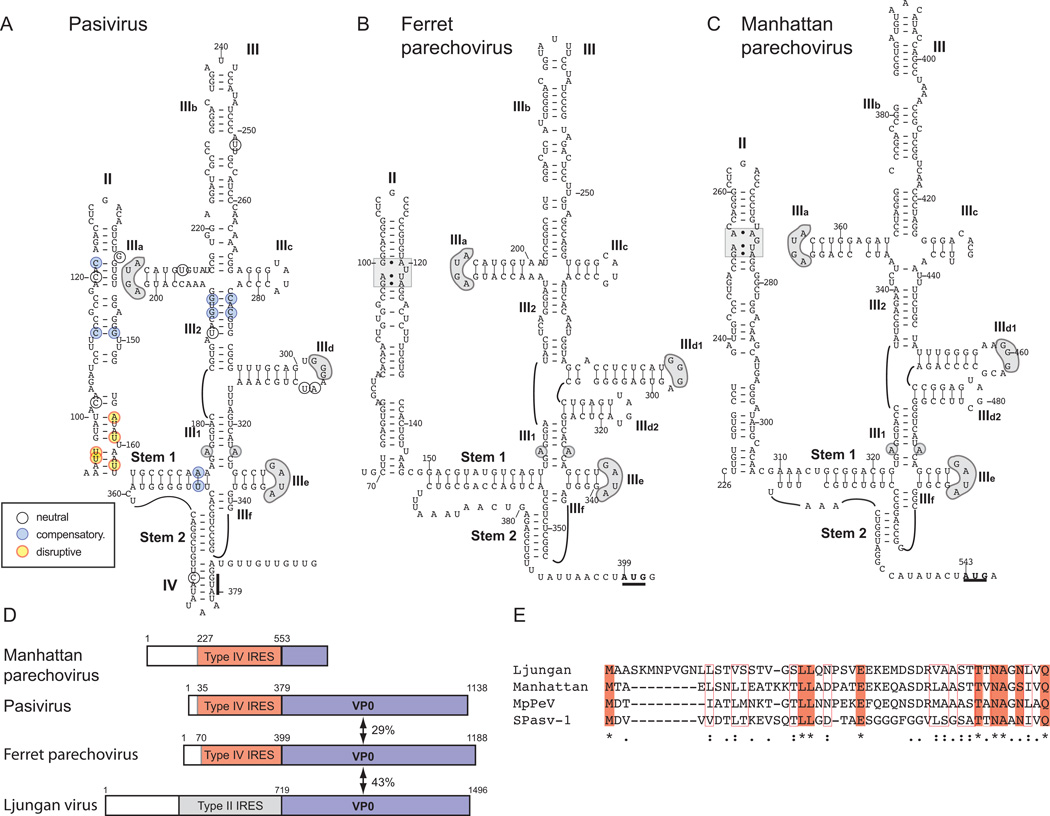
Figure 3. Related IRESs in Pasivirus A1, Ferret parechovirus and Manhattan parechovirus 5’UTRs.
Models of the structures of the IRESs of (A) Pasivirus A1 (PaV-1) isolate swine/France/2011, (B) Ferret parechovirus (FPV) isolate ferret/MpPeV1/NL and (C) Manhattan parechovirus (MPeV). The initiation codons for the viral polyproteins are bold and underlined, and the nomenclature of domains is as in [Hellen and de Breyne, 2007]. Nucleotide differences between the six current PaV-1 strains are assigned to different classes as indicated in the inset key, and as defined in the legend for Fig. 2. Sequence motifs in domain III of PaV-1, FPV and MPeV IRESs that are conserved in other Type IV IRESs (such as the conserved apical loops of domains IIId and IIIe and the unpaired purine residue in helix III1) are indicated by grey shading, as is an ‘E loop’ motif in domain II of FPV and MPeV IRESs. (D) Schematic representations of the 5’-UTRs and adjacent VP0 coding regions of MPeV, PaV-1 (isolate swine/France/2011), FPV (isolate ferret/MpPeV1/NL) and Ljungan virus strain 174F are divided to show the Type IV IRES in MPeV, PaV-1 and FPV (orange), the Type II IRES in Ljungan virus (grey) and the VP0 coding regions (blue). (E) Sequence alignment of the N-terminal regions of the VP0 proteins of PaV-1, FPV, MPeV and Ljungan virus. Identical amino acid residues are indicated by shading and asterisks, and related amino acid residues are indicated by open boxes and colons.