SUMMARY
Hepatocytes provide most liver functions, but they can also proliferate and regenerate the liver after injury. However, under some liver injury conditions, particularly chronic liver injury where hepatocyte proliferation is impaired, liver stem cells (LSCs) are thought to replenish lost hepatocytes. Conflicting results have been reported about the identity of LSCs and their contribution to liver regeneration. To address this uncertainty, we followed candidate LSC populations by genetic fate tracing in adult mice with chronic liver injury due to a choline-deficient, ethionine-supplemented diet. In contrast to previous studies, we failed to detect hepatocytes derived from biliary epithelial cells or mesenchymal liver cells beyond a negligible frequency. In fact, we failed to detect hepatocytes that were not derived from pre-existing hepatocytes. In conclusion, our findings argue against LSCs, or other nonhepatocyte cell types, providing a backup system for hepatocyte regeneration in this common mouse model of chronic liver injury.
INTRODUCTION
The adult liver is unique in its ability to efficiently regenerate after injury. Under most circumstances, liver function is restored through replacement of damaged hepatocytes by self-duplication of remaining hepatocytes. However, when hepatocyte proliferation is impaired—as under chronic injury conditions—other cells may contribute to liver regeneration by giving rise to hepatocytes (Itoh and Miyajima, 2014).
Liver stem cells (LSCs) have long been favored as the most likely alternative source of hepatocytes in the adult liver. In the classical view, LSCs are nonhepatocyte precursors of highly proliferative progenitor cells that can differentiate into both hepatocytes and biliary epithelial cells (BECs), thereby providing a backup system for liver regeneration (Duncan et al., 2009). In support of this view, cells that are bipotential in vitro can be isolated from the adult mouse liver (Dorrell et al., 2011; Huch et al., 2013; Shin et al., 2011). These cells exhibit markers of BECs, which accords with numerous studies locating LSCs in biliary structures, particularly at the interphase of bile ducts and hepatocyte plates (Itoh and Miyajima, 2014). However, specific markers of LSCs have not been identified, and therefore no direct evidence currently exists for a contribution from LSCs to hepatocytes in vivo.
In the absence of specific LSC markers, researchers have resorted to using broader lineage markers to delineate alternative cell sources of hepatocytes in vivo. Genetic fate-tracing studies in mice based on SRY (sex determining region Y) box 9 (Sox9), osteopontin (Opn), or hepatocyte nuclear factor 1 beta (Hnf1β) expression support that cells within the BEC population can differentiate into hepatocytes (Español-Suñer et al., 2012; Furuyama et al., 2011; Rodrigo-Torres et al., 2014). Other studies of fate tracing using a human glial fibrillary acidic protein (GFAP) promoter reported that stellate cells—a mesenchymal liver cell type at the center of liver fibrosis—can give rise to new hepatocytes (Michelotti et al., 2013; Swiderska-Syn et al., 2014; Yang et al., 2008). In addition, hematopoietic cells have been implicated as hepatocyte precursors, but these findings were later clarified to be due to cell fusion (Wang et al., 2003). Recent studies not only have challenged previous reports of stellate cells giving rise to hepatocytes (Mederacke et al., 2013), but also have raised doubt about the established concept of a subset of BECs being—or being able to act as—LSCs by giving rise to hepatocytes (Tarlow et al., 2014).
Because of these contradictory findings, the contribution of LSCs, or any nonhepatocyte cell type, to the formation of new hepatocytes in the chronically injured liver is uncertain. Here, we sought to resolve this uncertainty using our previously reported hepatocyte fate-tracing mouse model (Malato et al., 2011) and mouse models that afford highly specific labeling and therefore reliable fate tracing of BECs and mesenchymal liver cells.
RESULTS
Hepatocyte Fate Tracing in Choline-Deficient, Ethionine-Supplemented Diet-Induced Chronic Liver Injury
To study the contribution of LSCs or other nonhepatocytes to new hepatocytes, we chose a mouse model of chronic liver injury caused by a choline-deficient, ethionine-supplemented (CDE) diet. The CDE diet was originally observed in rats, and subsequently in mice, to cause emergence of liver progenitor cells—called oval cells in rodents—from portal tracts, thereby mimicking ductular reactions observed in chronic liver diseases in humans (Akhurst et al., 2001; Shinozuka et al., 1978). Although other chronic liver injury models exist, we focused on CDE diet feeding because, for mice, it is the only model with which multiple research groups obtained direct evidence for the conversion of nonhepatocytes into hepatocytes (Español-Suñer et al., 2012; Rodrigo-Torres et al., 2014).
