Abstract
Objective
Amyotrophic lateral sclerosis (ALS) is a fatal disorder of motor neuron degeneration. Most cases of ALS are sporadic (SALS), but about 5-10% of ALS cases are familial (FALS). Recent studies have shown that mutations in FUS are causal in approximately 4-5% of FALS and some apparent SALS cases. The pathogenic mechanism of the mutant FUS-mediated ALS and potential roles of FUS in non-FUS ALS remain to be investigated.
Methods
Immunostaining was performed on postmortem spinal cords from 78 ALS cases, including SALS (n=52), ALS with dementia (ALS/dementia, n=10) and FALS (n=16). In addition, postmortem brains or spinal cords from 22 cases with or without frontotemporal lobar degeneration (FTLD) were also studied. In total, 100 cases were studied.
Results
FUS-immunoreactive inclusions were observed in spinal anterior horn neurons in all sporadic and familial ALS cases, except for those with SOD1 mutations. The FUS-containing inclusions were also immunoreactive with antibodies to TDP43, p62 and ubiquitin. A fraction of tested FUS antibodies recognized FUS inclusions and an unusual antigen retrieval appeared to be important for detection of the skein-like FUS inclusions.
Interpretation
Although mutations in FUS account for only a small fraction of FALS and SALS, our data suggest that FUS protein may be a common component of the cellular inclusions in non-SOD1 ALS and some other neurodegenerative conditions, implying a shared pathogenic pathway underlying SALS, non-SOD1 FALS, ALS/dementia and related disorders. Our data also indicate that SOD1-linked ALS may have a distinct pathogenic pathway from SALS and other types of FALS.
INTRODUCTION
Amyotrophic lateral sclerosis (ALS) is a fatal paralytic disorder caused by degeneration of motor neurons in the brain and spinal cord. Most cases of ALS are of unknown etiology and appear as sporadic ALS (SALS). About 5-10% of ALS cases have a family history. Some forms of familial ALS (FALS) are linked to genetic mutations in specific genes. Mutations in the Cu/Zn superoxide dismutase gene (SOD1) account for approximately 20% of FALS cases 1, 2. Mutations in the TAR DNA-binding protein gene (TARDBP, or TDP43) may occur in about 3-4% of FALS 3-5. Rare genetic defects in several other genes have been found in ALS or ALS-like motor neuron diseases 6. Recently, mutations in FUS have been identified in 4-5% of FALS cases 7, 8 and some SALS cases 9-14.Altogether, mutations in specific genes have been identified in about 30% of the FALS cases. The causes for the remaining FALS cases have yet to be identified.
In contrast to FALS, the etiology and the pathogenic mechanisms underlying SALS, the most common form of ALS, remain largely unknown. Understanding the causes and pathogenic mechanisms of SALS remains to be a major challenge.
Protein aggregation has been recognized as a pathological hallmark in several neurodegenerative disorders, such as extracellular amyloid-beta plaques and intracellular tau neurofibrillary tangles in Alzheimer disease, and alpha-synuclein containing Lewy bodies in Parkinson disease 15. In ALS, protein aggregates are most common in spinal motor neurons and the ubiquitin-immunoreactive and skein-like aggregates are thought to be pathognomic of ALS 16, 17. SOD1-immunoreactive aggregates are a prominent pathology in SOD1-mediated ALS, but not in SALS 18. On the other hand, TDP43-immunoreactive aggregates have been identified in SALS, but not in SOD1-ALS 19, 20. Both TDP43 and FUS are DNA/RNA-binding proteins. They are predominantly present in cell nuclei in physiological condition. However, in neurons with cytoplasmic TDP43 aggregates, nuclear TDP43 is diminished 19, 20. It is currently unknown if FUS is involved in the pathogenesis of SALS and other forms of FALS without FUS mutations.
In this study, we analyzed postmortem tissues using immunostaining to explore post-translational involvement of FUS in different types of ALS and other neurodegenerative disorders, as well as neurological disease-free controls.
PATIENTS AND METHODS
Cases
Spinal cord sections from a total of 78 clinically and pathologically diagnosed ALS cases were analyzed using immunostaining in this study. The analyzed ALS cases included SALS (n=52), ALS with dementia (ALS/dementia, n=10) and FALS (n=16). We also analyzed frontal lobe or hippocampus sections from patients with frontotemporal lobar degeneration (FTLD-TDP) with (n=4) or without progranulin mutations (n=5), atypical FTLD with neuronal inclusions composed of an unidentified ubiquitinated protein (aFTLD-U or FTLD-FUS) (n=2), and controls without history of apparent neurological disorders (n=5). Spinal cord controls without motor neuron diseases and other neurological disorders (n=6) were also included in this study. In total, 100 cases were analyzed.
Genetic analysis
Genomic DNA was extracted from whole blood, transformed lymphoblastoid cell lines, or tissues by standard methods (Qiagen, Valencia, CA, USA). Exonic DNA fragments of the TDP43, FUS and SOD1 genes were directly sequenced for 24 cases of SALS, six cases of ALS/dementia and 16 cases of FALS, using a CEQ™ 8000 Genetic Analysis System (Beckman Coulter, CA, USA).
