Abstract
Tumor necrosis factor (TNF) and the members of the interferon (IFN) family are major inducible cytokines that function to counteract viral infections or cellular transformation. Recently, our lab has characterized a novel antiviral state which is induced in primary human fibroblasts by co-treatment with TNF plus IFNβ. Here, we demonstrate that this synergistic state is both antiviral and cytostatic for primary human cells. Significantly, we observed that a wide spectrum of transformed human cancer cells have universally lost the ability to induce the TNF/IFNβ synergistic state, as defined by three separate criteria. We hypothesize that the ability to induce the TNF/IFNβ synergistic state is a unique feature of primary cells and is incompatible with cellular immortalization and/or transformation.
Keywords: TNF, IFN, Synergy, Cancer, Cytostasis
1. Introduction
Complex organisms have evolved many innate defense mechanisms to respond to pathogen invasion or aberrant cellular states such as tumorigenic transformation. Some of these response mechanisms involve the induction and release of signaling cytokines such as interleukins, interferons (IFNs), and tumor necrosis factor (TNF) [1]. These cytokines bind to their cognate receptors on target cells and activate signaling cascades leading to the induction of novel gene products which mediate the antiviral and/or cytostatic states [1]. The capacity of individual cytokines, such as TNF or IFN, to induce an antiviral state or to mediate a cytostatic response in somatic cells has been studied extensively [1,2], however, it is far more likely that these self-defense cytokines are co-induced and act synergistically in virus-infected tissues or within complex tumor beds.
The rabbit-specific poxvirus myxoma (MV) is an example of a nonhuman virus which is unable to productively infect any animal except its natural host, in this case lagomorphs, but freely replicates in a variety of transformed cancer cells in vitro and in vivo [3–5]. Due to these characteristics, MV is currently being studied as a potential oncolytic therapeutic against cancer cells [6]. Recently, our lab has shown that treatment of primary human fibro-blasts with either TNF or IFNβ alone leads to a partial inhibition of MV [7]. Simultaneous treatment of primary human fibroblasts with TNF plus IFNβ, however, induces a novel synergistic antiviral state which completely abrogates MV replication and spread [7,8]. Microarray analysis revealed that the TNF/IFNβ synergistic state induced in primary human fibroblasts is highly distinct from the anti-viral states induced by addition of either cytokine alone and is characterized by significantly higher upregulation of several hundred cellular genes which were co-induced by the individual cytokines, plus induction of approximately 850 novel genes which were not upregulated by either cytokine alone [8].
In an effort to better understand how the TNF/IFNβ synergistic state might influence MV as a oncolytic virus for cancer therapy, we investigated whether co-treatment with these cytokines synergistically inhibited MV replication in a spectrum of normal primary human cells and transformed human cancer cells. Our results unexpectedly demonstrated that while treatment of all tested primary human cells with TNF plus IFNβ synergistically inhibits MV replication and spread, the identical cytokine treatment of a diverse spectrum of adherent and nonadherent transformed human cancer cell lines uniformly failed to abrogate MV amplification beyond the effect seen with either individual cytokine. Further examination revealed that treatment of transformed human cancer cell lines with TNF plus IFNβ failed to upregulate the canonical gene expression events associated with the TNF/IFNβ synergistic state in normal primary human fibroblasts or endothelial cells. Additionally, treatment of primary human cells with TNF plus IFNβ is uniformly cytostatic, but treatment of a wide spectrum of human cancer cell lines with both cytokines fails to synergistically block cellular proliferation beyond the levels observed using either cytokine alone. Taken together, using three independent criteria to measure induction of TNF/IFNβ synergy, we conclude that the ability to induce the synergistic TNF/IFNβ antiviral and cytostatic state is a unique property of primary human cells which is uniformly and uniquely defective in a wide spectrum of human cancer cells.
