Abstract
Guanine (G)-rich DNA sequences can adopt four-stranded quadruplex conformations that may play a role in the regulation of genetic processes. To explore the possibility of targeted molecular recognition of DNA sequences with short G-rich peptide nucleic acids (PNA) and to assess the strand arrangement in such complexes, we used PNA and DNA with the Oxytricha nova telomeric sequence d(G4T4G4) as a model. PNA probes were complexed with DNA targets in the following forms: single-stranded oligonucleotides, a loop of DNA in a hairpin conformation, and as supercoiled plasmid with the (G4T4G4)/(C4A4C4) insert. Gel-shift mobility assays demonstrated formation of stable hybrid complexes between the homologous G4T4G4 PNA and DNA with multiple modes of binding. Chemical and enzymatic probing revealed sequence-specific and G-quadruplex dependent binding of G4T4G4 PNA to dsDNA. Spectroscopic and electrophoretic analysis of the complex formed between PNA and the synthetic DNA hairpin containing the G4T4G4 loop showed that the stoichiometry of a prevailing complex is three PNA strands per one DNA strand. We speculate how this new PNA–DNA complex architecture can help to design more selective, quadruplex-specific PNA probes.
Introduction
There is a growing body of evidence that G-quadruplex structures play roles in various genetic processes [1,2]. Stabilization of such structures may result in alteration of gene expression and could potentially bring therapeutic benefits [3,4]. For this purpose, numerous small planar molecules that bind specifically to G-quadruplexes have been developed [5–7]; however, most of these small molecules bind G-tetrads via intercalation and base stacking that is nonselective among different types of quadruplexes. In addition, most of these molecules bind duplex DNA that is present in the genome. Therefore, the problem of selectivity between various quadruplexes has yet to be solved.
Another strategy to stabilize quadruplexes involves targeting the complementary strand of G-rich sequences with short peptide nucleic acids (PNA) [8–10] (reviewed in [3]). This approach is based on the earlier observation that PNA invade plasmid DNA more effectively at the target sequences that can form secondary structures, such as cruciforms [11]. We demonstrated the stabilization of a Bcl2 quadruplex induced by the formation of PNA–DNA duplex with the stand complementary to the G-rich quadruplex-forming strand [12]. We also demonstrated that there is no formation of a PNA/DNA quadruplex [12,13].
An alternative double-invasion approach was proposed when G-rich PNA binds both the complementary C-rich strand and participates in quadruplex formation with the G-rich strand [8]. The formation of mixed DNA–PNA quadruplexes was first demonstrated using mixtures of short, single-stranded PNA and DNA sequences [8]. The resulting quadruplexes were found to consist of two PNA and two DNA molecules (i.e., the stoichiometry of DNA to PNA was 1:1). Later the double invasion was used for sequence-specific scission of a double-stranded plasmid and genomic DNA [14]. However, the strand arrangements and stoichiometry of the DNA–PNA quadruplex complexes were not determined in these studies. Knowledge of the molecular structure of DNA–PNA quadruplex upon the double invasion would lead to the design of more efficient and specific PNA probes.
In this paper, we describe the quadruplex formation between PNA and its DNA target sequence presented in the form of single-stranded oligonucleotides, a single-stranded loop in a DNA hairpin, and as a duplex insert into plasmid DNA. For a model system, we used the well-characterized quadruplex-forming sequence of G4T4G4 the Oxytricha nova telomere [15]. The quadruplex structures formed by this sequence are quite stable and amenable to study. In addition, the use of this simple symmetrical sequence reveals structural features that are common in other quadruplexes.
Methods
PNA and DNA synthesis
PNA oligomers were synthesized by Boc-mediated solid phase synthesis on a 433A Automated Peptide Synthesizer from Applied BioSystems [16]. Boc-protected PNA monomers were purchased from Applied Biosystems. PNA containing 8-aza-7-deaza-guanine (ppG) were synthesized as described in [13]. PNA oligomer purity and identity were tested using an HP 1050 high-performance liquid chromatography (Agilent Technologies) and high-resolution mass spectra were obtained on a liquid chromatography-Time of Flight Mass Spectrometry System (Agilent Technologies). PNA concentration was measured at 260 nm on a HP 8452A Diode Array Spectrophotometer (Agilent Technologies), and was calculated with extinction coefficient calculator software (www.basic.northwestern.edu/biotools/oligocalc.html). Prior to all experiments, PNAs were incubated in a shaker for 15 min at 42°C and their concentration was measured before mixing with DNA samples.
The DNA oligonucleotides were synthesized on an ABI394 DNA synthesizer (Applied Biosystems), and purified by denaturing polyacrylamide gel electrophoresis (PAGE) as described in detail in [17]. The ppG phosphoramidite was purchased from Glen Research. The concentration of single-stranded oligonucleotides was measured at 260 nm on a HP 8452A spectrophotometer and was calculated with extinction coefficient calculator software. DNA oligonucleotides were 5′-32P labeled with [γ-32P]-ATP (Perkin Elmer) by T4 polynucleotide kinase using standard protocol followed by purification on G-25 microcolumns (GE Healthcare, UK).