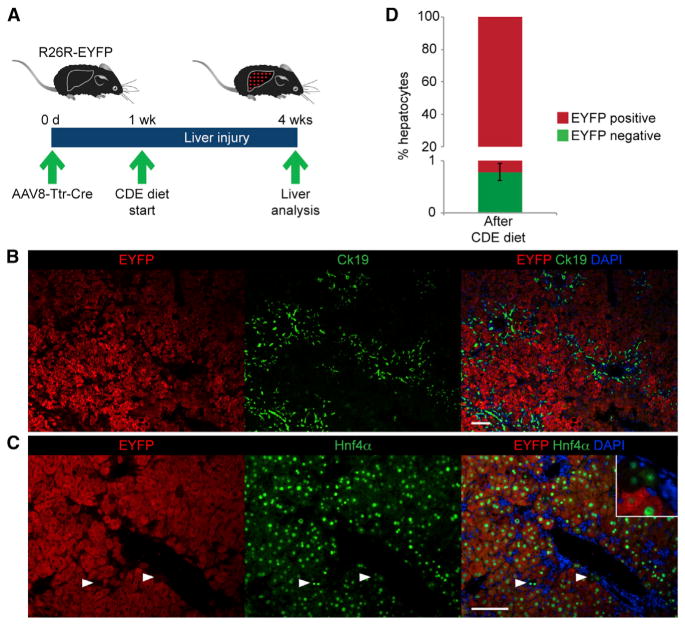
To determine the frequency at which new hepatocytes are formed from nonhepatocytes in CDE-diet-fed mice, we performed hepatocyte fate tracing. For this, we injected Cre recombinase reporter (R26R-EYFP) mice with an adenoassociated viral vector expressing Cre from the transthyretin promoter (AAV8-Ttr-Cre; Figure 1A). We showed previously that this nonintegrating vector affords specific and efficient reporter gene activation in hepatocytes but does not label BECs, stellate cells, macrophages, or endothelial cells in livers of R26R-EYFP mice (Malato et al., 2011). One week after genetically labeling hepatocytes with AAV8-Ttr-Cre, we started feeding mice the CDE diet. As previously reported (Español-Suñer et al., 2012; Rodrigo-Torres et al., 2014), the CDE diet was used for 3 weeks, after which we confirmed that a characteristic oval cell response was present—identified by the expansion of cytokeratin 19 (Ck19)-positive cells or Opn-positive cells in periportal regions (Figures 1B and S1A). Next, we analyzed the injured livers by coimmunostaining for EYFP and the hepatocyte markers hepatocyte nuclear factor 4 alpha (Hnf4α) and fumarylacetoacetate hydrolase (Fah) for the presence of EYFP-negative hepatocytes (Figures 1C and S1B). We found these non-fate-traced hepatocytes at a frequency of 0.76% ± 0.16% (Figure 1D). This result was in accord with results from previous studies showing that new hepatocytes originate—in small numbers—from a nonhepatocyte source, presumably LSCs, in mice with CDE-diet-induced chronic liver injury (Español-Suñer et al., 2012; Rodrigo-Torres et al., 2014).
Figure 1. Presence of Unlabeled Hepatocytes in a Hepatocyte Fate-Tracing Mouse Model after Chronic Liver Injury.
(A) The hepatocyte fate-tracing model was generated by injecting R26R-EYFP mice with AAV8-Ttr-Cre. Liver injury was induced by CDE diet feeding 1 week later. Livers were analyzed after 3 weeks of CDE diet feeding.
(B) Coimmunostaining for EYFP and Ck19 shows oval cell expansion characteristic for livers of mice after CDE diet feeding.
(C) Coimmunostaining for EYFP and Hnf4α shows EYFP-negative, Hnf4α-positive cells; i.e., non-fate-traced hepatocytes (arrowheads and inset).
(D) Quantification of non-fate-traced hepatocytes. Data are shown as mean ± SEM.
Scale bars, 100 μm. Representative images and results from three mice are shown. See also Figures S1 and S4B.
Biliary Cell Fate Tracing in CDE-Diet-Induced Chronic Liver Injury
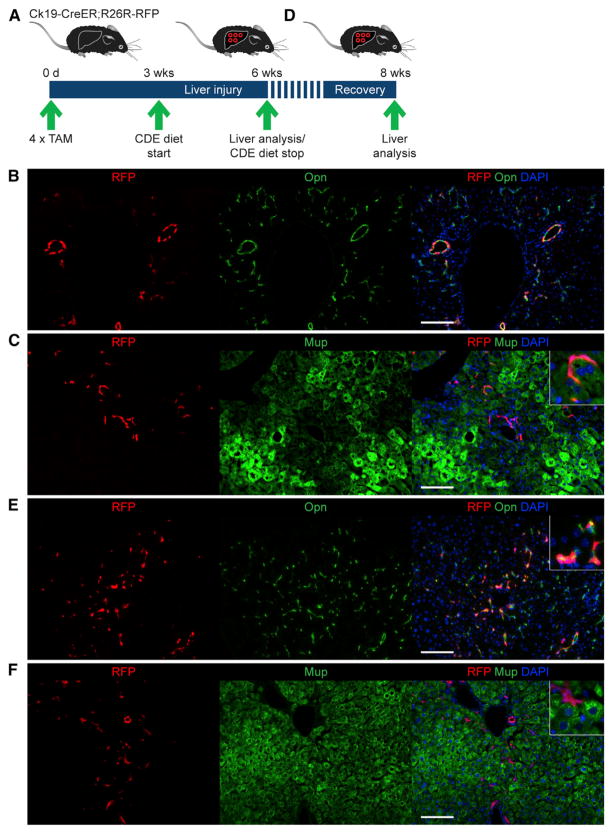
To determine the source of the observed non-fate-traced hepatocytes, we performed fate tracing of candidate cell populations. First, we tested the predominant view that LSCs capable of giving rise to hepatocytes reside in biliary structures (Español-Suñer et al., 2012; Furuyama et al., 2011; Rodrigo-Torres et al., 2014). Intriguingly, the frequency of non-fate-traced hepatocytes observed by us was similar to the frequency with which Opn-expressing cells were previously reported to give rise to hepatocytes in the same liver injury model (Español-Suñer et al., 2012). However, these results have been called into question by a recent Sox9-based fate-tracing study in which conversion of BECs into hepatocytes was not observed (Tarlow et al., 2014). To resolve this contradiction, we used a different BEC fate-tracing model, Ck19-CreER;R26R-RFP mice (Figures 2A). Direct RFP fluorescence combined with immunostaining for Opn after four tamoxifen (TAM) injections showed an overall BEC labeling efficiency