2. Materials and methods
2.1. Cell culture, reagents, and infections
GM02504 (Coriell Institute), 1059sk (ATCC# CRL-2072), Bud8 (ATCC# CRL-1554), Hek293 (ATCC# CRL-1573), 786-0 (ATCC# CRL-1932), T47D (ATCC# HTB-133), Caki (ATCC# HTB-46), Du145 (ATCC# HTB-81), Panc1 (ATCC# CRL-1469), HeLa (ATCC# CCL-2), A549 (ATCC# CCL-185), Hos (ATCC# CRL-1543), Hs913T (ATCC# HTB-152), Vero (ATCC# CCL-81), BHK (ATCC# CCL-10), BGMK [7] and MDA231 (ATCC# HTB-129) were maintained in DMEM (Gibco). Human Vascular Endothelial Cells (HUVEC) (ATCC# PCS-100-010) were maintained in endothelial cell growth media (Lonza– Biowhittiker). Hct116 cells (ATCC# CCL-247) were maintained in McCoys 5a Media (Gibco). BJAB and Namalwa cells (a generous gift of Dr. Sankar Swaminathan), KG-1 cells (a generous gift of Dr. Christopher R. Cogle), and CCRF-CEM cells (ATCC# CCL-119) were maintained in RPMI (Gibco). All media was supplemented with 10% FBS, 1× pen/strep, and 2 mM l-glutamine. Recombinant human TNF (Biosource) was used at a final concentration of 20 ng/ml. Recombinant human IFNβ (PBL Biomedical laboratories) was used at a final concentration of 500 U/ml.
Infections were done by removing existing media and replacing with a minimal amount of complete DMEM containing MV-GFP [9]. The virus was allowed to adsorb at 37° for 1 h after which media containing virus was removed and replaced with fresh media containing the indicated cytokines.
2.2. Immuno-florescence and flow cytometry
Cells (2.5 × 104) were plated in each well of a 96 well dish. The following day, cells were infected with MV-GFP at an MOI = 0.1. At 24, 48, and 72 h after infection the size and shape of GFP+ foci were observed using a Leica DMI 6000B microscope. Cells were then harvested using trypsin, fixed in 2% paraformaldehyde, and the percent of GFP+ cells was quantified on a BD FACScaliber.
2.3. cDNA synthesis
Total RNA (1–2 lg) was for each cDNA synthesis. Genomic DNA was removed from total RNA using the DNA-free™ kit (Ambion) as per the manufacturers’ recommendations. Following removal of genomic DNA, 1 μl of dNTP's (100 mM) and 1 ll of random hexamer primers (50 μg/ml) were added and the mixture was incubated for 5 min at 65°. The tube was then allowed to cool and 6 μl 5× Reaction Buffer, 3 μl DTT (0.1 M), 1 ll RNAsin (Promega), and 1 μl Superscript III™ reverse transcriptase (Invitrogen) were added followed by incubations of 1 h at 42° and for 15 min at 72°. The final reaction was diluted 1:10 with sterile H2O.
2.4. Real-time PCR
Four microliter of diluted cDNA was added to 21 μl of PCR mix containing: 0.5 U Taq polymerase (NEB), 1× Thermo Pol Buffer, 0.1× Sybr-Green (Molecular Probes), 0.5× Rox reference dye (Invitrogen), 160 μM dNTPs (Invitrogen), 4 mM MgCl (Invitrogen), 4 ng forward primer, and 4 ng reverse primer. The resulting 25 μl reaction was run on an ABI 7300 real time PCR machine using the following conditions: 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 1 min. Primers used in real time PCR analysis are listed in Supplemental Table 2.
2.5. Cell growth assay
Cells (1 × 104) were plated in each well of a 12 well dish. The following day cells were either mock treated or treated with TNF, IFNβ, or TNF plus IFNβ. The cells were then allowed to grow until the mock treated well had reached 100% confluence refreshing media and cytokines every 48 h. When the mock treated samples reached confluence, all cells were trypsinized and the number of cells was determined using a hemocytometer.
3. Results
3.1. TNF plus IFNβ fails to synergistically inhibit MV in transformed human cancer cells
Previously, our lab has shown that untreated primary human GM02504 fibroblasts are fully permissive for MV [7,8]. Treatment of these cells with either TNF or IFNβ results in a partial restriction of MV replication and spread, whereas complete abrogation of viral growth occurs only following co-treatment with TNF plus IFNβ [8]. To determine if the antiviral phenotype caused by TNF plus IFNβ was specific to GM02504 fibroblasts, we expanded our analysis to include three additional primary human cells: Bud8 and 1059sk fibroblasts, and human vascular micro-endothelial cells (HUVEC), as well as a spectrum of transformed human cancer cell lines (Supplemental Table 1).
