Summary
Proteostasis is maintained by multiple cellular pathways, including protein synthesis, quality control and degradation. An imbalance of neuronal proteostasis, associated with protein misfolding and aggregation, leads to proteinopathies or neurodegeneration. While genetic variations and protein modifications contribute to aggregate formation, components of the proteostasis network dictate the fate of protein aggregates. Here we provide an overview of proteostasis pathways and their interplay (particularly autophagy) with the metabolism of disease-related proteins. We review recent studies on neuronal activity-mediated regulation of proteostasis and transcellular propagation of protein aggregates in the nervous system. Targeting proteostasis pathways therapeutically remains an attractive but challenging task.
Introduction
Newly synthesized proteins are scrutinized and triaged by protein quality control systems to ensure proper folding and a final conformation that fits their unique cellular functions. Considering that approximately 30% of newly synthesized proteins are misfolded (Princiotta et al., 2003), the protein quality control process must operate continuously to manage the influx of misfolded proteins. Quality control involves re-folding, clearance and recycling of misfolded proteins that otherwise pose a threat to cell survival. The cell indeed has a daunting task in maintaining the homeostasis of the entire intracellular pool of proteins (proteostasis).
In young and healthy cells, proteostasis is maintained by balancing the cellular pathways that are responsible for protein synthesis, folding, processing, assembly, trafficking, localization and degradation (Balch et al., 2008). The degradation system represents a particularly critical line of defense against misfolded proteins, which alleviates the risk of proteotoxicity. Many disease-associated proteins, however, are misfolded and aggregate-prone in nature, and refractory to protein quality control systems including degradation pathways. Such proteins often polymerize and form aggregates, which typically have poor solubility in aqueous or detergent solvents and have no physiological function per se. The aggregates can be further deposited into large inclusion bodies (IBs) or aggresomes (Kopito, 2000). The formation of IBs is in fact a cellular stress response that neutralizes – in some situations - the toxic intermediate species of aggregates by confining them within a highly compact structure. It is indicative, however, of an alteration or a breakdown of local (and perhaps ultimately global) proteostasis due to loss of the equilibrium between oligomerization and clearance; favoring the former precipitates aggregate formation.
The intrinsic ability of the cell to maintain proteostasis varies drastically among different cell types. Not all tissues or cell types expressing disease-associated proteins develop IBs or pathologies. Neurons are among the most vulnerable cell types in the face of disease-related aggregate formation. A number of neurodegenerative diseases, such as Alzheimer's disease (AD), Parkinson's disease (PD), Huntington's disease (HD) and Amyotrophic lateral sclerosis (ALS), are characterized by the aggregation and accumulation of misfolded proteins in insoluble inclusions in the affected neuronal populations, whereas many other tissues or cell types are spared, despite the wide distribution of disease-associated proteins.
The exact mechanism underlying the particular vulnerability of neurons is not well understood. The cellular networks and pathways that maintain proteostasis in response to intrinsic and environmental stressors are regulated differently in neurons and in other cell types. For instance, neuronal activity impacts the two major degradation systems, the ubiquitin-proteasome system (UPS) and the autophagy-lysosomal system. It is likely that the autophagy-lysosomal pathway in neurons is controlled by signaling pathways distinct from the nutrient starvation mechanism that regulates autophagy in other tissues. In addition, neuronal autophagy actively prevents the accumulation of ubiquitinated proteins and formation of inclusion bodies (Hara et al., 2006; Komatsu et al., 2006). Notably, mounting evidence demonstrates the selectivity of autophagy in the degradation of aggregate-prone proteins and injured organelles through autophagy receptors, which link the autophagic cargo to the autophagy machinery for degradation (Rogov et al., 2014; Stolz et al., 2014). We will review the progress in understanding the role of autophagy in the removal of protein aggregates (aggrephagy) and its implication in drug target development. Recent studies have revealed an alternative pathway for the elimination of the disease-related protein fibrils in the nervous system: they are secreted and internalized by neighboring cells. Our review will highlight the recent reports of the trans-cellular movement and seeding of protein aggregates, which serve as a potential mechanism for the propagation of pathologies in neurodegenerative diseases.
Protein misfolding/aggregation and the cellular response
Our current understanding of protein aggregates is based on studies of mutant proteins that are aggregate-prone and causal to neurodegenerative diseases, such as Alzheimer's disease (AD), Parkinson's disease (PD), Huntington's disease (HD), Amyotrophic lateral sclerosis (ALS) and prion disease, which are increasingly recognized as protein conformation diseases. Upon the completion of translation, disease-related proteins adopt an abnormal conformation. The misfolded proteins are assembled into β-sheets, a common structural feature shared by many disease-related proteins, and assembly of this structure serves as an initiation step in the formation of an amyloid fibril. For example, β-amyloid (Aβ) and α-synuclein proteins, which are associated with AD and PD, respectively, form amyloid fibrils in vitro consisting primarily of β-sheets (Benzinger et al., 1998; Der-Sarkissian et al., 2003). The formation of a β-sheet significantly enhances the proteins’ propensity to oligomerize and aggregate prior to the formation of fibril structures. In addition, β-sheets are key components of the fibrils deposited in aggregates or inclusion bodies (IBs) (Ross and Poirier, 2004). Amyloid fibrils or protein aggregates are largely associated with pathological processes; however, physiological amyloid fibrils have also been reported. For example, fibrils of the Pmel17 protein are important for the biogenesis of melanosomes, which are lysosome-related organelles responsible for color and photoprotection (Berson et al., 2003).
Multiple factors are known to affect the intrinsic ability of disease-related proteins to oligomerize and aggregate. Genetic mutations in disease-related genes, such as those encoding amyloid precursor protein (APP), α-synuclein, poly-glutamine expanded huntingtin (polyQ-Htt; in HD) and superoxide dismutase 1 (SOD1; in ALS), disrupt the natural folding of the proteins and enhance their tendency to aggregate. Posttranslational modifications, including phosphorylation, nitration, age-related oxidative modifications and ubiquitination, facilitate protein oligomerization. In addition, proteolytic fragments of APP, polyQ-Htt and α-synuclein are known to enhance the aggregation compared to their corresponding full length proteins (Ross and Poirier, 2004).
The formation of microscopically visible inclusion bodies is the result of continuous overproduction of aggregated proteins, which are collected and deposited into a tight structure, called an aggresome. The collection process requires dynein motors that transport cellular aggregates along the microtubule network to the microtubule organization center (MTOC), where the aggregates are packed into aggresomes (Kopito, 2000). The synthesis of aggresomes is a response to proteostatic stress, and is functionally analogous to a “triage” center, thus being neuroprotective. Longitudinal tracking of toxicity in neurons expressing polyQ-Htt protein revealed the improved survival of neurons producing inclusion bodies, whereas increased levels of diffuse mutant proteins correlated with toxicity (Arrasate et al., 2004). Therefore, it has become increasingly apparent that the oligomeric intermediates of protein aggregates rather than the inclusion bodies are the toxic species. The inclusion bodies instead serve to isolate the toxic form of aggregates, which otherwise would poison the intracellular environment. The IBs thus prevent cell or neuronal death. In addition, the formation of inclusion bodies may allow efficient clearance of protein aggregates via MTOC (Kopito, 2000).
The misfolding and aggregation of disease-associated proteins may trigger the activation of molecular chaperones and co-chaperones, which recognize and bind misfolded proteins, and can assist in refolding or disaggregating the oligomers (Morimoto, 2011). The critical players of this protective response are the heat shock response (HSR) and heat shock (HS) transcription factor HSF-1 that regulates the induction of HS chaperones, including the abundant HS cognate protein 70 (Hsc70). The chaperones may serve as the first line of defense to prevent the accumulation of aggregated proteins. In spite of the upregulation of HSF-1 in response to protein misfolding, disease protein aggregates often overwhelm proteostasis pathways by sequestering chaperones and causing a functional depletion of chaperones that are required for many cellular processes. The malicious competition for the limited cellular pools of chaperones by disease-related aggregates was proposed to result in complex pathologies of protein conformational diseases (Yu et al., 2014).
A recent study showed that 12-15% of nascent polypeptides are ubiquitinated in human cells via a mechanism termed co-translational ubiquitination (CTU). CTU mainly generates K48-linked poly-ubiquitin chains for subsequent proteasome-mediated degradation. Protein misfolding enhances CTU activity within active translation complexes (Wang et al., 2013). Therefore, CTU may represent one of the earliest quality control systems, marking misfolded nascent polypeptides, while they are being synthesized, for proteasomal degradation.
Moreover, the endoplasmic reticulum (ER) is an important organelle for the maintenance of proteostasis since it regulates protein synthesis, folding, modifications, processing and transport of membrane proteins. Accumulation of misfolded proteins at the ER lumen causes ER stress, which subsequently engages the adaptive stress response known as the unfolded protein response (UPR). The UPR increases ER-resident chaperone levels, suppresses protein translation and facilitates the degradation of unfolded proteins in order to ameliorate the accumulation of unfolded proteins. The initial UPR is an adaptive and protective response but, if unresolved, can lead to inhibition of neuronal function and even to neurodegeneration (Hetz, 2012). Postmortem analysis of AD, PD, HD and ALS brains indicated that the presence of UPR markers is spatiotemporally correlated with abnormal protein aggregation and onset of neuropathologies (Hetz and Mollereau, 2014). Studies of animal models for the above neurological diseases also provide evidence for the involvement of UPR; however, the activation of UPR can enhance or reduce neurodegeneration, depending on the disease context and the specific UPR signaling that is activated. Thus the role of UPR in neurodegeneration can be either beneficial or deleterious, likely determined by the cross-talk between the UPR signaling pathways and other neuroprotective responses, such as degradation systems and in particular the autophagy-lysosomal pathway (Hetz and Mollereau, 2014) (Figure 1).
