Abstract
Protein folding homeostasis in the lumen of the endoplasmic reticulum is defended by signal transduction pathways that are activated by an imbalance between unfolded proteins and chaperones (so called ER stress). Collectively referred to as the unfolded protein response (UPR) this homeostatic response is initiated by three known ER stress transducers: IRE1, PERK and ATF6. These ER-localised transmembrane (TM) proteins posses lumenal stress sensing domains and cytosolic effector domains that collectively activate a gene expression programme regulating the production of proteins involved in the processing and maturation of secreted proteins that enter the ER. However, beyond limiting unfolded protein stress in the ER the UPR has important connections to lipid metabolism that are the subject of this review.
Current Opinion in Cell Biology 2015 33:67–73
This review comes from a themed issue on Cell regulation
Edited by Jodi Nunnari and Johan Auwerx
For a complete overview see the Issue and the Editorial
Available online 25th December 2014
http://dx.doi.org/10.1016/j.ceb.2014.12.002
0955-0674/© 2014 The Authors. Published by Elsevier Ltd. This is an open access article under the CC BY license (http://creativecommons.org/licenses/by/3.0/).
Lipid regulation of the endoplasmic reticulum unfolded protein response is conserved in eukaryotes
Clues to a lipid connection were provided by the very earliest studies in which UPR components were first identified. The genes encoding what we now know to be the UPR transducer IRE1 (also known as ERN1, for ER to nucleus transducer 1) and its downstream transcription factor HAC1/IRE2 (the yeast ortholog of the metazoan XBP1) were first identified as required for growth in medium deprived of inositol [1,2], an essential building block of yeast phospholipids. Depletion of inositol from growth medium strongly activates IRE1 signalling [3], whereas IRE1 and HAC1 are required for full expression of genes involved in lipid metabolism in yeast [4]. Furthermore, deletion of genes regulating lipid metabolism strongly activates UPR signalling in yeast [5••].
Lipid-dependent activation of IRE1 was also observed following loading of yeast with saturated fatty acids and sterol [6•] and following deletion of the regulators of sphingolipid synthesis ORM1 and ORM2 [7]. These findings established firm links between lipids and UPR signalling in yeast: the UPR is activated by altered lipid metabolism whose consequences are mitigated by UPR signalling.
In mammalian cells, enhanced UPR signalling has been observed in cholesterol-loaded macrophages [8], in pancreatic beta cells exposed to saturated fatty acids [9] and in cells in which increased lipid saturation was achieved by genetic or pharmacological inhibition of the Δ9 desaturase, stearoyl-CoA desaturase 1 [10,11]. Perturbation of sphingolipid metabolism causing increased levels of ceramides also activated the UPR in mammalian cells [12,13]. Increased UPR markers have also been observed in the liver and adipose tissue of mice fed a high fat diet and in severely obese humans [14,15]. These observations indicate that the ability of UPR transducers to sense perturbations to lipid homeostasis is conserved in eukaryotes.
Linking lipid perturbation to activation of UPR transducers
Lipid composition could modulate protein folding in, or trafficking through, the ER, indirectly activating UPR transducers by changing the level of unfolded proteins. Changes in the lipid composition could, for example, perturb ER calcium homeostasis, inhibiting the function of calcium-dependent enzymes and chaperones. In support of this idea, ER stress signalling in the liver of obese mice correlated with perturbations of ER calcium homeostasis through an inhibition of the SERCA transporter caused by an increase in the ratio between phosphatidylcholine and phosphatidylethanolamine in membranes of the hepatocytes [14]. UPR in cholesterol-loaded macrophages was also linked to inhibition of the SERCA pump [16].
However, there are clues that lipid changes may affect UPR signalling independently of their effect on protein folding in the ER lumen. In yeast, depletion of the phospholipid building block inositol strongly activated IRE1 but had no effect on the mobility of the ER chaperone BiP/KAR2 (BiP mobility is strongly retarded by unfolded protein stress) [17]. This indicates that inositol depletion activates the UPR without causing lumenal unfolded protein stress. In C. elegans, deletion of mdt-15, a subunit of the transcriptional regulator complex Mediator, was associated with an increase in membrane lipid saturation and the activation IRE1 and PERK without evidence for concomitant formation of protein aggregates in the ER, suggesting that activation of the UPR stress transducers may have a component that is independent of unfolded protein stress [18•].