Immunohistochemistry and confocal microscopy
Immunohistochemistry and confocal microscopy were performed using previously described methods 21. In brief, 6μm sections were cut from formalin-fixed, paraffin-embedded spinal cord and brain regions containing frontal lobe or hippocampus. The sections were deparaffinized and rehydrated by passing the slides in serial solutions: three times for 10 min each in xylene, three timesfor 5 min each in 100% ethanol, three times for 3 min each in 95% ethanol, once for 5 min each in 75% ethanol, 50% ethanol, deionized water and phosphate buffered saline (PBS), respectively. The antigens in the sections were retrieved using a decloaking chamber with a 1X Antigen Decloaker solution (Biocare Medical, Walnut Creek, CA,USA) at 125oC for 20 min. The sections were cooled to room temperature for 30 min and rinsed with deionized water for 5 min. For immunohistochemistry, the endogenous peroxidase activities were blocked with 2% hydrogen peroxide. Non-specific background was blocked with 1% BSA in PBS for 20 min at room temperature. An affinity-purified rabbit anti-human FUS polyclonal antibody (11570-1-AP, Proteintech Group, Inc., Chicago, IL, USA) at a concentration of 0.5-3μg/ml was applied to the slides and incubated at room temperature for 1 h. After rinsing the slides three times for 5 min each, a biotinylated goat anti-rabbit IgG was applied to the slides as the secondary antibody (GR608H, Biocare Medical, Concord, CA, USA). The slides were incubated at room temperature for 30 min. The excess secondary antibody was removed by rinsing the slides three times, 5 min for each time. The signals were detected with peroxidase-conjugated streptavidin (BioGenex, San Ramon, CA, USA) using 3-amino-9-ethylcarbazole as a chromogen. The slides were counterstained with hematoxylin and sealed with Aqua Poly/Mount (Polyscience, Warrington, PA, USA). The slides were examined and photographed under a light microscope. In addition to the anti-FUS antibody 11570-1-AP, we also tested eight other anti-FUS antibodies, including A300-292A, A300-293A, A300-294A and A300-302A (Bethyl Laboratories, Inc., Montgomery, TX, USA), ab23439 (Abcam, Inc., Cambridge, MA, USA), sc-25540 and sc-47711 (Santa Cruz Biotechnology, Inc., CA, USA) and HPA008784 (Sigma-Aldrich, Saint Louis, MO, USA). For confocal microscopy, FUS monoclonal or polyclonal antibodies were used in combination with either mouse anti-TDP43 monoclonal antibody (60019-2-Ig, 6μg/ml, Protein Tech Group, Inc. Chicago, IL, USA), rabbit anti-TDP43 polyclonal antibody (10782-2-AP, 5μg/ml, Protein Tech Group, Inc. Chicago, IL, USA), mouse anti-ubiquitin monoclonal antibody (10R-U101B, 3μg/ml, Fitzgerald Industries International, Concord, MA, USA) or mouse anti-p62 monoclonal antibody (H00008878-M01, 6μg/ml, Abnova Coporation, Taipei, Taiwan) as primary antibodies. Fluorescence signals were detected with donkey anti-rabbit IgG conjugated with FITC (Thermo Scientific, Rockford, IL, USA) and goat anti-mouse IgG conjugated with rhodamine (R-6393, Invitrogen, Carlsbad, CA, USA) using an LSM 510 META Laser Scanning Confocal Microscope with the multitracking setting. The same pinhole diameter was used to acquire each channel.
Western Blot
Spinal cord tissues from the lumbar sections were washed with 1X PBS and incubated with ice cold lysis buffer containing 20 mM Tris pH 7.6, 150 mM Sodium Chloride, 1% NP40, 1% SDS, 10% glycerol, 10 mM Magnesium Chloride, 1X HaltTM phosphatase inhibitor cocktail (Pierce) and proteases inhibitor cocktail (Roche). The tissues were then homogenized, sonicated for 10 seconds and centrifugated at 800g for 10 minutes at 4°C. The supernatants were removed and were subject to total protein quantification using BCA kit (Pierce). Twenty micrograms of the whole lysate total protein were separated on 4-12% Bis-tris polyacrylamide gel electrophoresis and blotted on PVDF membrane (Millipore). Two duplicate membranes were incubated overnight with primary antibodies, either anti-FUS (Protein Tech 11570-1-AP) or anti-TDP-43 (Protein Tech. 10782-2-AP). After washing with TBS 0.1% Tween 20, the membrane was incubated with secondary antibody, anti Rabbit HRP conjugated (BioRad) and visualized using ECL kit (GE). The membranes were striped and blotted with an antibody against actin (Sigma). A FUS positive control was derived from lysate of THP-1, a human monocytic leukemia cell line (sc-2238, Santa Cruz Biotechnology, Santa Cruz, CA, USA). The Molecular weight standard was Novex Sharp Prestained Protein Standards from Invitrogen (LC5800).
RESULTS
FUS-immunoreactive aggregates/inclusions in SALS, ALS/dementia and non-SOD1 FALS
Sequencing analysis of the FUS, TDP43 and SOD1 genes in 46 ALS samples revealed three cases with SOD1A4V mutation, and one case each with SOD1G85R, FUSR521L or TDP43G298S mutation among the 16 FALS cases. No mutations in the SOD1, FUS and TDP43 genes were found in 24 SALS and 6 ALS/dementia cases (unpublished data). Nine antibodies against FUS were initially tested on spinal cord sections of 10 SALS cases (Supplementary Table 1). Among these antibodies, two affinity-purified FUS antibodies against human FUS (11570-1-AP and HPA008784) revealed FUS-immunoreactive inclusions (Fig. 1). Because the antibody 11570-1-AP yielded more robust signals (Fig. 1 and supplementary table 1), it was then used for further analysis throughout this study. Immunohistochemical analysis of the spinal cord sections revealed that FUS-immunoreactive inclusions were present in some of the surviving spinal anterior horn neurons from all 52 SALS cases, twelve SOD1-negative FALS cases and 10 ALS/dementia cases (Fig. 2 and supplementary figure 1), but not in the four FALS cases with SOD1 mutations. Previous studies have demonstrated that SOD1/ubiqutin-positive inclusions in spinal motor neurons are a pathological hallmark in ALS patients with SOD1 mutations and in the transgenic mice overexpressing mutant SOD1 18, 22. Supporting the notion that the FUS-positive inclusions are absent in SOD1-linked ALS, we found that the ubiquitin-positive inclusions in spinal neurons were FUS-negative in patients with SOD1G85R or SOD1A4V mutation and in the transgenic mice overexpressing SOD1G93A and SOD1L126Z 21, 23 (Supplementary figure 2). We observed FUS-immunoreactive inclusions in a case with FUSR521L mutation (Fig. 2D). Interestingly, we also observed FUS-immunoreactive inclusions in a patient with TDP43G298S mutation (Fig. 2E). Most of the inclusions were skein-like (Fig. 2A-E). Some compact inclusions were also observed (Fig. 2F). The FUS inclusions appeared to be present predominantly in large anterior horn motor neurons and their neuritic processes (Figs. 1, 2 and supplementary figure 1). Significantly increased FUS staining was also observed in some glial cells (Fig. 2G). In spinal cord sections with preserved dorsal root ganglia of one SALS patient, FUS inclusions were observed in the spinal anterior horn motor neurons (Fig. 2A), but not in the ganglionic primary sensory neurons (Fig. 2H). In the motor neurons, the FUS inclusions were located primarily in the cytoplasm and neurites. The shape and distribution pattern of the FUS inclusions were similar to those of TDP43 inclusions observed in non-SOD1 ALS patients 19 (Fig. 2I). The skein-like, cytoplasmic and neuritic FUS-positive inclusions appeared characteristic in these non-SOD1 ALS cases. Even in some neurons with abundant lipofuscin, these FUS inclusions were apparent (Supplementary figure 1U and 1V). But these inclusions were not observed in six spinal cord samples from control cases without ALS (Supplementary figure 1W-B’).