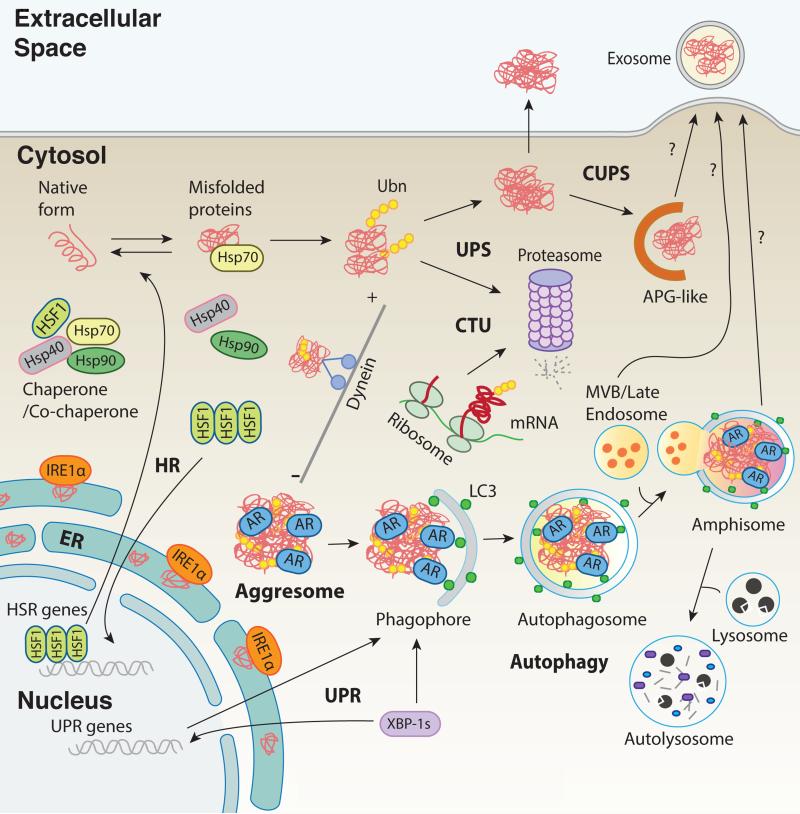
Figure 1. Proteostatic pathways regulate refolding, disaggregation, degradation and extracellular release of misfolded or disease related proteins.
Cells and neurons respond to the presence of misfolded proteins by activating heat shock transcription factor (HSF1), which regulates the levels and functions of molecular chaperones (including Hsp70) and co-chaperone systems such as the heat shock response (HR). Hsp70 recognizes and binds misfolded proteins, assisting in refolding or disaggregating oligomers. Accumulation of misfolded protein aggregates in the lumen of the endoplasmic reticulum (ER) also induces ER stress, which subsequently engages the adaptive stress response known as the unfolded protein response (UPR). For example, one of the three components of UPR, inositol-requiring enzyme 1 (IRE1), catalyzes the synthesis of transcription factor X-box binding protein 1 (XBP1), which in turn controls a subset of UPR genes related to protein folding, translocation, and degradation. The two degradation systems, the ubiquitin-proteasome system (UPS) and the autophagy-lysosomal system, are responsible for the degradation of diverse protein substrates. Nascent polypeptides can be ubiquitinated and degraded by the proteasome via a process known as co-translational ubiquitination (CTU). Protein aggregates can be further deposited into aggresomes via dynein-dependent retrograde transport along the microtubule network. Autophagy is the primary pathway for the clearance of protein aggregates or aggresomes. Protein aggregates, which are recognized by autophagy receptors (AR), are sequestered within autophagosomes, which are then fused with endosomes to form amphisomes, and delivered to lysosomes for degradation. However, the intracellular protein aggregates (or fibrils) can also be secreted outside of the cells or neurons via exosomes that are derived from multi-vesicular bodies (MVB). In addition, an unconventional secretory pathway (CUPS) associated with autophagosomes (APG) or amphisomes may provide an alternative path for extracellular release of protein fibrils (exophagy).
Degradation of misfolded proteins by the proteasome and autophagy
In addition to the chaperone system that helps refold or repair misfolded proteins, the cell may mobilize the degradation systems as the next line of defense to digest the compromised proteins in order to avoid the toxic accumulation. The ubiquitin-proteasome system (UPS) and the autophagy-lysosomal pathway are the two major protein degradation pathways and are critical components of the proteostasis network.
The UPS is responsible for the degradation of ubiquitinated, soluble and short-lived proteins. It requires three distinct enzymes, E1, E2 and E3 to covalently add ubiquitin (Ub) to a target protein (Goldberg, 2003). Activated ubiquitin forms a poly-ubiquitin chain on a target protein, serving as a signal for proteasome-mediated degradation. The UPS was shown to degrade several disease-related proteins, including α-synuclein, tau, SOD1, and polyQ-huntingtin (Dantuma and Bott, 2014). Inhibition of UPS activity results in the accumulation of poly-ubiquitinated proteins as well as disease-related proteins, causing their deposition into aggregates or inclusion bodies (See (Dantuma and Bott, 2014) for full references). Therefore, the UPS plays an important role in preventing protein aggregate formation. However, large protein complexes or aggregates, once formed, cannot penetrate the narrow proteasome barrels, and instead accelerate further the accumulation of protein aggregates via inhibition of proteasome activity (Verhoef et al., 2002). In contrast, autophagy is a lysosomal degradation pathway that targets long-lived proteins, large protein complexes, aggregates and cellular organelles. Since the autophagy machinery is physically compatible with large protein aggregates, autophagy is considered a major player in the clearance of disease–associated proteins.
Autophagy: the cardinal clearance pathway for aggregated proteins
Three types of autophagy have been described: macroautophagy, microautophagy and chaperone-mediated autophagy (CMA), which have distinct mechanisms for delivering substrates to lysosomes. Macroutophagy (hereafter autophagy) is the prevalent type of autophagy that has been extensively studied. It involves the synthesis, trafficking and fusion of autophagosomes with lysosomes. The formation of autophagosomes begins with an isolation membrane that emerges from the cellular organelle membrane network, followed by expansion of the isolation membrane and closing to form double-membrane vacuoles. As a result, part of the cytosol is engulfed inside the vesicle and subsequently degraded by lysosomes. The autophagy process is controlled by the coordination of a number of evolutionarily conserved autophagy-related (ATG) proteins (Xie and Klionsky, 2007), which can be categorized into four functional groups: the Atg1/unc-51-like kinase (ULK1) complex; the Vps34/class III phosphatidylinositol-3-phosphate (PI3P) kinase complex; mAtg9; Atg12 and Atg8/MAP1 light chain 3 (LC3) and their conjugation systems (Lamb et al., 2013).
One significant function of autophagy is to degrade protein aggregates. During early development of C. elegans, a number of maternally derived proteins exist in aggregates as components of P granules (specialized complexes of protein and RNA); these aggregates are selectively degraded with high efficiency by basal autophagy, which may provide a source of nutrients required for embryogenesis (Yang and Zhang, 2014). While it is unclear whether these aggregates would become harmful if not removed, autophagy is known to clear protein aggregates that are commonly associated with neurodegenerative diseases. Previous analyses of mouse models in which an autophagy gene was genetically deleted suggest that basal autophagy is required to fend off ubiquitinated protein aggregates in the nervous system (Hara et al., 2006; Komatsu et al., 2006). Existing evidence also suggests that autophagy compensates for impaired UPS function. For example, autophagy is up-regulated in response to UPS deficiency (Kageyama et al., 2014). Blocking UPS function induces phosphorylations of the autophagy receptor p62/SQSTM1, which promotes degradation of ubiquitinated cargos via selective autophagy (Matsumoto et al., 2011) (Lim et al., PLoS Genetics, in press).
Autophagy offers neuroprotection by clearing away disease-related proteins or their aggregates. Administration of chemicals that increase autophagy, such as rapamycin, trehalose and latrepirdine (dimebone), results in enhanced clearance of a variety of aggregate-prone disease proteins, including Aβ, tau, huntingtin and α-synuclein (Caccamo et al., 2010; Ravikumar et al., 2004; Sarkar et al., 2007; Steele et al., 2013). Furthermore, over-expression of beclin 1, an essential component of the VPS34 lipid kinase complex that is required for autophagy initiation, promotes clearance of disease-related proteins in PD, HD and AD animal models (Lonskaya et al., 2013; Nascimento-Ferreira et al., 2011; Shibata et al., 2006; Spencer et al., 2009). Recent studies show that over-expression of transcription factor EB (TFEB), a master regulator of lysosomal genes, promotes degradation of disease proteins, such as α-synuclein and polyQ-Htt, by enhancing autophagy-lysosomal activity (Decressac et al., 2013; Sardiello et al., 2009). These studies have provided compelling evidence that enhancing autophagic activity is an effective cellular defense mechanism against accumulation of disease-associated proteins and its toxicity.
