ABSTRACT
Sperm are highly differentiated cells characterized by their species-specific nuclear shapes and extremely condensed chromatin. Abnormal head shapes represent a form of teratozoospermia that can impair fertilization capacity. This study shows that poly(ADP-ribose) polymerase-11 (ARTD11/PARP11), a member of the ADP-ribosyltransferase (ARTD) family, is expressed preferentially in spermatids undergoing nuclear condensation and differentiation. Deletion of the Parp11 gene results in teratozoospermia and male infertility in mice due to the formation of abnormally shaped fertilization-incompetent sperm, despite normal testis weights and sperm counts. At the subcellular level, PARP11-deficient elongating spermatids reveal structural defects in the nuclear envelope and chromatin detachment associated with abnormal nuclear shaping, suggesting functional relevance of PARP11 for nuclear envelope stability and nuclear reorganization during spermiogenesis. In vitro, PARP11 exhibits mono(ADP-ribosyl)ation activity with the ability to ADP-ribosylate itself. In transfected somatic cells, PARP11 colocalizes with nuclear pore components, such as NUP153. Amino acids Y77, Q86, and R95 in the N-terminal WWE domain, as well as presence of the catalytic domain, are essential for colocalization of PARP11 with the nuclear envelope, but catalytic activity of the protein is not required for colocalization with NUP153. This study demonstrates that PARP11 is a novel enzyme important for proper sperm head shaping and identifies it as a potential factor involved in idiopathic mammalian teratozoospermia.
Keywords: acrosome, ADP-ribose, ARTD11, condensation, gametogenesis, infertility, male infertility, MAR, mono(ADP-ribosyl)ation, mouse, nuclear envelope, PAR, PARG, PARP, PARP11, poly(ADP-ribosyl)ation, sperm, sperm differentiation, spermatid, spermatogenesis, spermiogenesis, teratozoospermia, testis, transgenic/knockout model, WWE domain
INTRODUCTION
Spermatogenesis is the developmental process required to produce male gametes (spermatozoa). Following the meiotic cell division that reduces the genome to a haploid state, male germ cells, now called spermatids, undergo an extensive differentiation process called spermiogenesis that entails dramatic morphological changes—for example, formation of the flagellum and acrosome and, in mammals, packaging of the sperm genome into the highest condensed chromatin state known [1, 2]. The drastic size reduction of the spermatid nucleus requires extensive chromatin remodeling, which is accompanied by a transition from nucleosome- to protamine-based chromatin in mammals [3, 4]. In addition, during the elongation and condensation steps of spermatid differentiation in mid- to late-phase spermiogenesis, the nuclei adopt their final, species-specific shape. Proper shaping of the nucleus (and therefore the entire head, because spermatids shed most of their cytoplasm) is essential for sperm function and fertilization. Any significant deviation from the normal species-specific morphology (e.g., in teratozoospermic sperm) is associated with poor fertilization rates [5]. Spermatid nuclear elongation, which represents an essential part of the nuclear shaping process, is facilitated by well-orchestrated physical forces exerted by specialized cytoskeletal components within each spermatid, such as the microtubule manchette shaping the posterior nuclear end and the spermatid acroplaxome [1]. In addition, the adjacent Sertoli cells form F-actin hoops that are also involved in shaping the anterior part of the sperm nucleus [1, 6]. Molecular details of the unique process of spermatid nuclear shaping are not completely understood, but studies of mouse mutants with sperm head anomalies have demonstrated the importance of certain components of the Golgi apparatus giving rise to the acrosome as well as correct manchette formation [7–10].
Enzymes of the poly(ADP-ribose) polymerase (PARP) family cleave nicotinamide adenine dinucleotide (NAD+) into nicotinamide and ADP-ribose and then transfer the ADP-ribose unit onto specific amino acid side chains of target proteins as a posttranslational modification [11]. The enzyme family comprises both mono(ADP-ribosyl) transferases and “true” PARPs. Mono(ADP-ribosyl) transferases typically transfer only single ADP-ribose units to target proteins, whereas PARPs form and transfer ADP-ribose polymers of varying length (poly[ADP-ribose]). To date, at least 16 putative members of the ADP-ribosyltransferase family have been reported [11, 12]. Although most of those enzymes have historically been classified as PARPs or PARP-like proteins based on structural homology of their catalytic domains, true PARP activity could only be demonstrated for PARP members 1, 2, 4, 5, and 7. All others appear to be either mono(ADP-ribose) transferases [13], such as PARP10 and PARP14 [14] or PARP3 [15], or have no catalytic activity at all even though catalytic domains exist, like in PARP9 [16]. To more accurately reflect common enzymatic properties, it has been proposed to rename the PARP family of enzymes to ADP-ribose transferases, diphtheria (toxin-like) (ARTD), so that PARP11 should consequently be renamed to ARTD11 [11].
Both PARP1, the archetypal enzyme of the family, and PARP2 are well known to be involved in the regulation of DNA repair, chromatin dynamics, and many other cellular processes [17–21]. In contrast, the specific functions of most other PARPs/ARTDs have not yet been elucidated in such detail or, as in the case of PARP11, have so far remained completely unknown. Similar to other forms of posttranslational modifications, poly(ADP-ribosyl)ation alters protein-protein interactions, enzymatic activity, or other biochemical properties of the modified proteins. It has been shown that poly(ADP-ribosyl)ation affects various cellular processes, such as DNA repair and apoptosis, telomere homeostasis, mitotic segregation, cell-cycle regulation, cell differentiation, and epigenetic modifications [20, 22, 23]. However, much less is known about functions of physiological mono(ADP-ribosyl)ation in eukaryotic cells [24–27].
PARP11 is characterized by the presence of a globular WWE domain, which is named after the consensus signature containing two tryptophan residues (W) and a glutamate (E) residue [28]. This globular domain is also found in a number of other PARP proteins (PARP7/tiPARP, PARP12, PARP13/ZAP, and PARP14/BAL2), where it is accompanied by additional distinct domains. Structural analyses indicate that the WWE domain in PARP11 is capable of binding to a terminal ADP-ribose unit (i.e., a mono-ADP-ribosylated protein or the terminal moiety of a poly[ADP-ribose] chain) [29]. WWE domains have also been described in ubiquitin-conjugating enzymes, such as the ubiquitin E3 ligases DELTEX and TRIP12. It has been suggested that WWE domains function in specific protein-protein interactions and could thus help target the different kinds of E3-like and ADP-ribosylation activities to acceptor proteins participating in various signaling cascades [28].
In the present study, we analyzed overexpressed PARP11 protein in cultured cells and the phenotype of Parp11 knockout mice to characterize PARP11 functions in vivo. We demonstrate that PARP11 is a mono(ADP-ribosyl) transferase that localizes to the nuclear membrane in transfected cells. Parp11 is preferentially expressed in spermatids during nuclear condensation and differentiation in wild-type animals, and lack of Parp11 in knockout animals causes teratozoospermia with nuclear membrane abnormalities and results in male infertility.
MATERIALS AND METHODS
All chemicals used in the present study were purchased from Sigma except where noted otherwise.
Sequence Comparison Analysis
Protein sequences (Homo sapiens [NP_065100.2], Mus musculus [NP_852067.1], Xenopus laevis [NP_001086816.1], and Gallus gallus [P_416489.2]) were obtained from the National Center for Biotechnology Information (NCBI) database and were analyzed using the protein alignment program PRALINE available online (http://www.ibi.vu.nl/programs/pralinewww/).
ADP-Ribosylation Assay
Purified recombinant PARP11 protein was purchased (United States Biological) as an N-terminal GST fusion protein purified from baculovirus-infected SF9 cells. Per the manufacturer, the specific activity is 23 U/mg, where one unit is defined to incorporate 100 pmol of ADP-ribose from NAD+ into acid-insoluble form in 1 min. In our assays, a similar reaction buffer was used (50 mM Tris/HCl [pH 8.0], 5 mM MgCl2, and 2 μM ZnCl2) in 10-μl total reaction volume. Assays contained 10 ng of PARP11, and the final concentration of NAD+ was 50 μM, containing 0.925 μM [32P]NAD+. As a positive control, 2 ng of a PARP1 (E988K) mutant with severely reduced poly(ADP-ribosyl)ating activity were utilized [30, 31]. Proteins were separated by Standard SDS-PAGE with gradient gels (4%–15%; Bio-Rad), transferred onto Hybond-N+ membranes (GE Healthcare), and dried for autoradiography.
Cloning of Human Parp11 cDNA, Plasmid Construction, and Point Mutant Generation
The cDNA encoding human Parp11 was generated by full-length 5′ rapid amplification of cDNA ends with the Smart cDNA Synthesis Kit (Clontech) from testis total RNA (Stratagene), cloned into pcDNA3.1 TOPO (Invitrogen), sequenced, and their identity confirmed by comparison to NCBI reference sequences, including the reference sequence NM_020367.4 (human). Standard recombinant DNA techniques [32] were used to generate an in-frame fusion of the full-length Parp11 cDNA to the enhanced green fluorescent protein (EGFP) sequence in plasmid pEGFP-C1 (Invitrogen). The EGFP-Parp11 cDNA was excised from the resulting pEGFP-C1-Parp11 and inserted into pcDNA3.1, resulting in pcD-E-Parp11, which was used for expression of EGFP-PARP11 fusion proteins in eukaryotic cells.
For mutational analysis, targeted mutation of regions of interest were introduced in the Parp11 cDNA of plasmid pcD-E-Parp11 with the QuikChange Site-Directed Mutagenesis Kit (Agilent) according to manufacturer's instructions with the following primer pairs: Parp11_R95A_muta forward, GCT CTT TTT ATT AAG GCC TGC TTT CCA GTG GTG AG; Parp11_R95A_muta reverse, CTC ACC ACT GGA AAG CAG GCC TTA ATA AAA AGA GC; Parp11_Y77A muta forward, CTA CTT CCA AAT TCA GCG CTA AGA TAG ACT TTG C; Parp11_Y77A_muta reverse, GCA AAG TCT ATC TTA GCG CTG AAT TTG GAA GTA G; Parp11_Q86A_muta forward, GAC TTT GCA GAA ATG AAG GCA ATG AAT CTC AC; Parp11_Q86A_muta reverse, GTG AGA TTC ATT GCC TTC ATT TCT GCA AAG TC; Parp11_F41A_muta forward, GAA GTG GCA CAT GGC TCA GCC GGA TAC CAA C; Parp11_F41A_muta reverse, GTT GGT ATC CGG CTG AGC CAT GTG CCA CTT C; Parp11_Y31A_muta forward, CAG TGG GGC TGG TTT GCC TTG GCA GAA TGT G: Parp11_Y31A_muta reverse, CAC ATT CTG CCA AGG CAA ACC AGC CCC ACT G; Parp11_G198W_muta forward, ATG AAC AAA TGC TGT TTC ATT GGA CCA GCA GTG AA; Parp11_G198W_muta reverse, TTC ACT GCT GGT CCA ATG AAA CAG CAT TTG TTC AT.
