Abstract
Long non-coding RNAs (lncRNAs) are a large and diverse group of RNAs that are often lineage-specific and that regulate multiple biological functions. Many are nuclear and are essential parts of ribonucleoprotein complexes that modify chromatin segments and establish active or repressive chromatin states; others are cytosolic and regulate the stability of mRNA or act as microRNA sponges. This Review summarizes the current knowledge of lncRNAs as regulators of the endocrine system, with a focus on the identification and mode of action of several endocrine-important lncRNAs. We highlight lncRNAs that have a role in the development and function of pancreatic β cells, white and brown adipose tissue, and other endocrine organs, and discuss the involvement of these molecules in endocrine dysfunction (for example, diabetes mellitus). We also address the associations of lncRNAs with nuclear receptors involved in major hormonal signalling pathways, such as estrogen and androgen receptors, and the relevance of these associations in certain endocrine cancers.
Introduction
The traditional view of RNAs as merely message carriers from DNA to protein is fading. Pioneering conceptual work from several laboratories,1–4 together with early large-scale cloning undertakings such as FANTOM,5,6 revealed the existence of a large number of non-coding transcripts, most of which were regarded as transcriptional noise. Simultaneously, technologic advances such as DNA tiling arrays,2,7 next-generation sequencing8 and the availability of human genome sequences,9,10 expressed sequence tags11 and large datasets of genomic functional elements from the ENCODE consortium12–16 have enabled a more refined view of mammalian transcriptomes. Now we understand that a substantial proportion of the genome is transcribed, but only a small fraction of DNA encodes proteins.2,17,18 The non-protein-coding portion of the genome is transcribed to generate a vast array of non-coding RNAs, which include (but are not limited to) transfer RNAs (tRNAs), ribosomal RNAs (rRNAs), small nucleolar RNAs (snoRNAs), microRNAs (miRNAs), long non-coding RNAs (lncRNAs), small nuclear RNAs (snRNAs) and piwi-interacting RNAs (piRNAs).12,19 lncRNAs share many structural features with protein-coding mRNAs: lncRNAs are often transcribed by RNA polymerase II, are encoded by multiple exons and undergo canonical RNA splicing to yield RNA products that can be several kilobases in length.20 Mounting evidence is revealing regulatory roles for lncRNAs in governing gene expression during cellular development and homeostasis.21–24
In this Review, we summarize studies concerning the discovery and functions of lncRNAs in the endocrine system. As background information, we first describe the general features, functions and mechanisms of action of lncRNAs. Next, we discuss several studies from the past few years in which lncRNAs were identified in endocrine organs and their roles in the development and function of endocrine tissues were elucidated, as well as the potential links of lncRNAs to specific diseases such as diabetes mellitus and certain endocrine cancers.
General characteristics of lncRNAs
Definition
An operational definition of lncRNAs is that they are RNA transcripts longer than 200 bases and without evident protein-coding capacity. This definition is somewhat arbitrary, but this size cut-off can distinguish lncRNAs from many small RNAs, such as miRNAs, snRNAs, tRNAs, piRNAs and snoRNAs. The challenge in this definition is to test whether a transcript that apparently falls within this lncRNA classification can be translated into one or more small peptides. In early studies, the lack of coding potential was tested by analysing the degree of conservation of open reading frames across different species.25 New techniques such as ribosome footprint assays are being used to determine ribosome occupancy of specific RNAs and can be applied to determine if a lncRNA is being translated.26 However, some lncRNAs, such as H19, were observed to associate with ribosomes but no protein products were detected.25,26 Improved approaches are now being developed to accurately distinguish between real translation and non-productive ribosome occupancy.27–30
Categories
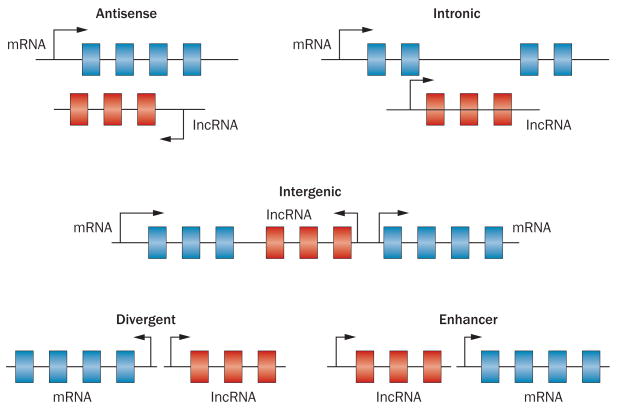
A detailed classification of lncRNAs can be very intricate. According to their relative genome position with respect to neighbouring protein-coding genes, lncRNAs can be categorized as antisense, intronic, intergenic, divergent and enhancer lncRNAs (Figure 1). Interestingly, numerous lncRNAs that are transcribed from enhancer regions (enhancer lncRNAs) mediate short-range and long-range interactions between the enhancers from which they are transcribed and other regulatory elements in the genome.31
Figure 1.
Categories of long non-coding RNAs classified on the basis of their genomic region.
According to their cellular localization, lncRNAs can be categorized as nuclear or cytosolic, but some lncRNAs can be found in both compartments. Nuclear lncRNAs such as Xist21are likely to exert their functions by modifying chromatin structure, thereby influencing gene transcription, whereas cytosolic lncRNAs regulate target mRNA stability and translational efficiency through RNA-RNA interactions (Figure 2). The observation that many nuclear lncRNAs can be depleted in cultured cells by expression of short hairpin RNAs suggests that lncRNAs probably shuttle between the nucleus and cytosol, although this hypothesis has not been tested.13,32
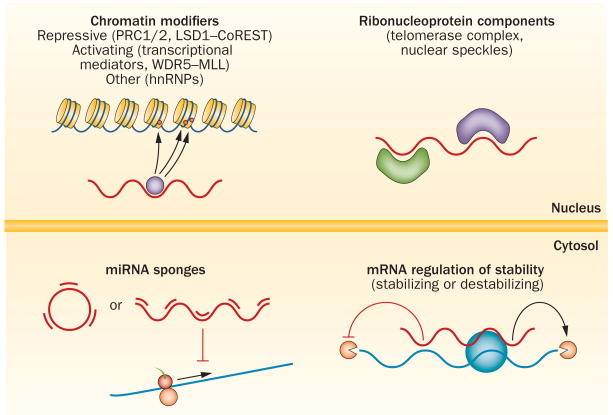
Figure 2.
Mechanisms of lncRNA action. Most lncRNAs are nuclear and their most common mechanism of action is via recruitment of chromatin modifiers to DNA. These chromatin modifiers can be repressive (such as the PRC1/2 or the LSD1–CoREST complexes), activating (such as transcriptional mediators or the WDR5–MLL complex) or other modifiers such as hnRNPs as nuclear organization factors. Some lncRNAs bind to specific proteins and act as scaffolds within ribonucleoprotein complexes. In the cytosol, circular or linear lncRNAs can act at the post-transcriptional level as sponges for miRNAs, therefore inhibiting the actions of miRNAs on mRNAs. A few examples of lncRNAs that affect the half-life of mRNAs by either destabilizing or stabilizing a specific subset of mRNAs have been described. Abbreviations: hnRNP, heterogeneous ribonucleoprotein; lncRNA, long non-coding RNA; miRNA, microRNA; PRC, polycomb repressor complex.
Tissue specificity and conservation
lncRNAs have lower sequence conservation and abundance than mRNAs, properties which were once used to argue against any biological function for lncRNAs. However, a striking feature of lncRNAs is that, as a group, they have greater tissue specificity than protein-coding RNAs,8,33 which suggests that lncRNAs might have a crucial role in the formation of multiple, if not all, cell types.