of 10.5% ± 2.0%, with some bile ducts showing >40% labeling; the frequency of labeled hepatocytes was negligible (defined as <0.1% of the hepatocyte population) (Figures S2A and S2B). Two weeks after the last TAM injection, mice were fed the CDE diet for 3 weeks. Their injured livers showed the expected fate-traced oval cells expressing RFP and Opn, but fate-traced hepatocytes—identified by immunostaining for major urinary protein (Mup)—were present at a negligible frequency (Figures 2B and 2C). The frequency of fate-traced hepatocytes remained negligible when we restricted our analysis to portal regions with >40% labeling efficiency (53.5% ± 2.6%) (Figure S2C), which suggested our results were not due to low labeling efficiency. Nevertheless, we reevaluated these results using a modified CDE diet model that was reported to yield higher numbers of BEC-derived hepatocytes (2.45% versus 0.78%) by allowing mice to recover from CDE-diet-induced chronic liver injury for 2 weeks before analysis (Español-Suñer et al., 2012) (Figure 2D). Using this CDE-Stop model, we still detected only a negligible number of fate-traced hepatocytes in Ck19-CreER;R26R-RFP mice (Figures 2E and 2F). These results argue against BECs, or LSCs contained in biliary structures, generating hepatocytes in mice with CDE-diet-induced chronic liver injury.
Figure 2. Absence of BEC-Derived Hepatocytes.
(A) The BEC fate-tracing model was generated by injecting Ck19-CreER;R26R-RFP mice with 4 mg TAM every other day for four injections total. Liver injury was induced by CDE diet feeding 2 weeks after the last TAM injection. Livers were analyzed after 3 weeks of CDE diet feeding.
(B) Direct fluorescence of RFP combined with immunostaining for Opn shows no RFP-positive, Opn-negative cells; i.e., non-BECs/nonoval cells derived from BECs.
(C) Direct fluorescence of RFP combined with immunostaining for Mup shows no RFP-positive, Mup-positive cells; i.e., hepatocytes derived from BECs.
(D) The CDE-Stop model was generated as in (A) with the difference that the mice were fed a normal diet between CDE diet feeding and analysis.
(E) Direct fluorescence of RFP combined with immunostaining for Opn shows no RFP-positive, Opn-negative cells; i.e., non-BECs/nonoval cells derived from BECs.
(F) Direct fluorescence of RFP combined with immunostaining for Mup shows noRFP-positive, Mup-positive cells; i.e., hepatocytes derived from BECs.
Scale bars, 100 μm. At least three mice were analyzed for each experiment. Representative images are shown. See also Figure S2.
Mesenchymal Cell Fate Tracing in CDE-Diet-Induced Chronic Liver Injury
Another potential alternative source of hepatocytes are mesenchymal liver cells, among which stellate cells are intriguing candidates because they undergo substantial changes in form and function when they become activated in chronic liver injury, including after CDE diet (Ueberham et al., 2010). Moreover, stellate cells are considered to be the liver’s pericytes, a cell type that has been implicated as a precursor of parenchymal cells in other tissues (Dellavalle et al., 2007). Previous GFAP-based fate-tracing studies reported that activated stellate cells—called myofibro-blasts—contribute up to 24% of hepatocytes in mice with liver injury (Michelotti et al., 2013; Swiderska-Syn et al., 2014; Yang et al., 2008). However, these results were contradicted by a recent fate-tracing study using lecithin-retinol acyltransferase as a more specific stellate cell/myofibroblast marker (Mederacke et al., 2013).