Plasmids expressing only the WWE domain or the catalytic domain of PARP11 fused to EGFP were generated by targeted PCR deletions as follows: To retain the WWE domain, only the sequences coding for the first 106 amino acids were maintained in frame with EGFP, and the rest of the Parp11 cDNA was replaced by a stop codon (pcD-E-WWE). To selectively keep only the PARP11 catalytic domain in the fusion protein, the sequence coding for the first 117 amino acids was deleted, and the sequence coding for the catalytic domain was fused in frame to the EGFP cDNA (pcD-E-CD).
All plasmids were sequence verified, and expression of fusion proteins of the expected sizes was verified detecting their EGFP tags in Western blot analyses of lysates from transfected cells.
Cell Culture and Transfections
Human cervix carcinoma cells (HeLa) and human embryonic kidney cells (HEK293; both from ATCC) were cultured under standard cell culture conditions (37°C, 5% CO2) in Dulbecco modified Eagle medium supplemented with 1% GlutaMAX, 10% fetal bovine serum (HyClone), and 1% Pen/Strep (Invitrogen). For transfection experiments, cells were seeded at a density of 1 × 105 cells/well into 35-mm cell culture dishes or six-well plates. After 18 h, cells were transfected using TransIT-LT1 Transfection Reagent (Mirus Bio) according to the manufacturer's instructions.
Antibodies
Primary antibodies against the following epitopes were used in the different experiments: mouse anti-tubulin-alpha (1:400; clone DM1a; Sigma), mouse anti-GFP (1:200; clone J18; Clontech), rabbit anti-GFP (1:200; Clontech), and mouse anti-nuclear pore protein 153 (NUP153; 1:200; clone QE5; Abcam). The following secondary antibodies (all from Jackson ImmunoResearch) were used: donkey anti-mouse AF488 (1:400), donkey anti-rabbit AF488 (1:400), donkey anti-mouse CY3 (1:400), and donkey anti-mouse CY5 (1:100). The acrosome was labeled with Alexa Fluor 488-conjugated peanut agglutinin (PNA-A488; Invitrogen).
Immunocytochemistry and Microscopy
Immunofluorescent staining of transfected cells and testis sections was performed as described previously [33–37]. Nuclei were visualized by staining with 4′,6-diamidine-2-phenylindole (Sigma). Sperm smears on glass slides were stained with hematoxylin and eosin. Microscopy was performed on a Leitz Ortholux fluorescent microscope (63×, 1.4 NA oil objective) equipped with a MagnaFire digital color camera (Model S99808; Optronics) and MicroFire software (Optronics, Inc.) and on a Zeiss Axioscope A1 with an AxioCam MR digital camera and AxioVision software (Zeiss). Confocal microscopy was performed on an Leica TCS SP5 as described previously [38]. To measure flagellum length of sperm, sperm smear preparations were stained with hematoxylin and eosin and imaged using a Zeiss Axioscope A1. Flagellum lengths were determined using the measurement function of the AxioVision software; statistical analysis of resulting values was performed using Mann-Whitney test in Prism 5.04 (GraphPad Software).
CMA3 staining of sperm chromatin condensation was performed as described previously [36, 38–40].
Generation of Parp11 Knockout Animals and Breeding
All animals were maintained according to the guidelines of the Institutional Animal Care and Use Committees of the University of Pennsylvania and of Utah State University. Mice with deletion of the mouse Parp11 gene were generated using frozen Parp11tm1(KOMP)Vlcg sperm (Project ID VG12032) obtained from the KOMP repository of the Knockout Mouse Project (www.komp.org). In these animals, the protein coding exons 4–8 was replaced by vector Zen-UB1, which abolished Parp11 gene expression and instead permitted expression of the reporter gene beta-galactosidase under the control of the endogenous Parp11 promoter (see Supplemental Fig. S3; Supplemental Data are available online at www.biolreprod.org). The vector utilizes the promoter of the human ubiquitin C gene to mediate expression of the neomycin phosphotransferase gene, permitting selective resistance to neomycin selection. Sperm were microinjected into oocytes from C57BL/6J female mice (Jackson Laboratories) and transferred into pseudopregnant ICR females using standard embryological techniques [41]. Offspring were genotyped, and heterozygous offspring were bred to generate homozygous Parp11−/− animals.
Quantitative PCR Analyses
Both RT-PCR and quantitative PCR were performed on total RNA from several tissues from Parp11−/− animals (Parp11tm1(KOMP)Vlcg) and from Parp11+/+ sibling control animals using QIAshredder spin column for tissue homogenization and RNeasy Mini Kit for total RNA isolation following the manufacturer's directions (both from Qiagen). The cDNA synthesis was carried out using the QuantiTect Reverse Transcription Kit (Qiagen), followed by quantitative real-time PCR with primers specific for Parp11 (Mm00619375_m1) and eEF-2 (Mm00833287_g1) or Gapdh (Mm99999915_g1) as endogenous controls (Applied Biosystems) in a reaction mixture including TaqMan Gene Expression Master Mix (Applied Biosystems) in MicroAmp Optical 96-Well Reaction Plates with barcode on an ABI PRISM 7000 cycler at default setting (1st cycle: 50°C for 2 min, 95°C, 10 min; subsequent 40 cycles: 95°C, 15 sec; 40 cycles, 60°C, 1 min). Expression levels of Parp11 were calculated using the comparative threshold cycle (Ct) method (ΔΔCt), using the formula FC = 2ΔΔCt, where FC is fold-change and ΔCt is the difference between the Ct values of the Parp11 gene and the internal control eEF-2. To compare expression levels between different tissues, testicular Parp11 gene expression was set to 100%, and relative expression in other tissues were calculated.
Assessment of Fertility and Embryo Development
To determine fertility of the animals, males of each genotype were mated with two females. Number of litters and number of pups born were recorded over the total breeding time (2–5 m), and as a measure of fertility of each male, the number of offspring produced per female and 30 days of breeding time was calculated.
Mean values for a minimum of five breeding cages per genotype were plotted. Statistical analysis using ANOVA and Mann-Whitney test was performed with Prism 5.04. To determine if the observed fertility defect was caused by inability of the sperm to fertilize or by early embryonic lethality, female mice (B6/C3 F1 hybrid females) were superovulated as described by Nagy [41] and naturally mated to either Parp11+/+ or to Parp11−/− males. At 1.5 days postcoitus, females were killed, embryos/oocytes flushed from the oviducts, and development of embryos determined microscopically by the progression to the 2-cell stage. Significance of the cleavage efficiency was calculated using Fisher exact test in Prism 5.04.
Beta-Galactosidase Staining of Testes
Isolated testes were fixed in 4% paraformaldehyde in PBS on ice for 1 h, washed in rinse buffer (0.2 M sodium phosphate [pH 7.3], 2 mM MgCl2, 0.02% NP40, and 0.01% sodium deoxycholate) three times for 30 min per wash, and incubated at 37°C in staining solution (5 mM K3Fe(CN)6, 5 mM K4Fe(CN)6, and 1 mg/ml of X-Gal). A postfixation step with 10% formalin in PBS for 1 h was performed. Stained testes were embedded in paraffin, cut into 8-μm sections, dewaxed, and counterstained with Nuclear Fast Red.
Histology and Electron Microscopy
Standard histological analysis was performed as described previously [36, 42]. For transmission-electron microscopy (TEM), testis and sperm samples of Parp11−/− and wild-type controls animals were fixed in 2% paraformaldehyde and 2% glutaraldehyde in deoxycholate buffer overnight. Sample preparation and processing was performed by the Electron Microscopy Resource Laboratory of the University of Pennsylvania. TEM imaging was performed on a JEM-1010 transmission-electron microscope (JEOL Ltd.).
RESULTS
PARP11 Is a Mono(ADP-Ribosyl) Transferase that Localizes to the Nuclear Envelope
PARP11 is one of the smallest members of the unique PARP protein family [12]. PARP11 comprises two distinct protein domains, the N-terminal WWE domain (amino acids 1–105) and a C-terminal catalytic PARP domain (amino acids 116–331) (Fig. 1A). The amino acid sequences of PARP11 homologs in human, mouse, chicken, and X. laevis show a very high level of conservation, with 78% sequence identity between vertebrate species (Supplemental Fig. S1). Human PARP11 shares 95% overall identity with mouse PARP11 protein and with all mammalian species analyzed here, with 100% conservation in the WWE domain (mouse amino acids 26–107) and PARP catalytic domain (mouse amino acids 194–330) (Supplemental Fig. S1) and with very few, conservative exchanges in the remaining regions of the protein.
FIG. 1.
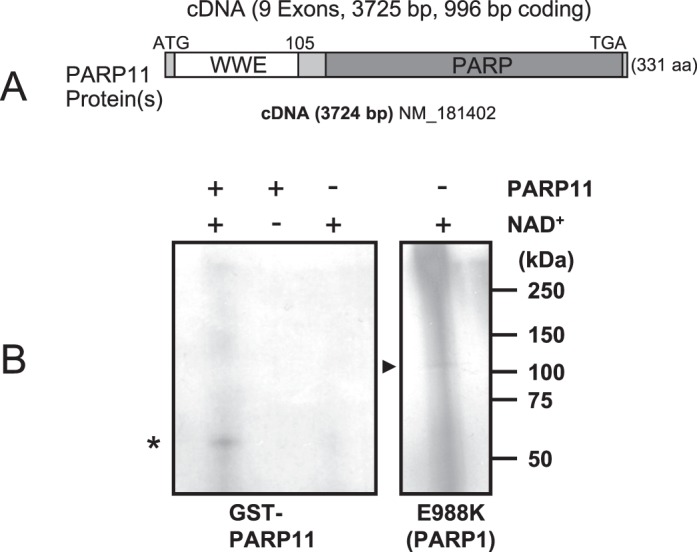
PARP11 is a mono(ADP-ribose) transferase. A) PARP11 protein contains two distinct domains, the WWE domain and the PARP domain. B) Automodification of PARP11 with monomeric ADP-ribose in the presence (+) or absence (−) of [32P]NAD+ as indicated by the presence of a defined band of the expected size of GST-PARP11 (*) and absence of an upward “smear” that would have indicated automodification with poly(ADP-ribose). As a positive control, an E988K mutant of the related protein PARP1 with known mono(ADP-ribosyl) transferase activity and low levels of poly(ADP-ribose) synthesis was used. It shows an equally sharp band of monomeric ADP-ribose (arrowhead) at the expected size of approximately 113 kDa (right), along with the typical upward smear indicating some residual poly(ADP-ribosyl)ation activity of the mutant. Note that overall acid-precipitable radioactivity was lower in PARP11, indicating a much lower specific activity of PARP11 compared to the PARP1 E988K mutant control.
