Summary
Progesterone drives mammary stem and progenitor cell dynamics through paracrine mechanisms that are currently not well understood. Here, we demonstrate that CXCR4, the receptor for stromal-derived factor 1 (SDF-1; CXC12), is a crucial instructor of hormone-induced mammary stem and progenitor cell function. Progesterone elicits specific changes in the transcriptome of basal and luminal mammary epithelial populations, where CXCL12 and CXCR4 represent a putative ligand-receptor pair. In situ, CXCL12 localizes to progesterone-receptor-positive luminal cells, whereas CXCR4 is induced in both basal and luminal compartments in a progesterone-dependent manner. Pharmacological inhibition of CXCR4 signaling abrogates progesterone-directed expansion of basal (CD24+CD49fhi) and luminal (CD24+CD49flo) subsets. This is accompanied by a marked reduction in CD49b+SCA-1− luminal progenitors, their functional capacity, and lobuloalveologenesis. These findings uncover CXCL12 and CXCR4 as novel paracrine effectors of hormone signaling in the adult mammary gland, and present a new avenue for potentially targeting progenitor cell growth and malignant transformation in breast cancer.
Graphical Abstract
Highlights
-
•
Progesterone induces distinct molecular programs in mammary cell compartments
-
•
CXCR4 induction occurs in lobuloalveoli and is progesterone dependent
-
•
CXCR4 inhibition abrogates luminal progenitor expansion and mammopoiesis
-
•
Targeting of the CXCL12-CXCR4 axis may limit mammary progenitor cell transformation
In this article, Khokha and colleagues use expression profiling and functional assays to identify CXCR4, the receptor for stromal-derived factor 1 (SDF-1; CXC12), as a crucial instructor of hormone-induced mammary stem and progenitor cell function. Pharmacological inhibition of CXCR4 signaling abrogates progesterone-directed expansion of basal and luminal subsets, resulting in a marked reduction in CD49b+SCA-1− luminal progenitors and their functional capacity, and lobuloalveologenesis.
Introduction
Cumulative lifetime hormone exposure as a result of natural menstrual cycles and exogenous hormone therapies is a strong determinant of increased breast cancer risk. Progesterone, an ovarian steroid hormone that peaks during the luteal phase of the menstrual cycle, promotes the expansion of stem and progenitor cells in the mammary gland (Joshi et al., 2010, 2012). Given their multipotency, self-renewal, proliferative properties, and shared molecular signatures with specific breast cancer subtypes, mammary stem and progenitor cells have been proposed to be cellular targets of transformation in breast cancer (Visvader, 2011). Since hormones play an integral role in breast cancer development and stem cell control, an in-depth understanding of the mechanisms responsible for hormone action on distinct cell types in the mammary gland is highly warranted. Through global transcriptomic analyses of purified mammary epithelial subsets (basal, luminal, and stromal) followed by phenotypic and functional studies, we identify and validate CXCR4 as a critical mediator of progesterone signaling in the adult mammary gland.
CXCR4, together with its ligand, CXCL12, is well known for its role in regulating the migration of hematopoietic stem cells to the bone marrow, and controlling their quiescence, proliferation, and recruitment to the circulation (Honczarenko et al., 2006; Lapidot and Kollet, 2002). Further, CXCR4 and CXCL12 promote metastasis in several cancers, including breast cancer, where they are associated with tamoxifen resistance and poor prognosis (Mukherjee and Zhao, 2013). Our study reveals a previously uncharacterized nexus between CXCR4 and progesterone signaling that drives the recruitment of mammary stem and progenitor cells during epithelial expansion and lobuloalveolar regeneration.
Results and Discussion
Progesterone Induces Distinct Molecular Programs in Mammary Cell Compartments
Mammary stem cells within the basal epithelium and CD49b+SCA-1− luminal progenitors are both hormone-receptor negative (HR−) (Asselin-Labat et al., 2006; Shehata et al., 2012), and we hypothesized that these cells respond to ovarian hormones via paracrine signals provided by HR+ luminal cells. To delineate the full spectrum of hormone-induced changes in the adult mammary gland, we generated microarray expression profiles of fluorescence-activated cell sorting (FACS)-purified basal (CD24−CD49fhi), luminal (CD24+CD49flo), and stromal (CD24−CD49f−) mammary cells from mice (Figure 1A) under defined hormone treatments (progesterone [P], 17β-estradiol [E], and 17β-estradiol plus progesterone [EP]) or vehicle controls. Unsupervised hierarchical clustering revealed a strong concordance within each cellular compartment, with tighter clustering among EP-treated samples compared with E, P, and vehicle treatments (Figure 1B). Visualization of hormone effects via volcano plots and Venn diagrams indicated that there were more differentially expressed genes in luminal and basal cells in the EP samples than in either the E or P samples alone, compared with vehicle (p < 2.2 × 10−16; proportions test; Figures 1C, 1D, S1A, and S1B). These data are tabulated in Figure S1C. Notably, treatment of mice with P alone had a limited effect on gene expression (Figures 1C and S1A–S1C). This is likely because estrogen is required to drive expression of the progesterone receptor (PR) in adult mice. Venn diagrams also illustrate greater overlap between basal and luminal cells after EP treatment versus E or P alone (Figures 1D and S1A). These data suggest that a combination of E and P generates more robust cellular responses, and these changes occur primarily in the epithelial subsets rather than in the stromal fraction.