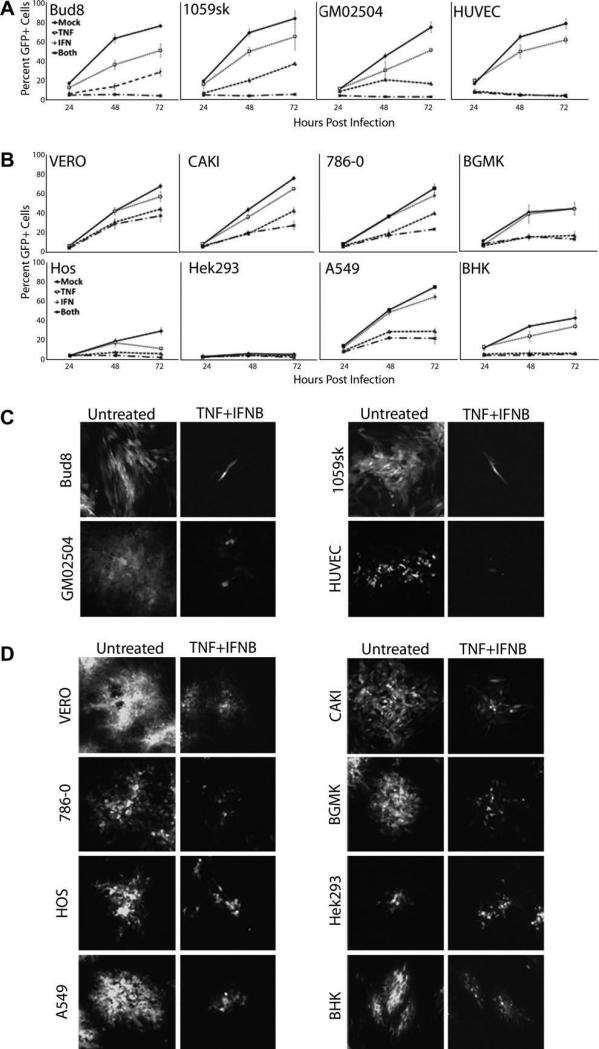
Cells were infected with MV that expresses GFP (MV-GFP) at an MOI = 0.1 and then either mock treated or treated with TNF, IFNβ, or TNF plus IFNβ. At 24, 48, and 72 h post infection, individual foci were examined by florescence microscopy and then cells were trypsinized and the percent of GFP+ cells analyzed using flow cytometry. Consistent with our previous results in GM02504 fibro-blasts, treatment of the three other primary human cells (Bud8, 1059sk, and HUVEC) with either TNF or IFNβ alone led to varying degrees of partial MV-GFP inhibition while treatment with both cytokines completely abrogated MV replication and spread in all of these cells (Fig. 1A and C). Similar to what was observed in primary cells, treatment of either human or primate transformed cell lines with either TNF or IFNβ usually led to a partial restriction of MV spread; however, in striking contrast to our results in primary human cells, none of the transformed cell lines were able to completely inhibit MV spread even following treatment with TNF plus IFNβ (Fig. 1B and D).
Fig. 1.
TNF plus IFNβ is unable to completely abrogate MV growth in transformed cells. The indicated primary (A) or transformed cell lines (B) were infected with MV-GFP at an MOI = 0.1 and then treated with TNF, IFNβ, or TNF plus IFNβ. At the indicated times post infection cells were trypsinized and the number of GFP+ cells was determined via flow cytometry. To determine the affect treatment with TNF, IFNβ, or TNF plus IFNβ had on MV-GFP foci formation the indicated primary (C) or transformed cell lines (D) were infected with MV-GFP at an MOI = 0.1 and then treated with TNF, IFNβ, or TNF plus IFNβ. Seventy-two hours post infection, MV foci formation was observed using fluorescent microscopy to track GFP+ cells.