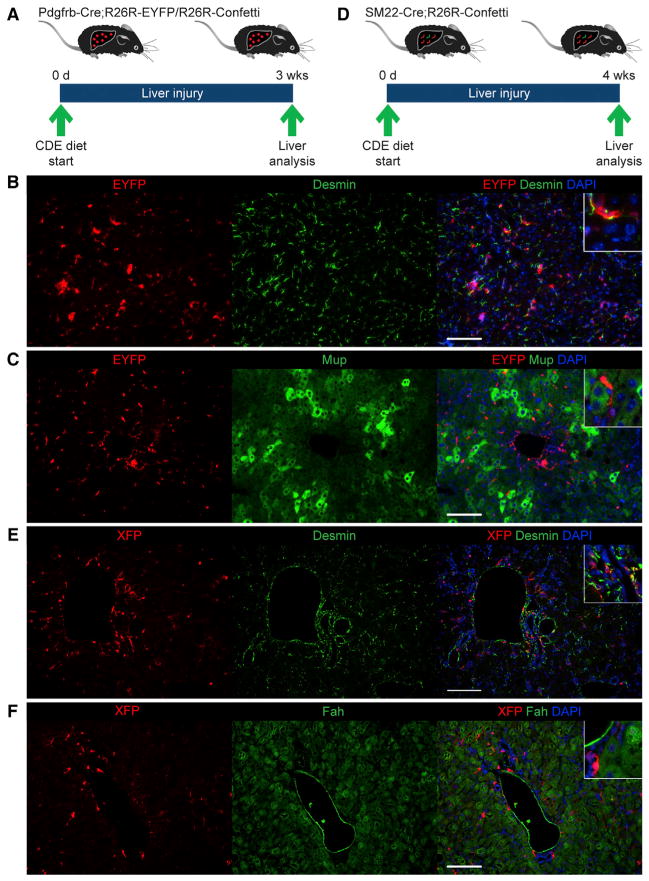
To independently assess the contribution of stellate cells/myofibroblasts to hepatocyte regeneration, we performed fate tracing based on platelet-derived growth factor receptor beta (Pdgfrβ) expression in mice fed CDE diet for 3 weeks (Figure 3A). For this, we generated R26R-EYFP and R26R-Confetti mice carrying a Pdgfrb-Cre allele that was recently shown to be specifically and efficiently expressed in stellate cells/myofibroblasts (Henderson et al., 2013). Combining immunostaining for EYFP or direct fluorescence of the Confetti markers (RFP, YFP, mCFP, and nGFP; data not shown) with immunostaining for the mesenchymal cell markers desmin and alpha smooth muscle actin (αSma) or the hepatocyte markers Mup and Fah showed fate tracing of stellate cells/myofibroblasts at >70% efficiency, whereas the number of fate-traced hepatocytes was negligible in both injured and noninjured livers (Figures 3B, 3C, and S3A–S3C). This result suggests that stellate cells/myofibroblasts are not a source of new hepatocytes in CDE-diet-induced chronic liver injury.
Figure 3. Absence of Mesenchymal Liver-Cell-Derived Hepatocytes.
(A) Pdgfrb-Cre;R26R-EYFP or Pdgfrb-Cre;R26R-Confetti mice were used for fate tracing of stellate cells/myofibroblasts. Livers were analyzed after 3 weeks of CDE diet feeding.
(B) Coimmunostaining for EYFP and desmin shows no EYFP-positive, desmin-negative cells; i.e., nonmesenchymal cells derived from stellate cells/myofibroblasts.
(C) Coimmunostaining for EYFP and Mup shows no EYFP-positive, Mup-positive cells; i.e., hepatocytes derived from stellate cells/myofibroblasts.
(D) SM22-Cre;R26R-Confetti mice were used for fate tracing of periportal mesenchymal cells. Livers were analyzed after 4 weeks of CDE diet feeding.
(E) Coimmunostaining for YFP/nGFP/mCFP (XFP) and desmin shows that desmin-positive periportal mesenchymal cells, but no cells with hepatocyte morphology, are XFP positive.
(F) Coimmunostaining for YFP/nGFP/mCFP (XFP) and Fah shows no XFP-positive, Fah-positive cells; i.e., hepatocytes derived from periportal mesenchymal cells.
Scale bars, 100 μm. At least three mice were analyzed for each experiment. See also Figure S3.
A previous study aimed at identifying LSCs based on label retention provided evidence for candidate cells not only residing in but also immediately adjacent to biliary structures (Kuwahara et al., 2008). The identity of these cells was not determined, but they appeared to be located within the periportal mesenchyme. To investigate the possibility that the periportal mesenchyme harbors cells capable of hepatocyte differentiation, we performed fate tracing using smooth muscle protein 22 alpha (SM22)-Cre mice, which were previously used for specific gene knockout in the periportal mesenchyme (Hofmann et al., 2010). For this, we generated SM22-Cre;R26R-Confetti mice and analyzed their livers after 4 weeks of CDE diet feeding (Figure 3D). We found >15% labeling efficiency of desmin-positive periportal mesenchymal cells, whereas the number of labeled hepatocytes was negligible in both injured and noninjured livers (Figures 3E, 3F, S3D, and S3E). Viewed together with our results obtained by stellate cell/myofibroblast fate tracing, these results argue against a contribution from mesenchymal liver cells to hepatocyte regeneration in chronic liver injury.