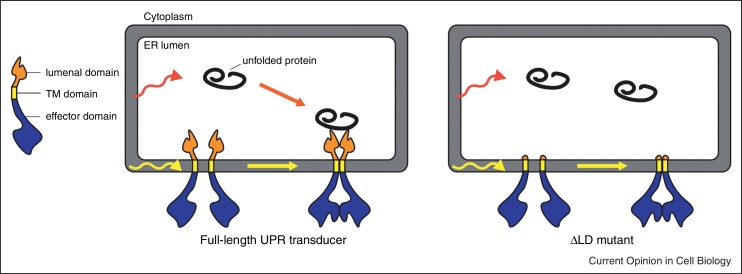
Direct evidence that lipids may activate the UPR independently of their effects on unfolded protein burden in the lumen was provided by the observation that IRE1 and PERK lacking their lumenal unfolded protein stress-sensing domains were activated in yeast deprived of inositol [19••] or mammalian cells exposed to saturated fatty acid [20••]. Activation of the mutant IRE1 and PERK lacking their lumenal domain required ER membrane tethering via a TM domain [20••]. Furthermore, sensitivity to the lipid composition of the membrane bilayer was observed in a reconstituted system composed only of liposomes and a truncated PERK lacking its lumenal domain, but retaining the TM and cytosolic effector domains [20••]. Thus, perturbations of ER membrane lipids can directly activate IRE1 and PERK independently of unfolded proteins in a process that requires TM domain insertion into the ER lipid bilayer (Figure 1).
Figure 1.
Mechanisms of lipid sensing by the UPR transducers. Perturbations of the ER lipid bilayer composition could impair the folding of ER proteins, thereby activating the full-length UPR transducers via their lumenal unfolded proteins sensing domain (left panel). However, mutant IRE1 and PERK lacking their lumenal domain (ΔLD mutants, right panel) are also activated by lipid perturbations in a process that requires ER membrane tethering via a TM domain [19••,20••]. Lipid-dependent activation of the ΔLD UPR transducers is independent of unfolded protein accumulation and likely proceeds by association of the TM domains in response to changes in the biophysical properties of the membrane. Dimerization of the effector domains of IRE1 and PERK (required for their allosteric activation) follows association of the TM domains. In the full-length proteins, stability of the activating effector-domain dimer would be further increased by the dimerization of the lumenal domain of IRE1 or PERK. Response of TM domains to changes in lipid composition of the ER membrane likely modulates a cooperative process involving the dimerization of both the effector and the lumenal domains.
Activation of the UPR signal transducers hinges on the transition from an inactive monomeric state to an active dimer/higher order oligomeric structure (reviewed in [21]). The recent findings obtained with mutant UPR signal transducers lacking their lumenal domains suggest that the TM domain of PERK and IRE1 could promote dimerization by responding to changes in the biophysical properties of the ER membrane [20••]. In comparison to other organelles, the ER lipid bilayer is a thin and fluid membrane, characterized by low cholesterol content (for a review of the key differences in the lipid composition of the different organelles, see [22]). Changes in the lipid composition, such as increased acyl chain saturation, are therefore likely to modify the ER membrane biophysical properties and influence the behaviour of TM peptides within the lipid bilayer.
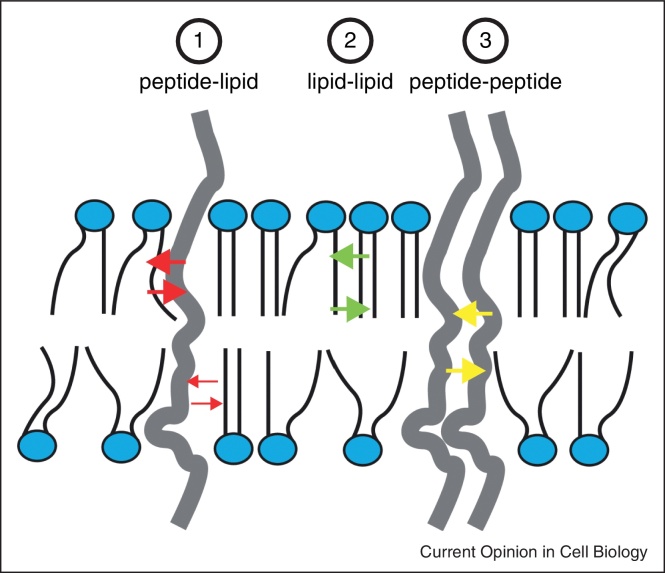
In the lipid bilayer, proteins and lipids are subject to three types of interactions: peptide–peptide, peptide–lipid and lipid–lipid (Figure 2). The relative strength of which influences the oligomeric state of a TM peptide in the lipid membrane [23]. The biophysical properties of membranes significantly influence the partitioning of TM peptides between their monomeric and dimeric/oligomeric state. This is played out through several defined mechanisms that might contribute to lipid regulation of UPR signalling, independently of unfolded proteins (Box 1).
Figure 2.
Molecular interactions in the plane of the lipid bilayer. Within the plane of the membrane, a TM helix can interact solely with neighbouring lipids and therefore remain as a monomer, or it can also interact with another TM helix and therefore form a dimer or higher order structure. The monomer/dimer equilibrium is modulated by the strength of the three types of interactions taking place in this simplified model of TM helices in a lipid bilayer: (1) peptide–lipid interaction, (2) lipid–lipid interaction and (3) peptide–peptide interaction. Changes to the lipid composition can affect the strength of these competing interactions and therefore modify the monomer/dimer equilibrium of the embedded TM [56].
Box 1. Biophysical principles regulating TM–TM dimerization.