The primary sequence of lncRNAs is often poorly, if at all, conserved across species. A study of the evolution of lncRNAs in 11 tetrapods indicated that many lncRNAs in humans evolved very late and only a very small fraction of these lncRNAs, as judged by primary sequence similarity, have conserved orthologous genes beyond primates.34 However, this criterion might be less appropriate for assessing conservation of lncRNAs than of mRNAs, so some researchers have proposed that lncRNA conservation should be assessed on the basis of synteny of their gene loci and on the basis of their structure, rather than relying on sequence similarity.8,35 Indeed, lncRNAs derived from syntenic loci (lncRNAs whose chromosome position in relation to surrounding genes is the same) tend to have higher sequence similarity8,36 and more conserved functions37 than nonsyntenic lncRNAs. For example, knockdown of two zebrafish lncRNAs that have low sequence conservation in human and mouse caused major developmental defects in fish embryos; these defects were rescued by adding either the mature fish lncRNAs or their human or mouse orthologs that were identified by chromosome synteny.37 In addition, as lncRNAs are likely to function through the formation of secondary and tertiary structures, the major evolutionary constraint for lncRNAs could be to maintain their functional structures.38-40 However, accurate prediction of lncRNA structures still remains a big challenge for the field.
Mechanisms
In contrast to miRNAs, which exert their functions via an RNA-RNA base-pairing mechanism,41 lncRNAs act through diverse mechanisms that probably rely on their secondary or tertiary structures.42 Most lncRNAs are located in the nucleus,13,18 where they can act as molecular scaffolds,43 aid alternative splicing44 or modify chromatin structures (Figure 2).45-47 However, emerging evidence indicates that some lncRNAs, of which TINCR, ½-sbsRNA and ciRS-7 are examples, have functions in the cytoplasm, such as modulating translation, promoting or inhibiting mRNA degradation, and acting as miRNA sponges (which can be circular or linear molecules).24,48-51 As the lncRNA field moves forward, new perspectives on the mechanisms of lncRNA function will be seen in the near future.
Association with human disease
Genome-wide association studies (GWAS) have been used to identify associations between DNA sequence variation and clinical phenotypes. Since the first GWAS in 2005,52 hundreds of reports have been published, which identified thousands of single nucleotide polymorphisms (SNPs) related to a variety of diseases including cancer, heart disease, neuron disorders, obesity and diabetes mellitus.53,54 However, most of these SNPs are found in non-coding regions, which poses an enigma for researchers attempting to connect functional DNA elements with diseases. The answer to this enigma lies, in part, in the comprehensive transcription process2,17,18,55 that produces thousands of species of non-coding RNAs, including lncRNAs. Several studies have linked disease-associated SNPs to lncRNAs and documented these associations in multiple databases.56–59 In one study, the association of SNPs with the expression levels (assessed through analysis of expression quantitative trait loci, eQTLs) of large intergenic non-coding RNAs was investigated using genome-wide gene expression and genotype data from five different tissues.56 The researchers found that 75% of the SNPs affected expression of lncRNAs (lncRNA cis-eQTLs) but not the neighbouring protein-coding genes. These results suggest a role of lncRNAs in human diseases.
lncRNAs in endocrine physiology and disease
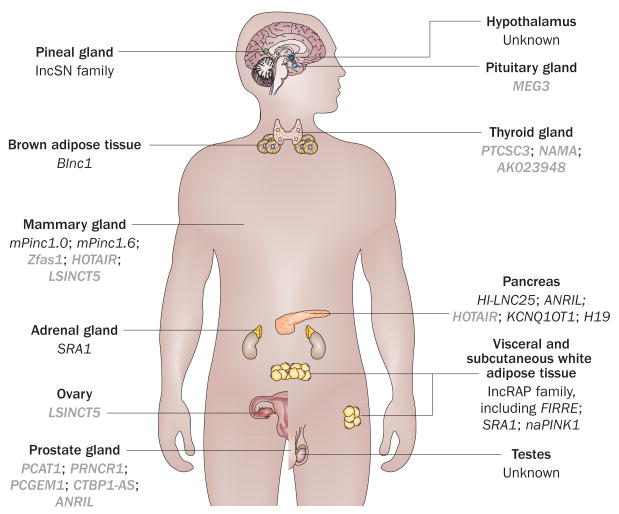
Until just a few years ago, the roles of lncRNAs in endocrine physiology had not been examined and this field is still in its infancy. Very little knowledge exists on the function of lncRNAs in many important endocrine organs, including the pituitary, thyroid and parathyroid glands, and the hypothalamus. Studies from our laboratories and other groups, however, have revealed crucial roles of lncRNAs in the normal development and function of several other endocrine organs, as well as showing their involvement in endocrine diseases such as diabetes mellitus and endocrine cancers (Figure 3, Table 1). The number of functional lncRNAs identified in a variety of biological systems is increasing, and we anticipate a rapid expansion of knowledge about the involvement of lncRNAs in endocrine physiology and disease in the near future.
Figure 3.
Endocrine organs and lncRNAs. lncRNAs known to be involved in the development, physiology and/or disease of each endocrine organ are indicated in the image. lncRNAs highlighted in bold grey are involved in cancer.
Table 1. lncRNAs in endocrine organs.
| Endocrine organ | lncRNA | Association with disease | Physiological function | Study |
|---|---|---|---|---|
| Hypothalamus | Unknown names | Unknown | Unknown | Roy et al.115 |
| Pineal gland | lncSN family | Unknown | Circadian rhythm regulation | Coon et al.90 |
| Pituitary gland | MEG3 | Pituitary adenomas | Unknown | Cheunsuchon et al.75; Zhou et al.104 |
| Thyroid gland | PTCSC3 | Thyroid cancer | Competing endogenous RNA for microRNAs | Gudmundsson et al.105,106; Jendrzejewski et al.107 |
| NAMA | Thyroid cancer | Unknown | Yoon et al.108 | |
| AK023948 | Thyroid cancer | Unknown | He et al.109 | |
| Mammary gland | mPinc1.0, mPinc1.6 | Unknown | Prevents alveolar differentiation | Ginger et al.87; Shore et al.88 |
| Zfas1 | Mammary cancer | Unknown | Askarian-Amiri et al.89 | |
| HOTAIR | Mammary cancer | Unknown | Bhan et al.93 | |
| LSINCT5 | Mammary cancer | Proliferation | Silva et al.96; Silva et al.97 | |
| Pancreas | HI-LNC25 | T2DM | Positively regulates GLIS3 mRNA levels | Morán et al.60 |
| HOTAIR | Pancreatic cancer | Enhances cell invasion | Kim et al.116 | |
| ANRIL | T2DM | In mouse, controls expression of Cdkn2a (also known as P16ink4a) | Krishnamurthy et al.71 | |
| KCNQ1OT1 | T2DM | Unknown | Voight et al.73 | |
| H19 | T2DM | Unknown | Ding et al.65 | |
| Adrenal gland | SRA1 | Unknown | Coactivator of steroidogenesis | Xu et al.85 |
| Adipose tissue | lncRAP family, including | Unknown | Promotes adipogenesis | Sun et al.78 |
| FIRRE | Unknown | Promotes adipogenesis by binding hnRNPU | Hacisuleyman et al.47 | |
| Sra1 | Unknown | Promotes adipogenesis by binding PPARγ | Xu et al.117; Liu et al.118 | |
| Blnc1 | Unknown | Thermogenic differentiation | Zhao et al.83 | |
| naPINK1 | T2DM | Suppressor of PINK1 | Scheele et al.72 | |
| Ovary | LSINCT5 | Ovarian cancer | Stimulates proliferation | Silva et al.96 |
| Testes | Unknown names | Unknown | Unknown | Sun et al.119 |
| Prostate gland | PCAT1 | Prostate cancer | Transcriptional repressor | Prensner et al.101 |
| PRNCR1, PCGEM1 | Prostate cancer | Ligand-independent activation of the androgen receptor | Chung et al.99; Yang et al.100; Li et al.120 | |
| CTBP1-AS | Prostate cancer | Represses CTBP1 expression | Takayama et al.103 | |
| ANRIL | Prostate cancer | Tumour suppressor | Yap et al.70 |
Abbreviations: hnRNPU, heterogeneous nuclear ribonucleoprotein U; lncRNA, long non-coding RNA; T2DM, type 2 diabetes mellitus.
