomic DNA. 5′ and 3′ PCR fragments were cloned into the plPBLP expression vector (Faix et al., 2004) using BamHI, PstI, NcoI and KpnI restriction sites, respectively, to incorporate a blasticidin-resistance cassette. The knockout cassette was linearised and transformed into Ax2 WT cells by electroporation (Gene Pulser Xcell, Bio-Rad, Hertfordshire, UK). Positive transformants were selected in nutrient medium containing blasticidin (10 µg/ml). Independent clones were screened for homologous integration by PCR, using a genomic and vector control as well as a diagnostic knockout band. Loss of gene transcription was confirmed using reverse-transcription PCR using a high pure isolation kit (Roche Diagnostics, Sussex, UK) according to the manufacturer's instructions.
An overexpression construct for grlJ was created using the full-length open reading frame amplified from cDNA with EcoRI on both 5′ and 3′ ends as flanking restriction sites. The full-length cDNA PCR product was digested and ligated into the Dictyostelium pDEX27 vector expressing GFP under the control of the actin 15 promoter (Müller-Taubenberger, 2006). Correct gene orientation was confirmed by digestion with BglII. Before cloning into Dictyostelium cells, plasmids were sequenced and compared with grlJ using reference strains on Dictybase.org. The human gene Q8NHA5 was manufactured using the Dictyostelium codon bias (GenScript) with restriction sites BglII and SpeI sites flanking either side of the gene. The full-length cDNA product was digested and ligated into the Dictyostelium PDM 323 and PDM320 vectors under the control of actin 15 promoters. All overexpression constructs were transformed into appropriate Dictyostelium cell lines by electroporation or calcium transformation and placed under G418 (20 µg/ml) selection.
Insertional mutagensis, selection for drug resistance and gene identification
AX4 cells were mutagenised by restriction-enzyme-mediated integration (REMI) of plasmid DNA using pBBC plasmids, which are derivatives of the pBSR1 plasmid (Adachi et al., 1994) that contain 60-mer DNA barcodes. REMI was performed using three combinations of restriction enzymes for plasmid linearization and electroporation (BamHI/DpnII, EcoRI/ApolI and SphI/NlaIII). Clonal transformants were propagated in 24-well culture plates and stored at −80°C in 10% DMSO for future recovery. The residual cells from these plates were inoculated onto bacterial lawns, allowed to grow for 2 days, and collected as pools (24 mutants per pool). A number of these pools of 24 (28–32 pools) were combined into 30 large pools of 672–768 mutants. 25 large pools were used in the enrichment experiments that were carried out for each pool, in triplicate, in 10 cm Petri dishes with 10 ml of HL-5 supplemented with 100 µg/ml streptomycin and 100 U/ml penicillin (Sussman, 1987). Phenylthiourea (1 mM final concentration) was added to 5×106 mutant cells in 10 ml of supplemented HL-5. After 3 days or when cell concentration reached ∼2×106 cells/ml, 1 ml of the initial culture medium was removed and added to 9 ml of fresh medium containing phenylthiourea. The same procedure was repeated for 21 days. Genomic DNA was purified from cells that were collected from each culture dish and plasmid insertion sites were cloned as described previously (Kuspa, 2006). After the insertion site insertion site was identified in a mutant by plasmid rescue, a clonal strain was recovered from the 24-well frozen stock to confirm resistance to growth inhibition by phenylthiourea and for further analysis.
Dictyostelium growth resistance with PTU
Dictyostelium WT and grlJ− cells were grown in axenic medium for 96 hours (to mid log-phase growth), divided into individual flasks, PTU added at the indicated concentrations and cell growth was counted at 24 and 48 hours. Growth rate comparison between the two strains at 3 mM was presented as the percentage of control growth from 96 hours (i.e. density at 96 hours 100%) with each experimental condition being analysed in triplicate.
Colony-forming assay
SM agar (Formedium) was heated and allowed to cool to ∼50°C before addition of desired quantities of DMSO (control) or phenylthiourea. Agar was then poured into Petri dishes and 500 µl of R. planticola was spread onto the plates upon setting. Approximately 30 cells suspended in phosphate buffer were spread onto the agar plates and cells allowed to grow over 4 days.