3.2. Transformed human cancer cells are unable to upregulate consensus cellular gene markers of TNF–IFNβ synergy
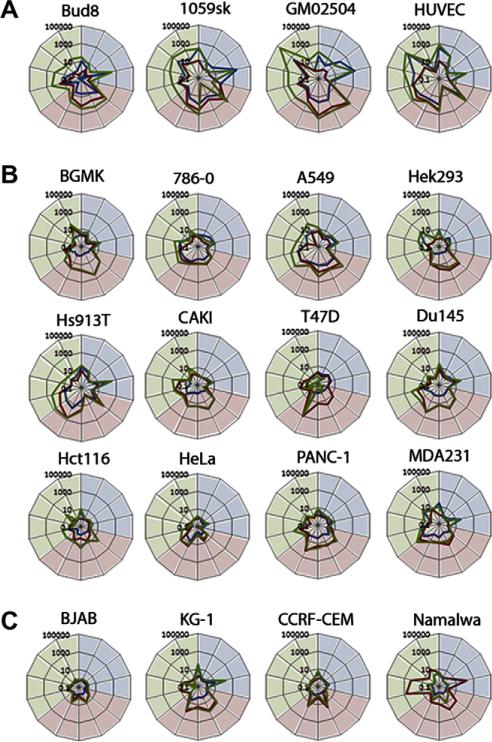
We next determined if the transformed human cancer cell lines also lacked the ability to induce the unique cellular genes associated with the TNF/IFNβ synergistic state. Our previous microarray analysis had demonstrated that, in primary human fibroblasts, the TNF/IFNβ synergistic state is characterized by increased fold-induction of a set of several hundred co-induced cellular genes as well as induction of a over 800 novel genes not induced by either cytokine alone [8]. To assess if we could use this characteristic profile of gene upregulation to evaluate induction of the TNF/IFNβ synergistic state, we selected 14 cellular marker genes predominantly induced in primary fibroblasts by either; TNF (BIRC3, CCL2, CXCL5, IL8), IFNβ (ISG15, ISG54, IFIT1, IFIT2, OAS1) or TNF plus IFNβ (INDO, GBP4, CXCL10, VCAM1, CCL5). The four primary human cells were treated with TNF, IFNβ, or TNF plus IFNβ, and at 24 h after treatment, RNA was extracted from each cell and the induction of each marker gene was measured using real-time PCR (rtPCR). The data is displayed with the genes predominantly induced by TNF (shaded in blue), those predominantly induced by IFNβ (shaded in red) and those induced only by TNF plus IFNβ (shaded in green). The fold-induction, compared to control cells which received no cytokine, of each gene is shown following treatment with TNF (blue line), IFNβ (red line), or TNF plus IFNβ (green line) (Fig. 2A). While the level of induction of each gene differed somewhat for each primary human cell tested, three responses were consistently observed: (1) all the TNF and IFNβ marker genes were induced following treatment with each cytokine alone (bulges in the red and blue lines in the correspondingly shaded areas), (2) an enhanced fold-increase in co-induced genes was uniformly observed following treatment with TNF plus IFNβ (i.e. the green line is consistently outside the red and blue lines), and (3) induction of those cellular genes characteristic of the TNF/IFNβ synergistic state is observed only following treatment with both cytokines (i.e. a bulge in the green line in the green shaded area).
Fig. 2.
Transformed cells are unable to induce markers of the TNF/IFNβ synergistic state. Cells (1 × 106) of the indicated type were plated in each well of a six well dish. The following day cells were either mock treated or treated with TNF, IFNβ, or TNF plus IFNβ. Twenty-four hours after treatment RNA was extracted and cDNA synthesized. rtPCR was used to track the induction of 14 marker genes in primary cells (A), transformed adherent cells (B), and transformed lymphoma cells (C). Each graph depicts the fold induction compared to mock of marker genes in response to TNF (blue line), IFNβ (red line), or TNF plus IFNβ (green line). Marker genes tested (clockwise from top): (induced by TNF and shaded in blue) BIRC3, CCL2, CXCL5, IL8, (induced by IFNβ and shaded in red) ISG15, ISG54, IFIT1, IFIT2, OAS1, (induced by TNF plus IFNβ and shaded in green) INDO, GBP4, CXCL10, VCAM1, and CCL5.
We next tested whether transformed human cancer cells were able to induce the same spectrum of characteristic gene upregulations. We measured the induction of the 14 defining cellular marker genes in 12 adherent cancer cell lines of epithelial or fibroblastic origin (Fig. 2B) and four nonadherent lymphoid cancer cell lines (Fig. 2C). Although defects in some of the single cytokine gene inductions were noted, the majority of tested cancer cells displayed induction of at least one marker gene associated with either TNF or IFNβ responsiveness in primary human cells. While the level of induction of these diagnostic genes was frequently lower than that observed in primary human cells, these data suggest that most of the human cancer cell lines tested retain at least some ability to both sense and respond to TNF and/or IFNβ. Thus, complete functional loss of the TNF or type I IFN receptors was relatively rare, and most cancer cells were capable of at least some downstream signaling and gene induction from both of these receptors. In contrast, treatment of 15 of the 16 cancer cell lines tested with TNF plus IFNβ failed to induce either enhanced upregulation of co-induced genes or induction of the unique TNF/IFNβ synergistic marker genes. In the one human cancer cell line which did display some partial synergistic characteristics of gene inductions, A549 lung carcinoma cells, both the levels of enhanced gene upregulation and induction of the unique TNF/IFNβ synergy marker genes was significantly reduced compared to all four primary human cells tested.