Hydrophobic mismatch
Stable membrane integration of single-pass proteins is favoured by matching TM length to the thickness of the lipid bilayer, as evidenced by the gradual increase in TM length of single pass resident proteins that matches the increase in the lipid bilayer thickness along the secretory pathway [43••]. However, a mismatch can occur when changes to lipid composition causes the thickness of the lipid bilayer to be different from the length of the hydrophobic TM peptide. The thickness of the lipid bilayer depends on the length of the acyl chains and on the degree of their unsaturation. The less flexible saturated acyl chains remain in an extended conformation and therefore tend to increase the thickness of the lipid bilayer [44].
As lipid bilayer thickness increases, polar amino acid side chains that would otherwise be located at the water–lipid interface are forced into the hydrophobic lipid bilayer. Their presence in the lipid bilayer creates a thermodynamically unfavourable situation, which can be relieved by tilting or by the formation of hydrogen bonds or salt bridges between polar side chains from adjacent TMs, favouring dimerization [45–47].
Acyl chain flexibility
The degree of phospholipid unsaturation can also influence TM–TM interactions, independently of effects on the thickness of the lipid bilayer. Saturated acyl chains are less flexible than unsaturated acyl chains. As a consequence saturated acyl chains interact less efficiently with the surface of the TM helices than unsaturated acyl chains. Weakening of TM–lipid interaction by acyl chain saturation favours the competing TM–TM interaction thereby promoting dimerization [48].
Lipid microdomains
Biological membranes composed of heterogeneous lipids can segregate into lipid microdomains, such as liquid ordered domains and liquid disordered domains [22,49]. TM proteins preferentially localize into different membrane domains where their local density increases [50,51]. Changes in lipid composition can modify the relative size of these membrane domains. As a consequence, the local concentration of a TM protein that partitions asymmetrically between lipid domains, may change, thereby affecting the monomer–dimer equilibrium [48,52]. In comparison to the plasma membrane, the relative low cholesterol and sphingolipid content of the ER makes the formation of liquid ordered microdomains unlikely. However, this observation does not exclude the possibility that microdomains with specific lipid composition are formed in the ER, either locally at the site of sphingolipid or cholesterol synthesis [53], or in the spatially distinct sub-compartments of the ER (sheets, tubules, ER exit sites, mitochondrial associated membrane) [54,55].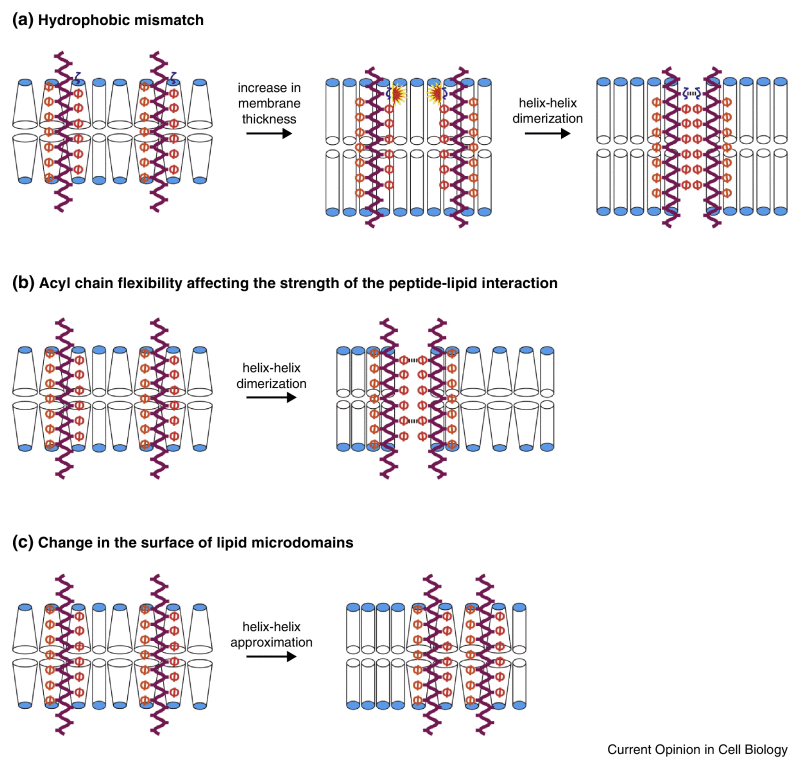
A measure of lipid-mediated activation of the mutant IRE1 lacking its lumenal domain was retained when the TM domain of IRE1 was swapped to that of calnexin, an unrelated ER protein, or when the TM peptide sequence was scrambled [20••]. These findings suggest that lipid-dependent activation of the UPR transducers has relaxed specificity with respect to protein–protein or protein–lipid interactions involving the TM amino acid side chains, but rather proceeds through generic biophysical mechanisms of dimerization and approximation that are shared by diverse TM domains, described in Box 1. As long as it allows insertion in the ER membrane, the TM domain of the UPR transducers can tolerate a range of amino acid substitutions in its sequence. However, the sensitivity of the assays used to measure the effects of TM swaps on intensity of UPR signalling is rather limited. It thus remains possible that more sensitive assays might reveal sequence constrains on TM domains of the UPR transducers driven by the need to respond to lipids.