Figure 1.
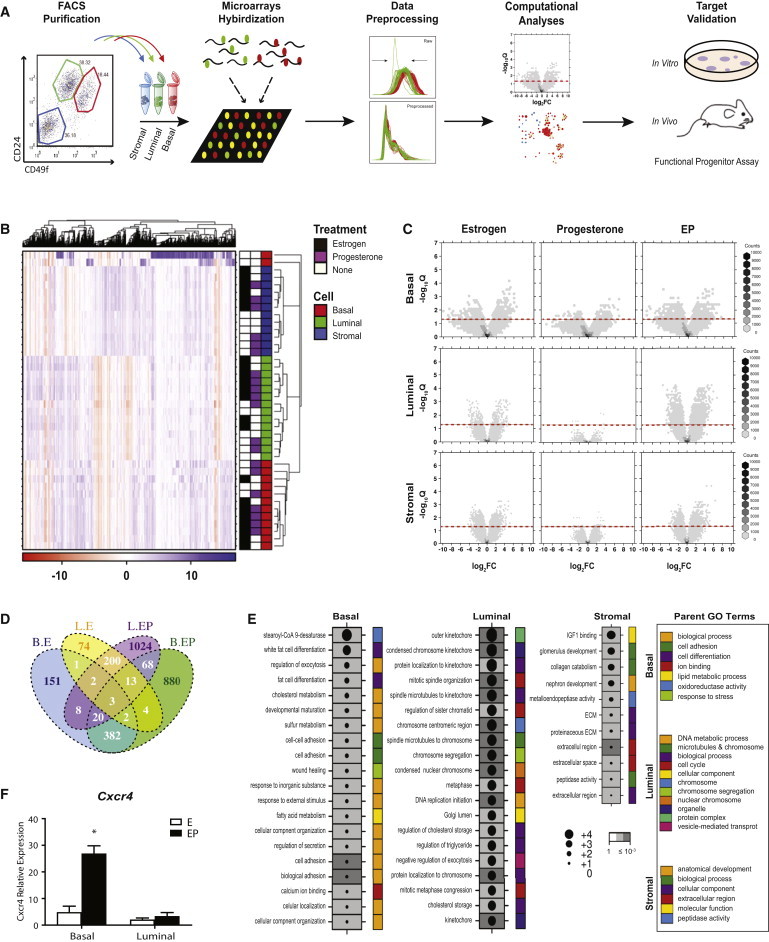
Computational Analyses of Microarray Expression Profiles of Mammary Subsets and mRNA Level Validation
(A) Schema of experimental pipeline.
(B) Heatmaps display levels of mRNA expression with a variance of >3 across all 40 samples.
(C) Volcano plots of differential mRNA abundance levels following hormone treatments in distinct cell compartments. X axis: coefficients from linear model in log2 scale; y axis: p values adjusted for multiple testing. The red dotted line depicts q value < 0.05.
(D) Venn diagram of significant genes with q value < 0.05 and |fold-change| ≥ 1.5.
(E) Dotmap of significant progesterone-responsive pathways in different cell populations as determined by GOMiner analyses. FDR values are represented by the shade of the gray boxes, and log2 enrichment scores are indicated by the size of the circle within each box.
(F) Independent qPCR validation of FACS-sorted basal and luminal samples. Mean ± SEM, EP n = 4, E n = 3 mice/group. Significance levels based on two-tailed unpaired t test, ∗p < 0.05.
See also Figure S1.
To identify progesterone-driven networks associated with mammary stem and progenitor cell expansion, we analyzed genes identified as significantly altered (q value < 0.05, |fold-change| ≥ 1.5) in specific compartments following EP treatment using GOMiner software (threshold of 10% false-discovery rate [FDR]; Figure 1E). The basal population was enriched for alterations in the cell-adhesion pathway, consistent with these cells expressing high levels of β1 (CD29) and α6 (CD49f) integrins (Shackleton et al., 2006; Stingl et al., 2006). Integrins facilitate adhesion, and β1 integrin is important for maintaining stem cell activity within specialized niches (Taddei et al., 2008). Interestingly, ovarian hormones regulate expression of integrins during mammary morphogenesis, with α5β1 integrin increasing following puberty, in early pregnancy, and in response to EP treatment (Haslam and Woodward, 2001). Luminal cells were enriched for genes involved in cell-cycle and chromosomal regulation, suggesting that EP exerts direct mitogenic effects on this population. This finding agrees with reports that progesterone elicits two waves of cellular proliferation in the mammary epithelia, with the first occurring in PR+ luminal cells (Beleut et al., 2010). In contrast, expression analyses of the stromal compartment indicated an enrichment of pathways involved in extracellular matrix (ECM) regulation in response to EP, consistent with data suggesting that progestin has the capacity to modify the ECM composition (Xie and Haslam, 1997). An examination of the top 25 common pathways altered by EP across all three cell populations suggested that more similar pathways are regulated by progesterone in basal and luminal cells compared with the stroma (Figure S1D). Gene set enrichment analysis (GSEA) was also performed and was largely consistent with the GOMiner results (Figure S1E). GSEA also showed basal cells to be enriched for metabolic and ATP synthesis pathways, luminal cells for DNA repair, and stromal cells for translation and receptor signaling pathway (Figure S1E).