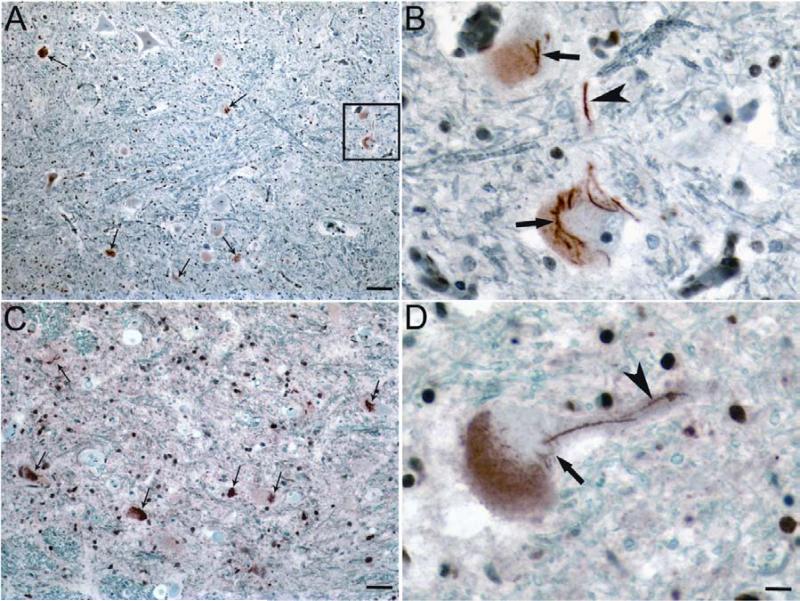
Figure 1.
FUS-immunoreactive aggregates/inclusions in spinal cord of SALS patients. (A, C) Representative FUS-immunoreactive inclusions detected with antibody 11570-1-AP in several spinal anterior horn neurons of SALS cases. (B) Higher magnification image of the boxed area in panel A. (D) Representative FUS-positive inclusions detected with antibody HPA008784 in SALS cases. Small arrows in (A) and (C) indicate representative neurons with FUS-positive inclusions. Large arrows in (B) and (D) indicate neuronal cytoplasmic FUS-positive inclusions. Arrowheads in (B) and (D) indicate neuritic FUS-positive inclusions. Bar: A, C=100μm; D=25 μm.
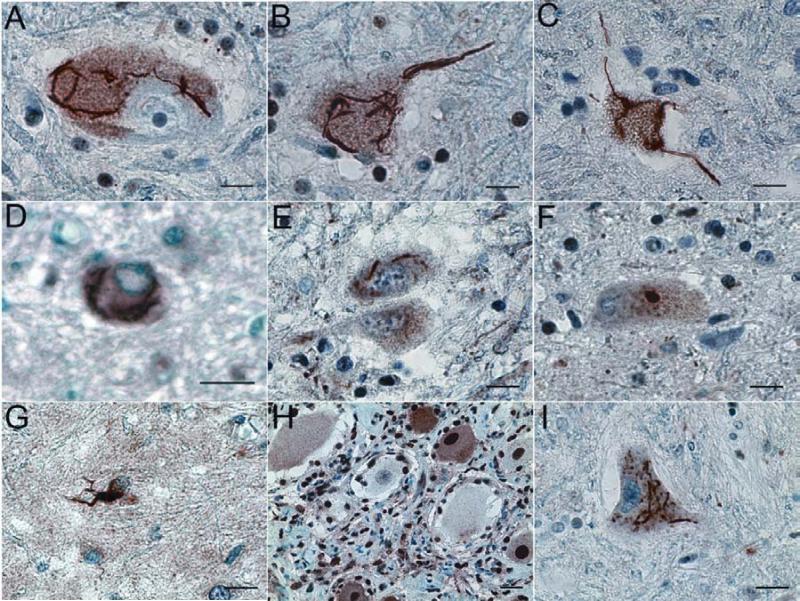
Figure 2.
Representative FUS-immunoreactive aggregates/inclusions in SALS, ALS/dementia and non-SOD1 FALS. Human spinal cord sections were analyzed with immunohistochemistry using an affinity purified anti-FUS antibody (11570-1-AP). Representative FUS-immunoreactive aggregates/inclusions are shown here in anterior horn neurons in patients with SALS (A), FALS without known genetic mutations in SOD1, FUS or TDP43 (B), ALS/dementia (C), FALS with FUSR521L mutation (D), and FALS with TDP43G298S mutation (E). Most of the inclusions were skein-like (A-E), but some of them appeared compact (F). The FUS inclusions were predominantly localized in the large neurons of the anterior horn (A-F). Some glial cells exhibited significantly increased FUS staining (G). No FUS inclusions were found in primary sensory neurons of the dorsal root ganglia (H) of a patient exhibiting FUS inclusions in spinal neurons (A). Representative TDP43-immunoreactive inclusions in spinal cord neurons of SALS patients are shown in (I). Bar, 50μm.
Co-localization of TDP43, p62 and ubiquitin with FUS in spinal neuron inclusions in non-SOD1 ALS cases
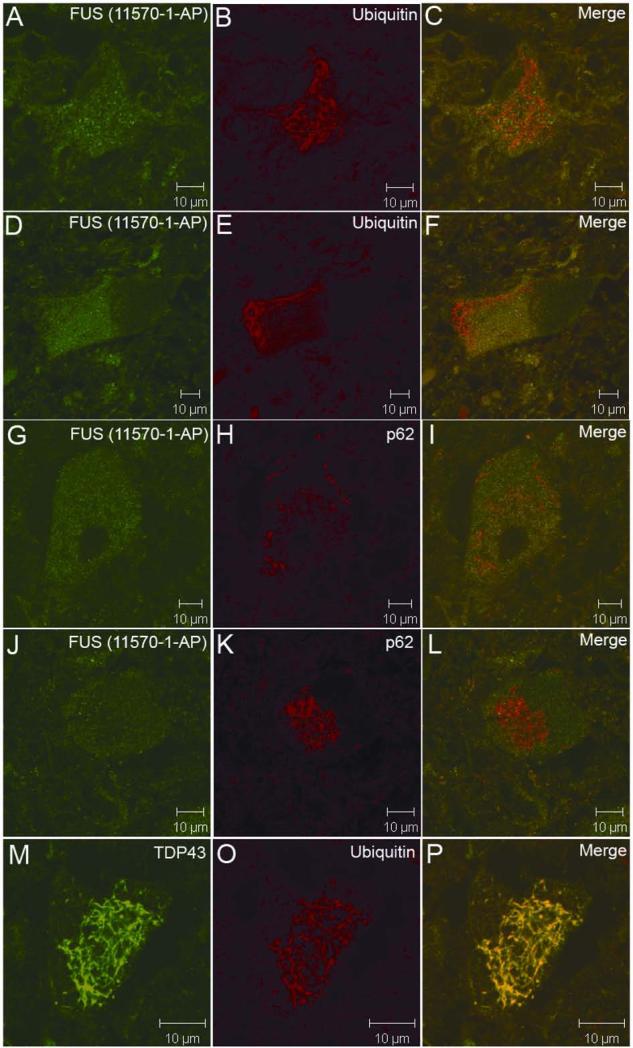
Since the FUS inclusions were similar in morphology and distribution pattern to the TDP43 inclusions observed in non-SOD1 ALS patients 19, we tested if FUS and TDP43 were co-localized. Confocal microscopy demonstrated that the FUS-positive inclusions were also immunorective with the same TDP43 antibody used to detect TDP43 inclusions in ALS 19 (Fig. 3A-3C).