It is noteworthy that the response of full-length IRE1 and PERK to lipid perturbation was considerably stronger than their lumenal domain-deleted derivatives [20••]. This observation is consistent with an important contribution of unfolded protein stress to lipid-mediated activation of IRE1 and PERK. Alternatively, as the lumenal domain stabilises the dimer [24,25], it may contribute to lipid-mediated activation of the UPR even in circumstances that are not associated with any further increase in unfolded protein stress. Cooperativity in dimerization suggests that perturbations in the ER lipid bilayer composition may lower the threshold for unfolded protein stress-mediated activation and that direct lipid-dependent regulation and conventional lumenal unfolded protein stress mediated activation of the UPR transducers are likely to modulate each other.
UPR modulation of lipid metabolism
IRE1 and PERK also modulate lipid metabolism, placing the UPR transducers as both sensors of primary lipid perturbations and regulators of lipid homeostasis. In yeast, the importance of IRE1 to lipid metabolism is stressed by the dependence of IRE1 mutant yeast on exogenous inositol for their survival [1,3]. In the absence of exogenous inositol, yeast IRE1 is required for the expression of INO1 encoding inositol-3-phosphate synthase, an enzyme catalysing a rate-limiting step in the synthesis of phosphatidylinositol [3,26]. Yeast genes controlling the expression of key enzymes in lipid metabolism are upregulated following induction of the UPR [4]. The role of IRE1 in regulating phospholipid synthesis is conserved in mammals, where activated splicing of its downstream effector XBP1 has been shown to contribute to ER membrane expansion through the stimulation of the expression of genes involved in phospholipid synthesis [27•]. In addition, IRE1 and PERK signalling have been shown to regulate lipid metabolism in vivo [28,29].
Remarkably, lipid perturbations in yeast triggered predominantly compensatory changes affecting protein quality control, contrasting with minimal adjustments to lipid metabolism [30••]. Restoration of protein quality control was dependent on IRE1, while expression of lipid metabolism genes previously identified as IRE1 targets remained largely unchanged. In this study, lipid disequilibrium was triggered by genetic deletion of CHO2 or OPI3, two enzymes catalysing respectively the initial and late steps of phosphatidylcholine synthesis from phosphatidylethanolamine. It should be noted that OPI3 expression is upregulated following IRE1 activation [4,31], raising the possibility that IRE1-dependent compensatory changes in lipid composition might have been blocked by the mutation in OPI3.
Following perturbation of the ER membrane lipid composition, compensatory changes to lipid homeostasis and protein homeostasis could be equally important to alleviate cellular stress. Indeed, as discussed earlier, lipid disequilibrium within the ER membrane is likely to affect protein folding within the ER lumen, for example by perturbing ER calcium homeostasis [14]. Lipid perturbations could also cause proteotoxicity by affecting protein folding within the membrane, protein translocation or trafficking, or by causing membrane protein aggregation. Activation of IRE1α and PERK by changes in ER membrane lipid composition may be a mechanism allowing the cell to adapt the flow of protein entering the ER, as well as the folding apparatus in response to changes in the lipid composition that might otherwise promote unfolded protein accumulation.
Physiological significance of lipid activation of the UPR
Activation of the UPR and perturbations in the ER lipid composition are observed in morbid obesity [15,32]. Moreover, UPR activation has been linked to the development of insulin resistance or beta-cell death in morbid obesity [9,14,33]. Altogether these observations suggest that lipid-dependent activation of the UPR transducers could contribute to the pathogenesis of morbid obesity.
Flaviviral non-structural proteins are ER membrane associated proteins triggering membrane rearrangements [34]. Though they lack substantial ER lumenal domains, they have been shown to activate the UPR, suggesting that lipid-dependent activation of the UPR could operate in Flavivirus infected cells [35–37]. Lipid-dependent activation of the UPR transducers could also occur during cellular processes marked by a discrepancy between the level of UPR activation measured and the level of unfolded proteins detected in the ER, such as B lymphocyte development [38–40]. In line with this hypothesis, modifications of the ER lipid membrane could also initiate what has been called anticipatory ER stress [41,42] in which the UPR is triggered independently of unfolded proteins, thereby allowing the cells to adapt their ER folding capacity in anticipation of unfolded protein stress.
Gaging the physiological or pathological significance of lipid-mediated activation of the UPR transducers represents a major challenge for the future. Currently, direct lipid-dependent activation is isolated from any affects of unfolded protein stress by the expression of mutant UPR transducers lacking their lumenal domain. Unfortunately this technique cannot be readily applied to study physiological circumstances, such as those listed above.