CXCR4 Induction Occurs in Lobuloalveoli and Requires PR
We next interrogated our significantly altered EP gene lists from basal, luminal, and stromal compartments using an online database (Database of Ligand-Receptor Partners [DLRP]) to seek ligand and receptor pairs that are likely involved in progesterone-driven stem and progenitor cell activation (Graeber and Eisenberg, 2001). CXCL12-CXCR4 was identified as a potential pathway involved in progesterone-initiated paracrine signaling. Although the CXCL12 cytokine is known to function in stem cell homeostasis and breast cancer progression, little is known about its role in mammary epithelial cell fate and control by steroid hormone signaling. To validate changes in the CXCL12-CXCR4 axis observed in microarray profiles, we performed quantitative RT-PCR (qRT-PCR) analyses in independently sorted samples of luminal and basal epithelial populations. Consistent with global effects on gene expression, we confirmed that Cxcr4 was significantly upregulated in the basal population in response to EP (Figure 1F), but not following treatment with E or P alone (Figure S1F).
Immunofluorescence demonstrated that CXCR4 protein localized to the nucleus in both the E and EP treatment groups (Figure 2A). CXCR4 is a G-protein-coupled receptor (GPCR) that is typically found on the cell surface; however, Wang et al., (2009) showed that activated CXCR4 can translocate to the nucleus and mediate transcriptional changes. Co-staining of CXCR4 with basal (K14) and luminal (K18) cytokeratin markers confirmed its expression in both luminal and basal cells (Figure 2A). Conversely, CXCL12 staining was observed in the cytoplasm of luminal cells (Figure 2A). This staining was heterogeneous where strong CXCL12 expression overlapped with high expression of the differentiation marker K8 and with PR (Figure S2). These data indicate that CXCL12 is primarily expressed in HR+ luminal subpopulations, as opposed to the CD49b+SCA-1− luminal subset, which is known to be ER−PR− and enriched for progenitor cells.
Figure 2.
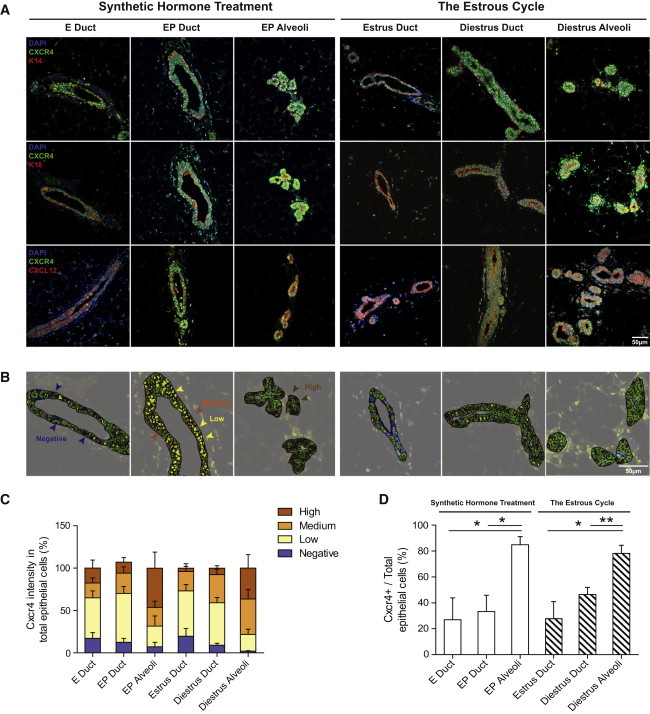
Mammary Epithelial Expression of CXCR4 and CXCL12 in Response to Hormones
(A) CXCR4 staining (green) was scored in ductal versus alveoli structures. Co-localization of CXCR4 with the basal marker K14 (red, top panels), luminal marker K18 (red, middle panels), and CXCL12 (red, bottom panels) is shown under synthetic hormone treatments and natural estrous cycle. DAPI (blue) stain indicates cell nucleus; scale bar, 50 μm.
(B) Intensity of CXCR4 staining was separated into four groups (high, medium, low, and negative) using Definiens Tissue Studio software.
(C) Bar graph shows the percentage of mammary epithelial cells that possess CXCR4 intensities in each group.
(D) Bar graph shows the percentage of CXCR4+ cells (high + medium group) in total mammary epithelia; mean ± SEM; EP n = 4, E n = 3, diestrus n = 3 and estrus n = 4 mice/group. Significance levels based on two-tailed unpaired t test; ∗p < 0.05, ∗∗p < 0.01.
See also Figure S2.