Refined Hepatocyte Fate Tracing in CDE-Diet-Induced Chronic Liver Injury
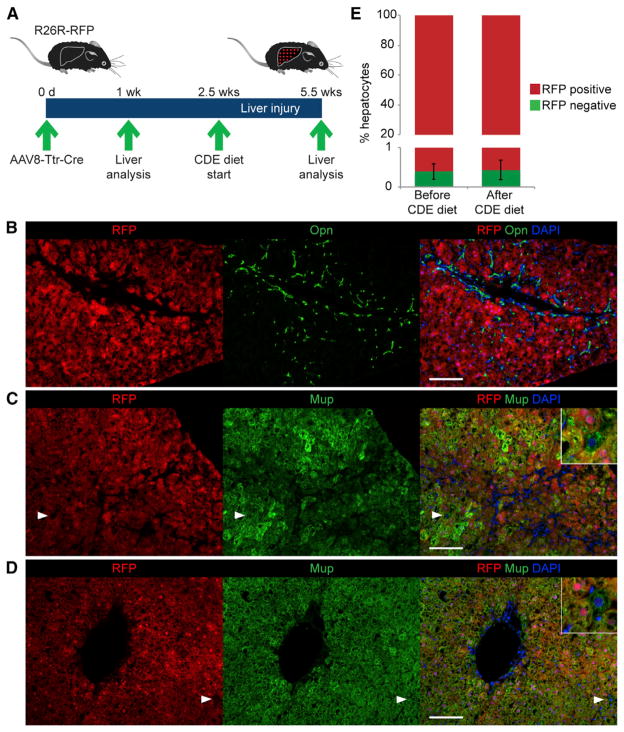
Prompted by our inability to confirm previous reports of hepatocyte formation from BECs or mesenchymal liver cells—not even at the low frequency observed in our hepatocyte fate-tracing studies (Figure 1D)—we revisited this model. We hypothesized that the small number of unlabeled hepatocytes observed in our hepatocyte fate-tracing model was due to less than 100% labeling efficiency of AAV8-Ttr-Cre, and not reflective of hepatocytes derived from nonhepatocytes. To address this possibility, we repeated hepatocyte fate tracing with AAV8-Ttr-Cre using different Cre reporter mice, R26R-RFP mice, which do not require immunostaining as R26R-EYFP mice do but can be analyzed by direct fluorescence (Figures 4A and S4A). In accord with improved ability to detect labeled hepatocytes, we found a slightly lower frequency of non-fate-traced hepatocytes of 0.40% ± 0.23% in R26R-RFP mice after 3 weeks of CDE diet feeding (Figures 4B, 4C, and 4E). More importantly, sampling of each mouse’s liver by one-third partial hepatectomy (PH) before CDE diet feeding revealed the same frequency (0.39% ± 0.18%) of non-fate-traced hepatocytes as after CDE diet feeding (Figures 4A, 4D, and 4E), confirming that the hepatocyte labeling efficiency of AAV8-Ttr-Cre is not 100%. Furthermore, the result indicates that non-fate-traced hepatocytes detected after CDE diet feeding are not newly generated by a nonhepatocyte source but are pre-existing hepatocytes that were not labeled by AAV8-Ttr-Cre. Thus, these results argue that new hepatocytes formed during CDE-diet-induced chronic liver injury derive from pre-existing hepatocytes and not other, nonhepatocyte cell types such as LSCs.
Figure 4. Origin of New Hepatocytes from Pre-Existing Hepatocytes.
(A) A refined hepatocyte fate-tracing model was generated by injecting AAV8-Ttr-Cre into R26R-RFP mice and analyzing the liver of each mouse before and after CDE diet feeding. For this, mice were subjected to one-third PH 1 week after AAV8-Ttr-Cre injection, followed by a 1.5-week-long recovery phase. Final liver analysis was carried out after 3 weeks of CDE diet feeding.
(B) Direct fluorescence of RFP combined with immunostaining for Opn shows the characteristic oval cell response after CDE diet feeding.
(C and D) Direct fluorescence of RFP combined with immunostaining for Mup shows in livers of mice after (C) and before (D) liver injury the presence of RFP-negative, Mup-positive cells; i.e., non-fate-traced hepatocytes (arrowheads and insets).
(E) Quantification of non-fate-traced hepatocytes in livers of mice before and after liver injury. Data are shown as mean ± SEM; p > 0.1.
Scale bars, 100 μm. Representative images and results from three mice are shown. See also Figure S4.
DISCUSSION
Conflicting results have been reported about the contribution of nonhepatocyte cell types to liver regeneration. To address the resulting uncertainty about the significance, or even existence, of LSCs, we used multiple fate-tracing strategies in mice with CDE-diet-induced chronic liver injury, all of which failed to provide convincing evidence for BECs or mesenchymal liver cells giving rise to new hepatocytes. Possibly, the labeling efficiency in our Ck19-based or the recent Sox9-based fate-tracing study by Tarlow et al. (2014) (10.5% ± 2.0% or ~7%, respectively, versus 69.1% in the Opn or 28.7% in the Hnf1β study) was not sufficient for capturing rare cells like LSCs. However, we also failed to find a significant number of fate-traced he-patocytes when we restricted our analysis to periportal regions with the highest labeling efficiency (53.5% ± 2.6%). Furthermore, we confirmed our Ck19 and our Pdgfrβ and SM22 expression-based fate-tracing results by using our refined hepatocyte fate-tracing approach, which was designed to detect even very rare events by excluding intermouse variability.