Conclusions and perspective
The proposed tuning of UPR signalling by lipids, mediated by simple biophysical principles, could represent an addition strand in the lipid-UPR dialectic; the physiological significance of which remains to be explored. Major lipid perturbations are found alongside UPR activation in important pathophysiological circumstances such as viral infection and severe obesity, and these would be good candidates for testing the biological role of the lipid-UPR dialectic.
The highly cooperative nature of IRE1 (and likely PERK) activation is poised to respond to subtle variation in the factors that alter the tendency of TM proteins to dimerize. Acting alone, each of these simple biophysical principles would probably have only weak effects on protein dimerization. This may account for the considerable redundancy in the sequence requirements for a functional IRE1 TM domain [20••]. However, we propose that in association with dimerization-competent lumenal and cytosolic domains that respond to other cues, these weak forces acting on the TM domains might allow lipids to tune UPR signalling.
Changes in membrane properties are likely to influence other TM proteins by such generic mechanisms. The identification of such proteins and a better understanding of the biophysical parameters of the membrane that govern the modulation of their function represent an interesting challenge for cell biology.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
Acknowledgements
This work was supported by a Wellcome Trust Intermediate Clinical Fellowship (to R.V.), a Wellcome Trust Principal Research Fellowship (to D.R.), and European Union Seventh Framework Programme Grant (Beta Bat, 277713).
Contributor Information
Romain Volmer, Email: r.volmer@envt.fr.
David Ron, Email: dr360@medschl.cam.ac.uk.
References
- 1.Nikawa J., Yamashita S. IRE1 encodes a putative protein kinase containing a membrane-spanning domain and is required for inositol phototrophy in Saccharomyces cerevisiae. Mol Microbiol. 1992;6:1441–1446. doi: 10.1111/j.1365-2958.1992.tb00864.x. [DOI] [PubMed] [Google Scholar]
- 2.Nikawa J., Akiyoshi M., Hirata S., Fukuda T. Saccharomyces cerevisiae IRE2/HAC1 is involved in IRE1-mediated KAR2 expression. Nucleic Acids Res. 1996;24:4222–4226. doi: 10.1093/nar/24.21.4222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cox J.S., Chapman R.E., Walter P. The unfolded protein response coordinates the production of endoplasmic reticulum protein and endoplasmic reticulum membrane. Mol Biol Cell. 1997;8:1805–1814. doi: 10.1091/mbc.8.9.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Travers K.J., Patil C.K., Wodicka L., Lockhart D.J., Weissman J.S., Walter P. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell. 2000;101:249–258. doi: 10.1016/s0092-8674(00)80835-1. [DOI] [PubMed] [Google Scholar]
- 5••.Jonikas M.C., Collins S.R., Denic V., Oh E., Quan E.M., Schmid V., Weibezahn J., Schwappach B., Walter P., Weissman J.S. Comprehensive characterization of genes required for protein folding in the endoplasmic reticulum. Science. 2009;323:1693-L1697. doi: 10.1126/science.1167983. [DOI] [PMC free article] [PubMed] [Google Scholar]; Using a comprehensive yeast genetic screen, this paper uncovers the diversity of gene deletions leading to upregulation of the UPR. Remarkably, a number of genes regulating lipid metabolism are identified as strong activatiors of the UPR.
- 6•.Pineau L., Colas J., Dupont S., Beney L., Fleurat-Lessard P., Berjeaud J.M., Berges T., Ferreira T. Lipid-induced ER stress: synergistic effects of sterols and saturated fatty acids. Traffic. 2009;10:673–690. doi: 10.1111/j.1600-0854.2009.00903.x. [DOI] [PubMed] [Google Scholar]; This paper provides evidence that UPR activation following both saturated fatty acid accumulation and ergosterol overload in yeast can be reverted by exogenous treatment with unsaturated fatty acids, suggesting that UPR activation is due to altered membrane fluidity.
