Summary
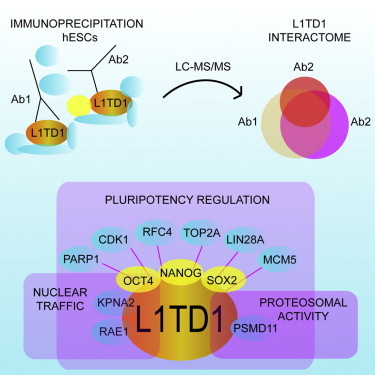
The RNA-binding protein L1TD1 is one of the most specific and abundant proteins in pluripotent stem cells and is essential for the maintenance of pluripotency in human cells. Here, we identify the protein interaction network of L1TD1 in human embryonic stem cells (hESCs) and provide insights into the interactome network constructed in human pluripotent cells. Our data reveal that L1TD1 has an important role in RNA splicing, translation, protein traffic, and degradation. L1TD1 interacts with multiple stem-cell-specific proteins, many of which are still uncharacterized in the context of development. Further, we show that L1TD1 is a part of the pluripotency interactome network of OCT4, SOX2, and NANOG, bridging nuclear and cytoplasmic regulation and highlighting the importance of RNA biology in pluripotency.
Graphical Abstract
Highlights
-
•
A protein interactome is constructed in human embryonic stem cells
-
•
L1TD1 interacts with U2af1 and Srsf3, which are vital for somatic cell reprogramming
-
•
L1TD1 is involved in the high proteasome activity of pluripotent cells
-
•
L1TD1 connects uncharacterized factors to the OCT4, SOX2, and NANOG network
In this work, Lahesmaa and colleagues constructed a protein interactome of a highly stem-cell-specific RNA-binding protein, L1TD1, in human embryonic stem cells. The L1TD1 interactome reveals uncharacterized factors that share the same network with OCT4, SOX2, and NANOG. In addition, the data show that RNA-binding proteins have an important role in the regulation of pluripotency.
Introduction
Pluripotent human embryonic stem cells (hESCs) can differentiate into all somatic cells of the human body and hold tremendous potential for developmental biology, drug screening, and regenerative medicine (Thomson et al., 1998). The importance of the core pluripotency transcription factors OCT4, NANOG, and SOX2 in the maintenance and induction of pluripotency has been well documented (Boyer et al., 2005; Takahashi and Yamanaka, 2006; Yu et al., 2007). However, a variety of post-transcriptional processes control and alter the original message after transcription. In addition, protein degradation by proteasomes has been suggested to have a vital role maintaining pluripotency (Buckley et al., 2012; Vilchez et al., 2012). The role of post-transcriptional regulators and protein networks in the maintenance of pluripotency is still largely unknown, especially in human cells.
L1TD1 is highly and specifically expressed in pluripotent cells (Iwabuchi et al., 2011; Mitsui et al., 2003; Närvä et al., 2012; Wong et al., 2011) under the control of OCT4, NANOG, and SOX2 (Boyer et al., 2005; Närvä et al., 2012; Wong et al., 2011). We previously reported that L1TD1 is essential for maintaining the pluripotent state in hESCs (Närvä et al., 2012). Like LIN28, which is one of the well-characterized post-translational regulators of pluripotency (Huang, 2012), L1TD1 is an RNA-binding protein (RBP). RBPs have a fundamental role in a wide variety of cellular processes, including RNA transcription, splicing, processing, localization, stability, and translation. Moreover, each RBP has unique and specific roles (Glisovic et al., 2008). Given its highly specific expression and vitality in pluripotent cells, we expect that L1TD1 regulates the maintenance of the pluripotent state.
Transcriptomic studies have provided a large amount of data concerning genes that are specifically expressed in the pluripotent state. However, this information can only be used to predict the behavior of proteins that are important for the maintenance of pluripotency. Combinations of mass spectrometry (MS) and bioinformatics studies have increased our understanding of the proteins and pathways that are active in ESCs (Jadaliha et al., 2012; Van Hoof et al., 2006, 2009). Only recently, affinity purifications combined with MS have shed light on the protein-protein interaction networks of individual pluripotency factors and the molecular networks that regulate pluripotency. The interactome for Oct4, Sox2, and Nanog, along with other individual transcription factors, has been reported in mouse ESCs (mESCs) (Ding et al., 2012; Gao et al., 2012; Liang et al., 2008; Pardo et al., 2010; van den Berg et al., 2010; Wang et al., 2006). However, the functional protein-protein interactions in hESCs have remained unexplored. Notably, the protein networks that are distinct from transcriptional-control networks in pluripotent cells remain unknown.
To elucidate interacting and functional components involved in the regulation of human pluripotency, we characterized proteins associated with L1TD1. Furthermore, we validated selected interactions in hESCs and human induced pluripotent stem cells (hiPSCs). We show that the pluripotency network of L1TD1 is shared with the OCT4, NANOG, and SOX2 interactomes, and contains uncharacterized protein players that are important for maintenance of pluripotency. Our data significantly expand the current knowledge about the biology of RBPs and provide an important resource of protein interactions in human pluripotent cells.
Results
L1TD1 Interactome
The biological function of a protein is determined by its structure. L1TD1 consists of a C-terminal coiled coil (CC) domain, an RNA recognition motif (RRM) domain, a C-terminal carboxyl tail domain (CTD) domain, and an N-terminal CTD domain (Figure 1A). The N-terminal RRM domain is exceptional in that only 30 of its C-terminal amino acids (K216–D248) are typical of RRM domains. This “unique sequence” possibly contains the CC domain and the rest of the RRM domain, but it clearly differs from the sequences found in typical CC and RRM domains. Based on these structural domains, we conclude that L1TD1 is an RBP. However, the middle region of L1TD1 contains an exceptional poly-E repeat region. Interestingly, nucleolin, whose structure has an unusual combination of Poly-E and RRM domains, has been shown to specifically recruit proteins that are important for its unique function, with the help of the acidic poly-E region (Edwards et al., 2000; Erard et al., 1988; Lapeyre et al., 1987). On the basis of these similarities, we speculated that by determining the proteins that interact with L1TD1, we could identify protein complexes that are important for the maintenance of pluripotency.