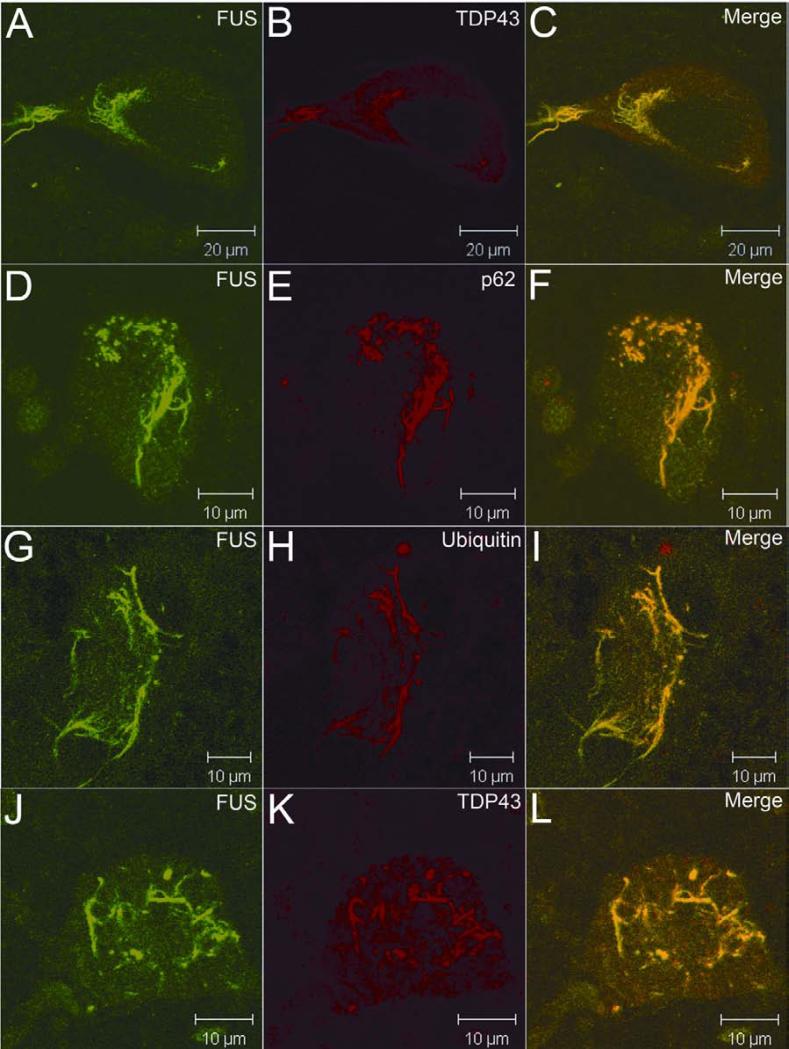
Figure 3.
Co-localization of TDP43, p62 and ubiquitin in FUS-positive inclusions. FUS-positive inclusions (green) were also immunoreactive to TDP43, p62 and ubiquitin (red) antibodies (A-I). In a patient with the TDP43G298S mutation, TDP43-positive inclusions (red) were also immunoreactive to the FUS antibody (green) (J-L).
Ubiquitin- and p62-immunoreactive inclusions in anterior horn motor neurons are hallmarks of ALS 24. To test if the FUS- and TDP43-containing inclusions were also immunoreactive to p62 and ubiquitin, we analyzed inclusions using confocal microscopy. We found that the FUS inclusions were also immunoreactive to p62 (Fig. 3D-3F) and ubiquitin (Fig. 3G-3I) antibodies. Consistent with the data seen in our immunohistochemical studies, these inclusions were present in cytosol and neurites. Using immunohistochemistry, we observed FUS-positive inclusions in spinal neurons of a patient with TDP43G298S mutation (Fig. 2E). Confocal microscopy demonstrated that TDP43-positive inclusions in this patient were also immunoreactive to the anti-FUS antibody (Fig. 3J-3L).
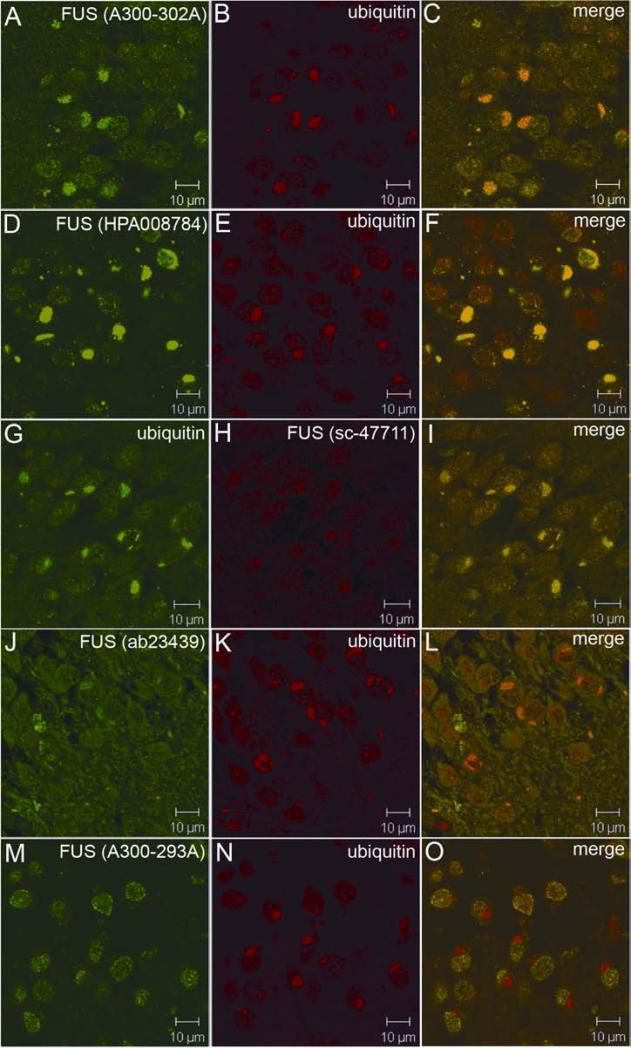
As the characteristic skein-like FUS inclusions detected with the FUS antibody 11570-1-AP were also immunoreactive to antibodies against TDP43, ubiquitin and p62, we further tested by confocal microscopy if the TDP43- and ubiquitin-positive inclusions could be detected with other commercially available FUS antibodies. Consistent with immunohistochemistry, HPA008784 also recognized the skein-like and TDP43-positive inclusions (Fig. 4A-4C). In addition, the TDP43-positive inclusions were also positive with FUS monoclonal antibody sc-47711 (Fig. 4D-4F). However, the 11570-1-AP antibody yielded apparently stronger FUS immunofluorescence (IF) intensity than the other two (Figs. 3 and 4, and supplementary figure 3). In approximately two-thirds of the cells with TDP43-positive inclusions, the FUS IF signals yielded by HPA008784 and sc-47711 were co-localized with strong TDP43 signals, but they were less robust when TDP43 signals were weak (supplementary figure 3D-3I). The other FUS antibodies, however, did not show apparent co-staining (supplementary figure 3J-3R and supplementary table 1). We also observed nuclear staining of FUS in some cells where FUS and TDP43 inclusions were present, although nuclear staining of the TDP43 were not apparent (supplementary figure 3D-3F). Notably, the antibodies HPA008784 and sc-47711, which recognize the inclusions, are raised using two separate polypeptides that are 61 amino acids apart (Supplementary figure 4).