PNA and DNA complexes were formed in TNE buffer containing 50mM Tris/HCl pH 7.4, 25 mM NaCl, 2.5 mM ethylenediaminetetraacetic acid (EDTA) by incubation at 37°C for 2 h. We did not use KCl in the binding buffer because both PNA and DNA oligonucleotides form strong quadruplex complexes in KCl, preventing them from interaction with each other.
Plasmids
The plasmid pGT4 was obtained by cloning G4T4G4/C4A4C4 synthetic duplex into pCR-Blunt vector using Zero Blunt PCR Cloning Kit (Invitrogen). Constructs were incorporated and amplified in One Shot TOP10 Chemically Competent Escherichia coli (Invitrogen) and selected with 50 μg/mL kanamycin. Plasmid DNA was purified using the Plasmid Maxi Kit (Qiagen). Plasmids were additionally purified by ultracentrifugation in CsCl gradient [18]. All plasmids were directly sequenced to prove the presence of the correct insert using Maxam-Gilbert sequencing procedure [19]. Incubation of plasmids with PNA was performed in TNE buffer at 37°C for 2 h. Contraction of pBcl2 plasmid was described previously [12]. The sequence of the pBcl2 insert is 5′-GGGCCAGGGAGCGGGGCGGAGGGGGCGGTCGG GTG-3′.
Gel shift assay
Native polyacrylamide gels (6% and 20%) were custom casted in 1 mm Novex cassettes and were run at room temperature in TBE buffer containing 12.5 mM sodium borate at 120V. Gels were quantified using BAS-2500 Bioimager (FUJI Medical Systems USA) for radioactivity signal and using FluorImager 595 (Molecular Dynamics) for fluorescent signal. For gel shift studies of plasmid fragments, following incubation with PNAs, plasmids were cut with AlwN1 or with HindIII and Xho1 restriction enzymes (Fermentas) for 1 at 37°C, followed by 3x′-32P labeling with [α-32P]-dCTP (Perkin Elmer) by Klenow fragment of DNA polymerase I (Fermentas). After purification on G-25 microcolumns (GE Healthcare), samples were analyzed with gel electrophoresis.
S1 nuclease and chemical probing
S1 nuclease cleavage of plasmid DNA complexes with PNA was performed in S1 nuclease buffer (30 mM NaAc pH 4.6, 10 mM ZnAc) with 10 units of S1 nuclease (Invitrogen) on ice for 2 min. Reactions were stopped with EDTA (25 mM final concentration), extracted with equal volume of phenol:chloroform:isoamyl alcohol (25:24:1, Life Technologies) and purified with G50 Microspin columns (GE Healthcare). The purified samples were cut with Fast Digest SpeI and Kpn1 restriction enzymes (Fermentas) for 20 min at 37°C followed by 3′-32P labeling with [α-32P]-dCTP (Perkin Elmer) by Klenow fragment of DNA polymerase 1 (Fermentas). Samples were analyzed by electrophoresis in 12% denaturing polyacrylamide gels allowing the shorter fragment to run off the gel.
Hairpin DNA was probed with 2.5 mM osmium tetroxide (OsO4) plus 2.5 mM 2.2′-dipyridyl disulfide for 5 min at room temperature, or with 2 μL 10% DMS in ethanol for 15 min at room temperature. Reactions were stopped with 5× stop solution on ice (1.5 M sodium acetate pH 5.2, 1 M β-mercaptoethanol, 100 μg/mL yeast tRNA) followed by ethanol precipitation and washing with 70% ethanol. After incubation in 10% piperidine at 95°C for 20 min and two repeated lyophilizations, samples were analyzed by electrophoresis in 12% denaturing PAGE [12].
Circular dichroism spectroscopy
Circular dichroism (CD) analyses were performed on Jasco J-810 spectrophotometer in a 2-mm pathlength cuvette and a wavelength scanning speed of 100 nm/min. DNA oligonucleotides were incubated for G-quadruplex formation as described above in a buffer containing 10 mM Na-phosphate, pH 7.4, 25 mM NaCl, 1 mM EDTA. Concentrations of DNA and PNA were 2μM and 6 μM respectively. CD spectra of samples were generated at 20°C in the range from 200 nM to 320 nM. The spectra (the averages of three scans) were corrected on blank buffer spectra and normalized on oligonucleotide concentration.
Results
G4T4G4 PNA and DNA form multiple complexes
An earlier study by Datta and coworkers found that G4T4G4 PNA and DNA oligonucleotides formed parallel G-quadruplexes with 1:1 DNA:PNA stoichiometry [20]. They also observed a drastic difference in CD spectra of their DNA and PNA complexes obtained by annealing at different temperatures. With these results in mind, we studied this system further by using gel-electrophoresis that allows separation of DNA-PNA complexes of different structures and stoichiometry, giving us a better understanding of the complexes formed.