To determine enzymatic activity of PARP11, ADP-ribosylation assays were performed in vitro using standard experimental conditions [43]. PARP11 protein showed mono(ADP-ribosyl)ation activity in an automodification reaction, albeit moderate, as indicated by a single signal band that was obtained (signal in Fig. 1B, left, asterisk). A similar single band was also detected in a parallel control reaction using a mutated PARP1 enzyme (E988K), which retains mainly mono- and not poly(ADP-ribosyl)ation activity (signal in Fig. 1B, right, arrowhead). However, in addition to the mono(ADP-ribosyl)ated PARP1 signal, significant levels of signals were also found upward of the single band of approximately 113 kDa, which appears to be due to the low level of residual poly(ADP-ribosyl)ation still found in this mutant. In summary, our results suggest that PARP11 does not possess PARP activity under the experimental conditions used but rather is a mono(ADP-ribosyl) transferase, which is in line with results obtained in a recent study focusing on PARP family-wide enzymatic activities [13, 14].
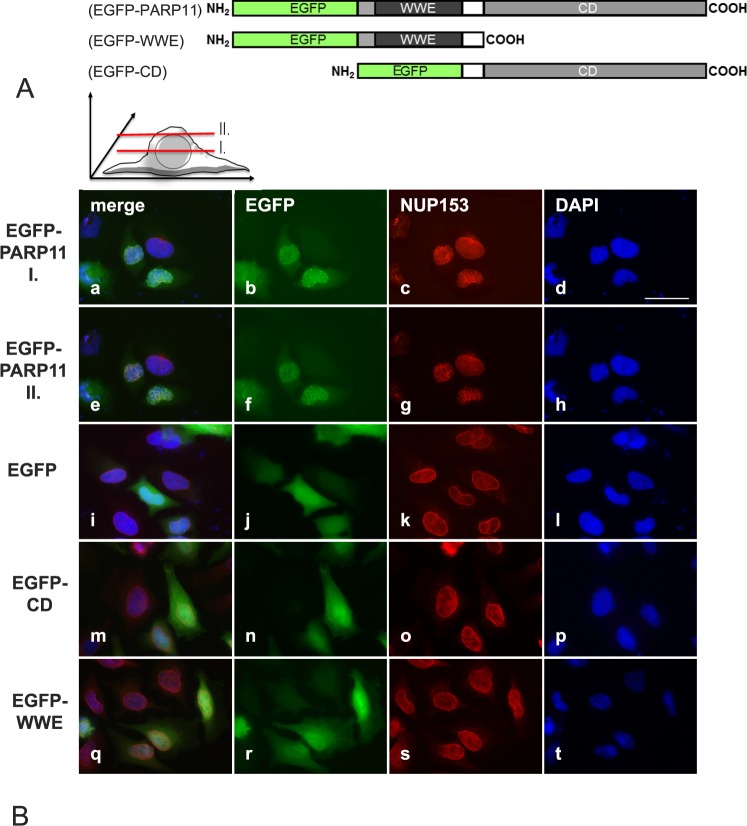
Utilizing transfected cultured cells, Parp11 expressing and knockout tissues, we were not able to identify any antibodies that recognize PARP11 in immunohistochemistry or in immunoblots. To study the subcellular localization of PARP11, with the aim to identify the functional relevance of PARP11's individual domains, we therefore generated eukaryotic vectors for expression of EGFP-tagged PARP11 proteins (full-length wild-type PARP11, the WWE domain alone, or the catalytic PARP domain by itself fused to EGFP) (Fig. 2A).
FIG. 2.
Overexpressed PARP11 preferentially localizes to the nuclear envelope in HeLa cells. A) PARP11-EGFP fusion protein structure. Eukaryotic expression vectors were generated that express fusion proteins consisting of either EGFP and the full-length PARP11 protein, EGFP fused to the PARP11 WWE domain only, or EGFP fused to the PARP domain only. B) Subcellular localization of the various EGFP-fusion proteins in transiently transfected HeLa cells. EGFP-PARP11 fusion were visualized with an EGFP-specific antibody (pseudo-colored green; b, f, j, n, and r). To localize the nuclear envelope, NUP153 was visualized by indirect immunofluorescence detections (pseudo-colored red; c, g, k, o, and s). DNA was counterstained with 4′,6-diamidine-2-phenylindole to visualize the nuclear area (pseudo-colored blue; d, h, l, p, and t). Merged images are shown in a, e, i, m, and q. EGFP-PARP11 fusion proteins comprising the full-length PARP11 protein preferentially localized to the nuclear envelope and colocalized with NUP153, a nuclear pore complex component. Images shown in rows EGFP-PARP11 I (a–d) and II (e–h) show the same cells imaged at slightly different focal planes, as illustrated in the schematic above images in B, to demonstrate colocalization of PARP11 with NUP153. Yellowish combined signals in the merged image (e) show colocalization. Images i–l reflect the signals observed for EGFP protein alone and show that the reporter EGFP alone did not exhibit any particular subcellular localization preference. Deletion of either the PARP11 WWE domain (EGFP-CD; m–p) or deletion of the catalytic domain (EGFP-WWE; q–t) completely abolished nuclear pore localization of PARP11. Bar = 40 μm.
Indirect immunofluorescence analysis of HeLa and HEK293 cells transfected with these PARP11 expression vectors revealed a striking localization of EGFP-tagged wild-type PARP11 at the nuclear envelope together with very weak, diffuse cytoplasmic and intranuclear localization of the protein (Fig. 2B, a, b, e, f). More specifically, EGFP-PARP11 colocalized with NUP153 at nuclear pores (Fig. 2B, c, g, and overlays in a and e). Confocal microscopy of transiently transfected HEK293 cells expressing EGFP-tagged PARP11 confirmed the colocalization of PARP11 with NUP153 (Supplemental Fig. S2). EGFP protein by itself did not show any particular subcellular localization preference (Fig. 2B, i–l).
Subcellular localization preferences of individual proteins are generally mediated by distinct protein domains or specific targeting peptides and posttranslational protein modifications, such as phosphorylation, sumoylation, glycosylation, ubiquitination, and ADP-ribosylation (for review, see [44–46]). The PARP11 protein comprises two distinct domains, the WWE domain and the catalytic PARP domain. Surprisingly, neither fusion protein of PARP11's WWE domain or the catalytic domain by themselves with EGFP was targeted to the nuclear envelope. Whereas the catalytic domain fusion protein appeared to be evenly distributed throughout the cell (Fig. 2B, m–p), the WWE containing fusion protein did, in addition to being cytoplasmic, exhibit a slight preference for nuclear localization (Fig. 2B, q–t). These results suggest that both domains must be present in PARP11 to allow its localization at nuclear pores.
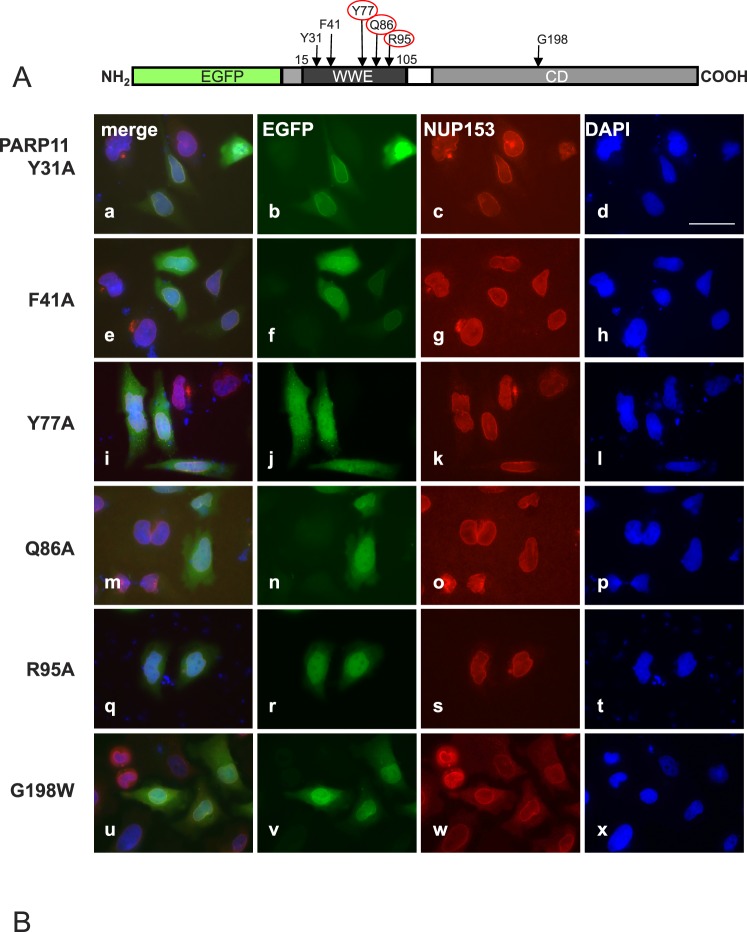
Recent studies demonstrated that the WWE domain in PARP11 is capable of binding ADP-ribose, and five amino acids within the WWE domain were identified as essential for this physical interaction in vitro [29]. We introduced point mutations to change all of these amino acid residues to alanine (A) in PARP11 to investigate a possible involvement of ADP-ribose binding in the subcellular localization of PARP11 (Fig. 3A). Substituting tyrosine in position 31 (Y31A) and phenylalanine in position 41 (F41A) did not visibly influence the subcellular localization observed for the mutant EGFP-PARP11 protein, and subcellular localization was indistinguishable from wild-type EGFP-PARP11 fusion protein (Fig. 3B, a–d and e–h). Replacing the tyrosine residue in position 77 (Y77A), glutamine in position 86 (Q86A), and arginine in position 95 (R95A) each abolished nuclear membrane colocalization (Fig. 3B, i–t), and subcellular distribution of the mutant proteins changed to a pattern similar to that observed for EGFP alone. These data indicate that binding of PARP11 to ADP-ribose units (e.g., as a modification of a target protein), produced by either PARP11 itself or a different PARP enzyme, may be involved in PARP11 binding to the nuclear pore.
FIG. 3.