Glucose homeostasis and diabetes mellitus
In a first attempt to identify lncRNAs in the β-cell transcriptome of mice, the expression of 1,359 potential lncRNAs was detected in mouse β cells; many of these lncRNAs are β-cell specific.53 At the same time, another group published a transcriptome analysis of human β cells and, through de novo assembly, identified 1,128 lncRNAs expressed in human pancreatic islets.60 Intriguingly, many of these lncRNA-encoding genes are located near genes encoding important regulators of β-cell function and the expression of some of these lncRNAs increased after addition of glucose to a β-cell culture, which suggested that these lncRNAs might be relevant for mature islet cell physiology. In an attempt to identify mouse counterparts of these lncRNAs, the researchers also analyzed mouse islet cells and tested whether the lncRNA orthologs are regulated in a similar manner. The investigators compared the expression pattern of eight mouse lncRNAs that were orthologous to human lncRNAs in the mouse embryonic pancreas at embryonic day 13.5 and in islet cells from adult mice. Five of the eight tested lncRNAs were not expressed in the embryonic pancreas but were expressed in the adult pancreas. This observation suggests that lncRNAs might be important for the development and cell identity of β cells in the adult pancreas. The researchers then focused on one of the identified lncRNAs, HI-LNC25, to determine its function. Knockdown of HI-LNC25 resulted in reduced mRNA levels of GLIS3, which encodes an important islet transcription factor for which genetic variants associated with risk of type 2 diabetes mellitus (T2DM) are known.61–63 However, the underlying mechanisms by which HI-LNC25 functions remain to be elucidated.
In an effort to identify novel genes influencing glucose metabolism, microarray, RNA sequencing and exome sequencing methods were combined to analyse pancreatic islets from 89 deceased individuals with or without T2DM.64 Besides the expression of many protein-coding genes, the researchers identified 493 lncRNAs expressed in the pancreatic islets, of which 54 were involved in regulating gene expression and exon usage, as well as being correlated to the HbA1c levels of donors.64 This study provides a comprehensive catalog of gene regulation in human pancreatic β cells, including lncRNA expression, and offers insight into how genetic variation can influence glucose metabolism.64 Of note, seven of the 54 lncRNAs identified in this study were also reported in one of the previous transcriptome analyses of human β cells of patients with T2DM.60
An imprinted lncRNA, H19, has also been shown to be involved in the intergenerational transmission of diabetes mellitus in a mouse model of gestational diabetes mellitus.65 The investigators showed that F2 offspring of mice with impaired glucose tolerance had increased risk of also developing impaired glucose tolerance through the paternal line that was accompanied by downregulated expression of Igf2 and H19 in pancreatic islets and abnormal DNA methylation status in differentially methylated regions in the Igf2–H19 locus.65
Three GWAS revealed that the locus encoding the lncRNA ANRIL is associated with risk of T2DM.66-69 ANRIL is thought to associate with chromobox protein homolog 7, a component of the polycomb repressive complex 1 (PRC1), and mediate transcriptional silencing of the CDKN2A locus.70 One possible link between ANRIL and T2DM is the tumour suppressor p16-INK4a, a 148 amino acid protein encoded in the CDKN2A locus in humans and the Cdkn2a (also known as P16ink4a) locus in mice, as the expression of Cdkn2a was observed to be upregulated with ageing of the endocrine pancreas in mice, which limited the regenerative capacity of β cells.71 ANRIL might contribute to glucose homeostasis by maintaining the repressive state of the Cdkn2a locus and thus promoting β-cell division.
Natural antisense to PINK1 (naPINK1), which is expressed in muscle and adipose tissue, has also been associated with obesity and T2DM.72 An association of KCNQ1OT1, which is expressed in the pancreas,60,73 with T2DM has also been found.67,73 However, the functional roles of naPINK1 and KCNQ1OT1 in these diseases, if any, remain to be established.
Not much is known about the roles of lncRNAs in the development of type 1 diabetes mellitus (T1DM), but genetic studies have identified associations between the imprinted DLK1-MEG3 genetic region and T1DM.74 The protective allele of MEG3, a lncRNA, was less common among fathers than among mothers of affected offspring.74 Of note, the DLK1-MEG3 imprinted locus is also implicated in the pathogenesis of nonfunctioning pituitary adenomas.75
Whereas these studies have begun to implicate lncRNAs in the development and physiology of β cells and in glucose metabolism, further studies in cell culture and in animal models are needed to clarify their roles in these processes and their mechanisms of action.
Adipose tissue function
Two principal types of fat tissue exist in mammals. White adipose tissue (WAT) is specialized in storing chemical energy in the form of triglycerides, whereas brown adipose tissue (BAT) is specialized in generating heat and consuming energy as a defence against cold and obesity.76 Although not primarily known as an endocrine organ, adipose tissue secretes about 30 different soluble factors, some of which act as hormones. For instance, adipose tissue secretes leptin, which binds to its receptor in the hypothalamus and regulates appetite, and adiponectin, which modulates a number of metabolic processes, including glucose metabolism and fatty acid oxidation.
In the first description of a role for a non-coding RNA in the development of adipocytes, the steroid receptor RNA activator 1 (Sra1) lncRNA was shown to bind to and coactivate PPARγ in mice.58 In a follow-up study, the same research group generated mice in which the Sra1 locus was deleted.59 These mice were resistant to developing obesity and glucose intolerance induced by a high-fat diet and had increased whole-body insulin sensitivity. However, whether the observed phenotype reflects an impaired development of adipocytes owing to decreased PPARγ function needs to be determined, as the Sra1 knockout might also affect organs other than adipose tissue. Moreover, Sra1 might not be an authentic non-coding RNA, as some isoforms of the Sra1 gene encode a protein called SRAP.77 Knocking out Sra1 would have diminished the expression of SRAP and the observed phenotype could possibly have been the result of SRAP loss.
Using massively parallel sequencing of polyadenylation-selected RNAs during adipogenesis, we identified more than 175 differentially regulated lncRNAs during adipogenesis in both WAT and BAT.78 In adipocytes, the promoters of 57 of these 175 genes were bound by PPARγ and C/EBPα, two major adipogenic transcription factors. Furthermore, loss-of-function studies using small interfering RNAs (siRNAs) characterized 10 lncRNAs that, when suppressed, inhibited the differentiation of adipocytes to different extents; we named these lncRNAs regulated in adipogenesis (RAP) 1–10.