Spatial differences between CXCR4 and its ligand CXCL12 in the mammary gland suggest that CXCR4 and CXCL12 are adequately positioned for autocrine and paracrine signaling in luminal and basal cells, respectively. CXCR4 signal intensities were quantified with the use of Definiens Tissue Studio software and categorized as high, medium, low, or negative; high- and medium-intensity groups were considered to represent a positive signal (Figures 2B and 2C). Since progesterone signaling leads to the formation of sac-like alveolar structures that functionally differentiate to secrete milk during pregnancy and lactation (Fata et al., 2001; Joshi et al., 2010), ductal and alveolar structures were individually scored in EP-treated glands. Only ducts were quantified in E-treated glands, because such ducts are devoid of alveoli. Significantly more CXCR4+ cells were present in EP alveoli structures (84.9% ± 6.3%, n = 3) compared with EP (33.3% ± 12.6%, n = 4, p = 0.022) and E (26.9% ± 17.0%, n = 3, p = 0.038) ductal structures, suggesting that progesterone-induced CXCR4 acts primarily within alveolar structures (Figure 2D).
The spatial relationship of CXCR4 and CXCL12 was then examined during the estrus (progesterone-low) and diestrus (progesterone-high) stages of the murine estrous cycle. Again, nuclear staining of CXCR4 was observed in luminal and basal cells, whereas CXCL12 expression was cytoplasmic and restricted to the luminal population (Figure 2A). More CXCR4+ cells were seen in diestrus alveoli structures (78.2% ± 6.2%, n = 3) compared with diestrus (46.5% ± 5.6%, n = 3, p = 0.007) and estrus (27.8% ± 13.2%, n = 4, p = 0.016) ductal structures, demonstrating lobuloalveoli enrichment of CXCR4 in this physiological state (Figure 2D). Thus, progesterone uniquely induces CXCR4+ cells in the luminal and basal compartments of lobuloalveoli, whereas CXCL12 remains confined to luminal cells.
To determine whether CXCL12-CXCR4 signaling within the luminal compartment involves an autocrine or paracrine mode of cellular crosstalk, we used the PR-LacZ reporter mouse to determine whether CXCR4 was preferentially expressed in HR+ or HR− luminal cells. We noted that all cells that were positive for β-galactosidase were CXCR4+, although CXCR4 positivity was not limited to this HR+ subset (Figure 3A), indicating the likely presence of both autocrine and paracrine mechanisms of CXCL12 action within luminal cells. We reasoned that if progesterone signaling through PR is essential for CXCR4 expression, depletion of this GPCR would be observed in the mammary glands of PR-null mice (PRKO). Compared with mammary tissues from wild-type (WT) EP-treated mice, minimal CXCR4 staining was seen in glands from EP-treated PRKO mice, confirming the requirement of a functional PR for CXCR4 expression in the adult mammary gland (Figure 3A).
Figure 3.
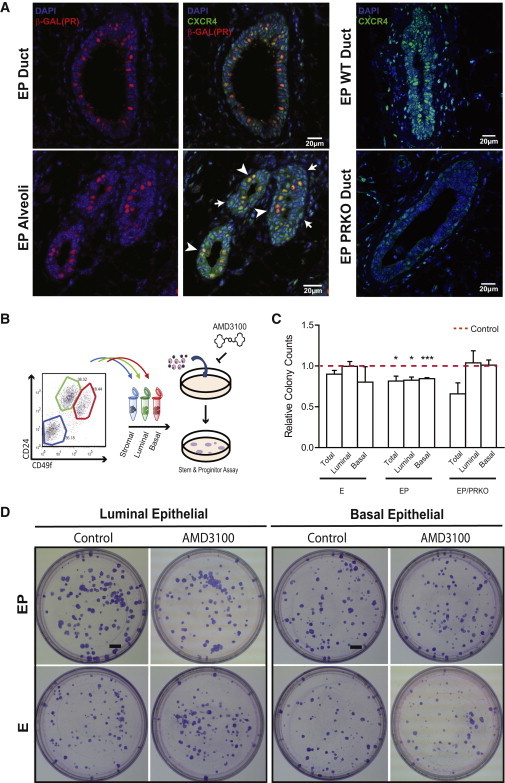
Effects of CXCR4 Inhibition on Total, Luminal, and Basal Clonogenic Capacity
(A) Left panel shows in situ staining of β-galactosidase (red) and CXCR4 (green) in PR-LacZ reporter mice. Arrowheads mark PR+ cells and arrows mark PR−CXCR4+ cells. Right panels compare CXCR4 expression in EP WT and PRKO tissue sections. DAPI (blue) indicates cell nucleus. Scale bars, 20 μm.
(B) Experimental outline for colony-forming assay.
(C) CXCR4 inhibition with AMD3100 (1 μm) of total, luminal, and basal colony-forming capacity in WT (EP n = 4, E n = 4 mice) and PRKO mice (EP n = 4 mice) compared with control. Mean ± SEM; significance levels based on two-tail paired-wise t test; ∗p < 0.05, ∗∗∗p < 0.001.
(D) Plates from luminal and basal colony-forming assays treated with vehicle or 1 μM AMD3100. Scale bar, 5 mm.
See also Figure S3.