- 7.Han S., Lone M.A., Schneiter R. Chang A: Orm1 and Orm2 are conserved endoplasmic reticulum membrane proteins regulating lipid homeostasis and protein quality control. Proc Natl Acad Sci U S A. 2010;107:5851–5856. doi: 10.1073/pnas.0911617107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feng B., Yao P.M., Li Y., Devlin C.M., Zhang D., Harding H.P., Sweeney M., Rong J.X., Kuriakose G., Fisher E.A. The endoplasmic reticulum as the site of cholesterol-induced cytotoxicity in macrophages. Nat Cell Biol. 2003;5:781–792. doi: 10.1038/ncb1035. [DOI] [PubMed] [Google Scholar]
- 9.Cunha D.A., Hekerman P., Ladriere L., Bazarra-Castro A., Ortis F., Wakeham M.C., Moore F., Rasschaert J., Cardozo A.K., Bellomo E. Initiation and execution of lipotoxic ER stress in pancreatic beta-cells. J Cell Sci. 2008;121:2308–2318. doi: 10.1242/jcs.026062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ariyama H., Kono N., Matsuda S., Inoue T., Arai H. Decrease in membrane phospholipid unsaturation induces unfolded protein response. J Biol Chem. 2010;285:22027–22035. doi: 10.1074/jbc.M110.126870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Minville-Walz M., Pierre A.S., Pichon L., Bellenger S., Fevre C., Bellenger J., Tessier C., Narce M., Rialland M. Inhibition of stearoyl-CoA desaturase 1 expression induces CHOP-dependent cell death in human cancer cells. PLoS ONE. 2010;5:e14363. doi: 10.1371/journal.pone.0014363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sorli S.C., Colie S., Albinet V., Dubrac A., Touriol C., Guilbaud N., Bedia C., Fabrias G., Casas J., Segui B. The nonlysosomal beta-glucosidase GBA2 promotes endoplasmic reticulum stress and impairs tumorigenicity of human melanoma cells. FASEB J. 2013;27:489–498. doi: 10.1096/fj.12-215152. [DOI] [PubMed] [Google Scholar]
- 13.Spassieva S.D., Mullen T.D., Townsend D.M., Obeid L.M. Disruption of ceramide synthesis by CerS2 down-regulation leads to autophagy and the unfolded protein response. Biochem J. 2009;424:273–283. doi: 10.1042/BJ20090699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fu S., Yang L., Li P., Hofmann O., Dicker L., Hide W., Lin X., Watkins S.M., Ivanov A.R., Hotamisligil G.S. Aberrant lipid metabolism disrupts calcium homeostasis causing liver endoplasmic reticulum stress in obesity. Nature. 2011;473:528–531. doi: 10.1038/nature09968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gregor M.F., Yang L., Fabbrini E., Mohammed B.S., Eagon J.C., Hotamisligil G.S., Klein S. Endoplasmic reticulum stress is reduced in tissues of obese subjects after weight loss. Diabetes. 2009;58:693–700. doi: 10.2337/db08-1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Y., Ge M., Ciani L., Kuriakose G., Westover E.J., Dura M., Covey D.F., Freed J.H., Maxfield F.R., Lytton J. Enrichment of endoplasmic reticulum with cholesterol inhibits sarcoplasmic–endoplasmic reticulum calcium ATPase-2b activity in parallel with increased order of membrane lipids: implications for depletion of endoplasmic reticulum calcium stores and apoptosis in cholesterol-loaded macrophages. J Biol Chem. 2004;279:37030–37039. doi: 10.1074/jbc.M405195200. [DOI] [PubMed] [Google Scholar]
- 17.Lajoie P., Moir R.D., Willis I.M., Snapp E.L. Kar2p availability defines distinct forms of endoplasmic reticulum stress in living cells. Mol Biol Cell. 2012;23:955–964. doi: 10.1091/mbc.E11-12-0995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18•.Hou N.S., Gutschmidt A., Choi D.Y., Pather K., Shi X., Watts J.L., Hoppe T., Taubert S. Activation of the endoplasmic reticulum unfolded protein response by lipid disequilibrium without disturbed proteostasis in vivo. Proc Natl Acad Sci U S A. 2014 doi: 10.1073/pnas.1318262111. [DOI] [PMC free article] [PubMed] [Google Scholar]; Using different reporters of misfolded protein accumulation, papers 17 and 18 provide evidence that activation of the UPR following perturbations of lipid metabolism does not correlate with significant ER unfolded protein stress.
- 19••.Promlek T., Ishiwata-Kimata Y., Shido M., Sakuramoto M., Kohno K., Kimata Y. Membrane aberrancy and unfolded proteins activate the endoplasmic reticulum stress sensor Ire1 in different ways. Mol Biol Cell. 2011;22:3520–3532. doi: 10.1091/mbc.E11-04-0295. [DOI] [PMC free article] [PubMed] [Google Scholar]; These two papers [19••,20••] show that the UPR transducers IRE1 and PERK can be activated by lipid preturbations independently of their ability to sense unfolded protein stress.
- 20••.Volmer R., van der Ploeg K., Ron D. Membrane lipid saturation activates endoplasmic reticulum unfolded protein response transducers through their transmembrane domains. Proc Natl Acad Sci U S A. 2013;110:4628–4633. doi: 10.1073/pnas.1217611110. [DOI] [PMC free article] [PubMed] [Google Scholar]; These two papers [19••,20••] show that the UPR transducers IRE1 and PERK can be activated by lipid preturbations independently of their ability to sense unfolded protein stress.