Spore shape assay
1×107 Dictyostelium cells were allowed to develop on nitrocellulose membrane filters for 24 hours (Robery et al., 2011). Fruiting bodies were harvested and suspended in phosphate buffer and added to coverslips. Images of spores were taken using light microscopy at 40× magnification.
Chemotaxis and random-cell-movement assays
To prepare Dictyostelium cells (Ax2) for chemotaxis assays, cells were grown in shaking suspension in Axenic medium (Formedium) for 48 hours, washed and resuspended in phosphate buffer at 1.7×106 cells/ml. Cells were then pulsed for 5 hours with 30 nM cyclic adenosine monophosphate (cAMP) at 6 minute intervals while shaking at 120 rpm. Cells were then washed in phosphate buffer, resuspended at 1×107 cells/ml and used in a Dunn chamber (Hawksley, Sussex, UK) assay (Robery et al., 2011) migrating toward 5 µM cAMP. A stable chemotactic gradient was allowed to form over a 30 minute period, before recording cell shape and position using an Olympus IX71 microscope at 40× magnification with a QImaging RetigaExi Fast1394 digital camera. Cell images were recorded every 6 seconds over a 15 minute period, with the initial 5 minute period recorded before the addition of test compounds (within a 10 µl aliquot diluted in 5 µM cAMP) to the outer well of the Dunn chamber. Subsequent images were recorded over the following 10 minute period for each compound, and at each concentration, with a minimum of three independent experiments for each drug and each concentration, and an average cell number of ∼30 cells quantified per experiment. Cell recordings were prepared in the second quadrant of the Dunn chamber, enabling cell angular movement to be recorded at around −50°. Solvent-only controls were carried out for all experiments to ensure readouts were based upon compounds alone.
For random-cell-movement assays, cells were prepared as described above but used in eight-well glass coverslips (Thermo Scientific, Northumberland, UK), containing 250 µl cell suspension. Cells were allowed to adhere and begin random movement before cell recordings were started, capturing one image every 18 seconds for ∼20 minutes. After 15 frames, 250 µl of double-concentrated tastant was added and cell behaviour observed over the remaining frames. A minimum of three independent experiments were performed for each compound, whereby an average of 10 cells were quantified per experiment.
PIP3 GFP global stimulation
Dictyostelium cells expressing PH-GFP (described in mutant construction) were prepared as described in random-cell-movement assays, whereby 200 µl cells were added to eight-well glass coverslips (Thermo Scientific) suspended in either KK2 or tastant. Cells were allowed to adhere for 10 minutes and cell fluorescence was subsequently recorded at 60× magnification, using a GFP filter, with images being taken every 2 seconds for 2 minutes. After 30 seconds, 50 µl of 5 µM cAMP was added to the cell suspension and changes in fluorescence were measured. A minimum of 10 cells were quantified for changes in fluorescence intensity over a minimum of five independent experiments.
PIP3 mass ELISA
To prepare Dictyostelium cells for a PIP3 mass ELISA, 6×108 cells were starved and pulsed with 30 nM cAMP for 5 hours in phosphate buffer. Endogenous cell signalling was removed by incubation with caffeine (3 mM final concentration) for 30 minutes before being washed and resuspended in phosphate buffer at 4×107 cells/ml with cells either incubated in phosphate buffer or phenylthiourea (3 mM final concentration) for 10 minutes. 2.5 ml of cells were added to a tube containing 250 µl cAMP (1 µM final concentration) and cell signalling was stopped by addition of 3 ml trichloriacetic acid (TCA, 0.5 M). Cells were then vortexed and incubated for 5 minutes on ice at time points 0, 5 and 20 seconds after addition of cAMP. Samples were then spun and washed with 5% TCA before following the lipid extraction protocol for mass ELISA as described in the kit (Echelon, Logan, UT).