CXCR4 Inhibition Abrogates Luminal Progenitor Expansion and Mammopoiesis
We next determined whether CXCR4 is important for mammary stem and progenitor cell function in the adult gland, using two CXCR4 inhibitors with distinct modes of action: AMD3100 and AMD3465. Previous studies found that only specific cell types in the mammary gland, termed progenitor cells, can contribute to colony formation in vitro (Shehata et al., 2012). Both CXCR4 inhibitors decreased mammary clonogenic capacity (total, luminal, and basal) by ∼20% following EP treatment, whereas the clonogenic capacity of E-treated mammary epithelial populations remained unchanged (Figures 3C, 3D, and S3). These data indicate that a proportion of progenitor cells in the luminal and basal populations that expand in response to progesterone are functionally dependent on CXCR4, which is consistent with the elevated CXCR4 expression observed in the progesterone-stimulated state (see above, Figure 2D). Application of CXCR4 inhibitors to cells derived from EP-treated PRKO mice (total, luminal, and basal) did not result in significant differences in colony-forming capacity, attesting to the importance of intact PR signaling for CXCR4-mediated effects on mammary progenitors (Figure 3C).
We then investigated whether CXCR4 signaling is essential for progesterone effects in the mammary gland in vivo. Mice were treated concurrently with EP and AMD3100 (18 mg/kg/day and 6 mg/kg/day) for 2 weeks, followed by mammary cell dissociation and phenotypic analyses of mammary epithelial subsets (Figure 4A). The effects of AMD3100 on peripheral white blood cell counts were measured as a control (Figure S4). CXCR4 inhibition resulted in a significant attenuation in the number of CD24+CD49flo luminal cells and CD24−CD49fhi basal cells (at the higher concentration) compared with PBS-treated controls (p < 0.01 and p < 0.021, respectively; Figure 4B). The luminal epithelial population was further segregated using the cell-surface markers SCA-1 and CD49b to identify HR− progenitor cells (CD49b+SCA-1−), HR+ progenitor cells (CD49b+SCA-1+), and non-clonogenic luminal cells (CD49b−SCA-1+) (Shehata et al., 2012). CXCR inhibitor treatment led to a profound reduction in the number of CD49b+SCA-1− progenitor cells and a modest yet significant decline in the number of HR+ progenitor cells (p = 0.042 and p = 0.006, respectively; Figure 4B). A profound reduction in colony-forming capacity (up to 45%) was observed in total mammary cells taken from mice treated with CXCR4 inhibitor in vivo (p < 0.05; Figure 4C). Total mammary cells were also dissociated from mice treated with vehicle control or 18 mg/kg/day AMD3100 and assayed for their ability to repopulate a cleared mammary fat pad in vivo (100 injected cells/fat pad, n = 11 mice/group). Repopulation was observed in 8/11 control mice (72.7%) compared with 4/11 AMD3100-treated mice (36.4%), indicating that inhibition of CXCR4 impedes the mammary repopulating capacity. Finally, we noted that gross mammary morphology was compromised in response to CXCR4 inhibition, with a marked reduction in side branching and lobuloalveologenesis (Figure 4D). Taken together, these data suggest that CXCR4-CXCL12 signaling is required for progesterone-mediated effects on progenitors and lobuloalveolar generation in the adult mammary gland. Phenotypically, progesterone-stimulated expansion of epithelial subsets was blocked by CXCR4 inhibition; functionally, this translated to a decrease in mammary progenitor cell numbers and mammopoiesis in the adult gland.
Figure 4.
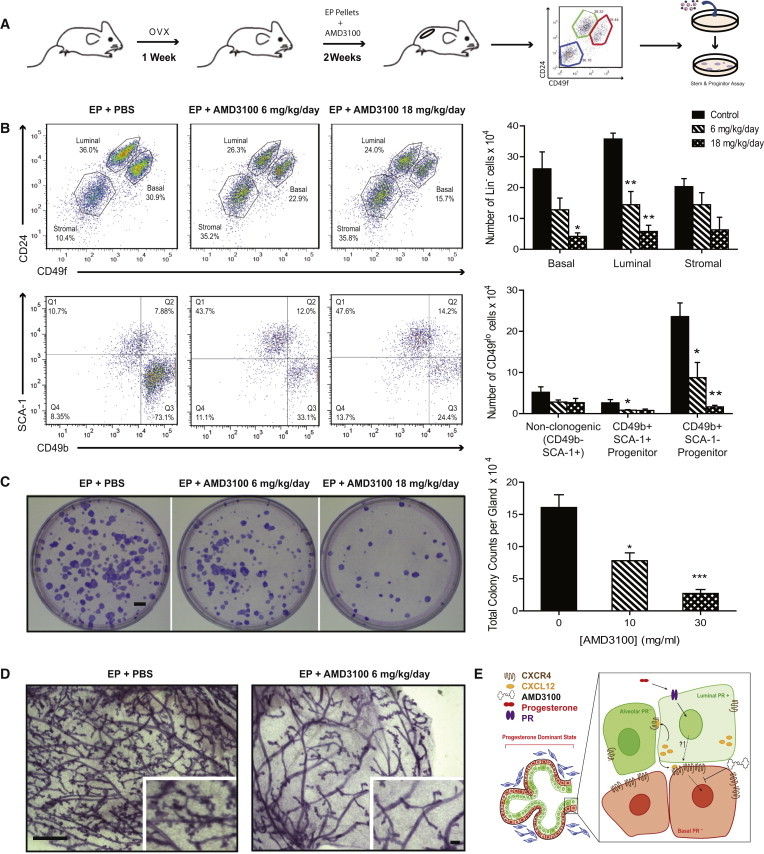
CXCR4 Signaling Inhibition Reduces Mammary Progenitors and Lobuloalveologenesis In Vivo
(A) Schema of in vivo experiments.