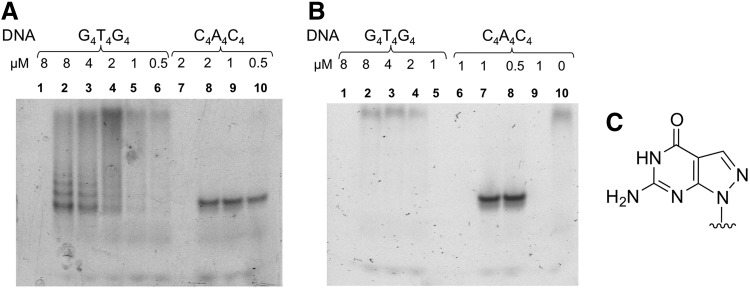
G4T4G4 PNA was fluorescently labeled and the concentration of PNA was kept constant at 1μM. PNA samples were mixed with G4T4G4 DNA in different molar ratios and incubated at 37°C for 2 h. These conditions were considered as physiologically relevant. The samples were then analyzed in 12% native PAGE. Figure 1A (lanes 2–6) shows multiple bands corresponding to DNA–PNA complexes of different mobilities in the gel. PNA oligomers are slightly positively charged; therefore, they do not enter the gel when uncomplexed with DNA. However, they do migrate in the gel as complexes with DNA. As the amount of DNA oligonucleotides diminishes from lane 2 to 6, so too does the intensity of the bands corresponding to the complexes. For comparison, formation of complementary complexes between G4T4G4 PNA and C4A4C4 DNA is shown in lanes 8 to 10. At least three separate bands are seen in the lanes 2 and 3. The most plausible explanation of these multiple bands is that they correspond to DNA–PNA quadruplexes with different stoichiometries. The bottom band with the fastest mobility corresponds to a quadruplex with a 3:1 DNA to PNA ratio as it carries the highest number of the negatively charged DNA strands, while the top of the three bands corresponds to a quadruplex that consists of 1 DNA and 3 PNA strands. The bands above that most likely correspond to higher order quadruplexes, or G-wire formation.
FIG. 1.
(A) Binding of fluorescent G4T4G4 peptide nucleic acids (PNA) to G4T4G4 and C4A4C4 DNA oligonucleotides. G4T4G4 PNA (1μM) was incubated with varying concentrations (shown on the top of the lanes in μM) of DNA oligonucleotides. Lanes 1 to 6 contain samples with G4T4G4 DNA oligonucleotides; lanes 7 to 10 contain samples with C4A4C4 DNA oligonucleotides; lanes 1 and 7 are samples with no PNA added. (B) Binding of fluorescent G4T4G4 PNA to ppG4T4ppG4 and C4A4C4 DNA oligonucleotides. G4T4G4 PNA (1μM) was incubated with varying concentrations (shown on the top of the lanes in μM) of DNA oligonucleotides. Lanes 1 to 5 contain samples with ppG4T4ppG4 DNA oligonucleotides; lanes 6 to 10 contain samples with C4A4C4 DNA oligonucleotides. Lanes 1, 6, and 9 are samples with no PNA added. (C) Chemical structure of 8-aza-7-deazaguanine (ppG).
A PNA binding experiment with DNA oligonucleotides carrying 8-aza-7-deaza-guanine (ppG) nucleotides instead of guanine that are unable to form G-quartets [13,21] is shown in Fig. 1B. The fact that the ppG containing oligonucleotides do not form complexes with G4T4G4 PNA demonstrates that complexes observed in Fig. 1A between G4T4G4 PNA and DNA were indeed stabilized by quadruplex formation. CD spectra of the PNA–DNA complexes are also consistent with the formation of anti-parallel quadruplexes (Supplementary Fig. S1; Supplementary Data are available online at www.liebertpub.com/nat). Under similar conditions without thermal annealing, Datta et al. observed very similar CD spectra, which they interpreted to indicate the presence of antiparallel quadruplexes. At the same time, they demonstrated that fully equilibrated quadruplexes contained 2 PNA and 2 DNA molecules in a parallel orientation [20]. Therefore, DNA–PNA quadruplex complexes may not be limited to those with 1:1 stoichiometry, but can consist of different proportions of DNA and PNA strands with different strand orientation.