The recent Sox9 study suggests unspecific fate tracing of hepatocytes as an explanation for the discrepant results obtained in previous BEC or stellate cell/myofibroblast fate-tracing studies (Español-Suñer et al., 2012; Furuyama et al., 2011; Michelotti et al., 2013; Rodrigo-Torres et al., 2014; Tarlow et al., 2014). The authors show that TAM, which was needed to activate fate tracing in all of these studies, remains active in mouse livers for a week and potentially longer. Initiating CDE diet feeding or other injuries at this time—as done in the original Sox9, Opn, Hnf1β and GFAP studies, but not in the recent Sox9 study or our Ck19 study—may have resulted in marker gene activation in injured hepatocytes that unspecifically expressed the genes whose promoters were used to drive Cre.
Our results also appear to be at odds with previous reports of isolation of cells from adult mouse liver that express BEC markers and can be expanded and differentiated into both hepatocytes and BECs in vitro (Dorrell et al., 2011; Huch et al., 2013; Shin et al., 2011). These differences suggest that removing liver cells from their in vivo environment may confer properties of plasticity not representative of in vivo biology. An alternative explanation, at least in liver injury models, is that these cells are hepatocytes that underwent biliary differentiation, but retained the ability to convert back into hepatocytes (Michalopoulos, 2014). Being derived from hepatocytes, such cells would not have been identified as nonhepatocytes by our approach to hepatocyte fate tracing.
An important implication of our inability to confirm previous findings of nonhepatocytes giving rise to new hepatocytes is that it calls into question whether adult mice have LSCs, which—in the classical view—are defined as a hepatocyte-independent backup system for liver regeneration. Despite many published studies about LSCs, no direct in vivo evidence exists supporting their existence, and conceivably the regenerative capabilities of hepatocytes—and their ability to undergo biliary differentiation—have rendered LSCs expendable. Because our approach to hepatocyte fate tracing allows investigation of the hepatocyte population only as a whole, it remains to be determined whether LSC-like subsets of hepatocytes exist that are more regenerative and/or plastic than others.
Another potential explanation for our findings is that current mouse models of liver injury are insufficient for LSC activation. Because LSCs are thought to serve as a backup system when hepatocytes fail to proliferate, LSC activation may require a true hepatocyte cell-cycle arrest. None of the currently available mouse models of liver injury—including CDE diet feeding (Figures S4B and S4C)—block hepatocyte proliferation. Although new mouse models could be developed, this question could be addressed in rats where intoxication with 2-acetylaminofluor-ene followed by two-thirds PH not only induces an oval cell response, but also blocks hepatocyte proliferation (Trautwein et al., 1999). Combining this protocol with tools for genetic fate tracing in rats may reveal the existence of LSCs or BECs capable of hepatocyte differentiation in vivo as recently reported for zebrafish (Choi et al., 2014; He et al., 2014).
In conclusion, our findings provide no evidence for hepatocytes deriving from LSCs, or other nonhepatocyte cell types, in mice with CDE-diet-induced chronic liver injury.
EXPERIMENTAL PROCEDURES
Mice
R26R-EYFP (Srinivas et al., 2001), R26R-RFP (Luche et al., 2007), R26R-RFP (Ai14) (Madisen et al., 2010), R26R-Confetti (Snippert et al., 2010), Ck19-CreER (Means et al., 2008), Pdgfrb-Cre (Henderson et al., 2013), and SM22-Cre (Hofmann et al., 2010) mice were used. All mice were kept under barrier conditions. All procedures were approved by the Institutional Animal Care and Use Committee at UCSF.
Adenoassociated Virus
AAV8-Ttr-Cre was generated as previously described (Malato et al., 2011). Purified virus was injected via the tail vein in <150 μl volume to avoid hydrodynamic effects.
Choline-Deficient, Ethionine-Supplemented Diet
Mice 5.5–8.5 weeks of age and ~20 g in body weight received choline-deficient chow (MP Biomedicals) supplemented with 0.15% (w/v) ethionine (E5139, Sigma-Aldrich) in drinking water.
Partial Hepatectomy
Removal of the left liver lobe (one-third of liver mass) was performed under isoflurane anesthesia, buprenorphine analgesia, sterile conditions, and homeothermia.
Tamoxifen
Tamoxifen (Sigma) was dissolved in sterile corn oil to a concentration of 40 mg/ml. Mice received a series of four intraperitoneal injections of 4 mg at 2 day intervals.
Immunostaining
Tissue samples were fixed either by 4% PFA (Sigma) perfusion (R26R-RFP and Confetti) or drop fixed overnight in 4% PFA (R26R-RFP), neutral-buffered formalin containing zinc (Anatech), or 10% PBS-buffered formalin (R26R-EYFP). Samples were embedded in OCT (Tissue-Tek) and cryosectioned into 6-μm-thick sections or embedded in paraffin and cut into 5-μm-thick sections. Paraffin sections were treated with Antigen Retrieval Citra Solution (Bio-Genex) before antibody staining. Sections were blocked in 10% serum for 1 hr and incubated with primary antibodies at 4°C overnight and secondary antibodies at room temperature for 1 hr (Tables S1 and S2). DAPI (Millipore) was used to stain nuclear DNA. Hnf4α immunostaining of paraffin-embedded sections was done using the M.O.M. kit (Vector Laboratories).