(B) Top panels show flow cytometry analysis of mammary subsets: luminal (CD24+CD49flo), basal (CD24−CD49fhi), and stromal (CD24−CD49f−). Bottom panel shows luminal population subdivided by the CD49b and SCA-1 cell-surface markers. Bar graph depicts absolute cell numbers for specific populations (EP + PBS, n = 3; EP + AMD3100 10 mg/ml, n = 4; EP + AMD3100 30 mg/ml, n = 3 mice). Significance levels based on two-tailed unpaired t test.
(C) Colony-forming capacity of total mammary epithelial cells from the indicated mouse groups (scale bar, 5 mm). The bar graph shows the absolute numbers of colony-forming cells/gland/treatment group. Significance levels based on one-way ANOVA. Sample sizes as in (B). All bar graphs show mean ± SEM; ∗p < 0.05, ∗∗p < 0.01.
(D) Whole mounts of the indicated mouse groups (scale bars, 100 μm and 20 μm).
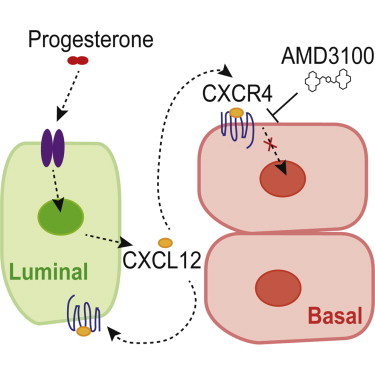
(E) Model of progesterone-mediated CXCR4-CXCL12 autocrine/paracrine effects within luminal and basal compartments.
See also Figure S4.
Conclusions
CXCL12 is a homeostatic chemokine that is integral to the homing and retention of adult hematopoietic stem cells in the bone marrow microenvironment, as well as lymphocyte trafficking (Honczarenko et al., 2006; Lapidot and Kollet, 2002). The CXCL12-CXCR4 pathway also operates in cancer metastasis, with CXCL12 providing a distal chemotactic signal for metastatic dissemination of tumor cells to tissues such as the lung, bone, and lymph node (Mukherjee and Zhao, 2013). Our study demonstrates a hitherto unknown role of the CXCL12-CXCR4 signaling pathway in hormone-driven mammary epithelial progenitor cell function. Previous work addressing the roles of CXCR4 in normal breast and breast cancer cell lines found that CXCR4 affected in vitro progenitor activity (Ablett et al., 2014). We show that CXCL12 acts in the local mammary microenvironment in vivo, with CXCL12-CXCR4 signaling occurring both within (luminal) and across (luminal to basal) mammary epithelial subsets in response to progesterone. In vivo models capture the heterogeneity of mammary epithelial cell populations while also preserving the numerous cell-cell and cell-matrix interactions that constitute the mammary stem/progenitor niche (Joshi et al., 2012). This study positions CXCL12 as a critical progesterone-stimulated effector whose signaling is enabled by PR-dependent upregulation of its cognate receptor, CXCR4, to result in the generation of progenitors that are indispensable for alveolar development. Understanding the programs required for normal adult mammopoiesis can provide insight into cellular events and signaling axes that become deregulated in breast cancer. Since hormones are known risk factors for breast cancer development, the discovery of hormone-regulated signaling processes that impinge on mammary stem and progenitor cells can potentially be harnessed to develop molecular therapies aimed at uncoupling the crosstalk between hormones and cellular precursors involved in breast cancer.
Experimental Procedures
Mice
All mouse experiments were conducted according to guidelines from the Canadian Council for Animal Care under protocols approved by the Animal Care Committee of the Princess Margaret Cancer Centre, Toronto, Canada. FVB mice were used for all experiments except those involving PRKO mice and WT littermate controls (Ismail et al., 2002). For microarray analyses, mice were ovariectomized and treated with vehicle or hormones as previously described (Joshi et al., 2010). Briefly, pellets of 0.14 mg 17β-estradiol (E) or 0.14 mg 17β-estradiol + 14 mg progesterone (EP) (Innovative Research of America) were inserted subcutaneously for 14 days. Alzet osmotic pumps (model 2002; Alza) were used to deliver AMD3100 (or PBS control) and inserted with an EP pellet in vivo according to the manufacturer’s instructions.
FACS Analyses and Colony Assays
Mammary glands were minced for 2 min followed by enzymatic digestion (Dulbecco’s modified Eagle’s medium: F12 [1:1], 750 Uml−1 collagenase/hyaluronidase, 1.5 hr at 37°C, samples vortexed briefly after 1 and 1.5 hr). Single-cell-suspension preparation and FACS analyses were performed as described by Joshi et al. (2010) and Shehata et al. (2012), using single color controls for compensation and established gating strategies based on Fc isotype-negative controls. Colony-forming assays were performed with irradiated NIH 3T3 cells according to Shehata et al., (2012) and cultured at 3% oxygen to allow basal colony growth.