3.3. Failure to induce the TNF/IFNβ synergistic state in human cancer cells is not simply a kinetic defect in target gene upregulations
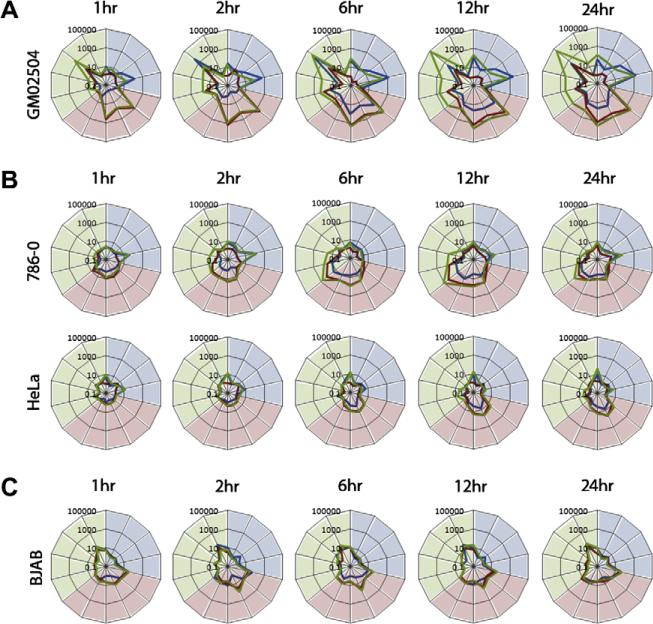
Since various cell types can induce gene upregulations in response to TNF and/or IFNβ with different kinetics, we wanted to rule out the possibility that the apparent failure of transformed cells to induce the TNF/IFNβ synergistic state was due to an alteration of marker gene upregulation kinetics between primary and transformed human cells. To address this, we tested the cytokine response kinetics to TNF, IFNβ, or TNF plus IFNβ in primary GM02504 human fibroblasts and three different transformed human cancer cell lines that we characterized as TNF/IFNβ synergy-defective (HeLa, 786-0, and BJAB). Cells were either mock treated or treated with TNF, IFNβ, or TNF plus IFNβ. At the indicated time points, RNA was extracted and cDNA was synthesized for rtPCR analysis. In primary GM02504 human fibroblasts, the diagnostic marker genes for TNF, IFNβ, and TNF plus IFNβ were all upregulated within 1 h following cytokine addition, continued to increase until 12 h and were maintained through 24 h (Fig. 3A). In contrast, 786-0 human renal carcinoma cells did not respond to addition of cytokines until 2 h and their response peaked at 12 h then subsided (Fig. 3B). At no point did the robustness of the 786-0 cell gene inductions in response to cytokine compare to that observed in the primary GM02504 fibroblasts. HeLa and BJAB cells showed only minimal responses to TNF, IFNβ, or TNF plus IFNβ at any time point analyzed (Fig. 3B and C).
Fig. 3.
The inability of transformed cells to respond to TNF plus IFNβ is not due to altered kinetics. Cells (1 × 106) were plated in each well of a six well dish. The following day cells were either mock treated or treated with TNF, IFNβ, or TNF plus IFNβ. At each indicated time point, cells were harvested and frozen. After all time points were collected, RNA was extracted and used for synthesis of cDNA. rtPCR was then used to measure the response of each cell to each treatment at each time point. Each graph depicts the fold induction compared to mock of 14 chosen marker genes in response to TNF (blue line), IFNβ (red line), or TNF plus IFNβ (green line). Marker genes are (clockwise from top): (induced by TNF and shaded in blue) BIRC3, CCL2, CXCL5, IL8, (induced by IFNβ and shaded in red) ISG15, ISG54, IFIT1, IFIT2, OAS1, (induced by TNF plus IFNβ and shaded in green) INDO, GBP4, CXCL10, VCAM1, and CCL5.