Binding G4T4G4 PNA to G4T4G4 loop in DNA hairpin
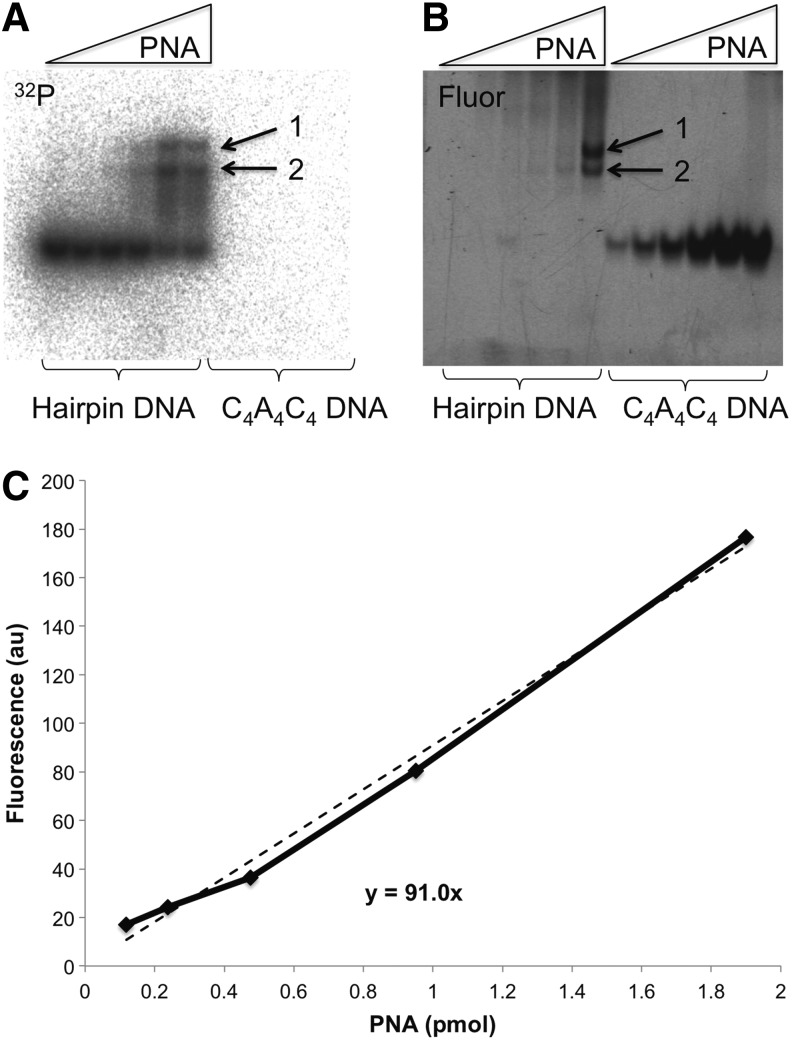
Next, we studied G4T4G4 PNA–DNA binding when G4T4G4 DNA is presented as a loop in the DNA hairpin shown in Fig. 2. In our design the loop contained two extra Ts on each side to increase its size and facilitate G-quadruplex formation. This model is more relevant to PNA invasion into duplex DNA than binding of PNA to an oligonucleotide with two free ends. This situation can happen, for example, when the complimentary DNA strands are separated and the complementary C-strand forms the i-motif structure [22]. In these experiments, the DNA hairpin was labeled with 32P, while G4T4G4 PNA was labeled with fluorescein. A fixed amount of DNA hairpin was incubated with increasing amounts of PNA, followed by analysis with PAGE. Autoradiography and fluorescent images of the gel are shown in Fig. 3A and B, respectively. As the amount of PNA in the samples increases, two shifted bands appear in the gel corresponding to two PNA–DNA hairpin complexes. Chemical probing of the complexes with OsO4 indicates that DNA preserves the compact G-rich loop without unwinding the hairpin stem after PNA binding (Supplementary Fig. S2).
FIG. 2.
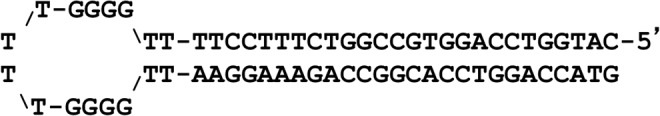
Nucleotide sequence of DNA hairpin.
FIG. 3.
Binding of fluorescent G4T4G4 PNA to 32P-labeled DNA hairpin and nonlabeled C4A4C4 DNA oligonucleotides. Two images of the same gel are shown: (A) 32P autoradiography and (B) fluorescent image. A fixed concentration (0.75 μM) of 32P-labeled DNA hairpin (left lanes on the gel), or nonlabeled C4A4C4 DNA (right lanes on the gel) were incubated with increasing concentrations (values on x-axis in C) of G4T4G4 PNA. Increasing amounts of fluorescently labeled G4T4G4 PNA were bound to the excess of C4A4C4 DNA oligonucleotide (B, right lanes). Fluorescence expressed in arbitrary units (au); PNA amount expressed in picomoles. Linear regression Y=91*X was used to calculate amounts of PNA in bands corresponding to PNA-hairpin complexes 1 and 2 (B, left lanes). Arrows 1 and 2 indicate the two shifted bands. (C) Calibration curve of the fluorescence of the bands in the gel versus amount of G4T4G4 PNA.
To assess the stoichiometry of these complexes, we titrated PNA with known amounts of complementary C4A4C4 DNA and produced a calibration curve of the total fluorescent signal of the bands versus the amount of PNA with the assumption that PNA binding to DNA was complete (Fig. 3C). Using this calibration curve and knowing the amount of 32P-labeled hairpin DNA, we calculated the molar ratio of PNA to DNA in complex 1 (upper band) as 3:1, and 1:1 in complex 2 (lower band). These estimations are based on the assumption that the quantum yields of the fluorescein dye are not considerably different for PNA–DNA duplex and PNA–DNA quadruplex complexes.