Aggrephagy: selective autophagy for degradation of protein aggregates
Morphological analysis showed indiscriminate sequestration of a portion of the cytosol during autophagy induction, suggesting a non-selective process. However, recent studies demonstrate that autophagy degrades its cargos in a selective manner. Aggrephagy, a term originally coined for selective degradation of protein aggregates by autophagy (Overbye et al., 2007), has now emerged as an important mechanism for aggregate clearance that is relevant to the understanding of proteinopathies. The selectivity towards specific cargos is mediated largely by a class of proteins known as autophagy receptors, which recognize cargos tagged with degradation signals. The autophagy receptors connect the cargos and the autophagosome apparatus, which also functions in non-selective autophagy induced by starvation (Stolz et al., 2014).
The role of autophagy receptors in aggrephagy
Although more than twenty autophagy receptors have been identified, only a few are characterized, including p62/Sequestosome 1 (SQSTM1), NBR1, Optineurin (OPTN), NIX, Autophagy linked FYVE protein (ALFY/WDFY3) and NDP52 (Stolz et al., 2014). These receptors typically contain an LC3-interaction region or recognition sequence (LIR or LRS) and a ubiquitin association (UBA) domain. The autophagy receptors directly interact with ubiquitinated cargos via UBA and LC3 (the mammalian homolog of Atg8) through LIR, leading to the engulfment and subsequent degradation of ubiquitinated cargos. Similar to the “tagging” of substrates during the UPS, ubiquitination is used by autophagy as a degradation signal. It was proposed that Lys11-, 29- or 48-Ub is mainly targeted to the UPS and Lys63-Ub to autophagy (Kirkin et al., 2009). Interestingly, p62 has a preference for K63-Ub over K48-Ub (Seibenhener et al., 2004). However, results from another study suggest that substrate oligomerization rather than specific Ub-chains may direct cargos towards autophagic degradation (Riley et al., 2010). Indeed, autophagy receptors can oligomerize and form aggregates with cargoes, which may contribute to efficient clearance through autophagy. The above receptors may work in a cooperative manner in the process of selective autophagy, but they may have redundant roles in selecting specific cargos (Stolz et al., 2014). Furthermore, autophagy receptors need to be tightly regulated through degradation or post-translational modifications. More recently, two new classes of autophagy receptors have been reported, including ubiquitin-binding CUE domain targeting adaptors (CUET) (Lu et al., 2014) and TRIMs, which are a large family of pattern recognition receptors (PRRs) (Mandell et al., 2014). Specific members of these two classes mediate selective autophagy by binding autophagy core components and cargos. Identification of CUET and TRIMs as autophagy receptors has expanded our understanding of the selective autophagy process. It is likely that each receptor recognizes distinct sets of cargos, which accounts for their specificities; however, the functional relationship among different receptors and the exact mechanism by which they detect and degrade cargos through selective autophagy remains to be elucidated.
p62/SQSTM1
p62 is the prototypical autophagy receptor with a role in the clearance of protein aggregates (Komatsu et al., 2007a; Pankiv et al., 2007), damaged mitochondria (Geisler et al., 2010), peroxisomes (Kim et al., 2008) and invading microbes (Zheng et al., 2009) through selective autophagy. Notably, p62 possesses several distinct domains that enable it to function as an autophagy receptor: a UBA domain for ubiquitin binding (Seibenhener et al., 2004), a PB1 domain for oligomerization and aggregation with cargos (Bjorkoy et al., 2005) and an LIR sequence for LC3 binding (Pankiv et al., 2007). In addition, p62 has nuclear localization signals (NLSs) and nuclear export signals (NESs) that regulate its nucleocytoplasmic shuttling. In the nucleus, p62 may direct misfolded nuclear proteins to proteasomes for degradation (Pankiv et al., 2010). Interestingly, p62 has a rather low ubiquitin binding affinity, requiring a conformational switch in the UBA domain from dimer to monomer of p62's UBA domain (Long et al., 2008) (Lim, et al. PLoS Genetics, in press). One study previously found that casein kinase-2 (CK2)-mediated phosphorylation of the p62 UBA domain at S403 enhances the affinity of p62 for ubiquitin (Matsumoto et al., 2011). Our data shows that autophagy kinase ULK1 phosphorylates both S403 and S407 (equivalent to S409 in murine p62) in the UBA domain, and that phosphorylation of S407 critically regulates S403 phosphorylation, destabilizes the UBA dimer interface, and increases the affinity of p62 for ubiquitin. Interestingly, inhibition of the proteasome or expression of polyQ-Htt induces p62 phosphorylation at both S403 and S407; however, nutrient starvation fails to trigger modification of S407. Therefore, ULK1-mediated phosphorylation of the UBA domain of p62 may regulate selective autophagy of poly-ubiquitinated proteins or aggregated disease proteins through a non-canonical autophagy signaling pathway (Figure 2) (Lim, et al. PLoS Genetics, in press). Recently, genetic p62 mutations were linked to familial and sporadic ALS (Fecto et al., 2011); it remains to be shown whether the disease-linked mutations alter p62 function in selective autophagy.
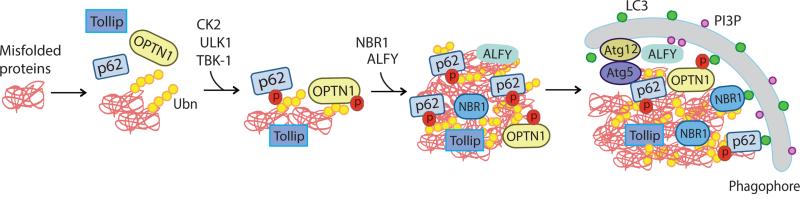
Figure 2. Regulation of aggrephagy: selective autophagy degrades protein aggregates via autophagy receptors.
Autophagy receptors, such as p62/SQSTM1, NBR1, OPTN1, ALFY and Tollip, mediate degradation of protein aggregates via selective autophagy by coupling autophagic cargos (ubiquitinated protein aggregates) and autophagy protein LC3. However, the binding affinity of certain autophagy receptors to either cargos (e.g. p62) or LC3 (e.g. OPTN1) is weak under normal condition. Phosphorylation of p62 by CK2, TBK1 and/or ULK1 enhances the affinity of p62 for ubiquitinated proteins (cargos), while phosphorylation of OPTN1 by TBK1 increases the binding of OPTN1 to autophagy modifier LC3. These modifications of p62 promote the selective degradation of protein aggregates by recruiting the autophagy machinery to the proximity of the aggregates.
ALFY (WDFY3)
Autophagy receptor ALFY is a highly conserved member of the Beige and Chediak-Higashi syndrome (BEACH) domain protein family. The C-terminal region of ALFY contains a FYVE domain, WD40 repeats and a Pleckstrin homology (PH)- BEACH domain, which mediate binding to PI3P, Atg5 and p62, respectively (Isakson et al., 2013). ALFY colocalizes with ubiquitin and p62-positive aggregates and is required for the clearance of polyQ-Htt aggregates. ALFY stabilizes the interaction between LC3 and p62 and also recruits Atg5 to enable the formation of autophagosomes adjacent to protein aggregates, leading to the clearance of polyQ-Htt (Filimonenko et al., 2010).
OPTN
Autophagy receptor OPTN binds ubiquitinated proteins through its UBA and NF-κB essential modulator (NEMO) domains. Genetic mutations of OPTN are associated with ALS, Paget disease of the bone and glaucoma (Kachaner et al., 2012). OPTN also binds LC3 via the OPTN LIR domain, thus regulating the degradation of protein aggregates (Kachaner et al., 2012). As with p62, phosphorylation of OPTN enhances its binding affinity for LC3 and degradation of selective autophagy cargos (Wild et al., 2011).
Cue5/Tollip
Cue5 is a newly identified yeast autophagy receptor that binds Atg8 and ubiquitin. The mammalian homologue of Cue5 is Tollip, which binds ubiquitin through a CUE domain and binds Atg8/LC3 through the AIM1 and AIM2 motifs, separately. The functions of Cue5 and Tollip as autophagy receptors are highly conserved: both play a critical role in autophagic clearance of cytotoxic protein aggregates such as polyQ-Htt. Under normal conditions, Tollip binds poly-ubiquitin chains with a higher affinity compared to p62 and may function cooperatively rather than sequentially with p62 in selective autophagy (Lu et al., 2014).
TRIM5α
Nearly half of the TRIM family members regulate autophagy. TRIM5α is one of the TRIM members that also functions as a selective autophagy receptor. TRIM5α interacts with several autophagy-related proteins, such as LC3, p62, ULK1 and Beclin 1, but not with ubiquitin. Instead, TRIM5α recognizes and delivers specific viral capsid proteins for selective autophagy (Mandell et al., 2014). The role of TRIM5α in the clearance of disease-related protein aggregates has yet to be shown.
Regulation of proteostasis by neural activity and misfolded proteins
Neurons are highly polarized cell types characterized by the production of electrical signals and the presence of synapses, which are the basis for transmission of the signals from one cell to another. Maintenance of proteostasis in neurons is affected by neuronal signaling and synaptic activity. Mounting evidence suggests that neurotransmitters, signaling molecules and neurotransmitter receptors all play regulatory roles in neuronal proteostasis. N-methyl-d-aspartate (NMDA) treatment was shown to cause accumulation of autophagosomes, providing early evidence for the regulation of autophagy by neural activity (Borsello et al., 2003). Another study reported that NMDA-induced long-term depression (LTD) enhances autophagy in spines and dendrites, along with increased degradation of GluR1, an α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptor subunit (Otabe et al., 2014; Shehata et al., 2012). In addition, neuronal activity affects levels of ubiquitinated proteins in the postsynaptic density (Ehlers, 2003), as well as the subcellular localization and biochemical composition of proteasomes (Bingol and Schuman, 2006; Tai et al., 2010). Thus, neuronal activity alters the cellular degradation system and therefore likely affects global neuronal proteostasis and the turnover of disease-related proteins.