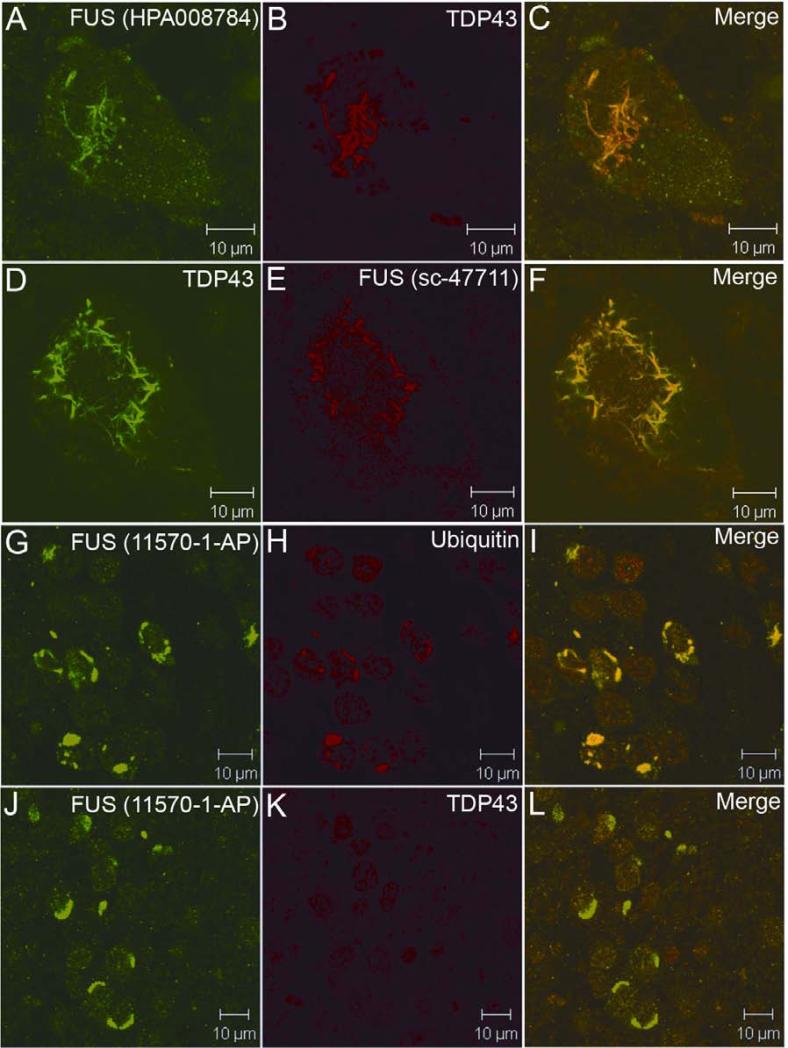
Figure 4.
Confocal analysis of the FUS pathology with different FUS antibodies. The representative FUS pathology is shown with FUS antibodies HPA008784 (A-C) and sc-47711 (D-F) in spinal neurons of SALS cases. The ubiquitin-positive inclusions in aFTLD-U were immunoreactive to FUS antibody (G-I), but not to TDP43 antibody (J-L).
FUS pathology is independent of TDP43 pathology in aFTLD-U
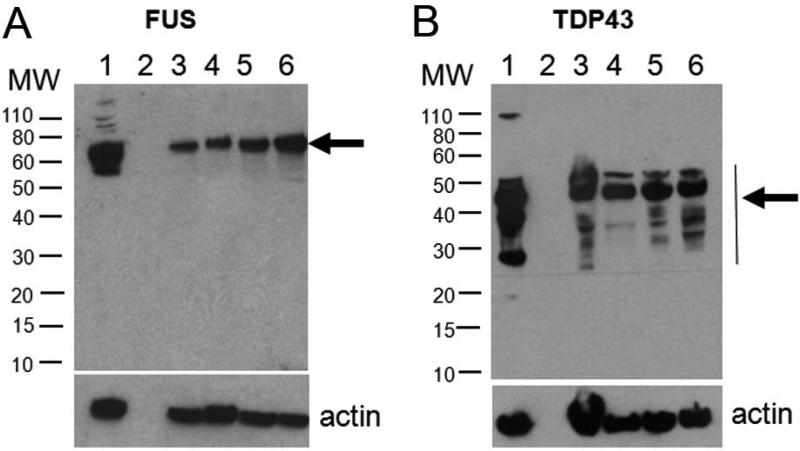
Most cases of frontotemporal dementia (FTD) are associated with inclusions containing TDP43. Recently, a subgroup of approximately 10% FTD cases has been described, which presents an unusual clinical phenotype and pathology characterized by frontotemporal lobar degeneration with neuronal inclusions composed of an unidentified ubiquitinated protein (atypical FTLD-U, aFTLD-U)25. The inclusions in aFTLD-U are positive for ubiquitin and FUS, but are negative for TDP43 (aFTLD-U or FTLD-FUS) 25. To test if FUS-positive inclusions are the result of cross-reactivity of tested FUS antibodies to TDP43, we analyzed hippocampus sections of two patients with aFTLD-U. Consistent with previous data 25, we found that pronounced oval or crescentic ubiquitin-positive inclusions in the dentate granule cells of the hippocampus were strongly immunoreactive with the FUS antibody 11570-1-AP (Fig. 4G-4I). However, these FUS-positive inclusions were negative for TDP43 (Fig. 4J-4L). Western blot using antibody 11570-1-AP showed a single band of the expected size that did not overlap with the TDP43 bands (Fig. 5).