Microarray Preparation and qPCR Analyses
RNA isolation, cDNA conversion, and amplification were performed as described in Joshi et al. (2010). For microarray analyses, three to four biological replicates were generated for each subset and hormone treatment. Samples were hybridized to Agilent mouse GE 8 × 60 K two-color platforms with mouse universal reference RNA. qPCR was performed as described in Joshi et al. (2010) or using SYBR Green (Quanta). Primer sequences are provided in Supplemental Experimental Procedures.
Data Preprocessing and Identification of Signature Genes
Preprocessing and statistical analyses of raw Agilent Feature Extraction data files were performed within the R statistical environment (v2.15.2) using the limma library (v3.14.3) (Smyth, 2004; Smyth and Speed, 2003) packages of the BioConductor open-source project (Gentleman et al., 2004). Control probes were filtered before background correction (normexp with offset 50) was conducted. Log ratios from Cy3 and Cy5 were loess normalized within each array, and scale normalized between arrays. After preprocessing, mRNA expression was analyzed using the univariate linear model. All model-based t tests were corrected using empirical Bayes moderation of the SE, followed by FDR adjustment for multiple testing (Storey and Tibshirani, 2003). Genes that showed significant differential expression were filtered based on q < 0.05 and |fold-change| ≥ 1.5.
Pathway and Receptor-Ligand Pair Analyses
Genes that were significantly affected by EP treatment were analyzed using GOMiner (Zeeberg et al., 2003) (build 328, database build 2011-01) and GSEA (v2.0.13) (Subramanian et al., 2005) to determine enriched pathways. GSEA results were visualized using Cytoscape (v.2.8.1) (Shannon et al., 2003). DLRP was downloaded from http://dip.doe-mbi.ucla.edu/dip/DLRP.cgi, the Database of Interacting Proteins (Graeber and Eisenberg, 2001), and human Entrez gene IDs of the receptor and ligand pairs were converted into mouse Entrez gene IDs using the HomoloGene database from NCBI (data modified 12/12/2012).
Immunofluorescence and Whole Mount
Immunofluorescence of paraformaldehyde-fixed, paraffin-embedded tissue sections was performed as described in Joshi et al. (2010). Antibodies are provided in Supplemental Experimental Procedures. Images were taken on an LSM700 confocal microscope and processed with Definiens Tissue Studio software to compare the intensities of the antibody signals.
Author Contributions
Y.S. performed bioinformatics analyses and experiments confirming functional validation of key molecules in vitro and in vivo, and wrote the manuscript. P.T. facilitated in vivo experiments, flow analyses, and drug administration, and performed sample generation from PrlacZ reporter mice. A.E.C. aided in immunofluorescence and writing of the manuscript. P.A.J. isolated cells for microarray, obtained tissue samples for independent validation, and facilitated writing of the manuscript. T.D.M. performed imaging quantification analyses. A.G.B. obtained samples for microarray analyses. H.W.J. and P.D.W. assisted with mammary repopulation assays. M.A.C. helped with microarray preprocessing. G.D.B. facilitated network analyses. J.P.L. provided PrLacZ mice. P.C.B. oversaw all bioinformatics analyses. R.K. conceptualized the study and supervised its execution and writing of the manuscript.
Acknowledgments
P.T. and P.A.J. hold fellowships from the CBCF. This work was supported by grants from the Canadian Cancer Society Research Institute (CCSRI) and Canadian Breast Cancer Foundation (CBCF). The authors thank F. Tong of the OCI FACS facility for cell sorting, S. Yousef of the UHN Animal Resources Centre for performing ovariectomies, and V. Voisin for advice on GSEA. The authors would also like to acknowledge the Spatio-Temporal Targeting and Amplification of Radiation Response (STTARR) program and its affiliated funding agencies.
Footnotes
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/).
Accession Number
The GEO accession number for the data reported in this paper is GSE59558.