Data analysis and statistics
For Dictyostelium chemotaxis assays, changes in cell velocity and aspect (the ratio between the major and minor axes of an elliptical shape such as a cell) were monitored for every cell within each of the 150 frames recorded over the 15 minute period and analysed by ImagePro Plus software (Media Cybernetics, Buckinghamshire, UK). Compound effects were compared using the mean velocity and aspect of cells between the first 5 minutes and the final 5 minutes, and significance was determined using a two-tailed paired Student t-test (P≤0.05). Cell outlines of chemotaxing cells were mapped using MATLAB (MathWorks, USA).
Random cell movement was analysed using the Quimp 11b software package for ImageJ (Dormann et al., 2002; Tyson et al., 2010) and accompanying scripts for analysis in MATLAB. Cells were segmented and behaviour quantified before and after compound addition using QuimP measures of circularity, displacement of the cell centroid and local membrane velocities between frames (motility maps). Specifically, motility maps were used to compute average membrane velocity at each time point to quantify the level at which cells are actively deforming. Furthermore, protrusions were defined within motility maps as regional peaks exceeding an average speed of 0.1 µm/second, and counted automatically over a running window of 10 frames. Protrusion counts therefore represent protrusive activity within short time periods, centred around discrete time points.
Measures were compared using one-way ANOVA with Tukey's post-hoc test to assess significant difference between the WT and mutant under control conditions. The same tests were performed in the final 288 seconds of the assay comparing mutant responses to WT cells in the presence of the desired drug.
Fluorescence from global stimulation experiments was also quantified using Quimp 11b software and MATLAB as described above. For this, fluorescence at the cell membrane was monitored and quantified. Membrane fluorescence was normalised for each individual cell and compound effects were compared using the mean membrane fluorescence before, immediately after and during the final 6 seconds of the assay, and significance was determined using a two-tailed paired Student t-test (P≤0.05).
Supplementary Material
Footnotes
Author contributions
The experimental work was carried out by S.R., with data analysis support provided by R.T. and T.B. The REMI screen was carried out by C.D. and A.K. and mutants were provided by A.A.N. and Dictybase. The research was supervised by P.A. and R.S.B.W., and the paper was written by S.R., P.A. and R.S.B.W.
Funding
This work was supported by a Universities Federation for Animal Welfare PhD studentship to R.S.B.W. and P.A., and a SWan small project grant to R.S.B.W. R.T. was funded by an EPSRC PhD studentship through the Systems Biology Doctoral Training Centre, Warwick University.
Supplementary material available online at http://jcs.biologists.org/lookup/suppl/doi:10.1242/jcs.136440/-/DC1
References
- Adachi H., Hasebe T., Yoshinaga K., Ohta T., Sutoh K. (1994). Isolation of Dictyostelium discoideum cytokinesis mutants by restriction enzyme-mediated integration of the blasticidin S resistance marker. Biochem. Biophys. Res. Commun. 205, 1808–1814 10.1006/bbrc.1994.2880 [DOI] [PubMed] [Google Scholar]
- Anjard C., Loomis W. F. (2006). GABA induces terminal differentiation of Dictyostelium through a GABAB receptor. Development 133, 2253–2261 10.1242/dev.02399 [DOI] [PubMed] [Google Scholar]
- Brunert D., Klasen K., Corey E. A., Ache B. W. (2010). PI3Kgamma-dependent signaling in mouse olfactory receptor neurons. Chem. Senses 35, 301–308 10.1093/chemse/bjq020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bufe B., Hofmann T., Krautwurst D., Raguse J. D., Meyerhof W. (2002). The human TAS2R16 receptor mediates bitter taste in response to beta-glucopyranosides. Nat. Genet. 32, 397–401 10.1038/ng1014 [DOI] [PubMed] [Google Scholar]