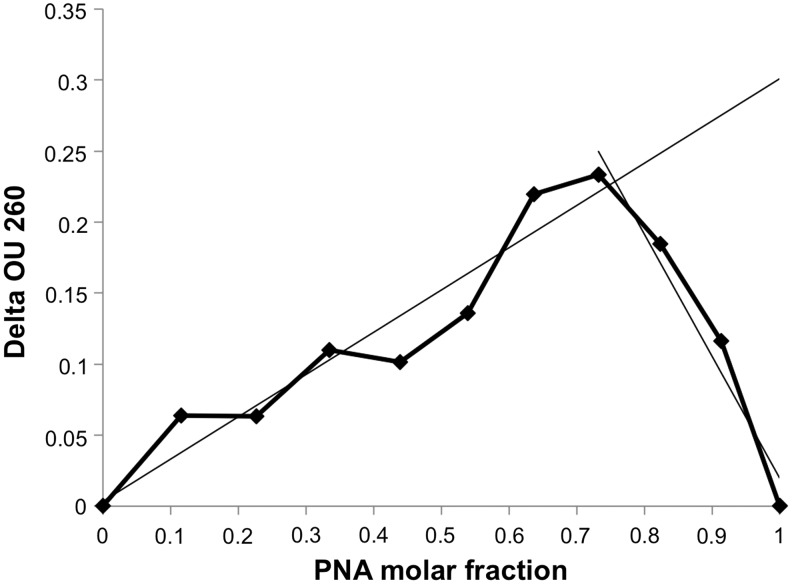
Using ultraviolet spectroscopy, we also studied PNA–DNA hairpin binding at various molar ratios but constant total concentration of PNA and DNA. The results of these experiments are shown in Fig. 4 as a Job plot. They confirm that the major complex between PNA and DNA hairpin has a 3:1 stoichiometry. Examples of such quadruplex complexes are shown in Fig. 5. CD spectra of PNA–hairpin complexes (Supplementary Fig. S1) show an increase in the maximum at 265 nm and decrease in the minimum at 240 nm compared with the CD spectrum of the duplex. This is consistent with a parallel quadruplex formation. Therefore, we conclude that the major quadruplex complex between PNA and DNA hairpin consists of three PNA and one DNA molecules most likely in a parallel orientation (3:1-P, Fig. 5), while the minor complex is an antiparallel quadruplex with 1:1 PNA to DNA ratio.
FIG. 4.
Job plot of G4T4G4 PNA and DNA hairpin mixtures. Difference in the absorbance at 260 nm in optical units (Delta OU 260) between PNA–DNA mixtures and the sum of the absorbance of individual PNA and DNA solutions plotted versus the PNA to DNA molar ratio. Total concentration of PNA and DNA was kept constant at 3 μM.
FIG. 5.
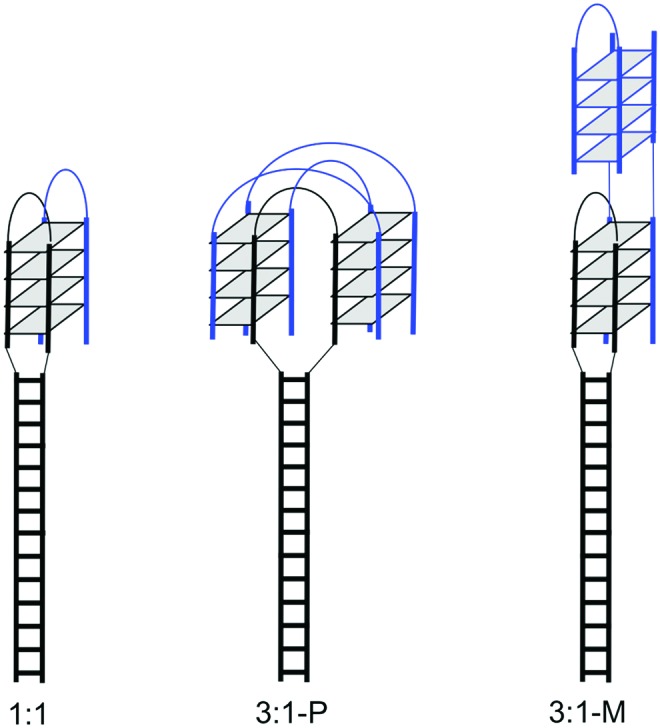
Strand arrangements in PNA–hairpin DNA complexes with 1:1 and 3:1 stoichiometries. Color images available online at www.liebertpub.com/nat
Binding G4T4G4 PNA and to a plasmid DNA containing G4T4G4 insert
We then studied G4T4G4 PNA binding to pGT4 plasmid DNA with a G4T4G4/C4A4C4 insert. The construction of the plasmid is described in the Materials and Methods. Due to the difficulty of synthesizing a plasmid containing ppG residues, we used ppG-substituted PNA to probe quadruplex formation in these experiments. Plasmid–PNA mixtures were analyzed in agarose gel (Fig. 6). Lane 1 on all panels shows binding with G4T4G4 PNA, and lane 2, ppG4T4ppG4 PNA. Both PNAs were labeled with fluorescein. Gels on the top and middle panels were not stained; only the fluorescent signal of PNA is visible. Clearly, G4T4G4 PNA binds considerably better to the plasmid than ppG4T4ppG4 PNA that cannot form quadruplexes (top panel). After PNA–plasmid complexes were cut with AlwN1 restriction enzyme producing two fragments (1995 bp and 1533 bp), the fluorescence of the bottom fragment containing the G4T4G4 insert is considerably higher (middle panel, lane 1). This reflects the G4T4G4 PNA binding specificity to the G4T4G4 insert. However, some nonspecific binding to the top fragment without the insert also occurred. The absence of fluorescent signal in lane 2 (middle panel) after AlwN1 digestion shows that ppG4T4ppG4 PNA has weaker affinity to the plasmid compared to G4T4G4 PNA and that the binding was stabilized by supercoiling. We believe that ppG–PNA binds only to the C4A4C4 strand of the insert, but this invasion is not stabilized by quadruplex formation with the G4T4G4 strand.