Misfolding of disease proteins and the formation of protein aggregates or inclusion bodies may lead to the enhanced vulnerability of neurons. The mechanism underlying the increased toxicity is likely related to the failure of the neuronal proteostasis network to handle the accumulating misfolded proteins. Why are neuronal proteostasis systems particularly vulnerable to the presence of the misfolded or aggregated proteins? As postmitotic cells, neurons cannot dissipate protein aggregates simply by cell division, and the age-related decline of the degradation systems further contributes to the vulnerability of neurons. These unique features of neurons may also contribute to their reduced ability to prevent the accumulation of protein aggregates. As mentioned earlier, specific neuronal signaling pathways and synaptic activity regulate proteostatic capacity. For example, the heat shock response and autophagy elements of proteostasis are differentially regulated in neurons (Skibinski and Finkbeiner, 2013). Proteostatic capacity is also likely to vary among different neuronal cell types due to differential expression levels of proteostasis components. Furthermore, different neuronal populations have different vulnerabilities to reduced proteostasis. For instance, Purkinje cells and midbrain dopaminergic (DA) neurons show a drastically different response to genetic inactivation of autophagy in mice. Mutant Purkinje cells exhibit severe degeneration by postnatal week eight, with no sign of ubiquitinated protein aggregates. In contrast, mutant DA neurons exhibit protracted and moderate death at 9 months but with extensive ubiquitinated inclusion bodies in the soma and dendrites (Friedman et al., 2012; Komatsu et al., 2007b). The presence of large inclusions leads to marked expansion of dendrite shafts and dendritic atrophy. Interestingly, the inclusions are enriched with autophagy receptor p62. While the results may suggest a more critical role of autophagy in the survival of Purkinje cells compared to DA neurons, they also show the importance of p62-mediated autophagy in preventing protein aggregate formation in DA neurons. It is likely that autophagy - via different autophagy receptors - targets diverse substrates in different types of neurons under physiological conditions. Moreover, the function of autophagy in specific subcompartments of neurons may also vary. In Purkinje cells, loss of autophagy seems to have little effect in dendrites but causes remarkable axonal swelling and degeneration. In contrast, DA neurons lacking autophagy exhibit severe dendritic atrophy.
Thus the unique polarized structures of neurons, i.e., the axons and dendrites, may additionally attribute to the particular vulnerability to impaired proteostasis that is associated with protein aggregates and damaged organelles. Indeed, the proteostasis pathway in neurons is differentially regulated in each subcompartment. Notably, autophagosome initiation is a constitutive and spatially restricted process in the distal ends of cultured dorsal root ganglion (DRG) neurons (Maday et al., 2012; Yue et al., 2008). Following synthesis, autophagosomes mature while undergoing retrograde transport to the soma along the axons in a dynein/dynactin-dependent manner (Wong and Holzbaur, 2014). Given the important role of axonal transport in supporting various neuronal activities, alteration in proteostasis and subsequent accumulation of protein aggregates or cargos can cause a “traffic jam” in the axon, leading to axonal dystrophy and neurodegeneration.
Cell non-autonomous regulation of protein aggregates in the nervous system
Early studies on proteostasis pathways focused mostly on cell-autonomous events. New work now shows that cell-cell communication also regulates proteostasis systems. It is known that chaperones and heat shock proteins can be activated in response to protein misfolding upon exposure to stressful environmental conditions. However, up-regulation of chaperones and activation of HSF-1 are rare in animal models and human tissues producing disease-related protein aggregates (Muchowski, 2002; Prahlad and Morimoto, 2011; Zourlidou et al., 2007). The lack of consistent HSR in the face of disease-related protein aggregates is perplexing, raising the question of the underlying mechanisms.
A number of studies in the nervous system of metazoan C. elegans provide evidence that the regulation of the quality control chaperone system involves cell-cell communication. One study showed that worm neurons exert an inhibitory and cell non-autonomous control over the organismal response to protein misfolding (Garcia et al., 2007). For example, the AFD sensory neurons negatively regulate the HSR of somatic cells ectopically expressing the polyQ expansion protein or the SOD-1 G93A mutant. On the other hand, the inhibitory effect of AFD can be relieved by down-regulating thermosensory neuron activity or by inhibiting calcium-mediated dense core vesicle neurosecretion from AFD. Thus, it is likely that thermosensory neurons function as a homeostatic switch for the control of chaperone expression in worms, serving to maintain a basal level of chaperones within the organism for normal function, yet responding to acute stressful conditions by increasing chaperone levels (Prahlad and Morimoto, 2011). The neuronal control of proteostasis and chaperone activity in a non-autonomous manner may provide a partial explanation for the absence of consistent HSR in degenerating neurons that produce disease protein aggregates associated with AD, HD and PD (Gidalevitz et al., 2011).
An interplay between neurons and glia during the cellular response to disease protein aggregates contributes an additional layer to the regulation of proteostasis, which in turn may modify the process of disease aggregate accumulation and the progression of neuropathologies. Although many disease-associated protein aggregates, such as tau, α-synuclein, TDP-43 and SOD1, are found intracellularly in various subcompartments, there is increasing evidence that they are secreted to the extracellular space. The aggregated form of disease proteins may adopt a specific conformation containing so-called danger-associated molecular patterns (DAMPs) that are known to induce robust immune responses (Rubartelli and Lotze, 2007). The DAMPs-mediated responses lead to posttranslational modifications that facilitate further aggregation and consequent toxicity of disease-associated proteins (Kummer et al., 2011; Saido et al., 1995; Wang et al., 2010). Recent studies have shown that disease-associated protein aggregates, when applied to glial cells exogenously, elicit innate immune and proinflammatory responses (Czirr and Wyss-Coray, 2012; Golde and Miller, 2009). Furthermore, LPS administration, which is commonly used to induce inflammation, results in enhanced aggregate deposition and exacerbated neuropathologies in AD, ALS and prion disease models. This observation suggests that proinflammatory responses modulate proteostasis status and contribute to neurodegeneration. Thus, extracellular aggregates, recognized by pattern recognition receptors in either neurons or glial cells, trigger innate immune responses and secretion of protein aggregates, which in turn induces further inflammation, creating a feed forward loop (Golde et al., 2013). Future studies are needed to determine whether intracellular disease protein aggregates also act as DAMPs in neurons to trigger an immune response.
In fact, an important function of microglia is related to the phagocytosis of cellular debris or protein aggregates in the extracellular space. This notion is supported by the finding that activation of microglia led to a decrease in Aβ load and plaque formation in APP transgenic mice (Grathwohl et al., 2009; Herber et al., 2004; Wilcock et al., 2004; Wyss-Coray et al., 2001). However, a definitive role for microglia in the clearance of extracellular Aβ load has yet to be firmly established (Grathwohl et al., 2009). Thus cross-talk between glial cells (particularly microglia) and neurons may play a significant role in modulating proteostasis pathways and the consequent alteration of intracellular and extracellular aggregate levels. The precise process and mechanism, however, remain to be characterized.
Seeding and trans-cellular propagation of protein aggregates
Over the past several years, an influx of studies has revealed the cell-to-cell transmissibility of disease-related protein aggregates, which has led to a paradigm-shift in terms of our understanding of the pathological progression mechanism for several major neurodegenerative diseases. Increasing evidence demonstrates the “prion-like” self-propagation property of a wide range of disease-related proteins, including Aβ, tau, α-synuclein, polyQ-Htt, SOD1 and TDP-43 (Guo and Lee, 2014; Holmes and Diamond, 2012). These observations not only help explain the Braak staging that describes the spatiotemporal pattern of neuropathologies in neurodegenerative diseases such as AD and PD, but also open up new research avenues for understanding the molecular mechanisms underlying a disease progression common to diverse proteinopathies.
It has been well established that the normal cellular prion protein (PrPc) can be converted into a disease-causing prion (PrPsc) (Prusiner, 1998; Prusiner and Scott, 1997). Similarly, lysates containing aggregates of disease causing proteins or their synthetic fibrils act as “seeds” that rapidly induce the extension of fibrils by recruiting the corresponding normal, soluble proteins (Guo and Lee, 2014). However, the nature of the seeds that template the fibrillization is poorly defined. In addition, lack of knowledge of the biochemical and structural characteristics of the seeds remains a major hurdle to the understanding of the self-propagating mechanism of disease proteins.
A stereotypic progression pattern of neuropathologies in various brain regions of AD and PD was histologically attributed to neuronal subtype or region-specific vulnerability. Recent evidence that fibrillar tau and α-synuclein transmit transcellularly along defined circuits or projections suggests a hypothesis regarding the spatiotemporal distribution of pathologies. For instance, injection of fibrillar species of disease proteins at different sites of rodent brains results in distinct patterns of neuropathology progression, consistent with the idea that the sequentially affected brain areas are synaptically connected. Therefore, it is likely that a combination of brain region- or neuronal subtype-specific vulnerability and neural circuits along which disease aggregates transmit transcellularly, contribute to the stereotypic disease staging as described by Braak and others.