We further characterized lncRAP-1 in detail; lncRAP-1 was particularly interesting because this lncRNA has a conserved human ortholog on the X chromosome, contains multiple 156 bp repeating RNA domains and has numerous alternatively spliced isoforms.47 Because of these features, lncRAP-1 was renamed functional intergenic repeating RNA element (FIRRE). FIRRE is a strictly nuclear lncRNA. It localizes across a 5 Mb domain near its site of transcription and is in close proximity to five distinct transchromosomal loci,47 four of which were previously described as having regulatory roles in adipogenesis.79-82 RNA-pulldown assays showed that the heterogeneous nuclear ribonucleoprotein U (hnRNPU) binds to FIRRE and that the distinct 156 bp RNA repeats are necessary for this interaction.47 Importantly, the FIRRE–hnRNPU interaction is needed for colocalization of the transchromosomal loci contacted by FIRRE, indicating that this lncRNA is an important nuclear organization factor that brings loci encoding known regulators of adipogenesis into close proximity within the nucleus and enables their co-regulation.
A lncRNA named Blnc1 is important for thermogenic differentiation of brown and beige adipocytes in mice.83 Blnc1 expression is regulated by the transcription factor COE2 (also known as EBF-2). Blnc1, in turn, binds to COE2, which forms a feed-forward loop that results in enhanced expression of COE2, as well as enhanced expression of UCP 1 and mitochondrial genes, which are important for thermogenesis. Enhancing BAT activity or inducing BAT features in WAT have been associated with beneficial metabolic phenotypes, and thus are promising therapeutic strategies for metabolic diseases.84
Adrenal gland function
The nuclear receptor steroidogenic factor 1 (SF-1) recruits another nuclear receptor, DAX-1 (dosage sensitive sex reversal–adrenal hypoplasia congenita critical region on the X chromosome protein 1, also known as nuclear receptor subfamily 0 group B member 1) to the promoter of the gene that encodes the adrenocorticotropic hormone receptor (also known as MC2-R), which induces transcription of this gene. Transcriptional activation of MC2R is dependent on the lncRNA SRA1, as knockdown of SRA1 in human JEG-3 and Y1 cells (placental and adrenal cell lines, respectively) reduced the expression of MC2R.85 These results suggest that SRA1 plays an important part in adrenal gland function and sex determination.
Mammary gland function
The expansion and regression of the mammary gland during pregnancy is regulated by hormones, including estrogens and progesterone. In a screen for persistently upregulated RNAs after estrogen and progesterone treatment in rats, the lncRNA Gb7 was identified.86 This lncRNA was then renamed pregnancy-induced non- coding RNA (Pinc). Homology searches in different species revealed several conserved homologs in mammals but not in fugu (pufferfish), zebrafish or Xenopus genomes.87 In mouse, the expression of two splice variants, mPinc1.0 and mPinc1.6, increases in lobuloalveolar structures of the mammary gland during pregnancy, drops during lactation and rises again at day 5 of involution, which suggests that mPinc prevents alveolar differentiation before parturition.87 Knockdown of mPinc1.0 promoted apoptotic cell death, whereas knockdown of mPinc1.6 facilitated the G1–S phase transition, and knockdown of both lncRNAs resulted in enhanced lactogenic differentiation. On the other hand, overexpression of mPinc1.0 inhibited activation of alveolar cells to produce and secrete milk proteins.88 RNA immunoprecipitation experiments revealed that mPinc binds to polycomb repressive complex 2 (PRC2), which suggests that mPinc facilitates the maintenance of a repressive chromatin state.88 In a similar manner, the mouse zinc finger antisense 1 (Zfas1) lncRNA was characterized as downregulated between pregnancy and lactation, and upregulated between lactation and involution.89 Of note, knockdown of Zfas1 promoted proliferation and differentiation, which suggests Zfas1 acts as a tumour suppressor. This hypothesis was further substantiated by the observation that the human ortholog ZFAS1 is downregulated in invasive ductal carcinoma tissue compared with normal breast tissue.89
Regulation of circadian rhythms
The pineal gland is an important endocrine organ that controls circadian periodicity of gene expression, mainly by the production of melatonin at night. In a search for lncRNAs whose expression oscillates throughout the day, a group of researchers analysed RNA expression profiles in the rat pineal gland. The group identified 112 lncRNAs that were differentially expressed during day and night, which were named lncSNs (lncRNAs, Section on Neuroendocrinology), with 59% of them having their expression increased during the night.90 Eight of these lnSNs, whose size ranged from <1 kb to >50 kb, were chosen for further analysis. The rhythmic oscillation of their expression depended on the suprachiasmatic nucleus (SCN)–pineal pathway, as this rhythmic oscillation was blocked after surgical removal of the SCN but not when the rats were housed in constant darkness.90 The precise mechanisms and effects of these lncRNAs on circadian rhythm systems needs to be further addressed.
Endocrine cancer
LncRNAs have been associated with several endocrine cancers, including breast, prostate, ovarian and thyroid cancers, as well as nonfunctioning pituitary tumours. The involvement of several lncRNAs in the estrogen and androgen signalling pathways underlies at least part of their role in breast and prostate cancers. Many other lncRNAs have also been implicated in these two types of endocrine cancers, although whether they affect any nuclear receptor pathway remains unknown. More work is needed to identify lncRNAs and annotate their function in these and other endocrine cancers, which might reveal new therapeutic targets.
Breast cancer
A global run-on and sequencing study performed in MCF-7 cells, a human breast cancer cell line, revealed a rapid, robust and transient induction of a large fraction of genes immediately after estrogen treatment, including many lncRNAs as well as other types of non-coding transcripts.91 A large number of previously undetected lncRNA transcripts was found in enhancer regions and intergenic regions proximal to estrogen-receptor binding sites, which suggests these transcripts have a role in the estrogen-dependent transcriptional response. In another study in the MCF-7 cell line, stimulation of the estrogen receptor by 17β-estradiol increased global transcription of enhancer lncRNAs near estrogen-activated coding genes.92 Knocking down a set of enhancer lncRNAs using siRNAs attenuated the expression of target genes. Furthermore, using a three-dimensional DNA selection and ligation assay, the researchers showed that blocking enhancer lncRNA expression could destabilize 17β-estradiol-induced promoter–enhancer interactions, at least at one of the 17β-estradiol targets, the Nrip1 locus.92 These findings show that enhancer lncRNA expression is functionally important for estrogen-dependent transcription activation.
The lncRNA HOTAIR is upregulated by estradiol binding to estrogen receptors ERα and ERβ. Co-regulators including histone methyltransferases MLL1 and MLL3 and histone acetylases of the p300–CBP family are recruited together with estrogen receptors to bind estrogen response elements in the HOTAIR promoter in response to 17β-estradiol treatment and are necessary for the upregulation of HOTAIR.93 HOTAIR was highly expressed in metastatic breast cancers with poor survival outcomes.23 Enforced expression of HOTAIR in epithelial cancer cells drives a genome-wide shift of the binding pattern of PRC2 towards a pattern that is typically seen in embryonic fibroblasts, which is associated with a PRC2-dependent alteration in gene expression and increased cancer invasiveness. Conversely, siRNA-induced loss of HOTAIR expression decreased matrix invasiveness in the MCF-7 cell line.23 Thus, HOTAIR is of functional importance for the progression of estrogen-supported breast cancers.
One of the best studied lncRNAs that interacts with the estrogen receptor is SRA1. SRA1 functions as a nuclear receptor corepressor in estrogen signalling by binding to SHARP-194 and SLIRP,95 two general corepressors of nuclear receptor signalling. SRA1 associates with NCoA-1 (also known as SRC-1) upon estrogen induction, which leads to activation of estrogen-induced gene transcription. Estrogen signalling then induces expression of the corepressor SHARP-1, which, together with SLIRP, competes for the binding of SRA1 and NCoA-1. This competition leads to repression of estrogen-induced genes, which is a possible regulatory mechanism whereby estrogen hormone responses are attenuated.