FIG. 6.
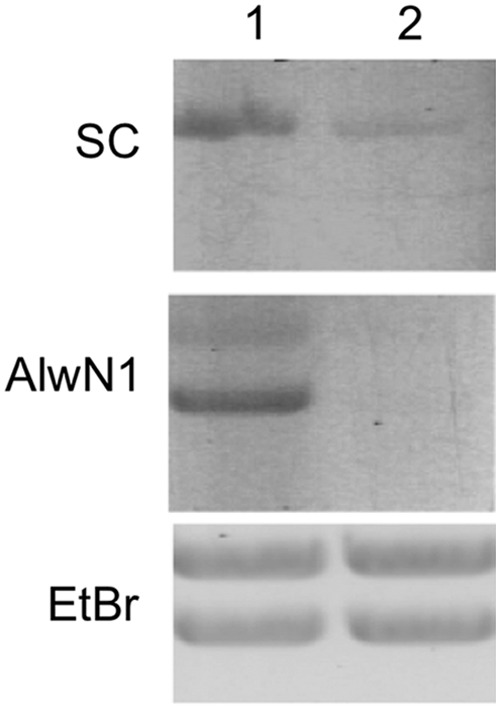
Binding fluorescently labeled G4T4G4 PNA and ppG4T4ppG4 PNA to pGT4 plasmid: Lane 1, binding with G4T4G4 PNA; lane 2, binding with ppG4T4ppG4 PNA. Top panel: native supercoiled plasmid DNA (SC), gel was unstained, PNA fluorescence was detected. Middle panel: pGT4 digested with AlwN1 restriction enzyme following PNA binding, gel was unstained, only PNA fluorescence was detected. Bottom panel: same gel as in the middle panel stained with ethidium bromide (EtBr).
To further assess the binding specificity of G4T4G4 PNA, the plasmid was cut by HindIII and Xho1 restriction enzymes resulting in the two fragments; the 110-bp fragment (containing the G4T4G4 insert) and the 3418-bp fragment. As a control, the binding was performed with another plasmid, pCRBcl2, based on the same pCRblunt vector with an insert of a quadruplex-forming sequence from the Bcl2 gene. This insert contains sequence GGGGCGGAGGGG of two G4 repeats separated by 4 nucleotides but not four Ts as in pGT4 [12]. The resulting fragments were end-labeled with 32P-dATP and separated in 6% PAGE (Fig. 7). The 110-bp G4T4G4 insert-containing fragment of pGT4 was partially shifted in the sample that was preincubated with PNA (lane 2). The fragment of pCRBcl2 containing Bcl2 quadruplex showed no band-shift, demonstrating the sequence-specificity of G4T4G4 PNA binding. Notably, complexes of PNA with the target sequence were stable during the reactions with the restriction enzymes, 32P labeling, and electrophoresis. Interestingly, there appears to be two shifted bands, possibly corresponding to the two binding modes of PNA to DNA loops. Sequence-specific binding of G4T4G4 PNA to the target sequence in the plasmid was also confirmed by S1 nuclease probing experiments (Supplementary Fig. S3).
FIG. 7.