Supplemental Information
References
- Ablett M.P., O’Brien C.S., Sims A.H., Farnie G., Clarke R.B. A differential role for CXCR4 in the regulation of normal versus malignant breast stem cell activity. Oncotarget. 2014;5:599–612. doi: 10.18632/oncotarget.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asselin-Labat M.-L., Shackleton M., Stingl J., Vaillant F., Forrest N.C., Eaves C.J., Visvader J.E., Lindeman G.J. Steroid hormone receptor status of mouse mammary stem cells. J. Natl. Cancer Inst. 2006;98:1011–1014. doi: 10.1093/jnci/djj267. [DOI] [PubMed] [Google Scholar]
- Beleut M., Rajaram R.D., Caikovski M., Ayyanan A., Germano D., Choi Y., Schneider P., Brisken C. Two distinct mechanisms underlie progesterone-induced proliferation in the mammary gland. Proc. Natl. Acad. Sci. USA. 2010;107:2989–2994. doi: 10.1073/pnas.0915148107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fata J.E., Chaudhary V., Khokha R. Cellular turnover in the mammary gland is correlated with systemic levels of progesterone and not 17beta-estradiol during the estrous cycle. Biol. Reprod. 2001;65:680–688. doi: 10.1095/biolreprod65.3.680. [DOI] [PubMed] [Google Scholar]
- Gentleman R.C., Carey V.J., Bates D.M., Bolstad B., Dettling M., Dudoit S., Ellis B., Gautier L., Ge Y., Gentry J. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graeber T.G., Eisenberg D. Bioinformatic identification of potential autocrine signaling loops in cancers from gene expression profiles. Nat. Genet. 2001;29:295–300. doi: 10.1038/ng755. [DOI] [PubMed] [Google Scholar]
- Haslam S.Z., Woodward T.L. Reciprocal regulation of extracellular matrix proteins and ovarian steroid activity in the mammary gland. Breast Cancer Res. 2001;3:365–372. doi: 10.1186/bcr324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honczarenko M., Le Y., Swierkowski M., Ghiran I., Glodek A.M., Silberstein L.E. Human bone marrow stromal cells express a distinct set of biologically functional chemokine receptors. Stem Cells. 2006;24:1030–1041. doi: 10.1634/stemcells.2005-0319. [DOI] [PubMed] [Google Scholar]
- Ismail P.M., Li J., DeMayo F.J., O’Malley B.W., Lydon J.P. A novel LacZ reporter mouse reveals complex regulation of the progesterone receptor promoter during mammary gland development. Mol. Endocrinol. 2002;16:2475–2489. doi: 10.1210/me.2002-0169. [DOI] [PubMed] [Google Scholar]
- Joshi P.A., Jackson H.W., Beristain A.G., Di Grappa M.A., Mote P.A., Clarke C.L., Stingl J., Waterhouse P.D., Khokha R. Progesterone induces adult mammary stem cell expansion. Nature. 2010;465:803–807. doi: 10.1038/nature09091. [DOI] [PubMed] [Google Scholar]
- Joshi P.A., Di Grappa M.A., Khokha R. Active allies: hormones, stem cells and the niche in adult mammopoiesis. Trends Endocrinol. Metab. 2012;23:299–309. doi: 10.1016/j.tem.2012.04.002. [DOI] [PubMed] [Google Scholar]
- Lapidot T., Kollet O. The essential roles of the chemokine SDF-1 and its receptor CXCR4 in human stem cell homing and repopulation of transplanted immune-deficient NOD/SCID and NOD/SCID/B2m(null) mice. Leukemia. 2002;16:1992–2003. doi: 10.1038/sj.leu.2402684. [DOI] [PubMed] [Google Scholar]
- Mukherjee D., Zhao J. The Role of chemokine receptor CXCR4 in breast cancer metastasis. Am J Cancer Res. 2013;3:46–57. [PMC free article] [PubMed] [Google Scholar]
- Shackleton M., Vaillant F., Simpson K.J., Stingl J., Smyth G.K., Asselin-Labat M.-L., Wu L., Lindeman G.J., Visvader J.E. Generation of a functional mammary gland from a single stem cell. Nature. 2006;439:84–88. doi: 10.1038/nature04372. [DOI] [PubMed] [Google Scholar]
- Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shehata M., Teschendorff A., Sharp G., Novcic N., Russell I.A., Avril S., Prater M., Eirew P., Caldas C., Watson C.J., Stingl J. Phenotypic and functional characterisation of the luminal cell hierarchy of the mammary gland. Breast Cancer Res. 2012;14:R134. doi: 10.1186/bcr3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth G.K. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004;3:e3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- Smyth G.K., Speed T. Normalization of cDNA microarray data. Methods. 2003;31:265–273. doi: 10.1016/s1046-2023(03)00155-5. [DOI] [PubMed] [Google Scholar]
- Stingl J., Eirew P., Ricketson I., Shackleton M., Vaillant F., Choi D., Li H.I., Eaves C.J. Purification and unique properties of mammary epithelial stem cells. Nature. 2006;439:993–997. doi: 10.1038/nature04496. [DOI] [PubMed] [Google Scholar]
- Storey J.D., Tibshirani R. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Mesirov J.P. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taddei I., Deugnier M.-A., Faraldo M.M., Petit V., Bouvard D., Medina D., Fässler R., Thiery J.P., Glukhova M.A. Beta1 integrin deletion from the basal compartment of the mammary epithelium affects stem cells. Nat. Cell Biol. 2008;10:716–722. doi: 10.1038/ncb1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visvader J.E. Cells of origin in cancer. Nature. 2011;469:314–322. doi: 10.1038/nature09781. [DOI] [PubMed] [Google Scholar]
- Wang L., Wang Z., Yang B., Yang Q., Wang L., Sun Y. CXCR4 nuclear localization follows binding of its ligand SDF-1 and occurs in metastatic but not primary renal cell carcinoma. Oncol. Rep. 2009;22:1333–1339. doi: 10.3892/or_00000572. [DOI] [PubMed] [Google Scholar]
- Xie J., Haslam S.Z. Extracellular matrix regulates ovarian hormone-dependent proliferation of mouse mammary epithelial cells. Endocrinology. 1997;138:2466–2473. doi: 10.1210/endo.138.6.5211. [DOI] [PubMed] [Google Scholar]
- Zeeberg B.R., Feng W., Wang G., Wang M.D., Fojo A.T., Sunshine M., Narasimhan S., Kane D.W., Reinhold W.C., Lababidi S. GoMiner: a resource for biological interpretation of genomic and proteomic data. Genome Biol. 2003;4:R28. doi: 10.1186/gb-2003-4-4-r28. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