3.4. TNF plus IFNβ synergistically inhibits cellular proliferation of primary but not transformed human cells
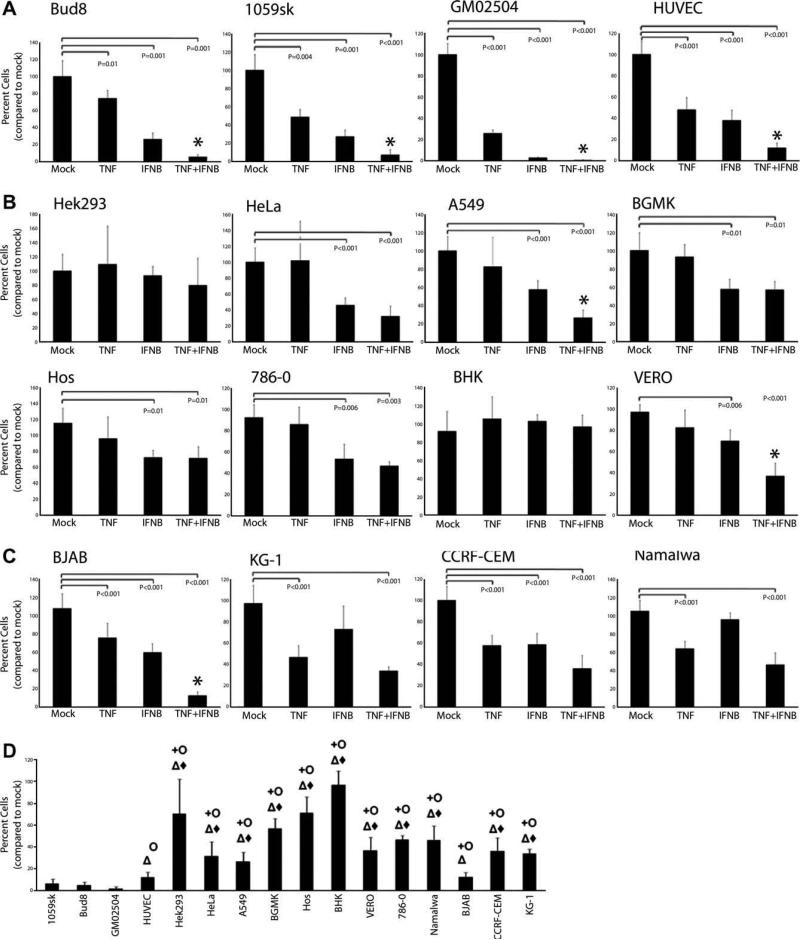
The failure of all transformed human cancer cells tested to induce the diagnostic genes associated with the TNF/IFNβ synergistic state suggests that this state might be incompatible with tumorigenic cell proliferation. To determine if the TNF/IFNβ synergistic state affects cellular proliferation, we tested the ability of both primary and transformed cells to propagate following treatment with TNF, IFNβ, or TNF plus IFNβ. 1 × 104 cells of each cell type were plated in a 12-well dish, 24 h later cells were mock treated or treated with TNF, IFNβ, or TNF plus IFNβ and allowed to grow to confluence as detailed in Section 2.5. When the control cells, which had received no cytokine, reached confluence the cell numbers were quantified using a hemocytometer. As expected, treatment with TNF or IFNβ alone significantly reduced the proliferation rates of all four primary human cells tested (Fig. 4A). Interestingly, treatment of all four cell primary cells with TNF plus IFNβ resulted in greater than 90% growth inhibition, a level which was significantly more profound than that caused by either TNF or IFNβ alone. Following the same treatment, replication of 10 of the 12 transformed human cancer cells tested was moderately reduced by treatment with either TNF or IFNβ alone (Fig. 4B and C). Interestingly, the adherent human cancer cell lines tended to be inhibited by IFNβ but not by TNF, while the human lymphoma cell lines tended to be inhibited by TNF but not by IFNβ. In contrast, only two transformed human cancer cells (A549, and BJAB) and one transformed monkey cell line (VERO) showed synergistically reduced cellular proliferation following treatment with TNF plus IFNβ. Even in these three transformed cell lines, however, the level of cytostasis was almost always significantly less than that observed in all four primary human cells tested (Fig. 4D).
Fig. 4.
TNF plus IFNβ synergistically inhibits cellular replication of primary but not transformed cells 1 × 104 cells were plated in each well of a 12 well dish. The following day cells were either mock treated or treated with TNF, IFNβ or TNF plus IFNβ. Cultures of primary cells (A) transformed adherent cells (B) or transformed lymphoma cells (C) were then incubated at 37° replenishing the media and cytokines every 48 hours until the mock treated sample had reached confluence. The number of cells in each treatment group was then determined using a hemocytometer. Each graph depicts the average percent of cells in each treatment group compared to mock (N = 8). Students T-test was used to determine statistically significant reductions (P < 0.01). Cells were considered to have a synergistic response (*) if the level of replication inhibition following treatment with TNF plus IFNβ was significantly greater than that observed following treatment with both TNF and IFNβ alone. (D) Depicts the replication inhibition of each transformed cell compared to all four primary cells in response to TNF plus IFNβ. Cells which displayed a significantly lower level of replication inhibition (students T-test P < 0.01) than each primary cell are marked as follows: 1059sk (+), Bud8 (J), GM02504 (Δ), HUVEC (◆).