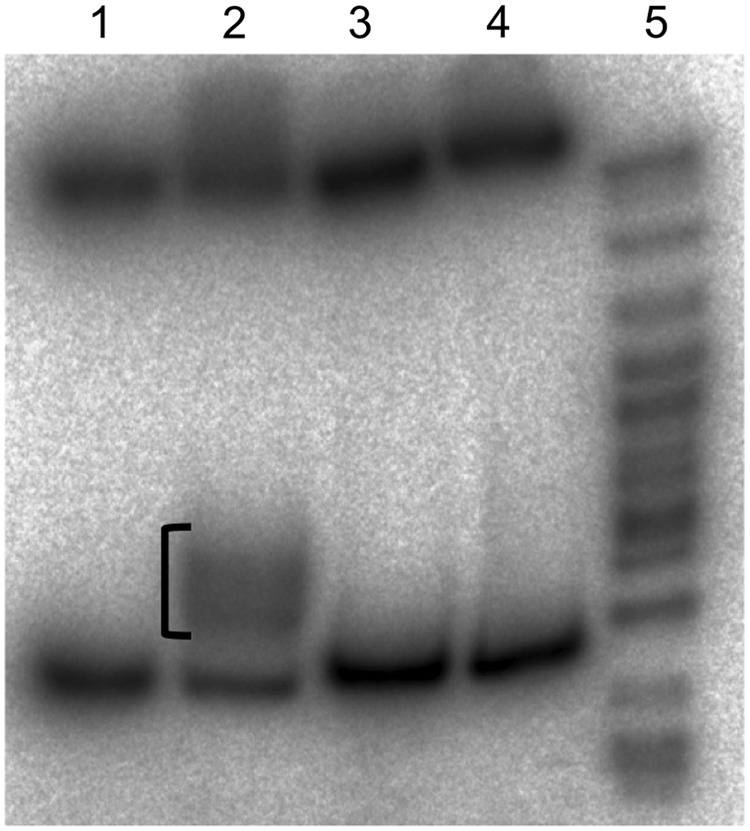
Binding G4T4G4 PNA to pGT4 and pCRBcl2. After binding plasmids were digested with HindIII and XhoI restriction enzymes and 32P-labeled. Autoradiography of 6% polyacrylamide gel is shown. Lane 1, pGT4 without PNA; lane 2, pGT4 with G4T4G4 PNA; lane 3, pCRBcl2 without PNA; lane 4, pCRBcl2 with G4T4G4 PNA; lane 5, pBR322/Msp1 32P-labeled molecular weight marker. PNA-shifted bands in lane 2 are marked with the bracket.
Discussion
Using three different DNA models we have demonstrated that G-quadruplex formation between PNA and DNA oligomers is more complicated than previously thought. In particular, the G4T4G4 PNA and DNA oligonucleotides form multiple complexes with different mobility in the gel most likely due to variations in PNA:DNA stoichiometry. Interaction of G4T4G4 PNA with G4T4G4 DNA placed in the loop of a hairpin structure resulted in formation of two distinct quadruplexes with 3:1 (major complex) and 1:1 (minor complex) stoichiometries. CD spectra implied that in the major quadruplex DNA and PNA strands are in parallel orientation. Chemical probing experiments showed that the invasion of PNA into the loop did not result in the melting of the hairpin. Based on these data, we propose a structure for this major PNA–DNA hairpin quadruplex (Fig. 5).
It has been shown that DNA supercoiling considerably enhances PNA invasion [23]. In our experiments we did not observe a significant invasion of PNA into the linear plasmid molecules (data not shown). However, PNA did invade supercoiled plasmid and the complexes remained stable even after linearization of the plasmid (Fig. 7). Our results also demonstrate that the G-quadruplex formation is required for the invasion of G4T4G4 PNA into the target G4T4G4/C4A4C4 sequence inserted into plasmid DNA. Gel-shift assays (Fig. 7) showed that at least two quadruplex complexes with different mobilities were formed. This could be 1:1 and 3:1 PNA:DNA complexes as shown in Fig. 8. However, we also cannot exclude 2PNA–DNA triplex formation in the C-strand [24].
FIG. 8.
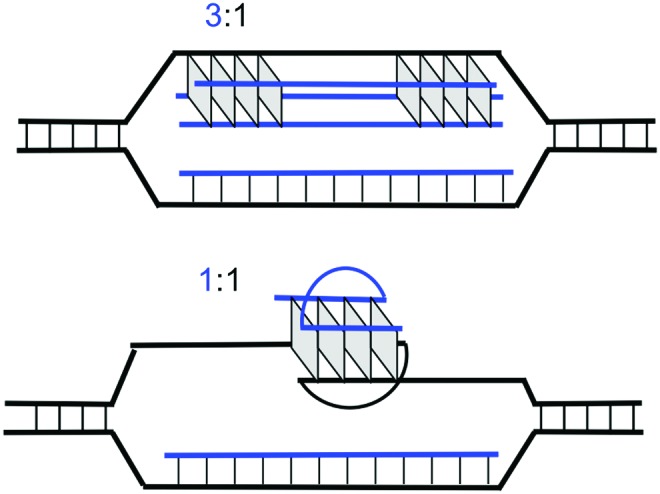
Two possible complexes of PNA invasion into plasmid DNA with 3:1 and 1:1 PNA:DNA stoichiometry. Color images available online at www.liebertpub.com/nat
At the same time, we observed PNA invasion into other locations in the plasmid. One of the major problems facing the design of DNA-targeting probes is the sequence selectivity (reviewed by Demidov et al. [25]). A short oligonucleotide probe will match numerous sequences in a large genome, while longer probes tend to have a lower selectivity due to the higher mismatch tolerance. Targeting DNA secondary structures (like G-quadruplexes) could significantly improve the sequence selectivity. However, this strategy faces the same probe length problem. Even in plasmid DNA, we observed some nonspecific binding, most likely because G4T4G4 PNA oligomers were able to invade numerous sequences containing three or more adjacent guanines. Three or four adjacent guanines in PNA oligomers can form a stable quadruplex with homologous sequences in DNA, diminishing sequence selectivity. In this case, the specificity of targeting is only reliant on the complementary pairing of the C-strand with the non-G-quadruplex-forming nucleotides between the runs of guanines in PNA (loop nucleotides). Because the complexes with 3:1 stoichiometry PNA do not fold back but rather stretch along DNA, such complexes can accommodate longer PNA. In addition, the interaction between loop nucleotides and DNA can be utilized for extra sequence specificity. Such interaction is prohibited in the complexes with 1:1 stoichiometry because the loops would lose needed flexibility. Of course, if the loop nucleotides are complementary to the G-quadruplex forming strand then they are not complementary to the C-rich strand. However, the nonspecific binding to the off-target sites that we observed in our experiments may indicate that only a portion of PNA (i.e. a run of Gs) is required for the initial invasion to the C-rich strand in order to initiate the heteroquadruplex formation.