Figure 5.
Western blots with antibodies to FUS and TDP43. Twenty micrograms of protein from spinal cord homogenates of ALS patients were loaded and blotted using affinity-purified rabbit anti-human FUS polyclonal antibody 11570-1-AP (A) and anti-TDP43 antibody (B). (A) Lane 1, lysate of THP-1, a human monocytic leukemia cell line (sc-2238, Santa Cruz Biotechnology, Santa Cruz, CA, USA); (B) lane 1, lysate of transiently transfected HEK293 cells overexpressing TDP43; (A and B) lane 2, blank; lanes 3-5, spinal cord homogenates from patients with SALS; lane 6, spinal cord homogenates from a patient with FUSR521L mutation. A single FUS band of the expected size (A) and multiple TDP43 bands (B) detected with each antibody are indicated with arrows. The sizes of molecular weight (MW) are indicated on the left (kd). Lower panel, actin control.
The robust immunoreactivity of the well-defined inclusions observed with 11570-1-AP in the FTLD-FUS led us to further evaluate the sensitivity of the other FUS antibodies to the inclusions in the FTLD-FUS cases. Consistent with the data obtained from spinal cord sections, the HPA008784 yielded strong signals (Fig. 6D-6F) and the sc-47711 generated moderate signals (Fig. 6G-6I). In addition, A300-292A also yielded moderate signals (Fig. 6A-6C) and the ab23439 revealed weak signals (Fig. 6J-6L). The other four antibodies (sc-25540, A300-292, A300-293 and A300294) did not show apparent signals on the ubiquitin-positive inclusions (Fig. 6M-O, and supplementary table 1).
Figure 6.
Sensitivity of FUS antibodies for detection of the FUS inclusions in aFTLD-U. Confocal microscopy of the hippocampal sections from two aFTLD-U cases stained with antibodies against ubiquitin and FUS.
Antigen retrieval-related detection of FUS pathology
FUS does not share homology with TDP43 at the protein level. We observed FUS-positive inclusions in SALS and non-SOD1 ALS using three out of the nine antibodies tested in this study. Although the epitopes of 11570-1-AP overlaps or partially overlaps with those of antibodies HPA008784 and sc-47711, the antibodies HPA008784 and sc-47711 have been raised using two separate polypeptides without any overlap and are 61 amino acids apart in different species of hosts (Supplementary figure 4). Moreover, sc-47711 is a monoclonal antibody. These FUS antibodies also detected FUS inclusions in FTLD-FUS, where TDP-43 inclusions were absent. Immunoblot using antibody 11570-1-AP, which is raised using the longest polypeptide of the FUS, showed a single band that did not overlap with the TDP43 bands. These data suggest that the presence of the inclusions immunoreactive to FUS antibodies is unlikely due to cross-reactivity of the FUS antibodies to TDP-43, but FUS is a component of the inclusions.
The FUS pathology in spinal cord has been previously evaluated using immunostaining and FUS-positive inclusions have not been identified in SALS and non-FUS ALS cases 8, 25, 26. In our previous studies of SOD1 inclusions in spinal cord of SOD1-linked FALS patients and SOD1 transgenic mice, we observed that antigen retrieval methodology played an essential role in detection of SOD1 inclusions in lower motor neurons 21, 22, especially in some human cases (unpublished data). We compared three antigen retrieval protocols: boiling and microwave that are most commonly used, and high pressure decloaking chamber. We found that antigen retrieval using the high pressure decloaking chamber was the most efficient in detection of SOD1 inclusions, and have therefore adopted this protocol as the routine method for antigen retrieval 21, 22. Because other studies failed to show FUS pathology in SALS using microwave protocol for antigen retrieval 8, 25, 26, we assumed that in addition to the sensitivity of the antibodies, antigen retrieval methodology may be an important factor for influencing the detestability of FUS-positive inclusions. To test this possibility, we compared boiling and microwave protocols to the high pressure decloaking chamber protocol using spinal cord sections from SALS cases. We observed that the FUS-positive inclusions were only observed when the high pressure decloaking chamber protocol was used and the ubiqutin- and p62-positive inclusions were consistently negative for FUS when boiling or microwave protocol was used (Fig. 7A-7L). However, the ubiquitin-, p62 and TDP43-positive inclusions were consistently observed with all three antigen retrieval protocols (Fig. 7A-7P).
Fig. 7.
Effect of the antigen retrieval protocols on detection of the inclusions. The representative ubiquitin- (A-F) and p62- (G-L) positive inclusions in spinal neurons of SALS cases were not apparently FUS-positive when antigen was retrieved using either boiling (A-C and G-I) or microwave (D-F and J-L) protocol. (M-O) Representative TDP43- and ubiquitin-positive inclusions are shown with microwave antigen retrieval protocol.
FUS pathology in the brain of FTLD-TDP43 with or without progranulin mutations, and ALS/dementia
We further tested frontal lobe sections from four FTLD cases with different progranulin mutations and another five FTLD cases without progranulin mutations. We observed that ubiquitin- and TDP43-immunoreactive inclusions were also positive for FUS, although the inclusions were rare in these cases (supplementary figure 5A-5F). In sections of the neocortex and hippocampus from three ALS/dementia patients, rare ubiquitin-positive inclusions were also positive for FUS (supplementary figure 5G-5L), but the FUS-immunoreactivity of the ubiquitin-positive inclusions in granule cells of hippocampal dentate gyrus appeared to be less intense compared to those in FTLD-FUS cases (supplementary figure 5J-5L). We also examined brain sections from frontal and hippocampal regions from five control cases without dementia, ranging in age from 56 to 75 years old. We did not observe FUS-positive inclusions in those cases.
DISCUSSION
In this study, we observed FUS-positive inclusions in anterior horn neurons and their neurites in spinal cord of 52 SALS cases, 10 ALS/dementia cases, one case each with TDP43 or FUS mutation and other 10 FALS cases with unknown genetic defects, but not in four cases with SOD1 mutations or SOD1-ALS transgenic mice. These data indicate that the FUS inclusions are involved not only in mutant FUS-linked FALS, but also in sporadic ALS and non-SOD1 FALS, representing a new finding with potentially significant implications 8, 27. These data suggest that the pathogenic pathway of mutant SOD1-mediated ALS is largely independent from other forms of ALS, including mutant FUS-linked ALS. We then explored possible explanations for the apparent discrepancy between our data and previous reports, in which FUS-positive inclusions were not observed in SALS or FALS without FUS mutation 8, 25, 26. We found that the high pressure decloaking chamber technique might be more effective than boiling and microwave methods for antigen retrieval of FUS in the inclusions. Although the discrepancy in the immunostaining remains to be further studied, our data suggest that the FUS in the inclusions of spinal motor neurons may have a conformation that requires optimized conditions such as efficient antigen retrieval or that FUS may be a less abundant component in the inclusions compared to TDP43.