4. Discussion
Our previous findings characterized a novel upregulated gene set associated with the synergistic antiviral state that is induced by co-treatment of primary human fibroblasts with TNF plus IFNβ [8]. Our current work expands the impact of this synergy by demonstrating that, while treatment of primary human cells with both TNF and IFNβ synergistically inhibits both viral and cellular replication, the ability to induce this state is either absent or severely compromised in all transformed human cancer cells we have tested. This failure of transformed human cancer cells to induce TNF/IFNβ synergy cannot be explained by defects or loss in the TNF or IFN receptors, since virtually all the human cancer cells tested induced at least some of the characteristic marker genes in response to either cytokine individually. We propose that this defect in the ability of human cancer cells to induce the TNF/IFNβ synergistic antiviral and cytostatic state is a genetic or phenotypic requirement for cellular transformation and/or immortalization.
Our data measures the induction of the TNF/IFNβ synergistic state using three different criteria: viral spread, unique cellular gene upregulations and cellular proliferation. The human cancer cell lines tested each display different levels of impairment of TNF/IFNβ synergy, depending on which criteria are used. These data suggest that the TNF/IFNβ synergistic response likely involves the induction of more than one unique signaling pathway, and that these pathways can be defective to different degrees in different human cancer cells. Importantly, however, the level of TNF/IFNβ synergy in all tested primary human cells is far more extensive than that observed in any of the transformed human cells, regardless of which criteria is used to measure synergy.
Consistent with the hypothesis that abrogation of the ability to induce the TNF/IFNβ synergistic state is a prerequisite for cellular immortalization and/or transformation, all previous reports describing TNF/IFNβ synergistic responses have been performed only in primary cells, such as human macrophages [10], human fibroblasts [8,11], or rat neuronal cells [12]. Interestingly, there is a large body of literature demonstrating that TNF can also synergize with IFNγ, but this synergistic state is demonstrably different from that induced by TNF plus type I IFN [13]. In contrast to our findings with TNF plus IFNβ, however, both primary and transformed cells can induce a synergistic state in response to TNF plus IFNγ [14–16]. These data suggest that the loss of the ability to induce synergy in response to TNF plus IFNβ plays a unique role in cellular transformation, either by specifically inhibiting oncogenesis in vivo or else by being functionally incompatible with cellular immortalization and/or transformation in general.
Interestingly, while many adherent human cancer cells displayed a strong correlation between marker gene induction and the antiviral or antiproliferative phenotype, several of the human lymphoma lines deviated significantly. For example, human Namalwa lymphoma cells display a dramatic induction of the classic IFN marker genes in response to IFNβ (Fig. 2) but this cytokine caused no obvious antiproliferative phenotype (Fig. 4). Conversely, human BJAB lymphoma cells exhibit a large antiproliferative phenotype in response to TNF, IFNβ, or TNF plus IFNβ, but display very little induction of any of the diagnostic marker genes that define these cytokine responses in primary human cells (Compare Fig. 2 to Figs. 1 and 4). We speculate that more detailed analysis of the genes induced by TNF plus IFNβ in a variety of primary and cancer cells types could lead to identification of the functional gene products mediating both the antiviral and antiproliferative affects of TNF/IFNβ synergy.
Finally, previous work has shown that the replication and spread of several candidate oncolytic viruses, including MV, can be dramatically inhibited by the TNF/IFNβ synergistic state in primary human cells [7,8,12]. Viruses which are strongly inhibited by this TNF/IFNβ synergistic state in primary human cells would also be especially safe candidates for oncolytic virotherapy in humans since either ectopic or induced TNF plus IFNβ would not impede virus spread through tumor tissue, in which the target cancer cells uniformly harbor defects in synergy, but would be potently antiviral in normal tissues.
Supplementary Material
Acknowledgment
GM was previously supported by an International Scholarship Award from the Howard Hughes Medical Institute (HHMI).
Footnotes
Publisher's Disclaimer: This article appeared in a journal published by Elsevier. The attached copy is furnished to the author for internal non-commercial research and education use, including for instruction at the authors institution and sharing with colleagues. Other uses, including reproduction and distribution, or selling or licensing copies, or posting to personal, institutional or third party websites are prohibited. In most cases authors are permitted to post their version of the article (e.g. in Word or Tex form) to their personal website or institutional repository. Authors requiring further information regarding Elsevier's archiving and manuscript policies are encouraged to visit: http://www.elsevier.com/copyright
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.cyto.2009.06.006.