- Bufe B., Breslin P. A., Kuhn C., Reed D. R., Tharp C. D., Slack J. P., Kim U. K., Drayna D., Meyerhof W. (2005). The molecular basis of individual differences in phenylthiocarbamide and propylthiouracil bitterness perception. Curr. Biol. 15, 322–327 10.1016/j.cub.2005.01.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrashekar J., Mueller K. L., Hoon M. A., Adler E., Feng L., Guo W., Zuker C. S., Ryba N. J. (2000). T2Rs function as bitter taste receptors. Cell 100, 703–711 10.1016/S0092-8674(00)80706-0 [DOI] [PubMed] [Google Scholar]
- Chaudhari N., Roper S. D. (2010). The cell biology of taste. J. Cell Biol. 190, 285–296 10.1083/jcb.201003144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung C. Y., Firtel R. A. (2002). Signaling pathways at the leading edge of chemotaxing cells. J. Muscle Res. Cell Motil. 23, 773–779 10.1023/A:1024479728970 [DOI] [PubMed] [Google Scholar]
- Dormann D., Libotte T., Weijer C. J., Bretschneider T. (2002). Simultaneous quantification of cell motility and protein-membrane-association using active contours. Cell Motil. Cytoskeleton 52, 221–230 10.1002/cm.10048 [DOI] [PubMed] [Google Scholar]
- Eichinger L., Pachebat J. A., Glöckner G., Rajandream M. A., Sucgang R., Berriman M., Song J., Olsen R., Szafranski K., Xu Q. et al. (2005). The genome of the social amoeba Dictyostelium discoideum. Nature 435, 43–57 10.1038/nature03481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faix J., Kreppel L., Shaulsky G., Schleicher M., Kimmel A. R. (2004). A rapid and efficient method to generate multiple gene disruptions in Dictyostelium discoideum using a single selectable marker and the Cre-loxP system. Nucleic Acids Res. 32, e143 10.1093/nar/gnh136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatemi S. H., Folsom T. D., Thuras P. D. (2011). Deficits in GABA(B) receptor system in schizophrenia and mood disorders: a postmortem study. Schizophr. Res. 128, 37–43 10.1016/j.schres.2010.12.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fey P., Dodson R., Basu S., Chisholm R. L. (2013). One stop shop for everything Dictyostelium: dictyBase and the Dicty Stock Cente. Methods Mol. Biol. 983, 59–92 10.1007/978-1-62703-302-2_4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerisch G., Schroth-Diez B., Müller-Taubenberger A., Ecke M. (2012). PIP3 waves and PTEN dynamics in the emergence of cell polarity. Biophys. J. 103, 1170–1178 10.1016/j.bpj.2012.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes J. E., Bartoshuk L. M., Kidd J. R., Duffy V. B. (2008). Supertasting and PROP bitterness depends on more than the TAS2R38 gene. Chem. Senses 33, 255–265 10.1093/chemse/bjm084 [DOI] [PubMed] [Google Scholar]
- Hilliard M. A., Bergamasco C., Arbucci S., Plasterk R. H., Bazzicalupo P. (2004). Worms taste bitter: ASH neurons, QUI-1, GPA-3 and ODR-3 mediate quinine avoidance in Caenorhabditis elegans. EMBO J. 23, 1101–1111 10.1038/sj.emboj.7600107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeller O., Kay R. R. (2007). Chemotaxis in the absence of PIP3 gradients. Curr. Biol. 17, 813–817 10.1016/j.cub.2007.04.004 [DOI] [PubMed] [Google Scholar]
- Holmes A. M., Rudd J. A., Tattersall F. D., Aziz Q., Andrews P. L. (2009). Opportunities for the replacement of animals in the study of nausea and vomiting. Br. J. Pharmacol. 157, 865–880 10.1111/j.1476-5381.2009.00176.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin T., Xu X., Fang J., Isik N., Yan J., Brzostowski J. A., Hereld D. (2009). How human leukocytes track down and destroy pathogens: lessons learned from the model organism Dictyostelium discoideum. Immunol. Res. 43, 118–127 10.1007/s12026-008-8056-7 [DOI] [PubMed] [Google Scholar]