Long stress-induced non-coding RNAs (LSINCTs) are highly expressed in breast cancers and were initially identified in a screen for genes that respond to the tobacco carcinogen nicotine-derived nitrosamine ketone.96 In a follow-up study, LSINCT5 was found to be overexpressed in most breast and ovarian cancer cell lines, and knockdown of this lncRNA resulted in decreased cell proliferation.97
Prostate cancer
Two lncRNAs, PCGEM1 and PRNCR1,98,99bind sequentially to the androgen receptor and strongly enhance both ligand-dependent and ligand-independent gene activation in prostate cancer cells.100 Binding of PRNCR1 is required for the recruitment of PCGEM1 to the androgen receptor. Each of these lncRNAs serves as a scaffold and forms a complex with a distinct set of proteins that enhances the looping of androgen-receptor-bound enhancers to target gene promoters, a mechanism similar to that observed in the enhancer lncRNAs interacting with the estrogen receptor.92 Interestingly, the expression of PCGEM1 and PRNCR1 is highly increased in aggressive prostate cancer, as well as castration- resistant prostate cancer cell lines. In castration- resistant prostate cancer cells, these overexpressed lncRNAs, through direct interactions with the androgen receptor, are required for ligand-independent activation of the androgen receptor and cell proliferation.100 Together, these data show that lncRNAs are required components of the regulatory network downstream of the androgen receptor.
In another study, 121 lncRNAs that were not annotated in the UCSC, Ensembl, Refseq, Vega or Encode genome databases as prostate-cancer-associated lncRNA transcripts (PCATs) were identified in a cohort of 102 prostate tissue specimens and cell lines.101 PCAT1 was specifically overexpressed in a subset of metastatic prostate cancers and functions as a transcriptional repressor in a complex with PRC2, which leads to the inhibition of some tumour-suppressor factors such as BRCA2, CENPE and CENPF, and the enhancement of cell proliferation.101
The androgen-responsive lncRNA CTBP1-AS was identified in a screen of androgen-sensitive prostate adenocarcinoma cells (LNCaP cells).102,103 CTBP1-AS is transcribed in antisense to the gene encoding CTBP1, a corepressor of androgen receptor signalling, and represses the expression of CTBP1, thereby promoting androgen-receptor signalling. These observations suggest CTBP1 has a role in the development of androgen-dependent and castration-resistant prostate cancers.103
Other endocrine cancers
The lncRNA MEG3 is implicated in the pathogenesis of nonfunctioning pituitary adenomas. MEG3 is expressed in normal pituitary cells but this expression is lost in non-functioning pituitary adenomas;75 this lncRNA has been proposed to act as a tumour suppressor.104 PTCSC3 was identified in a GWAS of papillary thyroid cancer.105,106 The expression of PTCSC3107 and that of NAMA and AK023948108,109 were found to be downregulated in thyroid cancer, but the contributions of these lncRNAs to thyroid cancer remain unclear.
Response to starvation
Induced expression of the lncRNA GAS5 represents a response to insufficient nutrient supply states, such as starvation and cell growth arrest.110 In these states, upregulated GAS5 binds the DNA-binding domain of the glucocorticoid receptor, thereby decoying the receptor away from its glucocorticoid response element targets in genomic DNA and inhibiting transcriptional responses to glucocorticoids.110
Conclusions and perspectives
On the basis of the pioneering studies reviewed above, several conclusions can be safely drawn. First, as in other cell types, lncRNAs have critical roles in the development of endocrine cells. Second, the regulated expression of many lncRNAs in response to hormones is required for these hormones to exert their full functions in target cells, including cells with aberrant functions such as cancer cells. Third, some SNPs in lncRNA loci have been linked with endocrine diseases as well as some endocrine cancers.
Our understanding of lncRNAs in the endocrine system is still at its infancy and many questions remain unanswered. First, because many lncRNAs are expressed in a cell-type-specific manner, currently annotated databases of lncRNAs that are based on previous RNA sequencing studies are likely to be incomplete or inaccurate for some cell types. This problem might be particularly important for the endocrine system, because many endocrine organs are composed of very specialized cell types that are likely to express functional lncRNAs that are unique to that cell type. Discovery and annotation of lncRNAs in these cell types remains important for future work. As shown by pioneering studies in this area,60 obtaining pure populations of different cell types and conducting in-depth RNA sequencing on both polyadenylated RNAs and non-polyadenylated RNAs that have been depleted of rRNAs is an essential first step, followed by detailed bioinformatic analysis to identify a comprehensive list of lncRNAs that is enriched in particular cell types.8,33
Second, the biological functions of lncRNAs in the endocrine system are still poorly understood. This problem will require developing genetic models to fully assess the role of lncRNAs on a variety of homeostatic processes in vivo. In addition, a large number of associations between endocrine diseases and SNPs affecting lncRNA expression have been established, but their cause–effect relationship should be carefully evaluated.
Third, we need to gain deeper insight into the molecular mechanisms used by lncRNAs to regulate gene expression. Numerous new techniques, developed to dissect the complicated interactions between RNA, proteins, and chromatin—including crosslinking immunoprecipitation, RNA antisense purification, chromatin isolation by RNA purification, capture–hybridization analysis of RNA targets and chromatin oligoaffinity precipitation—offer unprecedented opportunities to perform detailed mechanistic studies in the near future.
Fourth, a need exists to identify and classify the secondary and tertiary structures of lncRNAs according to their functions. Several dimethyl-sulphate-based methods have been developed for genome-wide profiling of the secondary structures of RNAs.111–113 The development of such methods might enable classification of lncRNA families on the basis of structural domains in ways similar to classification of protein families on the basis of structural domains.
Finally, lncRNAs can be potentially developed as novel diagnostic markers. For example, the expression levels of HOTAIR in primary breast tumours are powerful predictors of eventual metastasis and death.23 Several other lncRNAs have also been proposed as markers for endocrine malignancy.114 However, the sensitivity and reliability of these markers still need to be rigorously evaluated.
Key points.
A substantial proportion of the genome is transcribed into non-coding RNAs, including many long non-coding RNAs (lncRNAs) that are important regulators of endocrine cells
Many research findings point towards an important role of lncRNAs in regulating the development and maintenance of endocrine organs and hormonal signalling; misregulation of these processes can lead to disease
The biological functions of lncRNAs in endocrine organs are poorly understood; genetic models are needed to fully assess the role of lncRNAs in a variety of homeostatic processes in vivo
lncRNAs can be potentially developed as novel diagnostic markers to identify and classify certain tumours
Review criteria.
The PubMed search engine was used to identify articles relevant to the topic published from 1999 to September 2014. The following search terms were used: “long non-coding RNA”, “long non-coding RNAs endocrine”, “long non-coding RNAs cancer”, “long non-coding RNAs development”, “long non-coding RNAs nuclear receptors”, “long non-coding RNAs pancreas”, “long non-coding RNAs adipocytes”. Every search was done twice, using the filter “review” or not. Only full-text papers, reviews and resource articles in English were included. The lists of references of cited papers were searched for articles important for this Review and published before 1999. The authors regret that owing to space limitations not every relevant article could be cited.
Acknowledgments
M.K. is supported by the fellowship Kn1106/1-1 from the German Research Foundation. H.F.L. is supported by NIH grants DK047618-25 and DK068348-07. L.S. is supported by National Research Foundation grant NRF-2011NRF-NRFF 001-025. The authors thank J. Alvarez-Dominguez for critical discussions.