Three of the nine antibodies to FUS evaluated in the present study can be used for detection of FUS inclusions in spinal cord sections by immunostaining. The 11570-1-AP yielded the most robust signals. The HPA008784 also revealed FUS signals, but less robust than the 11570-1-AP. These two antibodies were generated using much longer polypeptides as immunogens (349 and 128 amino acids, respectively). The other polyclonal antibodies generated using much shorter polypeptides failed to detect FUS inclusions in ALS with immunostaining. A monoclonal antibody sc-47711 could also be used to detect FUS inclusions using immunofluorescence, but not by immunohistochemistry. This antibody was generated using the C-terminal half of FUS, but the epitope remains to be mapped. In ALS cases, both FUS and TDP43 were present in the inclusions. However, the presence of FUS in TDP43-negative inclusions in the aFTLD-U cases suggests that neuronal FUS pathology is independent of TDP43 pathology, at least in aFTLD-U. Although we did not observe apparent FUS-positive inclusions in non-SOD1 ALS cases with other six antibodies in this study, optimization of the immunostaining method may further increase the sensitivity for detection of the FUS pathology with other FUS antibodies.
A previous report demonstrated TDP43/ubiquitin inclusions in all evaluated cases of SALS, ALS/ dementia and SOD1-negative FALS 19. Our data support this observation. Confocal analysis demonstrated that the FUS-immunoreactive inclusions were also immunoreactive with antibodies to TDP43, ubiquitin and p62. Our study indicated that the FUS/TDP43/ubiquitin/p62-positive inclusions are a common feature in SALS, ALS/dementia, FUS or TDP43 mutation-linked FALS, and other SOD1-negative FALS. These data suggest that the SALS, ALS/dementia, FUS or TDP43 mutation-linked FALS and the other SOD1-negative FALS may share a common downstream pathway in the pathogenesis of motor neuron degeneration, although the initial cause of disease in each specific case may differ. Obviously, SOD1-linked ALS appears to have a pathogenic pathway that is independent, at least in part, from other forms of ALS.
The presence of FUS pathology in sporadic and most familial forms of ALS parallels that of TDP43 in ALS, β-amyloid in Alzheimer disease and α-synucle in Parkinson disease. Like TDP43, β-amyloid and α-synuclein, FUS is involved in the formation of inclusions in sporadic disease, even though genetic mutations only account for a small fraction of the familial form of these disorders. Moreover, FUS pathology has recently been identified in aFTLD-U 25, neuronal intermediate filament inclusion disease (NIFID) 26 and basophilic inclusion body disease (BIBD) 28. We showed FUS pathology in FTLD with or without progranulin mutations, as well as ALS/dementia. These data suggest that post-translational modification of FUS is involved in a wider range of neurodegenerative disorders in addition to motor neuron degeneration in ALS.
The TDP43 and FUS are multifunctional proteins with DNA/RNA binding properties. They have been implicated in transcription, splicing and transport of RNA, protein translation, microRNA biogenesis, apoptosis, and cell division 29, 30. The pathogenetic mechanisms by which FUS mutations cause motor neuron degeneration remain to be elucidated. It is not clear if these FUS/TDP43/ubiquitin/p62-positive inclusions per se present as toxic components to motor neurons, or if the formation of inclusions sequesters and thereby deprives neurons of other essential proteins, or RNA or if some other mechanism exists. Transgenic and knockout animal models may help address these issues. A previous study demonstrated that FUS-deficiency (FUS−/−) leads to perinatal death in mice, whereas heterozygous mice (FUS+/−) appear to have normal phenotype 31, favoring but not proving the hypothesis that mutations in FUS cause the disease through a gain of function mechanism. Co-localization of FUS and TDP43 in neuronal inclusions raises the possibility that interactions of FUS and TDP43 with other cellular components may be a common theme in motor neuron degeneration in most, if not all, of SOD1-negative forms of ALS. Investigation of the common pathway involving these post-translationally modified proteins may offer a novel avenue for developing therapies for most forms of ALS and related disorders. We believe that demonstration of FUS, along with TDP43 in SALS, non-SOD1 ALS and ALS/dementia is a major step forward in formulating a common pathogenetic pathway for these disorders.
Supplementary Material
ACKNOWLEDGEMENTS
The authors acknowledge the support of the National Institutes of Health (Grant NS050641), the Les Turner ALS Foundation, the Vena E. Schaff ALS Research Fund, the Harold Post Research Professorship, the Herbert and Florence C. Wenske Foundation, the David C. Asselin MD Memorial Fund and the Les Turner ALS Foundation/Herbert and Florence C. Wenske Professorship. We thank the following for retrieval of the autopsy tissues: Mary Myers, Ruth Macke, Patrick C. Allen, Christine M. Hulette, James S. Lewis and C. Ryan Miller. We thank Janice Caliendo for proofreading this manuscript.