References
- 1.Remick D, Friedland J. Cytokines in health and disease. Marcel Dekker, Inc.; New York, NY: 1997. [Google Scholar]
- 2.Weinberg R. The Biology of Cancer: Garland Science. 2007 [Google Scholar]
- 3.Kerr P, McFadden G. Immune responses to myxoma virus. Viral Immunol. 2002;15:229–46. doi: 10.1089/08828240260066198. [DOI] [PubMed] [Google Scholar]
- 4.Stanford MM, Werden SJ, McFadden G. Myxoma virus in the European rabbit: interactions between the virus and its susceptible host. Vet Res. 2007;38:299–318. doi: 10.1051/vetres:2006054. [DOI] [PubMed] [Google Scholar]
- 5.Fenner F, Poole WE, Marshall ID, Dyce AL. Studies in the epidemiology of infectious myxomatosis of rabbits. VI. The experimental introduction of the European strain of myxoma virus into Australian wild rabbit populations. J Hyg (Lond) 1957;55:192–206. doi: 10.1017/s0022172400037104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stanford MM, McFadden G. Myxoma virus and oncolytic virotherapy: a new biologic weapon in the war against cancer. Expert Opin Biol Ther. 2007;7:1415–25. doi: 10.1517/14712598.7.9.1415. [DOI] [PubMed] [Google Scholar]
- 7.Wang F, Gao X, Barrett JW, Shao Q, Bartee E, Mohamed MR, et al. RIG-I mediates the co-induction of tumor necrosis factor and type I interferon elicited by myxoma virus in primary human macrophages. PLoS Pathog. 2008;4:e1000099. doi: 10.1371/journal.ppat.1000099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bartee E, Mohamed MR, Lopez MC, Baker H, McFadden G. Addition of TNF plus IFN-{beta} induces a novel synergistic anti-viral state against poxviruses in primary human fibroblasts. J Virol. 2008;83:498–511. doi: 10.1128/JVI.01376-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnston JB, Barrett JW, Chang W, Chung CS, Zeng W, Masters J, et al. Role of the serine-threonine kinase PAK-1 in myxoma virus replication. J Virol. 2003;77:5877–88. doi: 10.1128/JVI.77.10.5877-5888.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yarilina A, Park-Min KH, Antoniv T, Hu X, Ivashkiv LB. TNF activates an IRF1-dependent autocrine loop leading to sustained expression of chemokines and STAT1-dependent type I interferon-response genes. Nat Immunol. 2008;9:378–87. doi: 10.1038/ni1576. [DOI] [PubMed] [Google Scholar]
- 11.Reis LF, Ho Lee T, Vilcek J. Tumor necrosis factor acts synergistically with autocrine interferon-beta and increases interferon-beta mRNA levels in human fibroblasts. J Biol Chem. 1989;264:16351–4. [PubMed] [Google Scholar]
- 12.Schijns VE, Van der Neut R, Haagmans BL, Bar DR, Schellekens H, Horzinek MC. Tumour necrosis factor-alpha, interferon-gamma and interferon-beta exert antiviral activity in nervous tissue cells. J Gen Virol. 1991;72(Pt. 4):809–15. doi: 10.1099/0022-1317-72-4-809. [DOI] [PubMed] [Google Scholar]
- 13.Bartee E, Mohamed MR, McFadden G. Tumor necrosis factor and interferon: cytokines in harmony. Curr Opin Microbiol. 2008;11:378–83. doi: 10.1016/j.mib.2008.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Watanabe N, Niitsu Y, Yamauchi N, Umeno H, Sone H, Neda H, et al. Antitumor synergism between recombinant human tumor necrosis factor and recombinant human interferon-r. J Biol Response Mod. 1988;7:24–31. [PubMed] [Google Scholar]
- 15.Sainz B, Jr, Halford WP. Alpha/beta interferon and gamma interferon synergize to inhibit the replication of herpes simplex virus type 1. J Virol. 2002;76:11541–50. doi: 10.1128/JVI.76.22.11541-11550.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yao Q, Qian P, Cao Y, He Y, Si Y, Xu Z, et al. Synergistic inhibition of pseudorabies virus replication by porcine alpha/beta interferon and gamma interferon in vitro. Eur Cytokine Netw. 2007;18:71–7. doi: 10.1684/ecn.2007.0088. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.