Figure 1.
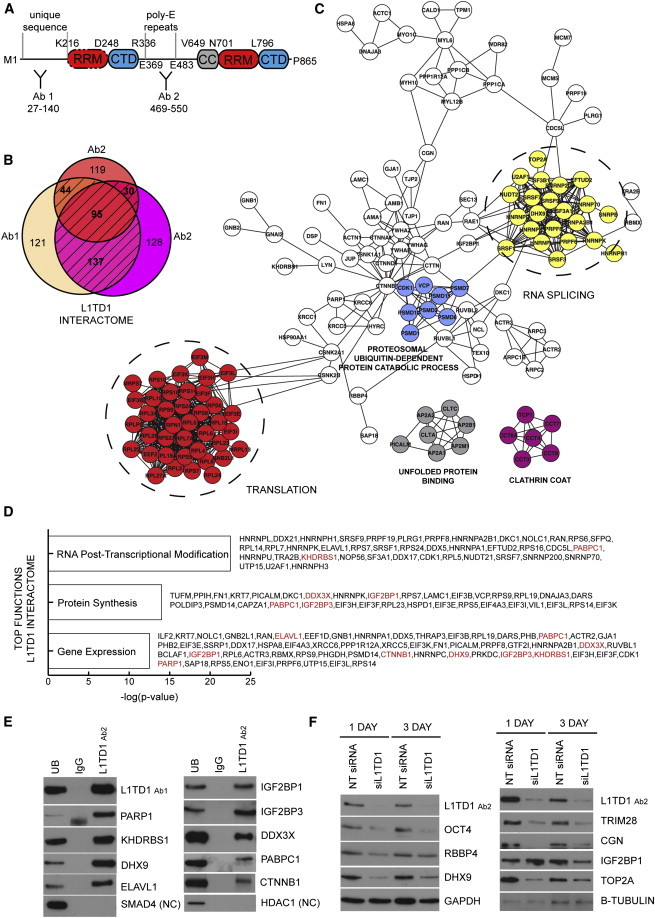
L1TD1-Interacting Proteins Are Involved in RNA Processing
(A) Schematic diagram of the human L1TD1 protein structure: coiled coil (CC) domain, RNA recognition motif (RRM) domain, and carboxyl tail domain (CTD). Regions targeted by antibodies 1 and 2 used in the IPs are shown. Protein domain borders are based on Khazina et al. (2011).
(B) Venn diagram of the proteins identified in the L1TD1 interactome (shaded area) based on MS analysis of three biological replicates.
(C) Clusters of the most enriched biological processes in the L1TD1 interactome. Data from the STRING interaction database (Szklarczyk et al., 2011) were visualized using Cytoscape 2.8.1 (Shannon et al., 2003).
(D) The three most significant functions of L1TD1 interactome proteins based on Ingenuity Pathway Analysis (IPA). Proteins validated experimentally are shown in red.
(E) Western blot (WB) validations of individual protein-protein interactions in L1TD1, IgG control, and unbound (UB) IP reactions. NC, negative control. hESC line HS360. CTNNB1 separate IP reaction, DDX3X and PABPC1 separate IP reaction, hESC line H9.
(F) Protein expression of the interacting proteins after L1TD1 silencing. NT, non-targeting. hESC line HS360. Quantifications can be found in the Supplemental Results.
To identify the L1TD1 protein network and to exclude secondary partners interacting through RNA, we carried out immunoprecipitation (IP) in the presence of RNase with two specific anti-L1TD1 antibodies (Figure S1) that recognize distinct regions of the L1TD1 protein (Figure 1A). The availability of highly specific antibodies against L1TD1 enabled IP of the endogenous protein, excluding possible overexpression effects. The strategy used to overexpress tagged fusion protein was unsuccessful, as the overexpressed fusion protein of L1TD1 was rapidly cleaved in living cells (data not shown). Liquid chromatography-tandem MS (LC-MS/MS) analysis of three biological replicates resulted in the detection of 306 proteins putatively belonging to the L1TD1 interactome (Figure 1B; Table S1). Immunoglobulin G (IgG) control IPs were used as a negative control. In addition, we carried out a CRAPome analysis (Mellacheruvu et al., 2013) to identify the specificity of the interactions (Table S2).
In order to identify biological processes through which L1TD1 mediates its function, we analyzed the L1TD1 interactome with several of the currently available Gene Ontology and interaction analysis tools. First, we investigated known interactions within the L1TD1 interactome based on the STRING interaction database (Szklarczyk et al., 2011). This revealed highly connected components of proteins involved in RNA-processing biology. In particular, RNA translation (21.6%) and splicing (10%) were identified as key functions (Figure 1C; Table S3). Among the other notable associated functions were intracellular transport (9.6%), protein folding (5.6%), and the proteosomal ubiquitin-dependent protein catabolic process (3%). Ingenuity Pathway Analysis (IPA) further confirmed that the top three functions of L1TD1-interacting proteins were gene expression, post-transcriptional modification, and protein synthesis (Figure 1D). Collectively, these results suggest that L1TD1 is involved in multiple steps ranging from RNA production to the production of a functional protein.
To establish the validity of the detected interaction network, we confirmed randomly selected individual interactions with specific antibodies (Figure 1E). In addition, silencing of L1TD1 resulted in reduced expression of most of the interacting partners (Figure 1F). Taken together with the previous observation that the silencing of L1TD1 has a rapid effect on pluripotency (Närvä et al., 2012), our results suggest that interacting proteins that respond to L1TD1 silencing participate in the maintenance of pluripotency along with L1TD1.
L1TD1 Interacts with Multiple RBPs
To gain further insight into the network revealed by these measurements, we classified the L1TD1 interactome on the basis of the structural domains of the interaction partners (Table S4). Based on this, 58 proteins (19%) were classified as ribosomal proteins, providing further evidence that L1TD1 is involved in translation. Interestingly, another major class (10%, 31 proteins) identified was proteins with the RNA-binding domain RRM or KH (K homology), suggesting that L1TD1 performs its function in a complex with other RBPs.