- Kenny E. E., Pe'er I., Karban A., Ozelius L., Mitchell A. A., Ng S. M., Erazo M., Ostrer H., Abraham C., Abreu M. T. et al. (2012). A genome-wide scan of Ashkenazi Jewish Crohn's disease suggests novel susceptibility loci. PLoS Genet. 8, e1002559 10.1371/journal.pgen.1002559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim U. K., Jorgenson E., Coon H., Leppert M., Risch N., Drayna D. (2003). Positional cloning of the human quantitative trait locus underlying taste sensitivity to phenylthiocarbamide. Science 299, 1221–1225 10.1126/science.1080190 [DOI] [PubMed] [Google Scholar]
- Kinnamon S. C. (2012). Taste receptor signalling - from tongues to lungs. Acta Physiol. (Oxf.) 204, 158–168 10.1111/j.1748-1716.2011.02308.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuspa A. (2006). Restriction enzyme-mediated integration (REMI) mutagenesis. Methods Mol. Biol. 346, 201–209. [DOI] [PubMed] [Google Scholar]
- Lilly P., Wu L., Welker D. L., Devreotes P. N. (1993). A G-protein beta-subunit is essential for Dictyostelium development. Genes Dev. 7, 986–995 10.1101/gad.7.6.986 [DOI] [PubMed] [Google Scholar]
- Manahan C. L., Iglesias P. A., Long Y., Devreotes P. N. (2004). Chemoattractant signaling in dictyostelium discoideum. Annu. Rev. Cell Dev. Biol. 20, 223–253 10.1146/annurev.cellbio.20.011303.132633 [DOI] [PubMed] [Google Scholar]
- McBride C. S., Arguello J. R., O'Meara B. C. (2007). Five Drosophila genomes reveal nonneutral evolution and the signature of host specialization in the chemoreceptor superfamily. Genetics 177, 1395–1416 10.1534/genetics.107.078683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller-Taubenberger A. (2006). Application of fluorescent protein tags as reporters in live-cell imaging studies. Methods Mol. Biol. 346, 229–246. [DOI] [PubMed] [Google Scholar]
- Pakes N. K., Veltman D. M., Rivero F., Nasir J., Insall R., Williams R. S. (2012). The Rac GEF ZizB regulates development, cell motility and cytokinesis in Dictyostelium. J. Cell Sci. 125, 2457–2465 10.1242/jcs.100966 [DOI] [PubMed] [Google Scholar]
- Pal S. K., Sharma K., Pathak A., Sawhney I. M., Prabhakar S. (2004). Possible relationship between phenylthiocarbamide taste sensitivity and epilepsy. Neurol. India 52, 206–209. [PubMed] [Google Scholar]
- Peyrot des Gachons C., Beauchamp G. K., Stern R. M., Koch K. L., Breslin P. A. Des Peyrot; Beauchamp; Stern; Koch; Breslin(2011). Bitter taste induces nausea. Curr. Biol. 21, R247–R248 10.1016/j.cub.2011.02.028 [DOI] [PubMed] [Google Scholar]
- Prabhu Y., Müller R., Anjard C., Noegel A. A. (2007). GrlJ, a Dictyostelium GABAB-like receptor with roles in post-aggregation development. BMC Dev. Biol. 7, 44 10.1186/1471-213X-7-44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pronin A. N., Tang H., Connor J., Keung W. (2004). Identification of ligands for two human bitter T2R receptors. Chem. Senses 29, 583–593 10.1093/chemse/bjh064 [DOI] [PubMed] [Google Scholar]
- Reichling C., Meyerhof W., Behrens M. (2008). Functions of human bitter taste receptors depend on N-glycosylation. J. Neurochem. 106, 1138–1148 10.1111/j.1471-4159.2008.05453.x [DOI] [PubMed] [Google Scholar]
- Robery S., Mukanowa J., Percie du Sert N., Andrews P. L., Williams R. S. (2011). Investigating the effect of emetic compounds on chemotaxis in Dictyostelium identifies a non-sentient model for bitter and hot tastant research. PLoS ONE 6, e24439 10.1371/journal.pone.0024439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogachevskaja O. A., Churbanov G. D., Bystrova M. F., Romanov R. A., Kolesnikov S. S. (2011). Stimulation of the extracellular Ca2+-sensing receptor by denatonium. Biochem. Biophys. Res. Commun. 416, 433–436 10.1016/j.bbrc.2011.11.095 [DOI] [PubMed] [Google Scholar]