In order to achieve transcellular transmission, disease causing protein aggregates must adapt themselves to cellular trafficking pathways for release and reuptake. The presence of extracellular α-synuclein and tau in interstitial and cerebrospinal fluid of healthy human and animals suggests the constitutive secretion of these two proteins under normal physiological conditions, despite the unknown significance of their release to the extracellular space. Cultured cells or neurons can release monomeric or oligomeric forms of α-synuclein and tau via exosomes (Danzer et al., 2012; Emmanouilidou et al., 2010; Saman et al., 2012). Whether fibrillar or seed-associated aggregates of the two proteins are released using the same mechanism is unclear. Alternative secretion routes for fibrillar α-synuclein have also been suggested (Kfoury et al., 2012; Poehler et al., 2014). In addition, the exosomal secretion of α-synuclein may involve calcium-dependent signaling (Emmanouilidou et al., 2010), recycling endosome associated small GTPase Rab11 (Chutna et al., 2014) and lysosome-related PD protein ATP13A2 (PARK9) (Tsunemi et al., 2014).
Each disease-related protein may adopt different mechanisms for entering recipient neurons in the form of oligomers. Fibrillar α-synuclein is likely to be taken up by the cells through receptor-mediated endocytosis that requires dynamin (Desplats et al., 2009; Hansen et al., 2011). The neuronal surface receptors that recognize fibrillar α-synuclein and initiate endocytosis have yet to be identified, although TLR2 was shown to be the relevant receptor for fibrillar α-synuclein in microglia (Kim et al., 2013). In addition, once internalized via endocytosis, fibrillar α-synuclein must escape from the lumen of endosomal vesicles to the cytosol in order to seed the polymerization of its normal counterparts. However, the mechanism by which the endocytosed α-synuclein exits the vesicles remains unknown. PolyQ expanded-protein was shown to enter cells through both cell surface receptor-mediated uptake (endocytosis) and direct penetration of the lipid bilayer (Ren et al., 2009; Trevino et al., 2012). Internalization of tau fibril was reported to go through a specific endocytic pathway known as macropinocytosis of fluid phase uptake (Frost et al., 2009; Wu et al., 2013). The internalized disease-related aggregates may then seed their counterpart cytosolic proteins, leading to the amplification of intracellular aggregates. Subsequently, the newly produced aggregates are secreted into the extracellular space to reinitiate the cycle.
Taken together, emerging evidence demonstrates the significance of transcellular propagation of pathogenic proteins fibrils or aggregates in understanding the progression of neuropathologies in neurodegenerative diseases; however, the molecular mechanisms delineating the process of fibrillar disease protein transmission remain poorly characterized at this stage.
Exocytosis of disease-related proteins: role of autophagy in unconventional secretory pathways?
The fate of intracellular protein aggregates is varied. For example, the aggregates associated with endosomes may be en route to the autophagy-lysosomal pathway for digestion; but some apparently can escape the digestion and exit the neurons. The control mechanism that determines the distinct fates of protein aggregates is unclear. Several studies showed that inhibition of autophagy alters exocytosis and intercellular transfer of oligomeric α-synuclein, suggesting that autophagy may also regulate the secretion of α-synuclein and consequently spreading of synucleinopathies (Danzer et al., 2012; Lee et al., 2013). Indeed autophagy was shown to be part of an unconventional secretory pathway (secretory autophagy or exophagy). Secretory autophagy participates in the extracellular delivery of α-synuclein monomers and aggregates through a mechanism involving impairment of the fusion between autophagosomes and lysosomes, and an increase in the pool of amphisomes (fusion product of autophagosomes and endosomes) (Ejlerskov et al., 2013).
In fact secretory autophagy participates in the release of several cytosolic proteins into the extracellular space. For example, proinflammatory cytokine IL-1β does not enter the conventional secretory pathway via the Golgi apparatus; instead it is delivered extracellularly via secretory autophagy (Dupont et al., 2011). It was shown in yeast that secretory autophagy involves a special cup-shaped structure named the compartment for unconventional protein secretion (CUPS) (Bruns et al., 2011). The formation of CUPS requires certain autophagy-related proteins; but CUPS differs from the pre-autophagosomal structure (PAS) or autophagosome in its composition and function (Rabouille et al., 2012). Interestingly, a block of autophagy through deleting the essential autophagy gene Atg7 results in aberrant accumulation of intraneuronal Aβ but reduced extracellular Aβ plaque burden in transgenic mouse brains due to inhibition of Aβ secretion, consistent with the role for autophagy in Aβ secretion (Nilsson et al., 2013). However, genetic inactivation of Atg7 enhances exocytosis of α-synuclein in cultured cells, suggesting that Atg7 or the early steps of autophagosome biosynthesis are not essential for the secretion of α-synuclein (Lee et al., 2013). Thus, it is likely that autophagy plays a distinct role in regulating the secretion of different disease-related proteins, though the exact mechanism by which autophagy contributes to their extracellular release remains to be elucidated. Future studies should identify the “CUPS” that functions in the exocytosis of disease-related proteins in mammalian cells and neurons.
Despite the unclear mechanism, current work suggests that autophagy intersects with the trafficking pathways of Aβ and α-synuclein, including secretion (Figure 1). Thus autophagy contributes to the elimination of neuronal protein aggregates by both degradative and secretory pathways.
Conclusions and outlook
Cells and neurons continuously produce misfolded proteins that must be monitored, repaired, and discarded. Proteostasis pathways that influence protein synthesis, stability, quality control and degradation coordinate to maintain the homeostasis and functionality of intracellular proteins and prevent the build-up of toxic misfolded proteins. However, disease causing proteins elude the tight control of proteostasis pathways, thus preventing the cells or neurons from mounting the effective defense response required to repair or degrade aggregated proteins. As a result, toxic intermediate species of oligomers accumulate and even dismantle the proteostasis systems, ultimately leading to cell death. Future studies should investigate in detail why multiple cellular systems including the chaperone quality control, the ER stress response (UPR), the UPS and the autophagy-lysosomal system fail to quickly detect, refold or dispose of misfolded and accumulated disease proteins. It is thus important to identify the molecular triggers and the specific signaling cascades (cell autonomous and non-autonomous) that lead to rapid activation of proteostasis systems that may be able to suppress the accumulation of misfolded oligomers or aggregates.
The toxicity of protein aggregates is more pronounced in neurons than in other cell types in part due to the highly polarized processes of neurons, including axons and dendrites, and neuron-associated synaptic activities. The recent finding that fibrillar α-synuclein and tau transmit through synaptic connections along neural circuits not only explains the spatiotemporal neuropathology pattern of the diseases, but also suggests a potential mechanism underlying the particular vulnerability of neurons to protein aggregates. The fibrils of disease proteins were shown to act as seeds that promote the conversion of intracellular wild-type proteins into oligomeric species. Despite this intriguing finding, many outstanding questions remain to be answered. The precise cellular process of internalization as well as the extracellular release of aggregated proteins in neurons awaits thorough characterization. The exact structures of disease protein fibrils that are transmitted in vivo are unclear. In addition, the molecular mechanism by which neurons decide to degrade intracellularly or release extracellularly aggregated proteins remains uncharacterized. Nonetheless, the discovery of cell-to-cell transmission of disease protein aggregates provides a promising target for therapeutic development.
Autophagy is emerging as a major clearance pathway for toxic protein aggregates and offers neuroprotection. The concept of selective autophagy is particularly attractive in light of the critical role of autophagy receptors in aggregate clearance or aggrephagy. Thus it is imperative to understand the regulation of selective autophagy, which is a promising therapeutic target for proteinopathies that are associated with major neurodenegerative diseases. In addition, emerging evidence suggests that the autophagy machinery intersects with trafficking pathways of disease-related proteins for their extracellular release, adding a new research direction for understanding the propagation of proteinopathies. Finally, there are clear differences in regulation of proteostasis between neurons and non-neuronal cells. To understand precisely the mechanism by which protein aggregates are formed, distributed, degraded or secreted in the context of neuronal environment or neuron-glia interaction, experimental systems recapitulating physiological conditions should be explored and constructed. Systems approaches combining longitudinal tracking and sophisticated analytical tools have been successfully employed to dissect the dynamic events of aggregate life cycle and associated proteostasis activities (Skibinski and Finkbeiner, 2013). Future studies should develop more robust, highly predictive and quantitative systems to explore the role of proteostasis networks in the regulation of aggregate formation, degradation, and transmission, and to harness this knowledge for therapeutic development.