Recently, Kwon et al. (2013) reported an RNA-binding repertoire of mESCs including 283 RBPs. A comparison of these RBPs with the L1TD1 interactome resulted in the identification of 89 shared RBPs, 72 of which had high significance based on the CRAPome (Figure 2A). Based on this comparison, 29% of the L1TD1-interacting proteins bind RNA. Moreover, two RBPs that interact with L1TD1, U2af1, and Srsf3 (Figure 2B) were recently shown to be vital for somatic cell reprogramming in mESCs (Ohta et al., 2013).
Figure 2.
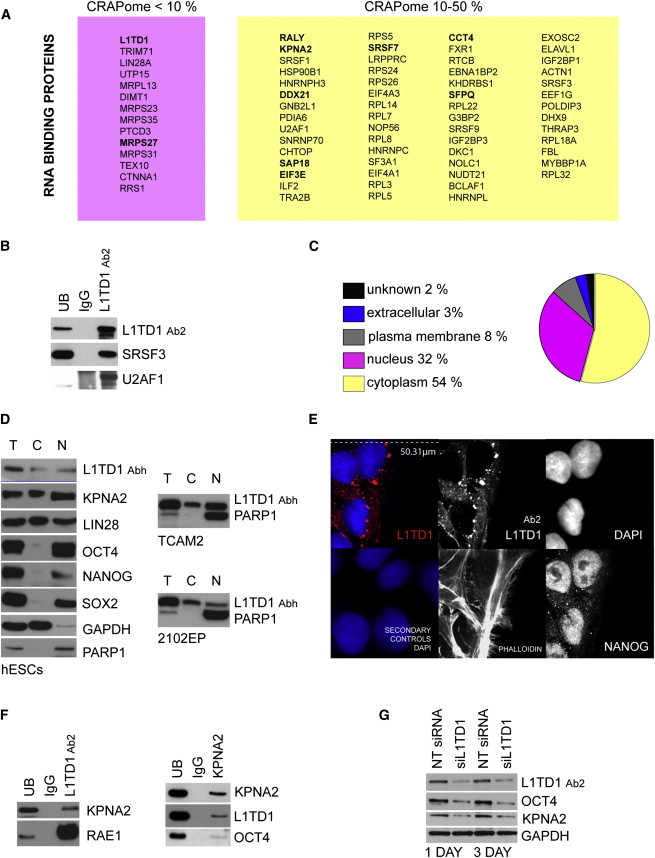
The L1TD1 Interactome Uncovers the Importance of RBPs and the Potential Role of L1TD1 in Nuclear Pore Traffic
(A) RBPs that were both identified here in the L1TD1 interactome in hESCs and previously reported in mESCs (Kwon et al., 2013). Proteins were grouped according to the CRAPome percentages (Workflow 1). Proteins with 0.8-1 SAINT probability are shown in bold (Workflow 3).
(B) WB validation of SRSF3 and U2AF1 interactions in L1TD1, IgG control, and UB IP reactions. hIPSC line HEL 11.4.
(C) Subcellular location of L1TD1-interacting proteins based on IPA software.
(D) Expression of L1TD1, KPNA2, LIN28, OCT4, NANOG, SOX2, GAPDH (cytoplasmic control), and PARP1 (nuclear control) in total (T), cytoplasmic (C), and nuclear (N) cell fractions of hESCs (H9), 2102EP embryoteratocarcinoma, and TCAM2 seminoma cell lines.
(E) Immunostaining of L1TD1 (red), DNA (DAPI staining, blue), NANOG, Phalloidin (actin), and secondary controls in hIPSCs (HEL 11.4).
(F) WB validation of KPNA2, L1TD1, and RAE1 interactions in L1TD1, KPNA2, IgG control, and UB IP reactions. Additional detections from the experiment shown in Figure 1E. KPNA2 IP hIPSC line HEL24.3.
(G) Effect of L1TD1 silencing on the expression of KPNA2 and OCT4. hESC line HS360. Quantifications can be found in the Supplemental Results.
See also Table S2.
L1TD1 Traffics between the Nucleus and Cytoplasm
Most of the L1TD1-interacting proteins identified were annotated as cytoplasmic in location (Figure 2C), although more than 30% were nuclear proteins. The presence of nuclear proteins in the L1TD1 interactome led us to reanalyze the cellular location of L1TD1. In a previous study, we localized L1TD1 in the cytoplasmic processing bodies (P-bodies) based on its co-location with AGO2 and LIN28 (Närvä et al., 2012). However, both of these proteins are able to traffic between the cytoplasm and nucleus (Balzer and Moss, 2007; Robb et al., 2005). In addition, a recent MS analysis identified L1TD1 in the nuclear fraction (Sarkar et al., 2012). To test whether L1TD1 is found in the nucleus, we carried out cell fractionation experiments. The results showed the presence of L1TD1 and LIN28 in both the nuclear and cytoplasmic fractions, whereas OCT4, NANOG, and SOX2 showed extensive nuclear expression (Figure 2D). The location was also validated in cancer cell lines expressing endogenous L1TD1 (Figure 2D).
Based on the immunostainings, we concluded that most of the L1TD1 complexes localize adjacent to the nuclear membrane (Figure 2E). This led us to analyze the L1TD1 interactome for the presence of possible binding partners associated with the nuclear membrane and nuclear pore traffic. In addition to identifying multiple proteins involved in cellular transport, we identified key factors in nuclear pore traffic, such as KPNA2, RAN, and RAE1. Interestingly, karyopherin KPNA2 is enriched in undifferentiated ESCs (Van Hoof et al., 2006) and has been shown to be responsible for the nuclear import of OCT4 (Young et al., 2011). Furthermore, RAE1 is a component of the nuclear pore complex involved in RNA export (Murphy et al., 1996). We validated the cytoplasmic and nuclear location of KPNA2 (Figure 2D), as well as interactions of KPNA2 and RAE1 with L1TD1 (Figure 2F). Further, by reciprocal pull-down of KPNA2, we were able to validate the interaction with L1TD1 and OCT4 (Figure 2F). In addition, we demonstrated that silencing of L1TD1 results in decreased KPNA2 and OCT4 levels (Figure 2G). These findings strongly suggest that L1TD1 is able to traffic between the nucleus and cytoplasm and participate in nuclear pore traffic.