- 21.Walter P., Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 2011;334:1081–1086. doi: 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
- 22.van Meer G., Voelker D.R., Feigenson G.W. Membrane lipids: where they are and how they behave. Nat Rev Mol Cell Biol. 2008;9:112–124. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cymer F., Veerappan A., Schneider D. Transmembrane helix–helix interactions are modulated by the sequence context and by lipid bilayer properties. Biochim Biophys Acta. 2012;1818:963–973. doi: 10.1016/j.bbamem.2011.07.035. [DOI] [PubMed] [Google Scholar]
- 24.Credle J.J., Finer-Moore J.S., Papa F.R., Stroud R.M., Walter P. On the mechanism of sensing unfolded protein in the endoplasmic reticulum. Proc Natl Acad Sci U S A. 2005;102:18773–18784. doi: 10.1073/pnas.0509487102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou J., Liu C.Y., Back A.H., Clark R.L., Peisach D., Xu Z., Kaufman R.J. The crystal structure of human IRE1 luminal domain reveals a conserved dimerization interface required for activation of the unfolded protein response. Proc Natl Acad Sci U S A. 2006;103:14343–14348. doi: 10.1073/pnas.0606480103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chang H.J., Jones E.W., Henry S.A. Role of the unfolded protein response pathway in regulation of INO1 and in the sec14 bypass mechanism in Saccharomyces cerevisiae. Genetics. 2002;162:29–43. doi: 10.1093/genetics/162.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27•.Sriburi R., Jackowski S., Mori K., Brewer J.W. XBP1: a link between the unfolded protein response, lipid biosynthesis, and biogenesis of the endoplasmic reticulum. J Cell Biol. 2004;167:35–41. doi: 10.1083/jcb.200406136. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper identifies the role of XBP1 in the regulation of phospholipid synthesis, indicating that the UPR transducer IRE1 regulates the size of the ER membrane in concert with the enzymatic equipment required for protein folding.
- 28.Lee A.H., Scapa E.F., Cohen D.E., Glimcher L.H. Regulation of hepatic lipogenesis by the transcription factor XBP1. Science. 2008;320:1492–1496. doi: 10.1126/science.1158042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oyadomari S., Harding H.P., Zhang Y., Oyadomari M., Ron D. Dephosphorylation of translation initiation factor 2 alpha enhances glucose tolerance and attenuates hepatosteatosis in mice. Cell Metab. 2008;7:520–532. doi: 10.1016/j.cmet.2008.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30••.Thibault G., Shui G., Kim W., McAlister G.C., Ismail N., Gygi S.P., Wenk M.R., Ng D.T. The membrane stress response buffers lethal effects of lipid disequilibrium by reprogramming the protein homeostasis network. Mol Cell. 2012;48:16–27. doi: 10.1016/j.molcel.2012.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]; Following lipid perturbations in yeast, the authors find that unexpectedly IRE1 activation triggers predominantly compensatory changes to the proteome, with only minimal changes to the lipidome.
- 31.Schuck S., Prinz W.A., Thorn K.S., Voss C., Walter P. Membrane expansion alleviates endoplasmic reticulum stress independently of the unfolded protein response. J Cell Biol. 2009;187:525–536. doi: 10.1083/jcb.200907074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pietilainen K.H., Rog T., Seppanen-Laakso T., Virtue S., Gopalacharyulu P., Tang J., Rodriguez-Cuenca S., Maciejewski A., Naukkarinen J., Ruskeepaa A.L. Association of lipidome remodeling in the adipocyte membrane with acquired obesity in humans. PLoS Biol. 2011;9:e1000623. doi: 10.1371/journal.pbio.1000623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ozcan U., Yilmaz E., Ozcan L., Furuhashi M., Vaillancourt E., Smith R.O., Gorgun C.Z., Hotamisligil G.S. Chemical chaperones reduce ER stress and restore glucose homeostasis in a mouse model of type 2 diabetes. Science. 2006;313:1137–1140. doi: 10.1126/science.1128294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Egger D., Wolk B., Gosert R., Bianchi L., Blum H.E., Moradpour D., Bienz K. Expression of hepatitis C virus proteins induces distinct membrane alterations including a candidate viral replication complex. J Virol. 2002;76:5974–5984. doi: 10.1128/JVI.76.12.5974-5984.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li S., Ye L., Yu X., Xu B., Li K., Zhu X., Liu H., Wu X., Kong L. Hepatitis C virus NS4B induces unfolded protein response and endoplasmic reticulum overload response-dependent NF-kappaB activation. Virology. 2009;391:257–264. doi: 10.1016/j.virol.2009.06.039. [DOI] [PubMed] [Google Scholar]
- 36.Yu C.Y., Hsu Y.W., Liao C.L., Lin Y.L. Flavivirus infection activates the XBP1 pathway of the unfolded protein response to cope with endoplasmic reticulum stress. J Virol. 2006;80:11868–11880. doi: 10.1128/JVI.00879-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ambrose R.L., Mackenzie J.M. West Nile virus differentially modulates the unfolded protein response to facilitate replication and immune evasion. J Virol. 2011;85:2723–2732. doi: 10.1128/JVI.02050-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gass J.N., Gifford N.M., Brewer J.W. Activation of an unfolded protein response during differentiation of antibody-secreting B cells. J Biol Chem. 2002;277:49047–49054. doi: 10.1074/jbc.M205011200. [DOI] [PubMed] [Google Scholar]
- 39.Hu C.C., Dougan S.K., McGehee A.M., Love J.C., Ploegh H.L. XBP-1 regulates signal transduction, transcription factors and bone marrow colonization in B cells. EMBO J. 2009;28:1624–1636. doi: 10.1038/emboj.2009.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Anken E., Romijn E.P., Maggioni C., Mezghrani A., Sitia R., Braakman I., Heck A.J. Sequential waves of functionally related proteins are expressed when B cells prepare for antibody secretion. Immunity. 2003;18:243–253. doi: 10.1016/s1074-7613(03)00024-4. [DOI] [PubMed] [Google Scholar]
- 41.Vitale A., Boston R.S. Endoplasmic reticulum quality control and the unfolded protein response: insights from plants. Traffic. 2008;9:1581–1588. doi: 10.1111/j.1600-0854.2008.00780.x. [DOI] [PubMed] [Google Scholar]
- 42.Rutkowski D.T., Hegde R.S. Regulation of basal cellular physiology by the homeostatic unfolded protein response. J Cell Biol. 2010;189:783–794. doi: 10.1083/jcb.201003138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43••.Sharpe H.J., Stevens T.J., Munro S. A comprehensive comparison of transmembrane domains reveals organelle-specific properties. Cell. 2010;142:158–169. doi: 10.1016/j.cell.2010.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper used large datasets of transmembrane protein sequences to demonstrate that the size of the transmembrane domain varies along the secretory pathway and correlates with the membrane thickness of the different organelles, suggesting that hydrophobic mismatch is under negative selection.