PARP11 nuclear envelope localization requires WWE domain amino acids (Y77, Q86, and R95) known to participate in ADP-ribose binding. A) Amino acids Y31, F41, Y77, Q86, and R95 in the WWE domain of PARP11 are involved in the binding to ADP-ribose [29]. To investigate the relevance of those amino acids for PARP11 subcellular localization, they were replaced with alanine (A) in expression vectors encoding the EGFP-PARP11 fusion protein. Positions of the particular amino acid exchanges within the EGFP-PARP11 protein are indicated by arrows. B) HeLa cells were transiently transfected to express the EGFP-PARP11 variants indicated on the left. Colocalization of mutant proteins with the nuclear envelope was evaluated by costaining with NUP153. EGFP fluorescence is visualized in green (b, f, j, n, r, and v) and NUP153 in red (c, g, k, o, s, and w). DNA was counterstained with 4′,6-diamidine-2-phenylindole (blue; d, h, l, p, t, and x). Merged images are shown in a, e, i, m, q, and u. Mutation of amino acids Y31 (a–d) and F41 (e–h) to A did not affect the observed subcellular localization pattern of the fluorescence signals compared to the original PARP11 fusion protein (see Fig. 2). In contrast, amino acid exchanges Y77A (i–l), Q86A (m–p), and R95A (q–t) ablated colocalization with NUP153 in the nuclear envelope and instead led to a fluorescence pattern comparable to that observed for EGFP only (see Fig. 2), indicating that presence of those residues (and likely ADP-ribose binding) is important for the association of PARP11 with the nuclear envelope. Mutation of an amino acid likely important for enzymatic activity (G198W; u–x) did not change EGFP-PARP11 subcellular localization. Bar = 40 μm.
To determine whether the enzymatic activity of PARP11 is necessary for colocalization with NUP153, an enzymatically inactive mutant of PARP11 with a point mutation in the catalytic center (G198W) was generated by site-directed mutagenesis. This amino acid was selected due to its important position in the catalytic domain and its analogy to the G888W mutation that renders PARP10 catalytically inactive [14]. Surprisingly, subcellular localization of the resulting mutant PARP11 protein was similar to that of the original PARP11 protein. This was unexpected, because in the absence of the catalytic domain, the residual PARP11 protein was unable to localize to the nuclear envelope, which we initially interpreted as merely due to the absence of PARP11 catalytic activity (Fig. 2B, q–t).
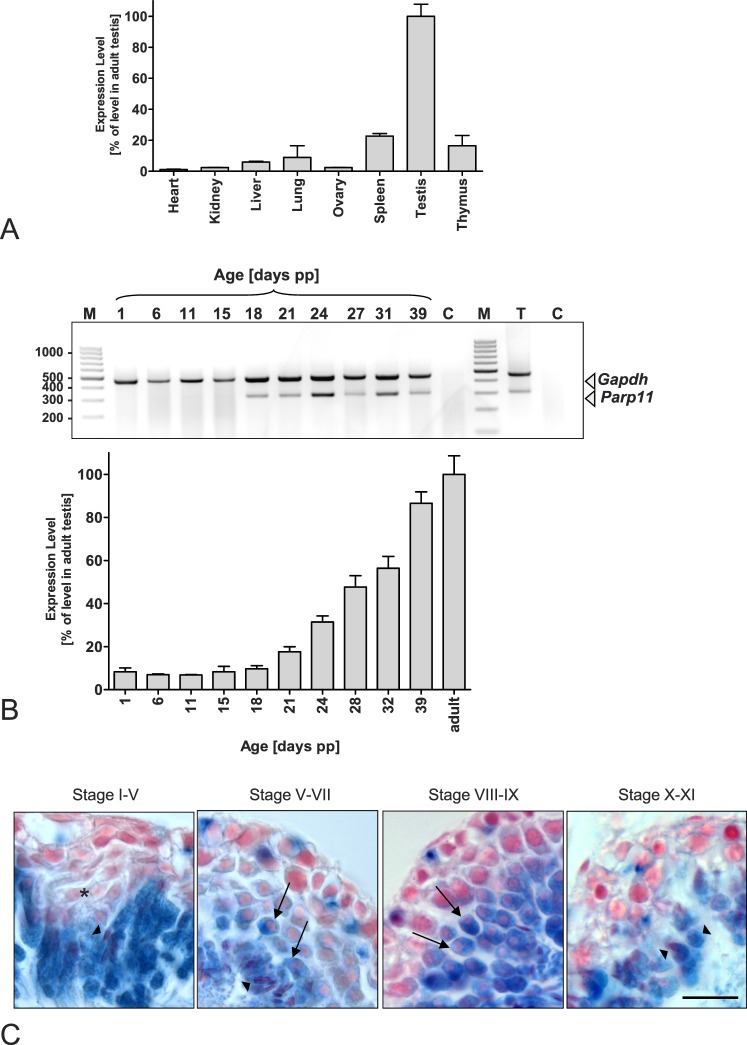
Parp11 Is Predominantly Expressed in Adult Testicular Tissues
Parp11 mRNA levels were analyzed in various tissues obtained from wild-type adult mice using quantitative and RT-PCR. Parp11 mRNA levels were highest in testicular tissue, but low-level expression was detectable also in other tissues, such as liver, lung, spleen, thymus, and brain (Fig. 4A and Supplemental Fig. S3C). To identify the exact time point when Parp11 expression is initiated during spermatogenesis, testes were collected from pubertal wild-type animals undergoing the first wave of spermatogenesis. Parp11 mRNA levels sharply increased at 18–21 days of age (Fig. 4B), which is consistent with the appearance of the first spermatids of the first wave of spermatogenesis just before initiation of elongation (spermatid steps 5–7 at spermatogenesis stage V).
FIG. 4.
Parp11 is predominantly expressed during spermatid condensation in the testis. Qualitative and quantitative analyses of mRNA expression by end-point RT-PCR and quantitative PCR (qPCR) of total RNA extracted from tissues of adult wild-type mice demonstrate a preferential expression of Parp11 in the testis, particularly in spermatid stages just before the initiation of nuclear condensation. A) Parp11 mRNA was detectable at high levels in testis, whereas lower expression levels were detected in liver, lung, spleen, and thymus tissues. Messenger RNA levels in heart, kidney, and ovary were near the detection threshold. Gapdh was amplified as an internal control, and qPCR analysis of wild-type tissues is shown. Data are presented as the mean ± SD (n = 3 samples). B) RT-PCR products of RNA isolated from murine testes collected from animals at the indicated time points after birth. Testicular Parp11 expression is detectable from Postnatal Day 18 to adulthood using qPCR (top) and end-point RT-PCR (bottom) analyses. Quantitative PCR data are presented as the mean ± SD (n = 3 samples). C) Beta-galactosidase staining of testicular tissue from adult mice expressing the LacZ gene under the control of the Parp11 promoter. Blue cellular staining indicates Parp11 promoter activity. Early round spermatids lack blue staining (asterisk). Round spermatids from developmental steps 5–6 onward (arrows) and subsequent elongating and condensing spermatids (arrowheads) express beta-galactosidase, representing endogenous Parp11 promoter activity. Bar = 30 μm.
We generated Parp11 gene-disrupted mice (Parp11tm1(KOMP)Vlcg, subsequently termed Parp11−/−). In these animals, exons 4–8 of the Parp11 gene, which encode the majority of the PARP11 protein, were replaced with a beta-galactosidase reporter gene that was subsequently expressed under the control of the endogenous Parp11 promoter (Supplemental Fig. S3, A and B). No Parp11 mRNA was detectable in the homozygous knockout tissues, confirming the gene deletion (Supplemental Fig. S3C).
In line with the observed sharp increase in testicular Parp11 mRNA levels at 18–21 days of age, Parp11−/− spermatids appeared blue after LacZ staining (Fig. 4C) from developmental steps 5 onward. In Parp11 gene-disrupted animals, beta-galactosidase is expressed from the endogenous Parp11 promoter in both heterozygous and Parp11−/− mice to mark the cell population that expresses the Parp11 gene in wild-type animals (Supplemental Fig. S3A). In conclusion, the expression analyses indicate that Parp11 is expressed preferentially in differentiating spermatids undergoing nuclear reorganization, elongation, and condensation.
Deletion of Parp11 Results in a Male Fertility Defect
When propagated by heterozygous mating, Parp11 gene-disrupted mice were viable, and both heterozygous and homozygous animals were born in normal mendelian ratios.
Parp11−/− males, but not females, exhibited a striking fertility defect, with the majority of males being sterile (five of nine animals), whereas a smaller portion of the analyzed males produced infrequent and small litters (four of nine animals) (Fig. 5A). Female Parp11−/− produced normal litter sizes when mated with wild-type males. Testis weights, body weights, and sperm counts did not significantly differ between wild-type, heterozygous, and homozygous Parp11−/− littermates (Supplemental Fig. S4, A–D). Mating behavior of the animals was normal, and the same frequency of copulation plugs was observed in all genotypes. To differentiate if the observed infertility was due to a primary fertilization defect of sperm or a potential early embryonic loss, we performed in vivo mating experiments. Homozygous Parp11−/− and wild-type males were mated with superovulated wild-type females, and early embryos and oocytes were retrieved at Day 1.5 after a copulation plug was observed. At this time point, most fertilized oocytes have undergone the first embryonic cleavage to the 2-cell stage and can reliably be distinguished microscopically from unfertilized and degenerating eggs. Seventy percent (166/237) of all embryos/oocytes retrieved after mating of wild-type females with wild-type males (two males and five females) had progressed to the 2-cell stage, whereas only 5% (16/312) of all embryos/oocytes retrieved from wild-type females after mating with Parp11−/− males (three males and eight females) developed normally (i.e., were at the 2-cell stage) (Fig. 5B and Table 1). Taken together, these data suggested a fertilization defect of Parp11−/− sperm.
FIG. 5.