L1TD1 Levels Are Rapidly Decreased by Proteasome Inhibitor
The LC-MS/MS analysis resulted in the identification of multiple members of the ubiquitin-dependent proteasome, such as PSMD1, PSMD2, PSMD7, PSMD8, PSMD11, and PSMD14. Of particular interest, PSMD11 was recently reported to be responsible for the increased proteasome activity in hESCs, which is important for the maintenance of pluripotency (Vilchez et al., 2012). The interaction between L1TD1 and PSMD11 was validated with immunoblotting (Figure 3A). Moreover, silencing of L1TD1 resulted in decreased levels of PSMD11 (Figure 3B).
Figure 3.
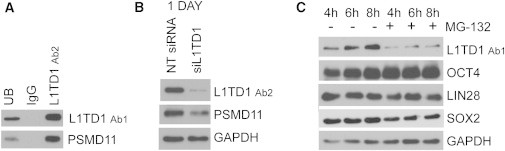
L1TD1 Participates in the High Proteasome Activity of Pluripotency
(A) WB detection of PSMD11 in L1TD1, IgG control, and UB IP reactions. hESC line HS360.
(B) Protein expression of PSMD11 after L1TD1 silencing. hESC line HS360. NT, non-targeting. Quantifications can be found in the Supplemental Results.
(C) Protein expression of L1TD1, OCT4, LIN28, SOX2, and GAPDH after inhibition of proteosomal activity with MG-132 inhibitor (10 μM) versus DMSO control. hESC line H9.
To further study the role of L1TD1 in the proteasome complex, we inhibited proteasome activity with MG-132, which is known to decrease the expression of SOX2 (Vilchez et al., 2012). Treatment with MG-132 resulted in an immediate reduction of L1TD1 and SOX2, but did not influence the levels of OCT4 or LIN28 at the early time points studied (Figure 3C). These results indicate that the protein levels of L1TD1 correlate with proteasome activity and that reduction of L1TD1 expression is an immediate indicator of a decrease in hESC proteasome activity and pluripotency.
L1TD1 Is a Member of the Pluripotency Network
To understand the role of L1TD1 in regulating pluripotency, we compared the L1TD1 interactome with lists of proteins (Jadaliha et al., 2012; Van Hoof et al., 2006) and genes (Assou et al., 2007) that are enriched in hESCs compared with their differentiated counterparts. Based on this analysis, 30% of the L1TD1 interaction partners are associated with pluripotency (Figures 4A and S2A). Nineteen of the interacting proteins had previously been shown to be enriched in hESCs at both the gene and protein levels. Interestingly, the panel contains several factors, such as DPPA4 (developmental pluripotency associated 4), RFC5 (replication factor C subunit 5), and TOP2A (topoisomerase-IIα), whose role in the regulation of pluripotency remains to be investigated. The interaction of L1TD1 with TOP2A was validated (Figure S2B). In addition, silencing of L1TD1 led to decreased TOP2A levels (Figure 1F).
Figure 4.
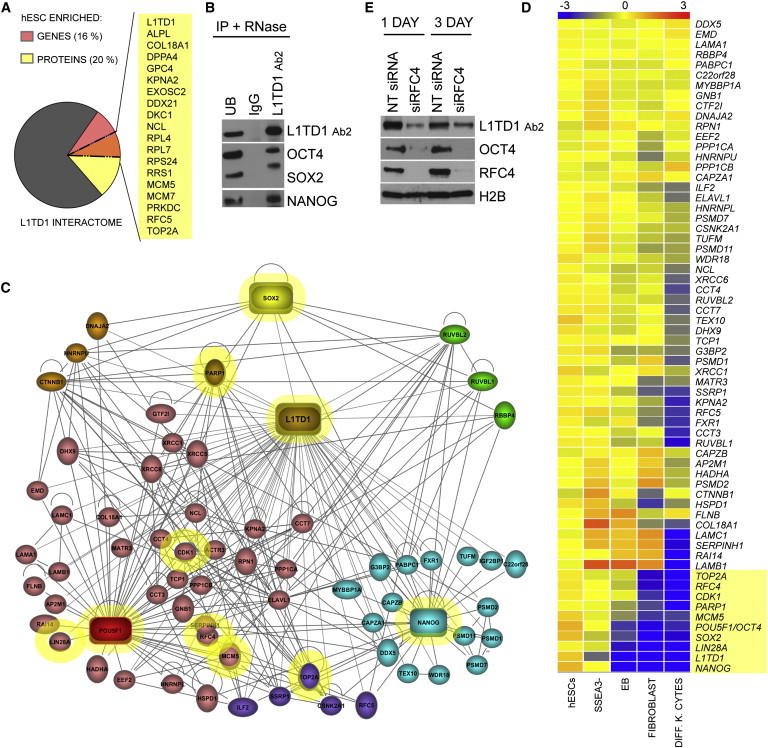
L1TD1 Connects Uncharacterized Factors to Core Pluripotency Regulation
(A) hESC-enriched genes (Assou et al., 2007) and proteins (Jadaliha et al., 2012; Van Hoof et al., 2006) in the L1TD1 interactome. Overlapping hits are listed.
(B) WB detection of OCT4, SOX2, and NANOG in L1TD1, IgG control, and UB IP reactions. hESC line HS360.
(C) Proteins that interact with L1TD1 and OCT4 (n = 45), NANOG (n = 25), or SOX2 (n = 8) (Ding et al., 2012; Nitzsche et al., 2011; van den Berg et al., 2010). The figure was constructed with the use of IPA software.