- 44.Lewis B.A., Engelman D.M. Lipid bilayer thickness varies linearly with acyl chain length in fluid phosphatidylcholine vesicles. J Mol Biol. 1983;166:211–217. doi: 10.1016/s0022-2836(83)80007-2. [DOI] [PubMed] [Google Scholar]
- 45.He L., Hristova K. Pathogenic activation of receptor tyrosine kinases in mammalian membranes. J Mol Biol. 2008;384:1130–1142. doi: 10.1016/j.jmb.2008.10.036. [DOI] [PubMed] [Google Scholar]
- 46.Weiner D.B., Liu J., Cohen J.A., Williams W.V., Greene M.I. A point mutation in the neu oncogene mimics ligand induction of receptor aggregation. Nature. 1989;339:230–231. doi: 10.1038/339230a0. [DOI] [PubMed] [Google Scholar]
- 47.Inda M.E., Vandenbranden M., Fernandez A., de Mendoza D., Ruysschaert J.M., Cybulski L.E. A lipid-mediated conformational switch modulates the thermosensing activity of DesK. Proc Natl Acad Sci U S A. 2014;111:3579–3584. doi: 10.1073/pnas.1317147111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Anbazhagan V., Schneider D. The membrane environment modulates self-association of the human GpA TM domain – implications for membrane protein folding and transmembrane signaling. Biochim Biophys Acta. 2010;1798:1899–1907. doi: 10.1016/j.bbamem.2010.06.027. [DOI] [PubMed] [Google Scholar]
- 49.Simons K., Sampaio J.L. Membrane organization and lipid rafts. Cold Spring Harb Perspect Biol. 2011;3:a004697. doi: 10.1101/cshperspect.a004697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nikolaus J., Scolari S., Bayraktarov E., Jungnick N., Engel S., Pia Plazzo A., Stockl M., Volkmer R., Veit M., Herrmann A. Hemagglutinin of influenza virus partitions into the nonraft domain of model membranes. Biophys J. 2010;99:489–498. doi: 10.1016/j.bpj.2010.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schafer L.V., de Jong D.H., Holt A., Rzepiela A.J., de Vries A.H., Poolman B., Killian J.A., Marrink S.J. Lipid packing drives the segregation of transmembrane helices into disordered lipid domains in model membranes. Proc Natl Acad Sci U S A. 2011;108:1343–1348. doi: 10.1073/pnas.1009362108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mall S., Broadbridge R., Sharma R.P., East J.M., Lee A.G. Self-association of model transmembrane alpha-helices is modulated by lipid structure. Biochemistry. 2001;40:12379–12386. doi: 10.1021/bi011075y. [DOI] [PubMed] [Google Scholar]
- 53.Muniz M., Riezman H. Intracellular transport of GPI-anchored proteins. EMBO J. 2000;19:10–15. doi: 10.1093/emboj/19.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sano R., Annunziata I., Patterson A., Moshiach S., Gomero E., Opferman J., Forte M., d‘Azzo A. GM1-ganglioside accumulation at the mitochondria-associated ER membranes links ER stress to Ca(2+)-dependent mitochondrial apoptosis. Mol Cell. 2009;36:500–511. doi: 10.1016/j.molcel.2009.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lynes E.M., Simmen T. Urban planning of the endoplasmic reticulum (ER): how diverse mechanisms segregate the many functions of the ER. Biochim Biophys Acta. 2011;1813:1893–1905. doi: 10.1016/j.bbamcr.2011.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li E., Wimley W.C., Hristova K. Transmembrane helix dimerization: beyond the search for sequence motifs. Biochim Biophys Acta. 2012;1818:183–193. doi: 10.1016/j.bbamem.2011.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]