Acknowledgement
This study is supported by NIH/NINDS grant R01NS060123 and CHDI; we thank Gary K Duberstein for critical reading and editing.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arrasate M, Mitra S, Schweitzer ES, Segal MR, Finkbeiner S. Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature. 2004;431:805–810. doi: 10.1038/nature02998. [DOI] [PubMed] [Google Scholar]
- Balch WE, Morimoto RI, Dillin A, Kelly JW. Adapting proteostasis for disease intervention. Science. 2008;319:916–919. doi: 10.1126/science.1141448. [DOI] [PubMed] [Google Scholar]
- Benzinger TL, Gregory DM, Burkoth TS, Miller-Auer H, Lynn DG, Botto RE, Meredith SC. Propagating structure of Alzheimer's beta-amyloid(10-35) is parallel beta-sheet with residues in exact register. Proc Natl Acad Sci U S A. 1998;95:13407–13412. doi: 10.1073/pnas.95.23.13407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berson JF, Theos AC, Harper DC, Tenza D, Raposo G, Marks MS. Proprotein convertase cleavage liberates a fibrillogenic fragment of a resident glycoprotein to initiate melanosome biogenesis. The Journal of cell biology. 2003;161:521–533. doi: 10.1083/jcb.200302072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bingol B, Schuman EM. Activity-dependent dynamics and sequestration of proteasomes in dendritic spines. Nature. 2006;441:1144–1148. doi: 10.1038/nature04769. [DOI] [PubMed] [Google Scholar]
- Bjorkoy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, Stenmark H, Johansen T. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. The Journal of cell biology. 2005;171:603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borsello T, Croquelois K, Hornung JP, Clarke PG. N-methyl-d-aspartate-triggered neuronal death in organotypic hippocampal cultures is endocytic, autophagic and mediated by the c-Jun N-terminal kinase pathway. The European journal of neuroscience. 2003;18:473–485. doi: 10.1046/j.1460-9568.2003.02757.x. [DOI] [PubMed] [Google Scholar]
- Bruns C, McCaffery JM, Curwin AJ, Duran JM, Malhotra V. Biogenesis of a novel compartment for autophagosome-mediated unconventional protein secretion. The Journal of cell biology. 2011;195:979–992. doi: 10.1083/jcb.201106098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caccamo A, Majumder S, Richardson A, Strong R, Oddo S. Molecular interplay between mammalian target of rapamycin (mTOR), amyloid-beta, and Tau: effects on cognitive impairments. The Journal of biological chemistry. 2010;285:13107–13120. doi: 10.1074/jbc.M110.100420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chutna O, Goncalves S, Villar-Pique A, Guerreiro P, Marijanovic Z, Mendes T, Ramalho J, Emmanouilidou E, Ventura S, Klucken J, et al. The small GTPase Rab11 co-localizes with alpha-synuclein in intracellular inclusions and modulates its aggregation, secretion and toxicity. Human molecular genetics. 2014;23:6732–6745. doi: 10.1093/hmg/ddu391. [DOI] [PubMed] [Google Scholar]
- Czirr E, Wyss-Coray T. The immunology of neurodegeneration. The Journal of clinical investigation. 2012;122:1156–1163. doi: 10.1172/JCI58656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dantuma NP, Bott LC. The ubiquitin-proteasome system in neurodegenerative diseases: precipitating factor, yet part of the solution. Frontiers in molecular neuroscience. 2014;7:70. doi: 10.3389/fnmol.2014.00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danzer KM, Kranich LR, Ruf WP, Cagsal-Getkin O, Winslow AR, Zhu L, Vanderburg CR, McLean PJ. Exosomal cell-to-cell transmission of alpha synuclein oligomers. Molecular neurodegeneration. 2012;7:42. doi: 10.1186/1750-1326-7-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decressac M, Mattsson B, Weikop P, Lundblad M, Jakobsson J, Bjorklund A. TFEB-mediated autophagy rescues midbrain dopamine neurons from alpha-synuclein toxicity. Proc Natl Acad Sci U S A. 2013;110:E1817–1826. doi: 10.1073/pnas.1305623110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Der-Sarkissian A, Jao CC, Chen J, Langen R. Structural organization of alpha-synuclein fibrils studied by site-directed spin labeling. J Biol Chem. 2003;278:37530–37535. doi: 10.1074/jbc.M305266200. [DOI] [PubMed] [Google Scholar]
- Desplats P, Lee HJ, Bae EJ, Patrick C, Rockenstein E, Crews L, Spencer B, Masliah E, Lee SJ. Inclusion formation and neuronal cell death through neuron-to-neuron transmission of alpha-synuclein. Proc Natl Acad Sci U S A. 2009;106:13010–13015. doi: 10.1073/pnas.0903691106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont N, Jiang S, Pilli M, Ornatowski W, Bhattacharya D, Deretic V. Autophagy-based unconventional secretory pathway for extracellular delivery of IL-1beta. The EMBO journal. 2011;30:4701–4711. doi: 10.1038/emboj.2011.398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlers MD. Activity level controls postsynaptic composition and signaling via the ubiquitin-proteasome system. Nature neuroscience. 2003;6:231–242. doi: 10.1038/nn1013. [DOI] [PubMed] [Google Scholar]
- Ejlerskov P, Rasmussen I, Nielsen TT, Bergstrom AL, Tohyama Y, Jensen PH, Vilhardt F. Tubulin polymerization-promoting protein (TPPP/p25alpha) promotes unconventional secretion of alpha-synuclein through exophagy by impairing autophagosome-lysosome fusion. J Biol Chem. 2013;288:17313–17335. doi: 10.1074/jbc.M112.401174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmanouilidou E, Melachroinou K, Roumeliotis T, Garbis SD, Ntzouni M, Margaritis LH, Stefanis L, Vekrellis K. Cell-produced alpha-synuclein is secreted in a calcium-dependent manner by exosomes and impacts neuronal survival. J Neurosci. 2010;30:6838–6851. doi: 10.1523/JNEUROSCI.5699-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fecto F, Yan J, Vemula SP, Liu E, Yang Y, Chen W, Zheng JG, Shi Y, Siddique N, Arrat H, et al. SQSTM1 mutations in familial and sporadic amyotrophic lateral sclerosis. Archives of neurology. 2011;68:1440–1446. doi: 10.1001/archneurol.2011.250. [DOI] [PubMed] [Google Scholar]
- Filimonenko M, Isakson P, Finley KD, Anderson M, Jeong H, Melia TJ, Bartlett BJ, Myers KM, Birkeland HC, Lamark T, et al. The selective macroautophagic degradation of aggregated proteins requires the PI3P-binding protein Alfy. Molecular cell. 2010;38:265–279. doi: 10.1016/j.molcel.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman LG, Lachenmayer ML, Wang J, He L, Poulose SM, Komatsu M, Holstein GR, Yue Z. Disrupted autophagy leads to dopaminergic axon and dendrite degeneration and promotes presynaptic accumulation of alpha-synuclein and LRRK2 in the brain. J Neurosci. 2012;32:7585–7593. doi: 10.1523/JNEUROSCI.5809-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost B, Jacks RL, Diamond MI. Propagation of tau misfolding from the outside to the inside of a cell. J Biol Chem. 2009;284:12845–12852. doi: 10.1074/jbc.M808759200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia SM, Casanueva MO, Silva MC, Amaral MD, Morimoto RI. Neuronal signaling modulates protein homeostasis in Caenorhabditis elegans post-synaptic muscle cells. Genes & development. 2007;21:3006–3016. doi: 10.1101/gad.1575307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler S, Holmstrom KM, Skujat D, Fiesel FC, Rothfuss OC, Kahle PJ, Springer W. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol. 2010;12:119–131. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- Gidalevitz T, Prahlad V, Morimoto RI. The stress of protein misfolding: from single cells to multicellular organisms. Cold Spring Harbor perspectives in biology. 2011;3 doi: 10.1101/cshperspect.a009704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg AL. Protein degradation and protection against misfolded or damaged proteins. Nature. 2003;426:895–899. doi: 10.1038/nature02263. [DOI] [PubMed] [Google Scholar]
- Golde TE, Borchelt DR, Giasson BI, Lewis J. Thinking laterally about neurodegenerative proteinopathies. The Journal of clinical investigation. 2013;123:1847–1855. doi: 10.1172/JCI66029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golde TE, Miller VM. Proteinopathy-induced neuronal senescence: a hypothesis for brain failure in Alzheimer's and other neurodegenerative diseases. Alzheimer's research & therapy. 2009;1:5. doi: 10.1186/alzrt5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grathwohl SA, Kalin RE, Bolmont T, Prokop S, Winkelmann G, Kaeser SA, Odenthal J, Radde R, Eldh T, Gandy S, et al. Formation and maintenance of Alzheimer's disease beta-amyloid plaques in the absence of microglia. Nature neuroscience. 2009;12:1361–1363. doi: 10.1038/nn.2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JL, Lee VM. Cell-to-cell transmission of pathogenic proteins in neurodegenerative diseases. Nature medicine. 2014;20:130–138. doi: 10.1038/nm.3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen C, Angot E, Bergstrom AL, Steiner JA, Pieri L, Paul G, Outeiro TF, Melki R, Kallunki P, Fog K, et al. alpha-Synuclein propagates from mouse brain to grafted dopaminergic neurons and seeds aggregation in cultured human cells. The Journal of clinical investigation. 2011;121:715–725. doi: 10.1172/JCI43366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, Yokoyama M, Mishima K, Saito I, Okano H, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- Herber DL, Roth LM, Wilson D, Wilson N, Mason JE, Morgan D, Gordon MN. Time-dependent reduction in Abeta levels after intracranial LPS administration in APP transgenic mice. Experimental neurology. 2004;190:245–253. doi: 10.1016/j.expneurol.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Hetz C. The unfolded protein response: controlling cell fate decisions under ER stress and beyond. Nature reviews. Molecular cell biology. 2012;13:89–102. doi: 10.1038/nrm3270. [DOI] [PubMed] [Google Scholar]