(D) RNA expression of proteins represented in Figure 4C in hESCs and differentiated cells. EB, embryoid body; DIFF.K.CYTES, differentiated keratinocytes. Microarray data collected from previous studies (Enver et al., 2005; Golan-Mashiach et al., 2005; Skottman et al., 2005). The figure was generated from normalized raw files using GeneSpring GX Software (Agilent Technologies Genomics). The top ten differentiation-responsive genes are highlighted in yellow.
(E) Effect of RFC4 silencing (siRNA1) on the expression of L1TD1 and OCT4. hESC line HS360. Quantifications can be found in the Supplemental Results.
See also Figure S2.
L1TD1 belongs to the interactomes of Nanog (Nitzsche et al., 2011) and Oct4 (Ding et al., 2012; van den Berg et al., 2010) in mESCs. However, NANOG and OCT4 were not detected as L1TD1-interacting proteins in our MS data. It has proved to be challenging to detect these pluripotency factors with MS (Ding et al., 2012; Liang et al., 2008; Pardo et al., 2010; van den Berg et al., 2010). Therefore, we tested the presence of these proteins by immunoblotting of the L1TD1 IPs. This approach led to the detection of OCT4, NANOG, and SOX2 as L1TD1-interacting proteins (Figure 4B). Successful optimization of SOX2-IP further verified L1TD1 interaction with SOX2 (Figure S2C). In addition to these transcription factors, we demonstrated that L1TD1 interacts with the pluripotency-related proteins DNMT3B and TRIM28 (Figure S2D). These interactions were initially identified only in individual MS replicates. In addition, silencing of L1TD1 led to decreased TRIM28 levels (Figure 1F).
Finally, we sought to determine which components of the L1TD1 interactome were also present in the mESC interactomes of OCT4, NANOG, and SOX2 (Figure 4C) (Ding et al., 2012; Nitzsche et al., 2011; van den Berg et al., 2010). We applied IPA and identified 45, 25, and 8 proteins that interact with OCT4, NANOG, and SOX2, respectively. We then analyzed the gene expression of these factors in human undifferentiated, early differentiated (SSEA3−), and differentiated (embryoid bodies [EBs], fibroblasts, and keratinocytes) cells (Figure 4D). This analysis resulted in the identification of proteins that form the key core of pluripotency at the protein level. In addition to NANOG, L1TD1, LIN28A, SOX2, and OCT4, this analysis suggested that MCM5 (minichromosome maintenance complex component 5), PARP1 (poly ADP-ribose polymerase 1), CDK1 (cyclin-dependent kinase 1), RFC4, and TOP2A could have important roles in the regulation of pluripotency. To evaluate the importance of these factors in terms of pluripotency, we selected one of them to determine the influence of its silencing. Depletion of RFC4 led to an immediate reduction of OCT4 and L1TD1 levels (Figure 4E), demonstrating that this factor is required for the maintenance of pluripotency.
To confirm our results, we repeated selected L1TD1 protein interactions in hiPSCs (Figure S2E) and tested the specificity of the L1TD1-bead-antibody complex by performing IP in differentiated HeLa control cells compared with pluripotent cells, including the RFC4 interaction (Figure S2F).
Discussion
RBPs are engaged in every aspect of RNA biology and are essential for carrying specific RNA products from the translation machinery to pre-mRNA splicing, transport, localization, translation, and turnover (Glisovic et al., 2008). Our study indicates that L1TD1 is a functional RBP in human pluripotent cells that is associated with proteins that participate in cellular RNA processes, thus shedding light on the RNA regulation of pluripotent cells.
Several observations support the idea that L1TD1 shuttles between the nucleus and cytoplasm. Based on our results and consistent with a previous report on the subcellular proteome of hESCs (Sarkar et al., 2012), we conclude that L1TD1 (like LIN28) can be found in cytoplasmic, nuclear, and membrane fractions. Based on the NetNES 1.1 server, the amino acids L811–L814 could be responsible for the nuclear location of L1TD1. As RBPs are known to participate in RNA traffic through nuclear pores, it is possible that L1TD1 has a role in that process. The interaction of L1TD1 with OCT4, SOX2, NANOG, and the nuclear pore traffic protein karyopherin KPNA2 suggests the intriguing possibility that L1TD1 is responsible for the nuclear import of these transcription factors after their translation to protein in the cytoplasm. Highly stem-cell-specific L1TD1 could be responsible for the nuclear localization, thereby controlling these transcription factors and providing an additional checkpoint for the maintenance of pluripotency.
The network shared among L1TD1, OCT4, SOX2, and NANOG is highly interesting. Our analysis suggests that the nuclear epigenetic regulator PARP1 and CDK1 are key proteins in the core pluripotency network. Recently, PARP1 expression and PARylation activity were shown to be increased in undifferentiated mESCs and to regulate nuclear reprogramming (Chiou et al., 2013; Doege et al., 2012; Lai et al., 2012). In addition, CDK1 was shown to affect the differentiation of mESCs through an interplay with Oct4 (Li et al., 2012). Our results also revealed a panel of proteins that were not previously reported in the context of self-renewal but may be involved in the regulation of pluripotency. Although the role of TOP2A, RCF4, and MCM5 in pluripotency remains uncharacterized, their contribution to this network and function in other cell types suggest they may have a role in regulation of the cell cycle and DNA replication in pluripotent cells. We chose to further investigate the role of RFC4 and demonstrated that it is vital for maintaining pluripotency.
Post-transcriptional regulators and protein turnover determine which factors are active in pluripotency. Recently, the deubiquitinating enzyme Psmd14 was shown to be essential for cellular reprogramming (Buckley et al., 2012), and PSMD11 was shown to regulate increased proteasome activity in hESCs (Vilchez et al., 2012). However, it is still unclear how proteins are selected for ubiquitination in pluripotent cells. The presence of multiple proteasome subunits in the highly pluripotent-specific L1TD1 interactome suggests that L1TD1 could participate in this selection.
One of the central functional categories detected in the L1TD1 interactome was splicing (Figure 1C). Cell-type-specific RBPs are able to regulate tissue-specific splicing (Licatalosi and Darnell, 2010), which has been shown to be unique in mESCs and reorganized during reprogramming by RBPs U2af1 and Srsf3 (Ohta et al., 2013). The interaction between L1TD1 (identified here in hESCs) and these two factors implies that U2AF1 and SRSF3 also have an important role in human pluripotency, and L1TD1 may play a role in stem-cell-specific splicing.