Cell Quantification
Periportal fields were randomly chosen from at least three lobes of each mouse, with the exception of analysis of samples collected by one-third PH. The overall number of hepatocytes per field was determined based on Mup or Hnf4α and DAPI staining and the overall number of BECs per field was determined based on Ck19 or Opn and DAPI staining. At least 10,000 hepatocytes were counted for each experiment except for analyses of highly labeled portal regions in Ck19-CreER;R26R-RFP mice and hepatocyte proliferation after CDE diet, where 2,500 hepatocytes were counted.
Statistics
Data are shown as mean ± SEM. Statistical significance was determined using the two-tailed Student’s t test.
Supplementary Material
Acknowledgments
H.W. was supported by funding from CIRM RN2-00950 and NIH P30 DK26743. J.R.S. was supported by NIH T32 DK060414. The authors thank Pamela Derish at UCSF for manuscript editing.
Footnotes
Supplemental Information includes four figures and two tables and can be found with this article online at http://dx.doi.org/10.1016/j.celrep.2014.07.003.
AUTHOR CONTRIBUTIONS
J.R.S. and H.W. designed the experiments. J.R.S., Y.M., and C.G. performed the experiments. J.R.S. and H.W. analyzed the experiments. J.R.S. and H.W. wrote the manuscript. All authors read and approved the final manuscript.
References
- Akhurst B, Croager EJ, Farley-Roche CA, Ong JK, Dumble ML, Knight B, Yeoh GC. A modified choline-deficient, ethionine-supplemented diet protocol effectively induces oval cells in mouse liver. Hepatology. 2001;34:519–522. doi: 10.1053/jhep.2001.26751. [DOI] [PubMed] [Google Scholar]
- Choi TY, Ninov N, Stainier DR, Shin D. Extensive conversion of hepatic biliary epithelial cells to hepatocytes after near total loss of hepatocytes in zebrafish. Gastroenterology. 2014;146:776–788. doi: 10.1053/j.gastro.2013.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellavalle A, Sampaolesi M, Tonlorenzi R, Tagliafico E, Sacchetti B, Perani L, Innocenzi A, Galvez BG, Messina G, Morosetti R, et al. Pericytes of human skeletal muscle are myogenic precursors distinct from satellite cells. Nat Cell Biol. 2007;9:255–267. doi: 10.1038/ncb1542. [DOI] [PubMed] [Google Scholar]
- Dorrell C, Erker L, Schug J, Kopp JL, Canaday PS, Fox AJ, Smirnova O, Duncan AW, Finegold MJ, Sander M, et al. Prospective isolation of a bipotential clonogenic liver progenitor cell in adult mice. Genes Dev. 2011;25:1193–1203. doi: 10.1101/gad.2029411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan AW, Dorrell C, Grompe M. Stem cells and liver regeneration. Gastroenterology. 2009;137:466–481. doi: 10.1053/j.gastro.2009.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Español-Suñer R, Carpentier R, Van Hul N, Legry V, Achouri Y, Cordi S, Jacquemin P, Lemaigre F, Leclercq IA. Liver progenitor cells yield functional hepatocytes in response to chronic liver injury in mice. Gastroenterology. 2012;143:1564–1575. doi: 10.1053/j.gastro.2012.08.024. [DOI] [PubMed] [Google Scholar]
- Furuyama K, Kawaguchi Y, Akiyama H, Horiguchi M, Kodama S, Kuhara T, Hosokawa S, Elbahrawy A, Soeda T, Koizumi M, et al. Continuous cell supply from a Sox9-expressing progenitor zone in adult liver, exocrine pancreas and intestine. Nat Genet. 2011;43:34–41. doi: 10.1038/ng.722. [DOI] [PubMed] [Google Scholar]
- He J, Lu H, Zou Q, Luo L. Regeneration of liver after extreme hepatocyte loss occurs mainly via biliary transdifferentiation in zebrafish. Gastroenterology. 2014;146:789–800. doi: 10.1053/j.gastro.2013.11.045. [DOI] [PubMed] [Google Scholar]
- Henderson NC, Arnold TD, Katamura Y, Giacomini MM, Rodriguez JD, McCarty JH, Pellicoro A, Raschperger E, Betsholtz C, Ruminski PG, et al. Targeting of αv integrin identifies a core molecular pathway that regulates fibrosis in several organs. Nat Med. 2013;19:1617–1624. doi: 10.1038/nm.3282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann JJ, Zovein AC, Koh H, Radtke F, Weinmaster G, Iruela-Arispe ML. Jagged1 in the portal vein mesenchyme regulates intrahepatic bile duct development: insights into Alagille syndrome. Development. 2010;137:4061–4072. doi: 10.1242/dev.052118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huch M, Dorrell C, Boj SF, van Es JH, Li VS, van de Wetering M, Sato T, Hamer K, Sasaki N, Finegold MJ, et al. In vitro expansion of single Lgr5+ liver stem cells induced by Wnt-driven regeneration. Nature. 2013;494:247–250. doi: 10.1038/nature11826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh T, Miyajima A. Liver regeneration by stem/progenitor cells. Hepatology. 2014;59:1617–1626. doi: 10.1002/hep.26753. [DOI] [PubMed] [Google Scholar]