- Hetz C, Mollereau B. Disturbance of endoplasmic reticulum proteostasis in neurodegenerative diseases. Nature reviews. Neuroscience. 2014;15:233–249. doi: 10.1038/nrn3689. [DOI] [PubMed] [Google Scholar]
- Holmes BB, Diamond MI. Cellular mechanisms of protein aggregate propagation. Current opinion in neurology. 2012;25:721–726. doi: 10.1097/WCO.0b013e32835a3ee0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isakson P, Holland P, Simonsen A. The role of ALFY in selective autophagy. Cell death and differentiation. 2013;20:12–20. doi: 10.1038/cdd.2012.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kachaner D, Genin P, Laplantine E, Weil R. Toward an integrative view of Optineurin functions. Cell cycle. 2012;11:2808–2818. doi: 10.4161/cc.20946. [DOI] [PubMed] [Google Scholar]
- Kageyama S, Sou YS, Uemura T, Kametaka S, Saito T, Ishimura R, Kouno T, Bedford L, Mayer RJ, Lee MS, et al. Proteasome Dysfunction Activates Autophagy and the Keap1-Nrf2 Pathway. J Biol Chem. 2014 doi: 10.1074/jbc.M114.580357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kfoury N, Holmes BB, Jiang H, Holtzman DM, Diamond MI. Trans-cellular propagation of Tau aggregation by fibrillar species. J Biol Chem. 2012;287:19440–19451. doi: 10.1074/jbc.M112.346072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C, Ho DH, Suk JE, You S, Michael S, Kang J, Joong Lee S, Masliah E, Hwang D, Lee HJ, et al. Neuron-released oligomeric alpha-synuclein is an endogenous agonist of TLR2 for paracrine activation of microglia. Nat Commun. 2013;4:1562. doi: 10.1038/ncomms2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim PK, Hailey DW, Mullen RT, Lippincott-Schwartz J. Ubiquitin signals autophagic degradation of cytosolic proteins and peroxisomes. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:20567–20574. doi: 10.1073/pnas.0810611105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkin V, McEwan DG, Novak I, Dikic I. A role for ubiquitin in selective autophagy. Molecular cell. 2009;34:259–269. doi: 10.1016/j.molcel.2009.04.026. [DOI] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, Ueno T, Koike M, Uchiyama Y, Kominami E, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Koike M, Sou YS, Ueno T, Hara T, Mizushima N, Iwata J, Ezaki J, Murata S, et al. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 2007a;131:1149–1163. doi: 10.1016/j.cell.2007.10.035. [DOI] [PubMed] [Google Scholar]
- Komatsu M, Wang QJ, Holstein GR, Friedrich VL, Jr., Iwata J, Kominami E, Chait BT, Tanaka K, Yue Z. Essential role for autophagy protein Atg7 in the maintenance of axonal homeostasis and the prevention of axonal degeneration. Proc Natl Acad Sci U S A. 2007b;104:14489–14494. doi: 10.1073/pnas.0701311104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopito RR. Aggresomes, inclusion bodies and protein aggregation. Trends in cell biology. 2000;10:524–530. doi: 10.1016/s0962-8924(00)01852-3. [DOI] [PubMed] [Google Scholar]
- Kummer MP, Hermes M, Delekarte A, Hammerschmidt T, Kumar S, Terwel D, Walter J, Pape HC, Konig S, Roeber S, et al. Nitration of tyrosine 10 critically enhances amyloid beta aggregation and plaque formation. Neuron. 2011;71:833–844. doi: 10.1016/j.neuron.2011.07.001. [DOI] [PubMed] [Google Scholar]
- Lamb CA, Yoshimori T, Tooze SA. The autophagosome: origins unknown, biogenesis complex. Nature reviews. Molecular cell biology. 2013;14:759–774. doi: 10.1038/nrm3696. [DOI] [PubMed] [Google Scholar]
- Lee HJ, Cho ED, Lee KW, Kim JH, Cho SG, Lee SJ. Autophagic failure promotes the exocytosis and intercellular transfer of alpha-synuclein. Experimental & molecular medicine. 2013;45:e22. doi: 10.1038/emm.2013.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long J, Gallagher TR, Cavey JR, Sheppard PW, Ralston SH, Layfield R, Searle MS. Ubiquitin recognition by the ubiquitin-associated domain of p62 involves a novel conformational switch. J Biol Chem. 2008;283:5427–5440. doi: 10.1074/jbc.M704973200. [DOI] [PubMed] [Google Scholar]
- Lonskaya I, Hebron ML, Desforges NM, Franjie A, Moussa CE. Tyrosine kinase inhibition increases functional parkin-Beclin-1 interaction and enhances amyloid clearance and cognitive performance. EMBO molecular medicine. 2013;5:1247–1262. doi: 10.1002/emmm.201302771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu K, Psakhye I, Jentsch S. Autophagic clearance of polyQ proteins mediated by ubiquitin-Atg8 adaptors of the conserved CUET protein family. Cell. 2014;158:549–563. doi: 10.1016/j.cell.2014.05.048. [DOI] [PubMed] [Google Scholar]
- Maday S, Wallace KE, Holzbaur EL. Autophagosomes initiate distally and mature during transport toward the cell soma in primary neurons. The Journal of cell biology. 2012;196:407–417. doi: 10.1083/jcb.201106120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandell MA, Jain A, Arko-Mensah J, Chauhan S, Kimura T, Dinkins C, Silvestri G, Munch J, Kirchhoff F, Simonsen A, et al. TRIM proteins regulate autophagy and can target autophagic substrates by direct recognition. Dev Cell. 2014;30:394–409. doi: 10.1016/j.devcel.2014.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto G, Wada K, Okuno M, Kurosawa M, Nukina N. Serine 403 phosphorylation of p62/SQSTM1 regulates selective autophagic clearance of ubiquitinated proteins. Molecular cell. 2011;44:279–289. doi: 10.1016/j.molcel.2011.07.039. [DOI] [PubMed] [Google Scholar]
- Morimoto RI. The heat shock response: systems biology of proteotoxic stress in aging and disease. Cold Spring Harbor symposia on quantitative biology. 2011;76:91–99. doi: 10.1101/sqb.2012.76.010637. [DOI] [PubMed] [Google Scholar]
- Muchowski PJ. Protein misfolding, amyloid formation, and neurodegeneration: a critical role for molecular chaperones? Neuron. 2002;35:9–12. doi: 10.1016/s0896-6273(02)00761-4. [DOI] [PubMed] [Google Scholar]
- Nascimento-Ferreira I, Santos-Ferreira T, Sousa-Ferreira L, Auregan G, Onofre I, Alves S, Dufour N, Colomer Gould VF, Koeppen A, Deglon N, et al. Overexpression of the autophagic beclin-1 protein clears mutant ataxin-3 and alleviates Machado-Joseph disease. Brain : a journal of neurology. 2011;134:1400–1415. doi: 10.1093/brain/awr047. [DOI] [PubMed] [Google Scholar]
- Nilsson P, Loganathan K, Sekiguchi M, Matsuba Y, Hui K, Tsubuki S, Tanaka M, Iwata N, Saito T, Saido TC. Abeta secretion and plaque formation depend on autophagy. Cell reports. 2013;5:61–69. doi: 10.1016/j.celrep.2013.08.042. [DOI] [PubMed] [Google Scholar]
- Otabe H, Nibuya M, Shimazaki K, Toda H, Suzuki G, Nomura S, Shimizu K. Electroconvulsive seizures enhance autophagy signaling in rat hippocampus. Progress in neuro-psychopharmacology & biological psychiatry. 2014;50:37–43. doi: 10.1016/j.pnpbp.2013.11.012. [DOI] [PubMed] [Google Scholar]
- Overbye A, Fengsrud M, Seglen PO. Proteomic analysis of membrane- associated proteins from rat liver autophagosomes. Autophagy. 2007;3:300–322. doi: 10.4161/auto.3910. [DOI] [PubMed] [Google Scholar]
- Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, Overvatn A, Bjorkoy G, Johansen T. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- Pankiv S, Lamark T, Bruun JA, Overvatn A, Bjorkoy G, Johansen T. Nucleocytoplasmic shuttling of p62/SQSTM1 and its role in recruitment of nuclear polyubiquitinated proteins to promyelocytic leukemia bodies. J Biol Chem. 2010;285:5941–5953. doi: 10.1074/jbc.M109.039925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poehler AM, Xiang W, Spitzer P, May V, Meixner H, Rockenstein E, Chutna O, Outeiro T, Winkler J, Masliah E, et al. Autophagy modulates SNCA/alpha-synuclein release, thereby generating a hostile microenvironment. Autophagy. 2014:e36436. doi: 10.4161/auto.36436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prahlad V, Morimoto RI. Neuronal circuitry regulates the response of Caenorhabditis elegans to misfolded proteins. Proc Natl Acad Sci U S A. 2011;108:14204–14209. doi: 10.1073/pnas.1106557108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Princiotta MF, Finzi D, Qian SB, Gibbs J, Schuchmann S, Buttgereit F, Bennink JR, Yewdell JW. Quantitating protein synthesis, degradation, and endogenous antigen processing. Immunity. 2003;18:343–354. doi: 10.1016/s1074-7613(03)00051-7. [DOI] [PubMed] [Google Scholar]
- Prusiner SB. Prions. Proc Natl Acad Sci U S A. 1998;95:13363–13383. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prusiner SB, Scott MR. Genetics of prions. Annual review of genetics. 1997;31:139–175. doi: 10.1146/annurev.genet.31.1.139. [DOI] [PubMed] [Google Scholar]
- Rabouille C, Malhotra V, Nickel W. Diversity in unconventional protein secretion. Journal of cell science. 2012;125:5251–5255. doi: 10.1242/jcs.103630. [DOI] [PubMed] [Google Scholar]