In addition to regulating pluripotency, L1TD1 may have an important function in cancer. We previously showed that L1TD1 is expressed in selected cancer types, such as colorectal carcinoma, ovarian germ cell tumors, and testis seminomas and non-seminomas (Närvä et al., 2012). Interestingly, the L1TD1 interactome reveals many proteins that have an important role or diagnostic value in cancer. For example, KPNA2 has been shown to affect malignant transformation and its elevated expression is correlated with poor diagnosis in multiple forms of cancer (Christiansen and Dyrskjot, 2012).
Of all the factors in the interactome, L1TD1 has the fastest kinetics in response to differentiation. This implies that L1TD1 has a regulatory role in the maintenance of pluripotency. Moreover, interaction with transcriptional, cell cycle, epigenetic, proteosomal, protein traffic, and splicing regulators further supports the regulatory role of L1TD1. The protein interactome constructed here can be used as an important resource for elucidating individual protein interactions in pluripotent stem cells and possibly in cancer stem cells. Moreover, this analysis sheds light on post-translational regulation in pluripotency and the importance of RBPs in pluripotent cells.
Experimental Procedures
All experiments were performed in feeder-free culture conditions on Matrigel (BD Biosciences) in mTeSR1 media (Stem Cell Technologies).
For IP, cells were lysed into NP-40 buffer. Lysates were preincubated with 10 μg/ml RNase A (QIAGEN) at +4°C. IP was carried out using M-280 sheep anti-rabbit IgG Dynabeads (11203D; Invitrogen) with L1TD1 HPA030064 (Ab1), HPA028501 (Ab2) (Sigma), SOX-2 #5024 (Cell Signaling), and normal rabbit IgG 12-370 (Millipore).
IP proteins were gel separated, digested, and submitted for LC-MS/MS analysis. The criteria used for inclusion as an L1TD1-interacting protein were (1) the presence of the protein in two out of three replicates, (2) more than one unique peptide was identified in the L1TD1 IP analysis, and (3) on/off or ≥3-fold enrichment of the identified peptides compared with the control IgG IP reaction.
Detailed methods are described in the Supplemental Experimental Procedures.
Author Contributions
M.R.E. and E.N. conceived and designed the study, performed experimental work, analyzed data, and wrote the manuscript. A.S., M.V., and N.R. performed experimental work. K.D. and D.C. analyzed data. A.R. performed mass spectrometry. R.M. provided scientific support and language editing. R.T. provided supervision for hIPSC cultures. R. Lund and L.L.E. performed data analysis and provided supervision. R. Lahesmaa was the leader and supervisor of the project from conception and design of the study to writing of the manuscript.
Acknowledgments
We thank Outi Hovatta (Karolinska Institute, Sweden) for the hESC lines and Timo Otonkoski (University of Helsinki) for hiPSC lines. We also thank Marjo Hakkarainen, Päivi Junni, Bogata Fezazi, and Krista Maurinen for excellent technical assistance, and Omid Rasool and Juha-Pekka Pursiheimo for fruitful discussions. We are grateful to the Turku Proteomics Facility, Cell Imaging Core, Turku Centre for Biotechnology (Turku, Finland), and Biomedicum Stem Cell Center (Helsinki, Finland) for their assistance. This study was supported by funding for the Juvenile Diabetes Research Foundation (JDRF), The Academy of Finland (the Centre of Excellence in Molecular Systems Immunology and Physiology Research, 2012-2017, Decision No. 250114 and Postdoctoral Researcher Decision No. 265723), the Finnish Cancer Organizations, the Sigrid Jusélius Foundation, the Turku Graduate School of Biomedical Sciences, the Finnish Cultural Foundation, European Commission Seventh Framework grant EC-FP7-SYBILLA-201106, and the Finnish Cancer Institute.
Footnotes
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/).
Supplemental Information
Workflow 1 (percentage found in other studies) and Workflow 3 (SAINT probability) for L1TD1 interactome proteins.
STRING9.0 and DAVID pathway analysis.
References
- Assou S., Le Carrour T., Tondeur S., Ström S., Gabelle A., Marty S., Nadal L., Pantesco V., Réme T., Hugnot J.P. A meta-analysis of human embryonic stem cells transcriptome integrated into a web-based expression atlas. Stem Cells. 2007;25:961–973. doi: 10.1634/stemcells.2006-0352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balzer E., Moss E.G. Localization of the developmental timing regulator Lin28 to mRNP complexes, P-bodies and stress granules. RNA Biol. 2007;4:16–25. doi: 10.4161/rna.4.1.4364. [DOI] [PubMed] [Google Scholar]