Footnotes
Competing interests: The authors declare no competing interests.
Author contributions: M.K. researched data for the article and wrote the article. All authors substantially contributed to discussing the content and reviewing and/or editing the manuscript before submission.
Contributor Information
Marko Knoll, Whitehead Institute for Biomedical Research, 9 Cambridge Center, MA 02142, USA.
Harvey F. Lodish, Whitehead Institute for Biomedical Research, 9 Cambridge Center, MA 02142, USA
Lei Sun, Cardiovascular and Metabolic Disorders, Duke-National University of Singapore Graduate Medical School, 8 College Road, 169857, Singapore.
References
- 1.Amaral PP, Dinger ME, Mercer TR, Mattick JS. The eukaryotic genome as an RNA machine. Science. 2008;319:1787–1789. doi: 10.1126/science.1155472. [DOI] [PubMed] [Google Scholar]
- 2.Kapranov P, et al. Large-scale transcriptional activity in chromosomes 21 and 22. Science. 2002;296:916–919. doi: 10.1126/science.1068597. [DOI] [PubMed] [Google Scholar]
- 3.Kapranov P, et al. Examples of the complex architecture of the human transcriptome revealed by RACE and high-density tiling arrays. Genome Res. 2005;15:987–997. doi: 10.1101/gr.3455305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wahlestedt C. Natural antisense and noncoding RNA transcripts as potential drug targets. Drug Discov Today. 2006;11:503–508. doi: 10.1016/j.drudis.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 5.Kawai J, et al. Functional annotation of a full-length mouse cDNA collection. Nature. 2001;409:685–690. doi: 10.1038/35055500. [DOI] [PubMed] [Google Scholar]
- 6.Okazaki Y, et al. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature. 2002;420:563–573. doi: 10.1038/nature01266. [DOI] [PubMed] [Google Scholar]
- 7.Rinn JL, et al. The transcriptional activity of human chromosome 22. Genes Dev. 2003;17:529–540. doi: 10.1101/gad.1055203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cabili MN, et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25:1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dunham I, et al. The DNA sequence of human chromosome 22. Nature. 1999;402:489–495. doi: 10.1038/990031. [DOI] [PubMed] [Google Scholar]
- 10.Lander ES, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 11.Schuler GD, et al. A gene map of the human genome. Science. 1996;274:540–546. [PubMed] [Google Scholar]
- 12.ENCODE Project Consortium et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Derrien T, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harrow J, et al. GENCODE: the reference human genome annotation for the ENCODE Project. Genome Res. 2012;22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hangauer MJ, Vaughn IW, McManus MT. Pervasive transcription of the human genome produces thousands of previously unidentified long intergenic noncoding RNAs. PLoS Genet. 2013;9:e1003569. doi: 10.1371/journal.pgen.1003569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jacquier A. The complex eukaryotic transcriptome: unexpected pervasive transcription and novel small RNAs. Nat Rev Genet. 2009;10:833–844. doi: 10.1038/nrg2683. [DOI] [PubMed] [Google Scholar]
- 18.Djebali S, et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bertone P, et al. Global identification of human transcribed sequences with genome tiling arrays. Science. 2004;306:2242–2246. doi: 10.1126/science.1103388. [DOI] [PubMed] [Google Scholar]
- 20.Guttman M, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Penny GD, Kay GF, Sheardown SA, Rastan S, Brockdorff N. Requirement for Xist in X chromosome inactivation. Nature. 1996;379:131–137. doi: 10.1038/379131a0. [DOI] [PubMed] [Google Scholar]
- 22.Yu W, et al. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature. 2008;451:202–206. doi: 10.1038/nature06468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gupta RA, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kretz M, et al. Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature. 2013;493:231–235. doi: 10.1038/nature11661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brannan CI, Dees EC, Ingram RS, Tilghman SM. The product of the H19 gene may function as an RNA. Mol Cell Biol. 1990;10:28–36. doi: 10.1128/mcb.10.1.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ingolia NT, Lareau LF, Weissman JS. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011;147:789–802. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guttman M, et al. Ribosome profiling provides evidence that large noncoding RNAs do not encode proteins. Cell. 2013;154:240–251. doi: 10.1016/j.cell.2013.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chew GL, et al. Ribosome profiling reveals resemblance between long non-coding RNAs and 5′ leaders of coding RNAs. Development. 2013;140:2828–2834. doi: 10.1242/dev.098343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bazzini AA, et al. Identification of small ORFs in vertebrates using ribosome footprinting and evolutionary conservation. EMBO J. 2014;33:981–993. doi: 10.1002/embj.201488411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ingolia NT, et al. Ribosome profiling reveals pervasive translation outside of annotated protein-coding genes. Cell Rep. 2014;8:1365–1379. doi: 10.1016/j.celrep.2014.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lam MT, Li W, Rosenfeld MG, Glass CK. Enhancer RNAs and regulated transcriptional programs. Trends Biochem Sci. 2014;39:170–182. doi: 10.1016/j.tibs.2014.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Heesch S, et al. Extensive localization of long noncoding RNAs to the cytosol and mono-and polyribosomal complexes. Genome Biol. 2014;15:R6. doi: 10.1186/gb-2014-15-1-r6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alvarez-Dominguez JR, et al. Global discovery of erythroid long noncoding RNAs reveals novel regulators of red cell maturation. Blood. 2014;123:570–581. doi: 10.1182/blood-2013-10-530683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Necsulea A, et al. The evolution of lncRNA repertoires and expression patterns in tetrapods. Nature. 2014;505:635–640. doi: 10.1038/nature12943. [DOI] [PubMed] [Google Scholar]
- 35.Diederichs S. The four dimensions of noncoding RNA conservation. Trends Genet. 2014;30:121–123. doi: 10.1016/j.tig.2014.01.004. [DOI] [PubMed] [Google Scholar]
- 36.Kutter C, et al. Rapid turnover of long noncoding RNAs and the evolution of gene expression. PLoS Genet. 2012;8:e1002841. doi: 10.1371/journal.pgen.1002841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ulitsky I, Shkumatava A, Jan CH, Sive H, Bartel DP. Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell. 2011;147:1537–1550. doi: 10.1016/j.cell.2011.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Torarinsson E, Sawera M, Havgaard JH, Fredholm M, Gorodkin J. Thousands of corresponding human and mouse genomic regions unalignable in primary sequence contain common RNA structure. Genome Res. 2006;16:885–889. doi: 10.1101/gr.5226606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Torarinsson E, et al. Comparative genomics beyond sequence-based alignments: RNA structures in the ENCODE regions. Genome Res. 2008;18:242–251. doi: 10.1101/gr.6887408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Johnsson P, Morris KV. Expanding the functional role of long noncoding RNAs. Cell Res. 2014;24:1284–1285. doi: 10.1038/cr.2014.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tsai MC, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tripathi V, et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell. 2010;39:925–938. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rinn JL, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang KC, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–124. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hacisuleyman E, et al. Topological organization of multichromosomal regions by the long intergenic noncoding RNA FIRRE. Nat Struct Mol Biol. 2014;21:198–206. doi: 10.1038/nsmb.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Karreth FA, et al. In vivo identification of tumor-suppressive PTEN ceRNAs in an oncogenic BRAF-induced mouse model of melanoma. Cell. 2011;147:382–395. doi: 10.1016/j.cell.2011.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yoon JH, et al. LincRNA-p21 suppresses target mRNA translation. Mol Cell. 2012;47:648–655. doi: 10.1016/j.molcel.2012.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gong C, Maquat LE. IncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3′ UTRs via Alu elements. Nature. 2011;470:284–288. doi: 10.1038/nature09701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hansen TB, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 52.Haines JL, et al. Complement factor H variant increases the risk of age-related macular degeneration. Science. 2005;308:419–421. doi: 10.1126/science.1110359. [DOI] [PubMed] [Google Scholar]