- Sibert J. R., Frude N. (1991). Bittering agents in the prevention of accidental poisoning: children's reactions to denatonium benzoate (Bitrex). Arch. Emerg. Med. 8, 1–7 10.1136/emj.8.1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern R. M., Koch K. L., Andrews P. L. R. (2011). The functions, identification and measurement of nausea and related behaviour in animals. Nausea: Mechanisms and Management 171–239New York, NY: Oxford University Press. [Google Scholar]
- Sussman M. (1987). Cultivation and synchronous morphogenesis of Dictyostelium under controlled experimental conditions. Methods Cell Biol. 28, 9–29 10.1016/S0091-679X(08)61635-0 [DOI] [PubMed] [Google Scholar]
- Takeda K., Sasaki A. T., Ha H., Seung H. A., Firtel R. A. (2007). Role of phosphatidylinositol 3-kinases in chemotaxis in Dictyostelium. J. Biol. Chem. 282, 11874–11884 10.1074/jbc.M610984200 [DOI] [PubMed] [Google Scholar]
- Tao W., Higgs M. H., Spain W. J., Ransom C. B. (2013). Postsynaptic GABAB receptors enhance extrasynaptic GABAA receptor function in dentate gyrus granule cells. J. Neurosci. 33, 3738–3743 10.1523/JNEUROSCI.4829-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tepper B. J., White E. A., Koelliker Y., Lanzara C., d'Adamo P., Gasparini P. (2009). Genetic variation in taste sensitivity to 6-n-propylthiouracil and its relationship to taste perception and food selection. Ann. N. Y. Acad. Sci. 1170, 126–139 10.1111/j.1749-6632.2009.03916.x [DOI] [PubMed] [Google Scholar]
- Terbach N., Shah R., Kelemen R., Klein P. S., Gordienko D., Brown N. A., Wilkinson C. J., Williams R. S. (2011). Identifying an uptake mechanism for the antiepileptic and bipolar disorder treatment valproic acid using the simple biomedical model Dictyostelium. J. Cell Sci. 124, 2267–2276 10.1242/jcs.084285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyson R. A., Epstein D. B., Anderson K. I., Bretschneider T. (2010). High resolution tracking of cell membrane dynamics in moving cells: an electrifying approach. Math. Model Nat. Phenom. 5, 34–55 10.1051/mmnp/20105102 [DOI] [Google Scholar]
- Ukhanov K., Brunert D., Corey E. A., Ache B. W. (2011). Phosphoinositide 3-kinase-dependent antagonism in mammalian olfactory receptor neurons. J. Neurosci. 31, 273–280 10.1523/JNEUROSCI.3698-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi B., Chen J., Yang L., Wang W., Fu M., Wang C. (2011). GABBR1 gene polymorphism(G1465A)isassociated with temporal lobe epilepsy. Epilepsy Res. 96, 58–63 10.1016/j.eplepsyres.2011.04.014 [DOI] [PubMed] [Google Scholar]
- Xu X., Müller-Taubenberger A., Adley K. E., Pawolleck N., Lee V. W., Wiedemann C., Sihra T. S., Maniak M., Jin T., Williams R. S. (2007). Attenuation of phospholipid signaling provides a novel mechanism for the action of valproic acid. Eukaryot. Cell 6, 899–906 10.1128/EC.00104-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarmolinsky D. A., Zuker C. S., Ryba N. J. (2009). Common sense about taste: from mammals to insects. Cell 139, 234–244 10.1016/j.cell.2009.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubare-Samuelov M., Peri I., Tal M., Tarshish M., Spielman A. I., Naim M. (2003). Some sweet and bitter tastants stimulate inhibitory pathway of adenylyl cyclase via melatonin and alpha 2-adrenergic receptors in Xenopus laevis melanophores. Am. J. Physiol. 285, C1255–C1262 10.1152/ajpcell.00149.2003 [DOI] [PubMed] [Google Scholar]
- Zubare-Samuelov M., Shaul M. E., Peri I., Aliluiko A., Tirosh O., Naim M. (2005). Inhibition of signal termination-related kinases by membrane-permeant bitter and sweet tastants: potential role in taste signal termination. Am. J. Physiol. 289, C483–C492 10.1152/ajpcell.00547.2004 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