- Boyer L.A., Lee T.I., Cole M.F., Johnstone S.E., Levine S.S., Zucker J.P., Guenther M.G., Kumar R.M., Murray H.L., Jenner R.G. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley S.M., Aranda-Orgilles B., Strikoudis A., Apostolou E., Loizou E., Moran-Crusio K., Farnsworth C.L., Koller A.A., Dasgupta R., Silva J.C. Regulation of pluripotency and cellular reprogramming by the ubiquitin-proteasome system. Cell Stem Cell. 2012;11:783–798. doi: 10.1016/j.stem.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou S.H., Jiang B.H., Yu Y.L., Chou S.J., Tsai P.H., Chang W.C., Chen L.K., Chen L.H., Chien Y., Chiou G.Y. Poly(ADP-ribose) polymerase 1 regulates nuclear reprogramming and promotes iPSC generation without c-Myc. J. Exp. Med. 2013;210:85–98. doi: 10.1084/jem.20121044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christiansen A., Dyrskjot L. The functional role of the novel biomarker karyopherin α 2 (KPNA2) in cancer. Cancer Lett. 2012;331:18–23. doi: 10.1016/j.canlet.2012.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding J., Xu H., Faiola F., Ma’ayan A., Wang J. Oct4 links multiple epigenetic pathways to the pluripotency network. Cell Res. 2012;22:155–167. doi: 10.1038/cr.2011.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doege C.A., Inoue K., Yamashita T., Rhee D.B., Travis S., Fujita R., Guarnieri P., Bhagat G., Vanti W.B., Shih A. Early-stage epigenetic modification during somatic cell reprogramming by Parp1 and Tet2. Nature. 2012;488:652–655. doi: 10.1038/nature11333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards T.K., Saleem A., Shaman J.A., Dennis T., Gerigk C., Oliveros E., Gartenberg M.R., Rubin E.H. Role for nucleolin/Nsr1 in the cellular localization of topoisomerase I. J. Biol. Chem. 2000;275:36181–36188. doi: 10.1074/jbc.M006628200. [DOI] [PubMed] [Google Scholar]
- Enver T., Soneji S., Joshi C., Brown J., Iborra F., Orntoft T., Thykjaer T., Maltby E., Smith K., Abu Dawud R. Cellular differentiation hierarchies in normal and culture-adapted human embryonic stem cells. Hum. Mol. Genet. 2005;14:3129–3140. doi: 10.1093/hmg/ddi345. [DOI] [PubMed] [Google Scholar]
- Erard M.S., Belenguer P., Caizergues-Ferrer M., Pantaloni A., Amalric F. A major nucleolar protein, nucleolin, induces chromatin decondensation by binding to histone H1. Eur. J. Biochem. 1988;175:525–530. doi: 10.1111/j.1432-1033.1988.tb14224.x. [DOI] [PubMed] [Google Scholar]
- Gao Z., Cox J.L., Gilmore J.M., Ormsbee B.D., Mallanna S.K., Washburn M.P., Rizzino A. Determination of protein interactome of transcription factor Sox2 in embryonic stem cells engineered for inducible expression of four reprogramming factors. J. Biol. Chem. 2012;287:11384–11397. doi: 10.1074/jbc.M111.320143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glisovic T., Bachorik J.L., Yong J., Dreyfuss G. RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett. 2008;582:1977–1986. doi: 10.1016/j.febslet.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golan-Mashiach M., Dazard J.E., Gerecht-Nir S., Amariglio N., Fisher T., Jacob-Hirsch J., Bielorai B., Osenberg S., Barad O., Getz G. Design principle of gene expression used by human stem cells: implication for pluripotency. FASEB J. 2005;19:147–149. doi: 10.1096/fj.04-2417fje. [DOI] [PubMed] [Google Scholar]
- Huang Y. A mirror of two faces: Lin28 as a master regulator of both miRNA and mRNA. Wiley Interdiscip. Rev. RNA. 2012;3:483–494. doi: 10.1002/wrna.1112. [DOI] [PubMed] [Google Scholar]
- Iwabuchi K.A., Yamakawa T., Sato Y., Ichisaka T., Takahashi K., Okita K., Yamanaka S. ECAT11/L1td1 is enriched in ESCs and rapidly activated during iPSC generation, but it is dispensable for the maintenance and induction of pluripotency. PLoS ONE. 2011;6:e20461. doi: 10.1371/journal.pone.0020461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jadaliha M., Lee H.J., Pakzad M., Fathi A., Jeong S.K., Cho S.Y., Baharvand H., Paik Y.K., Salekdeh G.H. Quantitative proteomic analysis of human embryonic stem cell differentiation by 8-plex iTRAQ labelling. PLoS ONE. 2012;7:e38532. doi: 10.1371/journal.pone.0038532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khazina E., Truffault V., Büttner R., Schmidt S., Coles M., Weichenrieder O. Trimeric structure and flexibility of the L1ORF1 protein in human L1 retrotransposition. Nat. Struct. Mol. Biol. 2011;18:1006–1014. doi: 10.1038/nsmb.2097. [DOI] [PubMed] [Google Scholar]
- Kwon S.C., Yi H., Eichelbaum K., Föhr S., Fischer B., You K.T., Castello A., Krijgsveld J., Hentze M.W., Kim V.N. The RNA-binding protein repertoire of embryonic stem cells. Nat. Struct. Mol. Biol. 2013;20:1122–1130. doi: 10.1038/nsmb.2638. [DOI] [PubMed] [Google Scholar]
- Lai Y.S., Chang C.W., Pawlik K.M., Zhou D., Renfrow M.B., Townes T.M. SRY (sex determining region Y)-box2 (Sox2)/poly ADP-ribose polymerase 1 (Parp1) complexes regulate pluripotency. Proc. Natl. Acad. Sci. USA. 2012;109:3772–3777. doi: 10.1073/pnas.1108595109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapeyre B., Bourbon H., Amalric F. Nucleolin, the major nucleolar protein of growing eukaryotic cells: an unusual protein structure revealed by the nucleotide sequence. Proc. Natl. Acad. Sci. USA. 1987;84:1472–1476. doi: 10.1073/pnas.84.6.1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Wang J., Hou J., Wu Z., Zhuang Y., Lu M., Zhang Y., Zhou X., Li Z., Xiao W., Zhang W. Cdk1 interplays with Oct4 to repress differentiation of embryonic stem cells into trophectoderm. FEBS Lett. 2012;586:4100–4107. doi: 10.1016/j.febslet.2012.10.030. [DOI] [PubMed] [Google Scholar]
- Liang J., Wan M., Zhang Y., Gu P., Xin H., Jung S.Y., Qin J., Wong J., Cooney A.J., Liu D., Songyang Z. Nanog and Oct4 associate with unique transcriptional repression complexes in embryonic stem cells. Nat. Cell Biol. 2008;10:731–739. doi: 10.1038/ncb1736. [DOI] [PubMed] [Google Scholar]