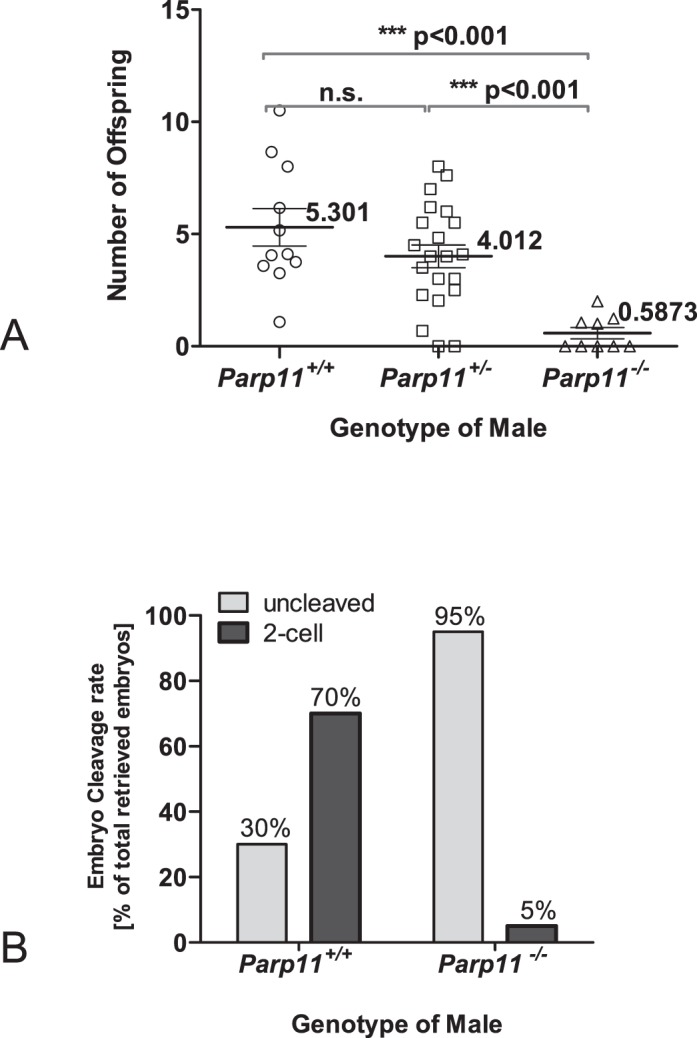
Fertility defect of male mice lacking Parp11. A) Mean number of live offspring produced by one male per female, normalized to 1 mo of breeding time. Mean values (numbers) with SEM are given for each genotype (with Parp11+/+, n = 11; Parp11+/−, n = 21; Parp11−/−, n = 9 individual males). Statistical significance was calculated by Mann-Whitney tests between each group as well as ANOVA followed by Bonferroni multiple comparison correction. Five of the nine Parp11−/− males were infertile, and the remaining four males sired very few and small litters. Average numbers of offspring born from control wild-type male/ Parp11−/− female pairings were not statistically different from those of wild-type control couples (not plotted). B) Reduced fertilization potential of Parp11−/− sperm. Percentages of 2-cell embryos, retrieved at 1.5 days postcoitum (dpc) after natural mating of either wild-type or Parp11−/− males with superovulated wild-type females are indicated above the graph bars; numbers of embryos and animals are listed in Table 1. Fisher exact test indicates a highly significant difference between the genotypes (P < 0.0001).
TABLE 1.
Reduced fertilization capacity of Parp11−/− sperm in vivo.a
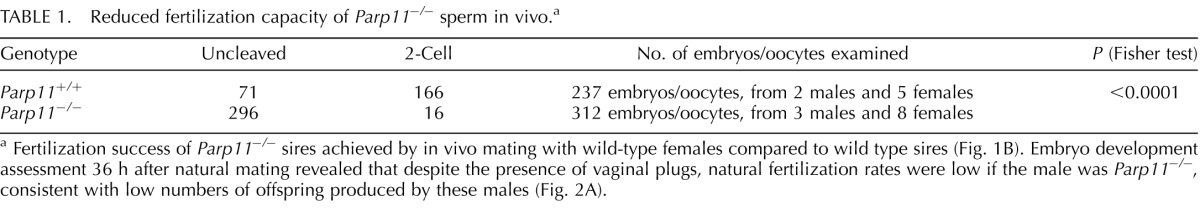
Fertilization success of Parp11−/− sires achieved by in vivo mating with wild-type females compared to wild type sires (Fig. 1B). Embryo development assessment 36 h after natural mating revealed that despite the presence of vaginal plugs, natural fertilization rates were low if the male was Parp11−/−, consistent with low numbers of offspring produced by these males (Fig. 2A).
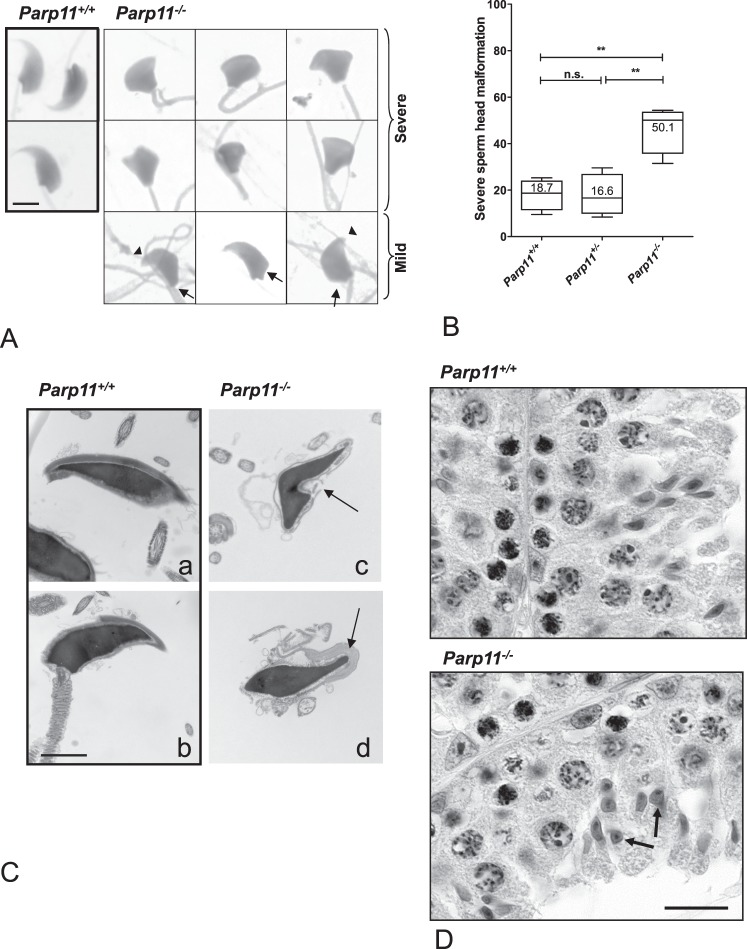
Sperm from Parp11−/− Mice Have Abnormally Shaped Nuclei
Microscopic evaluation of sperm morphology in homozygous Parp11 gene-disrupted mice (Parp11−/−) revealed that more than half of the sperm had malformed nuclei (Fig. 6A, “severe”), and the remaining sperm showed at least some type of subtle abnormalities (teratozoospermia, indicated by arrows in Fig. 6A, “mild”). Abnormal head shapes occurred at much higher frequency in sperm samples from Parp11−/− males compared to the wild-type and heterozygous Parp1+/− animals (Fig. 6B). The remaining spermatozoa exhibited at least some subtle form of teratozoospermia, such as blunt nuclear tips, no indentation at the tail attachment site, or concave indentations at the base of the sperm nucleus (arrows in Fig. 6A). Most of the severely abnormal sperm heads assume a hammer- or cupcake-like shape. Despite the abnormal nuclear shaping, the sperm chromatin appeared to be mostly condensed to normal levels, even in severely abnormally formed nuclei. TEM imaging of spermatozoa confirmed abnormal nuclear shaping (Fig. 6C, c and d, arrows) but also demonstrated the normal dark nuclear staining that is typical of fully condensed sperm nuclei (Fig. 6C). Further analysis of sperm chromatin condensation with CMA3 staining, in which positive staining indicates reduced protamine contents in sperm nuclei, showed that the frequency of CMA3-positive sperm in Parp11−/− was comparable to that of sperm from Parp11+/+ animals (3.8% positive in Parp11−/− sperm and 4% in Parp11+/+ sperm). Sperm with abnormal nuclear shape were not more frequent in the intensely CMA3-stained sperm fraction, and severely abnormal and only mildly affected nuclei were found in both the CMA3-positive and the CMA3-negative sperm groups (Supplemental Fig. S5). Frequencies of normal, mildly abnormal, and severely malformed sperm nuclei among the CMA3-positive fraction were not significantly different from frequencies of normal, mildly abnormal, and severely malformed nuclei in the total sperm population (P = 0.7, chi-square test, n = 315 sperm from four animals). Length of sperm flagella was not statistically significant between the genotypes (Supplemental Fig. S4E). Histological analysis of Parp11−/− testes did not show apparent signs of increased cell death, and epithelial cell composition was comparable between Parp11−/− and Parp11+/+ (stage X spermatogenic tubules are shown in Fig. 6D; additional stages are shown in Supplemental Fig. S6).
FIG. 6.
Teratozoospermia in Parp11−/− mice. A) More than 50% of Parp11−/− sperm cells have conspicuously abnormal head shapes (“severe”). The remaining spermatozoa display various subtle malformations (‘mild”), such as blunt sperm tips (arrowheads), abnormal formation of the nuclear base (arrows), and abnormal attachment of the tail (arrows). Bar = 5 μm. B) Box plots of severe head malformation frequencies (see also “severe” in A). The lines in the boxes indicate median values; whiskers indicate the SEM (5th to 95th percentiles). A total of n = 658–677 sperm from four animals per genotype were evaluated. One-way ANOVA followed by Bonferroni multiple comparison test as well as Whitney-Mann analyses indicate a significantly higher frequency of malformed sperm heads in the Parp11−/− sperm samples. C) TEM imaging of sperm indicates that Parp11−/− sperm heads are malformed (arrows) but contain normally condensed chromatin, with normal intense staining of the nuclear material. Bar = 2 μm. D) Histological staining confirmed abnormal spermatid nuclear shaping in Parp11−/− testes consistent with the observed teratozoospermia phenotype in absence of other overt histological changes (arrows indicate abnormal spermatids in stage X tubules of Parp11−/− but not Parp11+/+). Bar = 30 μm.
To identify the process that is perturbed in the absence of PARP11 and that causes the observed malformation of sperm nuclei in the Parp11−/− males, we next analyzed the ultrastructure of the differentiating spermatids in more detail.