- 53.Hindorff LA, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci USA. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.St Laurent G, Vyatkin Y, Kapranov P. Dark matter RNA illuminates the puzzle of genome-wide association studies. BMC Med. 2014;12:97. doi: 10.1186/1741-7015-12-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kapranov P, et al. The majority of total nuclear-encoded non-ribosomal RNA in a human cell is ‘dark matter’ un-annotated RNA. BMC Biol. 2010;8:149. doi: 10.1186/1741-7007-8-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kumar V, et al. Human disease-associated genetic variation impacts large intergenic non-coding RNA expression. PLoS Genet. 2013;9:e1003201. doi: 10.1371/journal.pgen.1003201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen G, et al. LncRNADisease: a database for long-non-coding RNA-associated diseases. Nucleic Acids Res. 2013;41:D983–D986. doi: 10.1093/nar/gks1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hrdlickova B, de Almeida RC, Borek Z, Withoff S. Genetic variation in the non-coding genome: Involvement of micro-RNAs and long non-coding RNAs in disease. Biochim Biophys Acta. 2014;1842:1910–1922. doi: 10.1016/j.bbadis.2014.03.011. [DOI] [PubMed] [Google Scholar]
- 59.Ning S, et al. LincSNP: a database of linking disease-associated SNPs to human large intergenic non-coding RNAs. BMC Bioinformatics. 2014;15:152. doi: 10.1186/1471-2105-15-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Morán I, et al. Human β cell transcriptome analysis uncovers lncRNAs that are tissue-specific, dynamically regulated, and abnormally expressed in type 2 diabetes. Cell Metab. 2012;16:435–448. doi: 10.1016/j.cmet.2012.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Senée V, et al. Mutations in GLIS3 are responsible for a rare syndrome with neonatal diabetes mellitus and congenital hypothyroidism. Nat Genet. 2006;38:682–687. doi: 10.1038/ng1802. [DOI] [PubMed] [Google Scholar]
- 62.Cho YS, et al. Meta-analysis of genome-wide association studies identifies eight new loci for type 2 diabetes in East Asians. Nat Genet. 2012;44:67–72. doi: 10.1038/ng.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nogueira TC, et al. GLIS3, a susceptibility gene for type 1 and type 2 diabetes, modulates pancreatic β cell apoptosis via regulation of a splice variant of the BH3-only protein Bim. PLoS Genet. 2013;9:e1003532. doi: 10.1371/journal.pgen.1003532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fadista J, et al. Global genomic and transcriptomic analysis of human pancreatic islets reveals novel genes influencing glucose metabolism. Proc Natl Acad Sci USA. 2014;111:13924–13929. doi: 10.1073/pnas.1402665111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ding GL, et al. Transgenerational glucose intolerance with Igf2/H19 epigenetic alterations in mouse islet induced by intrauterine hyperglycemia. Diabetes. 2012;61:1133–1142. doi: 10.2337/db11-1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Diabetes Genetics Initiative of Broad Institute of Harvard and MIT et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316:1331–1336. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- 67.Zeggini E, et al. Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science. 2007;316:1336–1341. doi: 10.1126/science.1142364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Scott LJ, et al. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science. 2007;316:1341–1345. doi: 10.1126/science.1142382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pasmant E, Sabbagh A, Vidaud M, Bièche I. ANRIL, a long, noncoding RNA, is an unexpected major hotspot in GWAS. FASEB J. 2011;25:444–448. doi: 10.1096/fj.10-172452. [DOI] [PubMed] [Google Scholar]
- 70.Yap KL, et al. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol Cell. 2010;38:662–674. doi: 10.1016/j.molcel.2010.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Krishnamurthy J, et al. p16INK4a induces an age-dependent decline in islet regenerative potential. Nature. 2006;443:453–457. doi: 10.1038/nature05092. [DOI] [PubMed] [Google Scholar]
- 72.Scheele C, et al. Altered regulation of the PINK1 locus: a link between type 2 diabetes and neurodegeneration? FASEB J. 2007;21:3653–3665. doi: 10.1096/fj.07-8520com. [DOI] [PubMed] [Google Scholar]
- 73.Voight BF, et al. Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nat Genet. 2010;42:579–589. doi: 10.1038/ng.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wallace C, et al. The imprinted DLK1–MEG3 gene region on chromosome 14q32.2 alters susceptibility to type 1 diabetes. Nat Genet. 2010;42:68–71. doi: 10.1038/ng.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cheunsuchon P, et al. Silencing of the imprinted DLK1–MEG3 locus in human clinically nonfunctioning pituitary adenomas. Am J Pathol. 2011;179:2120–2130. doi: 10.1016/j.ajpath.2011.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rosen ED, Spiegelman BM. Adipocytes as regulators of energy balance and glucose homeostasis. 2006;444:847–853. doi: 10.1038/nature05483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chooniedass-Kothari S, et al. The steroid receptor RNA activator is the first functional RNA encoding a protein. FEBS Lett. 2004;566:43–47. doi: 10.1016/j.febslet.2004.03.104. [DOI] [PubMed] [Google Scholar]
- 78.Sun L, et al. Long noncoding RNAs regulate adipogenesis. Proc Natl Acad Sci USA. 2013;110:3387–3392. doi: 10.1073/pnas.1222643110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nukitrangsan N, et al. Effect of Peucedanum japonicum Thunb on the expression of obesity-related genes in mice on a high-fat diet. J Oleo Sci. 2011;60:527–536. doi: 10.5650/jos.60.527. [DOI] [PubMed] [Google Scholar]
- 80.Seo J, et al. Atf4 regulates obesity, glucose homeostasis, and energy expenditure. Diabetes. 2009;58:2565–2573. doi: 10.2337/db09-0335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rubi B, del Arco A, Bartley C, Satrustegui J, Maechler P. The malate–aspartate NADH shuttle member Aralar1 determines glucose metabolic fate, mitochondrial activity, and insulin secretion in β cells. J Biol Chem. 2004;279:55659–55666. doi: 10.1074/jbc.M409303200. [DOI] [PubMed] [Google Scholar]
- 82.Lee EK, et al. miR-130 suppresses adipogenesis by inhibiting peroxisome proliferator-activated receptor γ expression. Mol Cell Biol. 2011;31:626–638. doi: 10.1128/MCB.00894-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhao XY, Li S, Wang GX, Yu Q, Lin JD. A long noncoding RNA transcriptional regulatory circuit drives thermogenic adipocyte differentiation. Mol Cell. 2014;55:372–382. doi: 10.1016/j.molcel.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kopecky J, Clarke G, Enerbäck S, Spiegelman B, Kozak LP. Expression of the mitochondrial uncoupling protein gene from the aP2 gene promoter prevents genetic obesity. J Clin Invest. 1995;96:2914–2923. doi: 10.1172/JCI118363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Xu B, et al. Dax-1 and steroid receptor RNA activator (SRA) function as transcriptional coactivators for steroidogenic factor 1 in steroidogenesis. Mol Cell Biol. 2009;29:1719–1734. doi: 10.1128/MCB.01010-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ginger MR, Gonzalez-Rimbau MF, Gay JP, Rosen JM. Persistent changes in gene expression induced by estrogen and progesterone in the rat mammary gland. Mol Endocrinol. 2001;15:1993–2009. doi: 10.1210/mend.15.11.0724. [DOI] [PubMed] [Google Scholar]