- Licatalosi D.D., Darnell R.B. RNA processing and its regulation: global insights into biological networks. Nat. Rev. Genet. 2010;11:75–87. doi: 10.1038/nrg2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellacheruvu D., Wright Z., Couzens A.L., Lambert J.P., St-Denis N.A., Li T., Miteva Y.V., Hauri S., Sardiu M.E., Low T.Y. The CRAPome: a contaminant repository for affinity purification-mass spectrometry data. Nat. Methods. 2013;10:730–736. doi: 10.1038/nmeth.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsui K., Tokuzawa Y., Itoh H., Segawa K., Murakami M., Takahashi K., Maruyama M., Maeda M., Yamanaka S. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell. 2003;113:631–642. doi: 10.1016/s0092-8674(03)00393-3. [DOI] [PubMed] [Google Scholar]
- Murphy R., Watkins J.L., Wente S.R. GLE2, a Saccharomyces cerevisiae homologue of the Schizosaccharomyces pombe export factor RAE1, is required for nuclear pore complex structure and function. Mol. Biol. Cell. 1996;7:1921–1937. doi: 10.1091/mbc.7.12.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Närvä E., Rahkonen N., Emani M.R., Lund R., Pursiheimo J.P., Nästi J., Autio R., Rasool O., Denessiouk K., Lähdesmäki H. RNA-binding protein L1TD1 interacts with LIN28 via RNA and is required for human embryonic stem cell self-renewal and cancer cell proliferation. Stem Cells. 2012;30:452–460. doi: 10.1002/stem.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitzsche A., Paszkowski-Rogacz M., Matarese F., Janssen-Megens E.M., Hubner N.C., Schulz H., de Vries I., Ding L., Huebner N., Mann M. RAD21 cooperates with pluripotency transcription factors in the maintenance of embryonic stem cell identity. PLoS ONE. 2011;6:e19470. doi: 10.1371/journal.pone.0019470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta S., Nishida E., Yamanaka S., Yamamoto T. Global splicing pattern reversion during somatic cell reprogramming. Cell Rep. 2013;5:357–366. doi: 10.1016/j.celrep.2013.09.016. [DOI] [PubMed] [Google Scholar]
- Pardo M., Lang B., Yu L., Prosser H., Bradley A., Babu M.M., Choudhary J. An expanded Oct4 interaction network: implications for stem cell biology, development, and disease. Cell Stem Cell. 2010;6:382–395. doi: 10.1016/j.stem.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robb G.B., Brown K.M., Khurana J., Rana T.M. Specific and potent RNAi in the nucleus of human cells. Nat. Struct. Mol. Biol. 2005;12:133–137. doi: 10.1038/nsmb886. [DOI] [PubMed] [Google Scholar]
- Sarkar P., Collier T.S., Randall S.M., Muddiman D.C., Rao B.M. The subcellular proteome of undifferentiated human embryonic stem cells. Proteomics. 2012;12:421–430. doi: 10.1002/pmic.201100507. [DOI] [PubMed] [Google Scholar]
- Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skottman H., Mikkola M., Lundin K., Olsson C., Strömberg A.M., Tuuri T., Otonkoski T., Hovatta O., Lahesmaa R. Gene expression signatures of seven individual human embryonic stem cell lines. Stem Cells. 2005;23:1343–1356. doi: 10.1634/stemcells.2004-0341. [DOI] [PubMed] [Google Scholar]
- Szklarczyk D., Franceschini A., Kuhn M., Simonovic M., Roth A., Minguez P., Doerks T., Stark M., Muller J., Bork P. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011;39:D561–D568. doi: 10.1093/nar/gkq973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Thomson J.A., Itskovitz-Eldor J., Shapiro S.S., Waknitz M.A., Swiergiel J.J., Marshall V.S., Jones J.M. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- van den Berg D.L., Snoek T., Mullin N.P., Yates A., Bezstarosti K., Demmers J., Chambers I., Poot R.A. An Oct4-centered protein interaction network in embryonic stem cells. Cell Stem Cell. 2010;6:369–381. doi: 10.1016/j.stem.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Hoof D., Passier R., Ward-Van Oostwaard D., Pinkse M.W., Heck A.J., Mummery C.L., Krijgsveld J. A quest for human and mouse embryonic stem cell-specific proteins. Mol. Cell. Proteomics. 2006;5:1261–1273. doi: 10.1074/mcp.M500405-MCP200. [DOI] [PubMed] [Google Scholar]
- Van Hoof D., Muñoz J., Braam S.R., Pinkse M.W., Linding R., Heck A.J., Mummery C.L., Krijgsveld J. Phosphorylation dynamics during early differentiation of human embryonic stem cells. Cell Stem Cell. 2009;5:214–226. doi: 10.1016/j.stem.2009.05.021. [DOI] [PubMed] [Google Scholar]
- Vilchez D., Boyer L., Morantte I., Lutz M., Merkwirth C., Joyce D., Spencer B., Page L., Masliah E., Berggren W.T. Increased proteasome activity in human embryonic stem cells is regulated by PSMD11. Nature. 2012;489:304–308. doi: 10.1038/nature11468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Rao S., Chu J., Shen X., Levasseur D.N., Theunissen T.W., Orkin S.H. A protein interaction network for pluripotency of embryonic stem cells. Nature. 2006;444:364–368. doi: 10.1038/nature05284. [DOI] [PubMed] [Google Scholar]
- Wong R.C., Ibrahim A., Fong H., Thompson N., Lock L.F., Donovan P.J. L1TD1 is a marker for undifferentiated human embryonic stem cells. PLoS ONE. 2011;6:e19355. doi: 10.1371/journal.pone.0019355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young J.C., Major A.T., Miyamoto Y., Loveland K.L., Jans D.A. Distinct effects of importin α2 and α4 on Oct3/4 localization and expression in mouse embryonic stem cells. FASEB J. 2011;25:3958–3965. doi: 10.1096/fj.10-176941. [DOI] [PubMed] [Google Scholar]
- Yu J., Vodyanik M.A., Smuga-Otto K., Antosiewicz-Bourget J., Frane J.L., Tian S., Nie J., Jonsdottir G.A., Ruotti V., Stewart R. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Workflow 1 (percentage found in other studies) and Workflow 3 (SAINT probability) for L1TD1 interactome proteins.
STRING9.0 and DAVID pathway analysis.