Elongating Spermatids in Parp11−/− Testes Have Nuclear Envelope Defects but Normal Acrosome and Manchette Formation
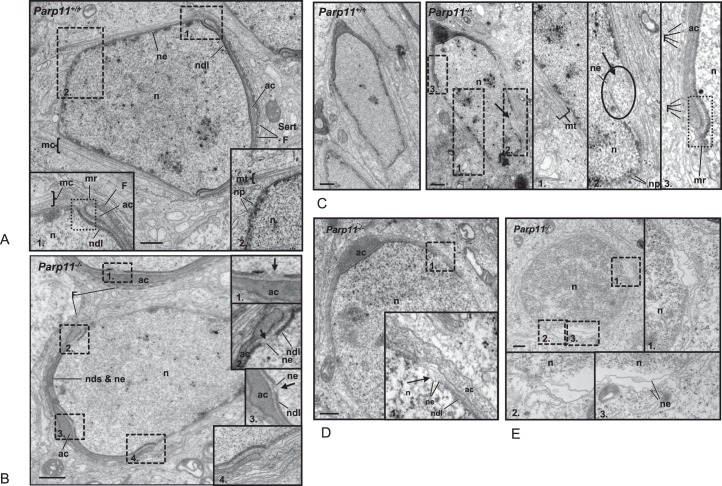
During spermiogenesis, the differentiation from round haploid spermatids to condensed elongated spermatozoa requires extensive reshaping of the nucleus and development of sperm-specific structures, including the acrosome and the flagellum. Achievement of the sperm nuclear shape is facilitated by several important cell-specific cytoskeletal molding structures (for review, see [1]), such as the manchette, which is a conical microtubule mantle that surrounds the distal part of the elongating spermatid nucleus (mc in Fig. 7A, inserts 1 and 2). Initiation of manchette formation occurs at a perinuclear ring structure that surrounds the nucleus adjacent to the marginal ring of the acroplaxome, which comprises a bent circular structure underlying the developing acrosome (mr in Fig. 7A, inset 1). The nuclear envelope subjacent to the acroplaxome appears thickened due to generation of a structure named the nuclear dense lamina (ndl in Fig. 7A, dark line underlying the subacrosomal space). The neighboring Sertoli cell forms F-actin hoops that cap the spermatid adjacent to the developing acrosome and thereby facilitate shaping of the anterior end of the spermatid nucleus (F in Fig. 7A). Nuclear pores in elongating spermatids are primarily detected at the very distal end of the nucleus (Fig. 7A, inset 2), in line with observations by others [47].
FIG. 7.
Nuclear shaping defects in Parp11 knockout spermatids. TEM analyses of elongating spermatids in Parp11+/+ and Parp11−/− testes confirm that wild-type Parp11+/+ early elongating spermatid present with normal structures typical of this developmental step. A) The electron-dense nuclear dense lamina (ndl) coats the nuclear envelope in the entire subacrosomal area. Extranuclear shaping structures are shown in the enlarged insets. Inset 1) F-actin rings inside the surrounding Sertoli cell (Sert) above the developing flat acrosomal area (ac) are visible as dark dots (F). The marginal ring of the acroplaxome (mr) marks the transitional region between the developing acrosome and the manchette (mc). Inset 2) The manchette is comprised of straight microtubule filaments (mt) that surround the central part of the nucleus (n). Nuclear pores (np) are located mainly at the distal end of the nucleus. B) Parp11−/− spermatid. The same basic shaping structures (manchette, F-actin rings, flat developing acrosome and acroplaxome [insets 2 and 4]) are also present in Parp11−/− spermatids of the same developmental step. However, in contrast to the wild-type, the nuclear dense lamia (ndl) coating the subacrosomal nuclear envelope appears interrupted at several positions in subacrosomal regions (arrows in Insets 1 [adjacent other spermatid] and Inset 3 [central region]) as well as under the marginal ring of the acroplaxome (Inset 2). C) Later stage (i.e., more elongated) Parp11+/+ (left) and Parp11−/− spermatids (right). Whereas formation of manchette (Inset 1) and marginal ring (mr; dotted line box in Inset 3) and caudal localization of nuclear pores (np; Inset 2) again appears normal in the Parp11+/+ spermatids, an abnormal invagination of the nuclear envelope (ne) into the nuclear lumen is apparent (arrow in oval of Inset 2). D and E) Round Parp11−/− spermatids have areas of abnormal separation of the nuclear envelope membranes from each other (D, inset 1, and E, inset 1) and detachment of the chromatin from the nuclear envelope (E, insets 2 and 3). adn, acrosome dense plaque. Bar = 500 nm.
Analysis of Parp11−/− testes by TEM showed that the cytoskeletal molding structures (i.e., the manchette in the spermatid and the F-actin hoops in the Sertoli cell) were present and of normal appearance (Fig. 7C, right, normal microtubules constituting the manchette in inset 1; also note the marginal ring of the acroplaxome as well as the F-actin hoops in the surrounding Sertoli cell shown in Fig. 7B, inset 3). Immunofluorescent staining of testis sections to visualize alpha-tubulin in the manchette (Supplemental Fig. S7A) and peanut agglutinin staining of the acrosome (Supplemental Fig. S7B) further supported normal development of these structures in Parp11−/− mice. Based on these results, we dismissed the hypothesis that an underlying gross abnormality of either the manchette or the acrosome contributes to the observed teratozoospermia in Parp11−/− animals.
In the TEM analyses, it also became apparent that Parp11−/− elongating spermatids display a focal failure to form the electron-dense lamina layer, which is normally visible as one distinct, dark line in the subacrosomal region. Instead, two thin and partially separated lines of flaccid appearance were detectable in the resulting gaps of the dense lamina, likely representing the two membrane bilayers of the nuclear envelope (Fig. 7B, insets 1–3, arrows). Other typical defects included the formation of large invaginations of the nuclear envelope into the nuclear lumen (Fig. 7C, inset 2, arrow in oval), separation and bulging of the outer or inner membrane of the nuclear envelope (Fig. 7D, inset 1, arrow), as well as an apparent detachment of adjacent chromatin from the inner nuclear membrane (Fig. 7E, insets 1–3). The portions of the nuclear envelope that underlie the acrosome and the acroplaxome, and which because of their position are most likely exposed to increased physical shaping forces, appeared most prone to display nuclear stretching effects (e.g., arrows in Fig. 7B, insets 1–3). In addition, similar abnormalities were observed in round spermatids at earlier developmental steps before formation of manchette and acroplaxome (Supplemental Fig. S8).
In summary, the present study shows, to our knowledge for the first time, that PARP11, the smallest WWE domain-containing PARP, is predominantly expressed in postmeiotic germ cells of the adult testis and that the enzyme has a subcellular localization at the nuclear envelope in transfected cells. Consistently, deletion of the Parp11 gene in mice results in nuclear envelope defects that occur during the nuclear reshaping process of spermatid elongation and condensation, teratozoospermia, and impaired male fertility. PARP11 showed mono-ADP-ribose transferase activity in vitro and colocalizes with nuclear pore complexes in the nuclear membrane in transfected cells. Localization at the nuclear envelope was dependent on the presence of both an intact WWE domain and the ADP-ribose transferase domain but not the catalytic activity of the enzyme. Taken together, these results suggest an important role of the ADP-ribose pathway and PARP11 in nuclear envelope stability and nuclear remodeling during spermiogenesis in mammals.
DISCUSSION
Sperm head shapes that deviate from the species-specific normal form are generally associated with fertility defects. In humans, the presence of a high percentage of abnormally shaped sperm (teratozoospermia) is a clinically relevant fertility problem, but underlying causes are mostly unknown.
In animal models, a small number of genetic defects have been shown to lead to abnormal sperm head morphology, but most are associated with a failure to properly form the acrosome or manchette [7, 9, 10, 48–51]. Other defects affect nuclear shaping by preventing proper nucleoprotein reorganization and thus sperm chromatin condensation [42, 52–55]. Spermatids possess a large array of nucleocytoplasmic transport factors and nuclear pore components, such as karyopherin A and B, NUP62, RAN, RANBP1, and RCC1 (regulator of chromosome condensation 1) [56, 57]. Mice lacking RANBP1 are infertile due to perturbed differentiation of elongating spermatids, underscoring the importance of effective nucleocytoplasmic transport for spermiogenesis [58]. The nuclear pores therefore appear to play a particularly important role in nuclear transport during early steps of spermatid maturation. Interestingly, they also seem to become obsolete once the nucleus is fully condensed and transcriptionally silent in the late spermatid. Accordingly, the position of nuclear pores in nuclear envelope changes from evenly distributed in round spermatids before elongation to an exclusively caudal accumulation of nuclear pores in condensed spermatids [47]. The processes associated with such remodeling of the nuclear envelope are not well understood, but based on the results of the present study, PARP11 functions could be part of them. In the present study, overexpressed PARP11 in cultured cells was found to colocalize with components of the nuclear pore complex (NUP153) (Figs. 2 and 3), and the enzyme has also previously been identified as an interaction partner of NUP98 [59, 60]. Taken together, the subcellular localization of PARP11, the deformation of sperm heads in Parp11−/− mice, and their defect of the nuclear envelope all suggest that PARP11 is a novel nuclear envelope/pore component in spermatids. The importance of PARP11 to these cells is readily apparent due to the dramatic infertility phenotype in the knockout; however, based on the RT-PCR results, we cannot fully exclude that Parp11 may also be expressed at relevant levels in other cells and tissues and that additional, less overt defects could exist in the knockout mouse.
Parp11−/− spermatids undergoing elongation and condensation show an intriguing lack of coherence of the inner and outer nuclear membranes, concomitant with a failure to generate the continuous nuclear dense lamina that normally forms underneath the developing acrosome (Fig. 7). The nuclear lamina appears stretched and seems to form abnormal gaps in the knockout, particularly in regions that likely are subject to strong physical forces exerted by the nuclear manchette and the forming acrosome during the nuclear shaping process. Inability of these membranes to accommodate reshaping of the nucleus might explain the resulting abnormal sperm head shapes prevalent in the gene-disrupted animals (Fig. 6). Parp11−/− males produce sperm in normal numbers and with normal tail length (Supplemental Fig. S4, A and E). Nevertheless, Parp11−/− males were largely infertile (Fig. 5), and only very few sperm were capable of fertilizing oocytes in vivo. Taken together, these data indicate that the abnormal sperm head shaping observed in knock-out sperm effectively prevents fertilization and that even a mild deviation from sperm nuclear shape, beyond the severe teratozoospermia present in approximately 50% of the sperm (Fig. 6, A and B), was sufficient to prevent efficient fertilization.
PARP11 protein function is expected to be mediated by its two protein domains, the WWE domain and a catalytic PARP domain (Fig. 1A). It has been suggested that WWE domains function in specific protein-protein interactions and could thus help to target the different kinds of E3-like and ADP-ribosylation activities to acceptor proteins participating in various signaling cascades [28]. In the PARP family, enzymes that have been shown to be capable of synthesizing polymeric ADP-ribose have an HYE triad of catalytic amino acid side chains (i.e. histidine [H], tyrosine [Y], and glutamate [E]), but in PARP11, homologs all of the acidic E residues are replaced by isoleucine (I in Supplemental Fig. S1). This conserved substitution is expected to limit of PARP11 to mono(ADP-ribosyl) transferase catalytic activity, similar to PARP10 [14]. In line with recent reports [13], our experimental analysis confirmed PARP11 as a mono(ADP-ribose) transferase (Fig. 1), and no PARP activity was detectable.