- Kuwahara R, Kofman AV, Landis CS, Swenson ES, Barendswaard E, Theise ND. The hepatic stem cell niche: identification by label-retaining cell assay. Hepatology. 2008;47:1994–2002. doi: 10.1002/hep.22218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luche H, Weber O, Nageswara Rao T, Blum C, Fehling HJ. Faithful activation of an extra-bright red fluorescent protein in “knock-in” Cre-reporter mice ideally suited for lineage tracing studies. Eur J Immunol. 2007;37:43–53. doi: 10.1002/eji.200636745. [DOI] [PubMed] [Google Scholar]
- Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malato Y, Naqvi S, Schürmann N, Ng R, Wang B, Zape J, Kay MA, Grimm D, Willenbring H. Fate tracing of mature hepatocytes in mouse liver homeostasis and regeneration. J Clin Invest. 2011;121:4850–4860. doi: 10.1172/JCI59261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Means AL, Xu Y, Zhao A, Ray KC, Gu G. A CK19(CreERT) knockin mouse line allows for conditional DNA recombination in epithelial cells in multiple endodermal organs. Genesis. 2008;46:318–323. doi: 10.1002/dvg.20397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mederacke I, Hsu CC, Troeger JS, Huebener P, Mu X, Dapito DH, Pradere JP, Schwabe RF. Fate tracing reveals hepatic stellate cells as dominant contributors to liver fibrosis independent of its aetiology. Nat Commun. 2013;4:2823. doi: 10.1038/ncomms3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalopoulos GK. The liver is a peculiar organ when it comes to stem cells. Am J Pathol. 2014;184:1263–1267. doi: 10.1016/j.ajpath.2014.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelotti GA, Xie G, Swiderska M, Choi SS, Karaca G, Krüger L, Premont R, Yang L, Syn WK, Metzger D, Diehl AM. Smoothened is a master regulator of adult liver repair. J Clin Invest. 2013;123:2380–2394. doi: 10.1172/JCI66904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigo-Torres D, Affò S, Coll M, Morales-Ibanez O, Millán C, Blaya D, Alvarez-Guaita A, Rentero C, Lozano JJ, Maestro MA, et al. The biliary epithelium gives rise to liver progenitor cells. Hepatology, Published online February. 2014;20:2014. doi: 10.1002/hep.27078. http://dx.doi.org/10.1002/hep.27078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin S, Walton G, Aoki R, Brondell K, Schug J, Fox A, Smirnova O, Dorrell C, Erker L, Chu AS, et al. Foxl1-Cre-marked adult hepatic progenitors have clonogenic and bilineage differentiation potential. Genes Dev. 2011;25:1185–1192. doi: 10.1101/gad.2027811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozuka H, Lombardi B, Sell S, Iammarino RM. Early histological and functional alterations of ethionine liver carcinogenesis in rats fed a choline-deficient diet. Cancer Res. 1978;38:1092–1098. [PubMed] [Google Scholar]
- Snippert HJ, van der Flier LG, Sato T, van Es JH, van den Born M, Kroon-Veenboer C, Barker N, Klein AM, van Rheenen J, Simons BD, Clevers H. Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell. 2010;143:134–144. doi: 10.1016/j.cell.2010.09.016. [DOI] [PubMed] [Google Scholar]
- Srinivas S, Watanabe T, Lin CS, William CM, Tanabe Y, Jessell TM, Costantini F. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiderska-Syn M, Syn WK, Xie G, Krüger L, Machado MV, Karaca G, Michelotti GA, Choi SS, Premont RT, Diehl AM. Myofibroblastic cells function as progenitors to regenerate murine livers after partial hepatectomy. Gut. 2014;63:1333–1344. doi: 10.1136/gutjnl-2013-305962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarlow BD, Finegold MJ, Grompe M. Clonal tracing of Sox9(+) liver progenitors in mouse oval cell injury. Hepatology. 2014;60:278–289. doi: 10.1002/hep.27084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trautwein C, Will M, Kubicka S, Rakemann T, Flemming P, Manns MP. 2-acetaminofluorene blocks cell cycle progression after hepatectomy by p21 induction and lack of cyclin E expression. Oncogene. 1999;18:6443–6453. doi: 10.1038/sj.onc.1203045. [DOI] [PubMed] [Google Scholar]
- Ueberham E, Böttger J, Ueberham U, Grosche J, Gebhardt R. Response of sinusoidal mouse liver cells to choline-deficient ethionine-supplemented diet. Comp Hepatol. 2010;9:8. doi: 10.1186/1476-5926-9-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Willenbring H, Akkari Y, Torimaru Y, Foster M, Al-Dhalimy M, Lagasse E, Finegold M, Olson S, Grompe M. Cell fusion is the principal source of bone-marrow-derived hepatocytes. Nature. 2003;422:897–901. doi: 10.1038/nature01531. [DOI] [PubMed] [Google Scholar]
- Yang L, Jung Y, Omenetti A, Witek RP, Choi S, Vandongen HM, Huang J, Alpini GD, Diehl AM. Fate-mapping evidence that hepatic stellate cells are epithelial progenitors in adult mouse livers. Stem Cells. 2008;26:2104–2113. doi: 10.1634/stemcells.2008-0115. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.