- Ravikumar B, Vacher C, Berger Z, Davies JE, Luo S, Oroz LG, Scaravilli F, Easton DF, Duden R, O'Kane CJ, et al. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nature genetics. 2004;36:585–595. doi: 10.1038/ng1362. [DOI] [PubMed] [Google Scholar]
- Ren PH, Lauckner JE, Kachirskaia I, Heuser JE, Melki R, Kopito RR. Cytoplasmic penetration and persistent infection of mammalian cells by polyglutamine aggregates. Nat Cell Biol. 2009;11:219–225. doi: 10.1038/ncb1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley BE, Kaiser SE, Shaler TA, Ng AC, Hara T, Hipp MS, Lage K, Xavier RJ, Ryu KY, Taguchi K, et al. Ubiquitin accumulation in autophagy-deficient mice is dependent on the Nrf2-mediated stress response pathway: a potential role for protein aggregation in autophagic substrate selection. The Journal of cell biology. 2010;191:537–552. doi: 10.1083/jcb.201005012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogov V, Dotsch V, Johansen T, Kirkin V. Interactions between autophagy receptors and ubiquitin-like proteins form the molecular basis for selective autophagy. Mol Cell. 2014;53:167–178. doi: 10.1016/j.molcel.2013.12.014. [DOI] [PubMed] [Google Scholar]
- Ross CA, Poirier MA. Protein aggregation and neurodegenerative disease. Nature medicine. 2004;10(Suppl):S10–17. doi: 10.1038/nm1066. [DOI] [PubMed] [Google Scholar]
- Rubartelli A, Lotze MT. Inside, outside, upside down: damage-associated molecular-pattern molecules (DAMPs) and redox. Trends in immunology. 2007;28:429–436. doi: 10.1016/j.it.2007.08.004. [DOI] [PubMed] [Google Scholar]
- Saido TC, Iwatsubo T, Mann DM, Shimada H, Ihara Y, Kawashima S. Dominant and differential deposition of distinct beta-amyloid peptide species, A beta N3(pE), in senile plaques. Neuron. 1995;14:457–466. doi: 10.1016/0896-6273(95)90301-1. [DOI] [PubMed] [Google Scholar]
- Saman S, Kim W, Raya M, Visnick Y, Miro S, Saman S, Jackson B, McKee AC, Alvarez VE, Lee NC, et al. Exosome-associated tau is secreted in tauopathy models and is selectively phosphorylated in cerebrospinal fluid in early Alzheimer disease. J Biol Chem. 2012;287:3842–3849. doi: 10.1074/jbc.M111.277061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardiello M, Palmieri M, di Ronza A, Medina DL, Valenza M, Gennarino VA, Di Malta C, Donaudy F, Embrione V, Polishchuk RS, et al. A gene network regulating lysosomal biogenesis and function. Science. 2009;325:473–477. doi: 10.1126/science.1174447. [DOI] [PubMed] [Google Scholar]
- Sarkar S, Davies JE, Huang Z, Tunnacliffe A, Rubinsztein DC. Trehalose, a novel mTOR-independent autophagy enhancer, accelerates the clearance of mutant huntingtin and alpha-synuclein. The Journal of biological chemistry. 2007;282:5641–5652. doi: 10.1074/jbc.M609532200. [DOI] [PubMed] [Google Scholar]
- Seibenhener ML, Babu JR, Geetha T, Wong HC, Krishna NR, Wooten MW. Sequestosome 1/p62 is a polyubiquitin chain binding protein involved in ubiquitin proteasome degradation. Molecular and cellular biology. 2004;24:8055–8068. doi: 10.1128/MCB.24.18.8055-8068.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shehata M, Matsumura H, Okubo-Suzuki R, Ohkawa N, Inokuchi K. Neuronal stimulation induces autophagy in hippocampal neurons that is involved in AMPA receptor degradation after chemical long-term depression. J Neurosci. 2012;32:10413–10422. doi: 10.1523/JNEUROSCI.4533-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata M, Lu T, Furuya T, Degterev A, Mizushima N, Yoshimori T, MacDonald M, Yankner B, Yuan J. Regulation of intracellular accumulation of mutant Huntingtin by Beclin 1. The Journal of biological chemistry. 2006;281:14474–14485. doi: 10.1074/jbc.M600364200. [DOI] [PubMed] [Google Scholar]
- Skibinski G, Finkbeiner S. Longitudinal measures of proteostasis in live neurons: features that determine fate in models of neurodegenerative disease. FEBS letters. 2013;587:1139–1146. doi: 10.1016/j.febslet.2013.02.043. [DOI] [PubMed] [Google Scholar]
- Spencer B, Potkar R, Trejo M, Rockenstein E, Patrick C, Gindi R, Adame A, Wyss-Coray T, Masliah E. Beclin 1 gene transfer activates autophagy and ameliorates the neurodegenerative pathology in alpha-synuclein models of Parkinson's and Lewy body diseases. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2009;29:13578–13588. doi: 10.1523/JNEUROSCI.4390-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele JW, Ju S, Lachenmayer ML, Liken J, Stock A, Kim SH, Delgado LM, Alfaro IE, Bernales S, Verdile G, et al. Latrepirdine stimulates autophagy and reduces accumulation of alpha-synuclein in cells and in mouse brain. Molecular psychiatry. 2013;18:882–888. doi: 10.1038/mp.2012.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolz A, Ernst A, Dikic I. Cargo recognition and trafficking in selective autophagy. Nat Cell Biol. 2014;16:495–501. doi: 10.1038/ncb2979. [DOI] [PubMed] [Google Scholar]
- Tai HC, Besche H, Goldberg AL, Schuman EM. Characterization of the Brain 26S Proteasome and its Interacting Proteins. Frontiers in molecular neuroscience. 2010;3 doi: 10.3389/fnmol.2010.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trevino RS, Lauckner JE, Sourigues Y, Pearce MM, Bousset L, Melki R, Kopito RR. Fibrillar structure and charge determine the interaction of polyglutamine protein aggregates with the cell surface. J Biol Chem. 2012;287:29722–29728. doi: 10.1074/jbc.M112.372474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsunemi T, Hamada K, Krainc D. ATP13A2/PARK9 regulates secretion of exosomes and alpha-synuclein. J Neurosci. 2014;34:15281–15287. doi: 10.1523/JNEUROSCI.1629-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhoef LG, Lindsten K, Masucci MG, Dantuma NP. Aggregate formation inhibits proteasomal degradation of polyglutamine proteins. Human molecular genetics. 2002;11:2689–2700. doi: 10.1093/hmg/11.22.2689. [DOI] [PubMed] [Google Scholar]
- Wang F, Durfee LA, Huibregtse JM. A cotranslational ubiquitination pathway for quality control of misfolded proteins. Molecular cell. 2013;50:368–378. doi: 10.1016/j.molcel.2013.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Garg S, Mandelkow EM, Mandelkow E. Proteolytic processing of tau. Biochemical Society transactions. 2010;38:955–961. doi: 10.1042/BST0380955. [DOI] [PubMed] [Google Scholar]
- Wilcock DM, Munireddy SK, Rosenthal A, Ugen KE, Gordon MN, Morgan D. Microglial activation facilitates Abeta plaque removal following intracranial anti-Abeta antibody administration. Neurobiology of disease. 2004;15:11–20. doi: 10.1016/j.nbd.2003.09.015. [DOI] [PubMed] [Google Scholar]
- Wild P, Farhan H, McEwan DG, Wagner S, Rogov VV, Brady NR, Richter B, Korac J, Waidmann O, Choudhary C, et al. Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science. 2011;333:228–233. doi: 10.1126/science.1205405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong YC, Holzbaur EL. The regulation of autophagosome dynamics by huntingtin and HAP1 is disrupted by expression of mutant huntingtin, leading to defective cargo degradation. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2014;34:1293–1305. doi: 10.1523/JNEUROSCI.1870-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JW, Herman M, Liu L, Simoes S, Acker CM, Figueroa H, Steinberg JI, Margittai M, Kayed R, Zurzolo C, et al. Small misfolded Tau species are internalized via bulk endocytosis and anterogradely and retrogradely transported in neurons. J Biol Chem. 2013;288:1856–1870. doi: 10.1074/jbc.M112.394528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyss-Coray T, Lin C, Yan F, Yu GQ, Rohde M, McConlogue L, Masliah E, Mucke L. TGF-beta1 promotes microglial amyloid-beta clearance and reduces plaque burden in transgenic mice. Nature medicine. 2001;7:612–618. doi: 10.1038/87945. [DOI] [PubMed] [Google Scholar]
- Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol. 2007;9:1102–1109. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- Yang P, Zhang H. You are what you eat: multifaceted functions of autophagy during C. elegans development. Cell research. 2014;24:80–91. doi: 10.1038/cr.2013.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu A, Shibata Y, Shah B, Calamini B, Lo DC, Morimoto RI. Protein aggregation can inhibit clathrin-mediated endocytosis by chaperone competition. Proc Natl Acad Sci U S A. 2014;111:E1481–1490. doi: 10.1073/pnas.1321811111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue Z, Wang QJ, Komatsu M. Neuronal autophagy: going the distance to the axon. Autophagy. 2008;4:94–96. doi: 10.4161/auto.5202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng YT, Shahnazari S, Brech A, Lamark T, Johansen T, Brumell JH. The adaptor protein p62/SQSTM1 targets invading bacteria to the autophagy pathway. J Immunol. 2009;183:5909–5916. doi: 10.4049/jimmunol.0900441. [DOI] [PubMed] [Google Scholar]
- Zourlidou A, Gidalevitz T, Kristiansen M, Landles C, Woodman B, Wells DJ, Latchman DS, de Belleroche J, Tabrizi SJ, Morimoto RI, et al. Hsp27 overexpression in the R6/2 mouse model of Huntington's disease: chronic neurodegeneration does not induce Hsp27 activation. Human molecular genetics. 2007;16:1078–1090. doi: 10.1093/hmg/ddm057. [DOI] [PubMed] [Google Scholar]