Detailed functional implications of PARP11 with its unique combination of functional domains are not currently known, but the sperm defect observed in the present study strongly supports a relevance for nuclear membrane stability during spermatid differentiation.
Intriguingly, the subcellular localization of PARP11 at the nuclear membrane and nuclear pore complexes requires the presence of both WWE and catalytic domains in the same protein (Fig. 2). Similarly, mutation of the amino acids in the WWE domain that mediate ADP-ribose binding abolished this preferential subcellular localization (Fig. 3). Because an enzymatically inactivating mutation (G198W) does not prevent PARP11 localization in the nuclear envelope (Fig. 3), the ADP-ribose moieties the PARP11 WWE domains binds to are unlikely to be conferred by the catalytic activity of PARP11 itself and instead might be generated by a different enzyme. A recent report demonstrated that PARP16/ARTD15 is capable of transferring mono(ADP-ribose) to karyopherin B1 (KPNB1, Importin B1) [61]. KPNB1 forms a heterodimeric complex with karyopherin A (KPNA), and this complex mediates transport of nuclear localization signal containing proteins through nuclear pores into the nucleus [62, 63]. During spermiogenesis, KPNA and KPNB1 were reported to be present in spermatids [57] as components of the subacrosomal nuclear lamina [56], which corresponds to the region of the nuclear envelope in which initial membrane defects in developing Parp11−/− spermatids were observed. Whereas these observations suggest that mono(ADP-ribosyl)ation and PARP11 might similarly be involved in nuclear transport processes during spermiogenesis, the identification of PARP11 interaction partners in ongoing studies should help in clarifying the exact biochemical and cellular mechanisms underlying PARP11 function.
ACKNOWLEDGMENT
We are grateful to Dr. Dewight Williams and Raymond Meade (University of Pennsylvania's Electron Microscopy Resource Laboratory) for support with the TEM analyses, to Ms. Stephanie Sterling for animal care and husbandry, and to Dr. Melissa Vadnais and Ms. Angel Lin (University of Pennsylvania, Center for Research on Reproduction and Women's Health).
Footnotes
Current address: Department of Gynecology and Obstetrics, Tohoku University Graduate School of Medicine, Sendai City, Miyagi, Japan.
Current address: Institute of Veterinary Pharmacology and Toxicology, University of Zurich, Zurich, Switzerland.
This work was supported by grants from the National Institutes of Health (NIH R01 HD48857 and U54HD068157) and the Utah Agricultural Experiment Station, Utah State University (UTA01166) to R.G.M. and by a Utah State University Research Catalyst Grant to M.M.F.
REFERENCES
- Kierszenbaum AL, Rivkin E, Tres LL. Molecular biology of sperm head shaping. Soc Reprod Fertil Suppl. 2007;65:33–43. [PubMed] [Google Scholar]
- Miller D, Brinkworth M, Iles D. Paternal DNA packaging in spermatozoa: more than the sum of its parts? DNA, histones, protamines and epigenetics. Reproduction. 2010;139:287–301. doi: 10.1530/REP-09-0281. [DOI] [PubMed] [Google Scholar]
- Ward WS, Coffey DS. DNA packaging and organization in mammalian spermatozoa: comparison with somatic cells. Biol Reprod. 1991;44:569–574. doi: 10.1095/biolreprod44.4.569. [DOI] [PubMed] [Google Scholar]
- Braun RE. Packaging paternal chromosomes with protamine. Nat Genet. 2001;28:10–12. doi: 10.1038/ng0501-10. [DOI] [PubMed] [Google Scholar]
- Szczygiel M, Kurpisz M. Teratozoospermia and its effect on male fertility potential. Andrologia. 1999;31:63–75. [PubMed] [Google Scholar]
- Kierszenbaum AL, Tres LL. The acrosome-acroplaxome-manchette complex and the shaping of the spermatid head. Arch Histol Cytol. 2004;67:271–284. doi: 10.1679/aohc.67.271. [DOI] [PubMed] [Google Scholar]
- Liska F, Gosele C, Rivkin E, Tres L, Cardoso MC, Domaing P, Krejci E, Snajdr P, Lee-Kirsch MA, de Rooij DG, Kren V, Krenova D. Rat hd mutation reveals an essential role of centrobin in spermatid head shaping and assembly of the head-tail coupling apparatus. Biol Reprod. 2009;81:1196–1205. doi: 10.1095/biolreprod.109.078980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivkin E, Eddy EM, Willis WD, Goulding EH, Suganuma R, Yanagimachi R, Kierszenbaum AL. Sperm tail abnormalities in mutant mice with neor gene insertion into an intron of the keratin 9 gene. Mol Reprod Dev. 2005;72:259–271. doi: 10.1002/mrd.20335. [DOI] [PubMed] [Google Scholar]
- Yao R, Ito C, Natsume Y, Sugitani Y, Yamanaka H, Kuretake S, Yanagida K, Sato A, Toshimori K, Noda T. Lack of acrosome formation in mice lacking a Golgi protein, GOPC. Proc Natl Acad Sci U S A. 2002;99:11211–11216. doi: 10.1073/pnas.162027899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang-Decker N, Mantchev GT, Juneja SC, McNiven MA, van Deursen JM. Lack of acrosome formation in Hrb-deficient mice. Science. 2001;294:1531–1533. doi: 10.1126/science.1063665. [DOI] [PubMed] [Google Scholar]
- Hottiger MO, Hassa PO, Luscher B, Schuler H, Koch-Nolte F. Toward a unified nomenclature for mammalian ADP-ribosyltransferases. Trends Biochem Sci. 2010;35:208–219. doi: 10.1016/j.tibs.2009.12.003. [DOI] [PubMed] [Google Scholar]
- Ame JC, Spenlehauer C, de Murcia G. The PARP superfamily. Bioessays. 2004;26:882–893. doi: 10.1002/bies.20085. [DOI] [PubMed] [Google Scholar]
- Vyas S, Matic I, Uchima L, Rood J, Zaja R, Hay RT, Ahel I, Chang P. Family-wide analysis of poly(ADP-ribose) polymerase activity. Nat Commun. 2014;5:4426. doi: 10.1038/ncomms5426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine H, Poreba E, Lesniewicz K, Hassa PO, Hottiger MO, Litchfield DW, Shilton BH, Luscher B. Substrate-assisted catalysis by PARP10 limits its activity to mono-ADP-ribosylation. Mol Cell. 2008;32:57–69. doi: 10.1016/j.molcel.2008.08.009. [DOI] [PubMed] [Google Scholar]
- Loseva O, Jemth AS, Bryant HE, Schuler H, Lehtio L, Karlberg T, Helleday T. PARP-3 is a mono-ADP-ribosylase that activates PARP-1 in the absence of DNA. J Biol Chem. 2010;285:8054–8060. doi: 10.1074/jbc.M109.077834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguiar RC, Takeyama K, He C, Kreinbrink K, Shipp MA. B-aggressive lymphoma family proteins have unique domains that modulate transcription and exhibit poly(ADP-ribose) polymerase activity. J Biol Chem. 2005;280:33756–33765. doi: 10.1074/jbc.M505408200. [DOI] [PubMed] [Google Scholar]
- Meyer RG, Meyer-Ficca ML, Jacobson EL, Jacobson MK. Enzymes in poly(ADP-ribose) metabolism In Burkle A. (ed.), Poly(ADP-Ribosyl)ation New York: Springer; 2004. 1 12 [Google Scholar]
- Meyer-Ficca ML, Meyer RG, Jacobson EL, Jacobson MK. Poly(ADP-ribose) polymerases: managing genome stability. Int J Biochem Cell Biol. 2005;37:920–926. doi: 10.1016/j.biocel.2004.09.011. [DOI] [PubMed] [Google Scholar]
- Oei SL, Keil C, Ziegler M. Poly(ADP-ribosylation) and genomic stability. Biochem Cell Biol. 2005;83:263–269. doi: 10.1139/o05-039. [DOI] [PubMed] [Google Scholar]
- Schreiber V, Dantzer F, Ame JC, de Murcia G. Poly(ADP-ribose): novel functions for an old molecule. Nat Rev Mol Cell Biol. 2006;7:517–528. doi: 10.1038/nrm1963. [DOI] [PubMed] [Google Scholar]
- Hassa PO, Hottiger MO. The diverse biological roles of mammalian PARPS, a small but powerful family of poly-ADP-ribose polymerases. Front Biosci. 2008;13:3046–3082. doi: 10.2741/2909. [DOI] [PubMed] [Google Scholar]
- Quenet D, El Ramy R, Schreiber V, Dantzer F. The role of poly(ADP-ribosyl)ation in epigenetic events. Int J Biochem Cell Biol. 2009;41:60–65. doi: 10.1016/j.biocel.2008.07.023. [DOI] [PubMed] [Google Scholar]
- Caiafa P, Guastafierro T, Zampieri M. Epigenetics: poly(ADP-ribosyl)ation of PARP-1 regulates genomic methylation patterns. FASEB J. 2008;23:672–678. doi: 10.1096/fj.08-123265. [DOI] [PubMed] [Google Scholar]
- Corda D, Di Girolamo M. Mono-ADP-ribosylation: a tool for modulating immune response and cell signaling Sci STKE 2002. 80. [DOI] [PubMed] [Google Scholar]
- Corda D, Di Girolamo M. Functional aspects of protein mono-ADP-ribosylation. EMBO J. 2003;22:1953–1958. doi: 10.1093/emboj/cdg209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Girolamo M, Dani N, Stilla A, Corda D. Physiological relevance of the endogenous mono(ADP-ribosyl)ation of cellular proteins. FEBS J. 2005;272:4565–4575. doi: 10.1111/j.1742-4658.2005.04876.x. [DOI] [PubMed] [Google Scholar]
- Yu M, Schreek S, Cerni C, Schamberger C, Lesniewicz K, Poreba E, Vervoorts J, Walsemann G, Grotzinger J, Kremmer E, Mehraein Y, Mersching J. PARP-10, a novel Myc-interacting protein with poly(ADP-ribose) polymerase activity, inhibits transformation. Oncogene. 2005;24:1982–1993. doi: 10.1038/sj.onc.1208410. [DOI] [PubMed] [Google Scholar]
- Aravind L. The WWE domain: a common interaction module in protein ubiquitination and ADP ribosylation. Trends Biochem Sci. 2001;26:273–275. doi: 10.1016/s0968-0004(01)01787-x. [DOI] [PubMed] [Google Scholar]