Footnotes
CONFLICT OF INTEREST STATEMENT. None
REFERENCES
- 1.Deng HX, Hentati A, Tainer JA, et al. Amyotrophic lateral sclerosis and structural defects in Cu,Zn superoxide dismutase. Science. 1993;261:1047–1051. doi: 10.1126/science.8351519. [DOI] [PubMed] [Google Scholar]
- 2.Rosen DR, Siddique T, Patterson D, et al. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;362:59–62. doi: 10.1038/362059a0. [DOI] [PubMed] [Google Scholar]
- 3.Kabashi E, Valdmanis PN, Dion P, et al. TARDBP mutations in individuals with sporadic and familial amyotrophic lateral sclerosis. Nat Genet. 2008;40:572–574. doi: 10.1038/ng.132. [DOI] [PubMed] [Google Scholar]
- 4.Sreedharan J, Blair IP, Tripathi VB, et al. TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science. 2008;319:1668–1672. doi: 10.1126/science.1154584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Van Deerlin VM, Leverenz JB, Bekris LM, et al. TARDBP mutations in amyotrophic lateral sclerosis with TDP-43 neuropathology: a genetic and histopathological analysis. Lancet Neurol. 2008;7:409–416. doi: 10.1016/S1474-4422(08)70071-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pasinelli P, Brown RH. Molecular biology of amyotrophic lateral sclerosis: insights from genetics. Nat Rev Neurosci. 2006;7:710–723. doi: 10.1038/nrn1971. [DOI] [PubMed] [Google Scholar]
- 7.Kwiatkowski TJ, Jr., Bosco DA, Leclerc AL, et al. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science. 2009;323:1205–1208. doi: 10.1126/science.1166066. [DOI] [PubMed] [Google Scholar]
- 8.Vance C, Rogelj B, Hortobagyi T, et al. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science. 2009;323:1208–1211. doi: 10.1126/science.1165942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Belzil VV, Valdmanis PN, Dion PA, et al. Mutations in FUS cause FALS and SALS in French and French Canadian populations. Neurology. 2009;73:1176–1179. doi: 10.1212/WNL.0b013e3181bbfeef. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Corrado L, Del Bo R, Castellotti B, et al. Mutations of FUS Gene in Sporadic Amyotrophic Lateral Sclerosis. J Med Genet. 2009 doi: 10.1136/jmg.2009.071027. [DOI] [PubMed] [Google Scholar]
- 11.Drepper C, Herrmann T, Wessig C, et al. C-terminal FUS/TLS mutations in familial and sporadic ALS in Germany. Neurobiol Aging. 2009 doi: 10.1016/j.neurobiolaging.2009.11.017. [DOI] [PubMed] [Google Scholar]
- 12.Groen EJ, van Es MA, van Vught PW, et al. FUS mutations in familial amyotrophic lateral sclerosis in the Netherlands. Arch Neurol. 2010;67:224–230. doi: 10.1001/archneurol.2009.329. [DOI] [PubMed] [Google Scholar]
- 13.Lai SL, Abramzon Y, Schymick JC, et al. FUS mutations in sporadic amyotrophic lateral sclerosis. Neurobiol Aging. 2010 doi: 10.1016/j.neurobiolaging.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tateishi T, Hokonohara T, Yamasaki R, et al. Multiple system degeneration with basophilic inclusions in Japanese ALS patients with FUS mutation. Acta Neuropathol. 2010;119:355–364. doi: 10.1007/s00401-009-0621-1. [DOI] [PubMed] [Google Scholar]
- 15.Lansbury PT, Lashuel HA. A century-old debate on protein aggregation and neurodegeneration enters the clinic. Nature. 2006;443:774–779. doi: 10.1038/nature05290. [DOI] [PubMed] [Google Scholar]
- 16.Leigh PN, Whitwell H, Garofalo O, et al. Ubiquitin-immunoreactive intraneuronal inclusions in amyotrophic lateral sclerosis. Morphology, distribution, and specificity. Brain. 1991;114(Pt 2):775–788. doi: 10.1093/brain/114.2.775. [DOI] [PubMed] [Google Scholar]
- 17.Lowe J. New pathological findings in amyotrophic lateral sclerosis. J Neurol Sci. 1994;124(Suppl):38–51. doi: 10.1016/0022-510x(94)90175-9. [DOI] [PubMed] [Google Scholar]
- 18.Shibata N, Hirano A, Kobayashi M, et al. Intense superoxide dismutase-1 immunoreactivity in intracytoplasmic hyaline inclusions of familial amyotrophic lateral sclerosis with posterior column involvement. J Neuropathol Exp Neurol. 1996;55:481–490. doi: 10.1097/00005072-199604000-00011. [DOI] [PubMed] [Google Scholar]
- 19.Mackenzie IR, Bigio EH, Ince PG, et al. Pathological TDP-43 distinguishes sporadic amyotrophic lateral sclerosis from amyotrophic lateral sclerosis with SOD1 mutations. Ann Neurol. 2007;61:427–434. doi: 10.1002/ana.21147. [DOI] [PubMed] [Google Scholar]
- 20.Neumann M, Sampathu DM, Kwong LK, et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–133. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- 21.Deng HX, Shi Y, Furukawa Y, et al. Conversion to the amyotrophic lateral sclerosis phenotype is associated with intermolecular linked insoluble aggregates of SOD1 in mitochondria. Proc Natl Acad Sci U S A. 2006;103:7142–7147. doi: 10.1073/pnas.0602046103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deng HX, Jiang H, Fu R, et al. Molecular dissection of ALS-associated toxicity of SOD1 in transgenic mice using an exon-fusion approach. Hum Mol Genet. 2008;17:2310–2319. doi: 10.1093/hmg/ddn131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gurney ME, Pu H, Chiu AY, et al. Motor neuron degeneration in mice that express a human Cu,Zn superoxide dismutase mutation. Science. 1994;264:1772–1775. doi: 10.1126/science.8209258. [DOI] [PubMed] [Google Scholar]
- 24.Nakano T, Nakaso K, Nakashima K, Ohama E. Expression of ubiquitin-binding protein p62 in ubiquitin-immunoreactive intraneuronal inclusions in amyotrophic lateral sclerosis with dementia: analysis of five autopsy cases with broad clinicopathological spectrum. Acta Neuropathol. 2004;107:359–364. doi: 10.1007/s00401-004-0821-7. [DOI] [PubMed] [Google Scholar]
- 25.Neumann M, Rademakers R, Roeber S, et al. Frontotemporal lobar degeneration with FUS pathology. Brain. 2009 doi: 10.1093/brain/awp214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Neumann M, Roeber S, Kretzschmar HA, et al. Abundant FUS-immunoreactive pathology in neuronal intermediate filament inclusion disease. Acta Neuropathol. 2009 doi: 10.1007/s00401-009-0581-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lagier-Tourenne C, Cleveland DW. Rethinking ALS: the FUS about TDP-43. Cell. 2009;136:1001–1004. doi: 10.1016/j.cell.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Munoz DG, Neumann M, Kusaka H, et al. FUS pathology in basophilic inclusion body disease. Acta Neuropathol. 2009 doi: 10.1007/s00401-009-0598-9. [DOI] [PubMed] [Google Scholar]
- 29.Andersson MK, Stahlberg A, Arvidsson Y, et al. The multifunctional FUS, EWS and TAF15 proto-oncoproteins show cell type-specific expression patterns and involvement in cell spreading and stress response. BMC Cell Biol. 2008;9:37. doi: 10.1186/1471-2121-9-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Buratti E, Baralle FE. Multiple roles of TDP-43 in gene expression, splicing regulation, and human disease. Front Biosci. 2008;13:867–878. doi: 10.2741/2727. [DOI] [PubMed] [Google Scholar]
- 31.Hicks GG, Singh N, Nashabi A, et al. Fus deficiency in mice results in defective B-lymphocyte development and activation, high levels of chromosomal instability and perinatal death. Nat Genet. 2000;24:175–179. doi: 10.1038/72842. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.