- 87.Ginger MR, et al. A noncoding RNA is a potential marker of cell fate during mammary gland development. Proc Natl Acad Sci USA. 2006;103:5781–5786. doi: 10.1073/pnas.0600745103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Shore AN, et al. Pregnancy-induced noncoding RNA (PINC) associates with polycomb repressive complex 2 and regulates mammary epithelial differentiation. PLoS Genet. 2012;8:e1002840. doi: 10.1371/journal.pgen.1002840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Askarian-Amiri ME, et al. SNORD-host RNA Zfas1 is a regulator of mammary development and a potential marker for breast cancer. RNA. 2011;17:878–891. doi: 10.1261/rna.2528811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Coon SL, et al. Circadian changes in long noncoding RNAs in the pineal gland. Proc Natl Acad Sci USA. 2012;109:13319–13324. doi: 10.1073/pnas.1207748109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hah N, et al. A rapid, extensive, and transient transcriptional response to estrogen signaling in breast cancer cells. Cell. 2011;145:622–634. doi: 10.1016/j.cell.2011.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Li W, et al. Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature. 2013;498:516–520. doi: 10.1038/nature12210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bhan A, et al. Antisense transcript long noncoding RNA (lncRNA) HOTAIR is transcriptionally induced by estradiol. J Mol Biol. 2013;425:3707–3722. doi: 10.1016/j.jmb.2013.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Shi Y, et al. Sharp, an inducible cofactor that integrates nuclear receptor repression and activation. Genes Dev. 2001;15:1140–1151. doi: 10.1101/gad.871201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hatchell EC, et al. SLIRP, a small SRA binding protein, is a nuclear receptor corepressor. Mol Cell. 2006;22:657–668. doi: 10.1016/j.molcel.2006.05.024. [DOI] [PubMed] [Google Scholar]
- 96.Silva JM, et al. Identification of long stress-induced non-coding transcripts that have altered expression in cancer. Genomics. 2010;95:355–362. doi: 10.1016/j.ygeno.2010.02.009. [DOI] [PubMed] [Google Scholar]
- 97.Silva JM, Boczek NJ, Berres MW, Ma X, Smith DI. LSINCT5 is over expressed in breast and ovarian cancer and affects cellular proliferation. RNA Biol. 2011;8:496–505. doi: 10.4161/rna.8.3.14800. [DOI] [PubMed] [Google Scholar]
- 98.Srikantan V, et al. PCGEM1, a prostate-specific gene, is overexpressed in prostate cancer. Proc Natl Acad Sci USA. 2000;97:12216–12221. doi: 10.1073/pnas.97.22.12216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chung S, et al. Association of a novel long non-coding RNA in 8q24 with prostate cancer susceptibility. Cancer Sci. 2011;102:245–252. doi: 10.1111/j.1349-7006.2010.01737.x. [DOI] [PubMed] [Google Scholar]
- 100.Yang L, et al. lncRNA-dependent mechanisms of androgen-receptor-regulated gene activation programs. Nature. 2013;500:598–602. doi: 10.1038/nature12451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Prensner JR, et al. Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression. Nat Biotechnol. 2011;29:742–749. doi: 10.1038/nbt.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Takayama K, et al. Integration of cap analysis of gene expression and chromatin immunoprecipitation analysis on array reveals genome-wide androgen receptor signaling in prostate cancer cells. Oncogene. 2011;30:619–630. doi: 10.1038/onc.2010.436. [DOI] [PubMed] [Google Scholar]
- 103.Takayama KI, et al. Androgen-responsive long noncoding RNA CTBP1-AS promotes prostate cancer. EMBO J. 2013;32:1665–1680. doi: 10.1038/emboj.2013.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zhou Y, et al. Activation of p53 by MEG3 non-coding RNA. J Biol Chem. 2007;282:24731–24742. doi: 10.1074/jbc.M702029200. [DOI] [PubMed] [Google Scholar]
- 105.Gudmundsson J, et al. Common variants on 9q22.33 and 14q13.3 predispose to thyroid cancer in European populations. Nat Genet. 2009;41:460–464. doi: 10.1038/ng.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Gudmundsson J, et al. Discovery of common variants associated with low TSH levels and thyroid cancer risk. Nat Genet. 2012;44:319–322. doi: 10.1038/ng.1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Jendrzejewski J, et al. The polymorphism rs944289 predisposes to papillary thyroid carcinoma through a large intergenic noncoding RNA gene of tumor suppressor type. Proc Natl Acad Sci USA. 2012;109:8646–8651. doi: 10.1073/pnas.1205654109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Yoon H, et al. Identification of a novel noncoding RNA gene, NAMA, that is downregulated in papillary thyroid carcinoma with BRAF mutation and associated with growth arrest. Int J Cancer. 2007;121:767–775. doi: 10.1002/ijc.22701. [DOI] [PubMed] [Google Scholar]
- 109.He H, et al. A susceptibility locus for papillary thyroid carcinoma on chromosome 8q24. Cancer Res. 2009;69:625–631. doi: 10.1158/0008-5472.CAN-08-1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kino T, Hurt DE, Ichijo T, Nader N, Chrousos GP. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci Signal. 2010;3:ra8. doi: 10.1126/scisignal.2000568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ding Y, et al. In vivo genome-wide profiling of RNA secondary structure reveals novel regulatory features. Nature. 2014;505:696–700. doi: 10.1038/nature12756. [DOI] [PubMed] [Google Scholar]
- 112.Rouskin S, Zubradt M, Washietl S, Kellis M, Weissman JS. Genome-wide probing of RNA structure reveals active unfolding of mRNA structures in vivo. Nature. 2014;505:701–705. doi: 10.1038/nature12894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wan Y, et al. Landscape and variation of RNA secondary structure across the human transcriptome. Nature. 2014;505:706–709. doi: 10.1038/nature12946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kentwell J, Gundara JS, Sidhu SB. Noncoding RNAs in endocrine malignancy. Oncologist. 2014;19:483–491. doi: 10.1634/theoncologist.2013-0458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Roy M, et al. Analysis of the canine brain transcriptome with an emphasis on the hypothalamus and cerebral cortex. Mamm Genome. 2013;24:484–499. doi: 10.1007/s00335-013-9480-0. [DOI] [PubMed] [Google Scholar]
- 116.Kim K, et al. HOTAIR is a negative prognostic factor and exhibits pro-oncogenic activity in pancreatic cancer. Oncogene. 2013;32:1616–1625. doi: 10.1038/onc.2012.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Xu B, et al. Multiple roles for the non-coding RNA SRA in regulation of adipogenesis and insulin sensitivity. PLoS ONE. 2010;5:e14199. doi: 10.1371/journal.pone.0014199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Liu S, et al. SRA gene knockout protects against diet-induced obesity and improves glucose tolerance. J Biol Chem. 2014;289:13000–13009. doi: 10.1074/jbc.M114.564658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Sun J, Lin Y, Wu J. Long non-coding RNA expression profiling of mouse testis during postnatal development. PLoS ONE. 2013;8:e75750. doi: 10.1371/journal.pone.0075750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Li L, et al. Association between polymorphisms in long non-coding RNA PRNCR1 in 8q24 and risk of colorectal cancer. J Exp Clin Cancer Res. 2013;32:104. doi: 10.1186/1756-9966-32-104. [DOI] [PMC free article] [PubMed] [Google Scholar]