- He F, Tsuda K, Takahashi M, Kuwasako K, Terada T, Shirouzu M, Watanabe S, Kigawa T, Kobayashi N, Guntert P, Yokoyama S, Muto Y. Structural insight into the interaction of ADP-ribose with the PARP WWE domains. FEBS Lett. 2012;586:3858–3864. doi: 10.1016/j.febslet.2012.09.009. [DOI] [PubMed] [Google Scholar]
- Beneke S, Scherr AL, Ponath V, Popp O, Burkle A. Enzyme characteristics of recombinant poly(ADP-ribose) polymerases-1 of rat and human origin mirror the correlation between cellular poly(ADP-ribosyl)ation capacity and species-specific life span. Mech Ageing Dev. 2010;131:366–3669. doi: 10.1016/j.mad.2010.04.003. [DOI] [PubMed] [Google Scholar]
- Rolli V, O'Farrell M, Menissier-de Murcia J, de Murcia G. Random mutagenesis of the poly(ADP-ribose) polymerase catalytic domain reveals amino acids involved in polymer branching. Biochemistry. 1997;36:12147–12154. doi: 10.1021/bi971055p. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- Ihara M, Stein P, Schultz RM. UBE2I (UBC9), a SUMO-conjugating enzyme, localizes to nuclear speckles and stimulates transcription in mouse oocytes. Biol Reprod. 2008;79:906–913. doi: 10.1095/biolreprod.108.070474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer-Ficca ML, Meyer RG, Coyle DL, Jacobson EL, Jacobson MK. Human poly(ADP-ribose) glycohydrolase is expressed in alternative splice variants yielding isoforms that localize to different cell compartments. Exp Cell Res. 2004;297:521–532. doi: 10.1016/j.yexcr.2004.03.050. [DOI] [PubMed] [Google Scholar]
- Meyer-Ficca ML, Scherthan H, Burkle A, Meyer RG. Poly(ADP-ribosyl)ation during chromatin remodeling steps in rat spermiogenesis. Chromosoma. 2005;114:67–74. doi: 10.1007/s00412-005-0344-6. [DOI] [PubMed] [Google Scholar]
- Meyer-Ficca ML, Ihara M, Lonchar JD, Meistrich ML, Austin CA, Min W, Wang Z-Q, Meyer RG. Poly(ADP-ribose) metabolism is essential for proper nucleoprotein exchange during mouse spermiogenesis. Biol Reprod. 2011;84:218–228. doi: 10.1095/biolreprod.110.087361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer-Ficca ML, Lonchar JD, Ihara M, Meistrich ML, Austin CA, Meyer RG. Poly(ADP-ribose) polymerases PARP1 and PARP2 modulate topoisomerase II beta (TOP2B) function during chromatin condensation in mouse spermiogenesis. Biol Reprod. 2011;84:900–909. doi: 10.1095/biolreprod.110.090035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer-Ficca ML, Lonchar JD, Ihara M, Bader JJ, Meyer RG. Alteration of poly(ADP-ribose) metabolism affects murine sperm nuclear architecture by impairing pericentric heterochromatin condensation. Chromosoma. 2013;122:319–335. doi: 10.1007/s00412-013-0416-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi PG, Manicardi G, Bizzaro D, Campana A, Bianchi U, Sakkas D. Use of the guanine-cytosine (GC) specific fluorochrome, chromomycin A3, as an indicator of poor sperm morphology. J Assist Reprod Genet. 1996;13:246–250. doi: 10.1007/BF02065944. [DOI] [PubMed] [Google Scholar]
- Singleton S, Mudrak O, Morshedi M, Oehninger S, Zalenskaya I, Zalensky A. Characterization of a human sperm cell subpopulation marked by the presence of the TSH2B histone. Reprod Fertil Dev. 2007;19:392–397. doi: 10.1071/rd06099. [DOI] [PubMed] [Google Scholar]
- Nagy A. Manipulating the Mouse Embryo: A Laboratory Manual Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2003. [Google Scholar]
- Meyer-Ficca ML, Lonchar J, Credidio C, Ihara M, Li Y, Wang ZQ, Meyer RG. Disruption of poly(ADP-Ribose) homeostasis affects spermiogenesis and sperm chromatin integrity in mice. Biol Reprod. 2009;81:46–55. doi: 10.1095/biolreprod.108.075390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beneke S, Meyer R, Burkle A. Isolation of cDNA encoding full-length rat (Rattus norvegicus) poly(ADP-ribose) polymerase. Biochem Mol Biol Int. 1997;43:755–761. doi: 10.1080/15216549700204571. [DOI] [PubMed] [Google Scholar]
- Bologna S, Ferrari S. It takes two to tango: Ubiquitin and SUMO in the DNA damage response. Front Genet. 2013;4:106. doi: 10.3389/fgene.2013.00106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine H, Herrmann A, Lamark T, Forst AH, Verheugd P, Luscher-Firzlaff J, Lippock B, Feijs LBH, Herzog N, Kremmer E, Johansen T, Mueller-Newen G. Dynamic subcellular localization of the mono-ADP-ribosyltransferase ARTD10 and interaction with the ubiquitin receptor p62. Cell Commun Signal. 2012;10:28. doi: 10.1186/1478-811X-10-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Offringa R, Huang F. Phosphorylation-dependent trafficking of plasma membrane proteins in animal and plant cells: phosphorylation-dependent trafficking of plasma membrane proteins. J Integr Plant Biol. 2013;55:789–808. doi: 10.1111/jipb.12096. [DOI] [PubMed] [Google Scholar]
- Ho HC. Redistribution of nuclear pores during formation of the redundant nuclear envelope in mouse spermatids. J Anat. 2010;216:525–532. doi: 10.1111/j.1469-7580.2009.01204.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza-Lujambio I, Burfeind P, Dixkens C, Meinhardt A, Hoyer-Fender S, Engel W, Neesen J. The Hook1 gene is non-functional in the abnormal spermatozoon head shape (azh) mutant mouse. Hum Mol Genet. 2002;11:1647–1658. doi: 10.1093/hmg/11.14.1647. [DOI] [PubMed] [Google Scholar]
- Cole A, Meistrich ML, Cherry LM, Trostle-Weige PK. Nuclear and manchette development in spermatids of normal and azh/azh mutant mice. Biol Reprod. 1988;38:385–401. doi: 10.1095/biolreprod38.2.385. [DOI] [PubMed] [Google Scholar]
- Meistrich ML, Trostle-Weige PK, Russell LD. Abnormal manchette development in spermatids of azh/azh mutant mice. Am J Anat. 1990;188:74–86. doi: 10.1002/aja.1001880109. [DOI] [PubMed] [Google Scholar]
- Oko R, Donald A, Xu W, van der Spoel AC. Fusion failure of dense-cored proacrosomal vesicles in an inducible mouse model of male infertility. Cell Tissue Res. 2011;346:119–134. doi: 10.1007/s00441-011-1248-9. [DOI] [PubMed] [Google Scholar]
- Martianov I, Brancorsini S, Catena R, Gansmuller A, Kotaja N, Parvinen M, Sassone-Corsi P, Davidson I. Polar nuclear localization of H1T2, a histone H1 variant, required for spermatid elongation and DNA condensation during spermiogenesis. Proc Natl Acad Sci U S A. 2005;102:2808–2813. doi: 10.1073/pnas.0406060102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzuk MM, McKeown MR, Filippakopoulos P, Li Q, Ma L, Agno JE, Lemieux ME, Picaud S, Yu RN, Qi J, Knapp S, Bradner JE. Small-molecule inhibition of BRDT for male contraception. Cell. 2012;150:673–684. doi: 10.1016/j.cell.2012.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka H, Iguchi N, Isotani A, Kitamura K, Toyama Y, Matsuoka Y, Onishi M, Masai K, Maekawa M, Toshimori K, Okabe M, Nishimune Y. HANP1/H1T2, a novel histone H1-like protein involved in nuclear formation and sperm fertility. Mol Cell Biol. 2005;25:7107–7119. doi: 10.1128/MCB.25.16.7107-7119.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu YE, Zhang Y, Unni E, Shirley CR, Deng JM, Russell LD, Weil MM, Behringer RR, Meistrich M. Abnormal spermatogenesis and reduced fertility in transition nuclear protein 1-deficient mice. Proc Natl Acad Sci U S A. 2000;97:4683–4688. doi: 10.1073/pnas.97.9.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrer M, Xu W, Oko R. The composition, protein genesis and significance of the inner acrosomal membrane of eutherian sperm. Cell Tissue Res. 2012;349:733–748. doi: 10.1007/s00441-012-1433-5. [DOI] [PubMed] [Google Scholar]
- Whiley PA, Miyamoto Y, McLachlan RI, Jans DA, Loveland KL. Changing subcellular localization of nuclear transport factors during human spermatogenesis. Int J Androl. 2012;35:158–169. doi: 10.1111/j.1365-2605.2011.01202.x. [DOI] [PubMed] [Google Scholar]
- Nagai M, Moriyama T, Mehmood R, Tokuhiro K, Ikawa M, Okabe M, Tanaka H, Yoneda Y. Mice lacking Ran binding protein 1 are viable and show male infertility. FEBS Lett. 2011;585:791–796. doi: 10.1016/j.febslet.2011.02.002. [DOI] [PubMed] [Google Scholar]
- BioGRID Database [Internet] CITY, STATE: Tyler Labs; http://thebiogrid.org/121365/summary/homo-sapiens/parp11.html. Accessed 20 June 2014. [Google Scholar]
- PARP11 [Internet] Bethesda, MD: National Center for Biotechnology Information, U.S. National Library of Medicine; http://www.ncbi.nlm.nih.gov/gene/57097. Accessed 20 June 2014. [Google Scholar]
- Di Paola S, Micaroni M, Di Tullio G, Buccione R, Di Girolamo M. PARP16/ARTD15 is a novel endoplasmic-reticulum-associated mono-ADP-ribosyltransferase that interacts with, and modifies karyopherin-ß1. PLOS ONE. 2012;7:e37352. doi: 10.1371/journal.pone.0037352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görlich D, Kostka S, Kraft R, Dingwall C, Laskey RA, Hartmann E, Prehn S. Two different subunits of importin cooperate to recognize nuclear localization signals and bind them to the nuclear envelope. Curr Biol. 1995;5:383–392. doi: 10.1016/s0960-9822(95)00079-0. [DOI] [PubMed] [Google Scholar]
- Görlich D, Vogel F, Mills AD, Hartmann E, Laskey RA. Distinct functions for the two importin subunits in nuclear protein import. Nature. 1995;377:246–248. doi: 10.1038/377246a0. [DOI] [PubMed] [Google Scholar]