We used G4T4G4 as a model sequence in our experiments because it forms stable quadruplexes that are easy to study with such methods as electrophoresis. Nonetheless, G4N4G4 sequences are very common among the potential quadruplex-forming sequences in human genome. We found total 8959 G[3-5]N[3-5]G[3-5]N[3-5]G[3-5]N[3-5]G[3-5] sequences within 2 kb of gene coding regions in human genome. Of these, 6372 contain G4N4G4 motif; G4T4G4 sequences are found in the GNAI3, SMARCC2, and PLA2G4D genes. It is worth noting that our target sequence has only 2 runs of 4 guanines and, therefore, cannot form an intramolecular quadruplex. In this case, the invasion of PNA results in G-quadruplex extrusion in genomic loci that do not contain potential quadruplex-forming sequences. This could be a useful tool for understanding the role of quadruplex structures in gene regulation and other genetic processes. Further studies will explore if these results extend to longer G-rich DNA sequences that are known to form intramolecular quadruplexes. In this case, more stable PNA–DNA invading complexes containing multiple PNA strands could be envisioned.
In conclusion, our findings extend the repertoire of the PNA-based DNA sequence specific probes, and may lead to the design of longer and more selective DNA-invading PNA.
Supplementary Material
Acknowledgments
This research was supported by the Intramural Research Program of the National Institutes of Health Clinical Center and National Institute of Diabetes and Digestive and Kidney Diseases.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Belotserkovskii BP, Mirkin SM. and Hanawalt PC. (2013). DNA sequences that interfere with transcription: Implications for genome function and stability. Chem Rev 113:8620–8637 [DOI] [PubMed] [Google Scholar]
- 2.Bochman ML, Paeschke K. and Zakian VA. (2012). DNA secondary structures: stability and function of g-quadruplex structures. Nat Rev Genet 13:770–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Panyutin IG, Onyshchenko MI, Englund EA, Appella DH. and Neumann RD. (2012). Targeting DNA g-quadruplex structures with peptide nucleic acids. Curr Pharm Des 18:1984–1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balasubramanian S, Hurley LH. and Neidle S. (2011). Targeting g-quadruplexes in gene promoters: A novel anticancer strategy? Nat Rev Drug Discov 10:261–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nielsen MC. and Ulven T. (2010). Macrocyclic g-quadruplex ligands. Curr Med Chem 17:3438–3448 [DOI] [PubMed] [Google Scholar]
- 6.Parrotta L, Ortuso F, Moraca F, Rocca R, Costa G, Alcaro S. and Artese A. (2014). Targeting unimolecular g-quadruplex nucleic acids: a new paradigm for the drug discovery? Expert Opin Drug Discov 9:1167–1187 [DOI] [PubMed] [Google Scholar]
- 7.Vy Thi Le T, Han S, Chae J. and Park HJ. (2012). G-quadruplex binding ligands: From naturally occurring to rationally designed molecules. Curr Pharm Des 18:1948–1972 [DOI] [PubMed] [Google Scholar]
- 8.Roy S, Tanious FA, Wilson WD, Ly DH. and Armitage BA. (2007). High-affinity homologous peptide nucleic acid probes for targeting a quadruplex-forming sequence from a myc promoter element. Biochemistry 46:10433–10443 [DOI] [PubMed] [Google Scholar]
- 9.Paul A, Sengupta P, Krishnan Y. and Ladame S. (2008). Combining g-quadruplex targeting motifs on a single peptide nucleic acid scaffold: a hybrid (3+1) PNA-DNA bimolecular quadruplex. Chemistry 14:8682–8689 [DOI] [PubMed] [Google Scholar]
- 10.Amato J, Pagano B, Borbone N, Oliviero G, Gabelica V, Pauw ED, D'errico S, Piccialli V, Varra M, Giancola C, Piccialli G, and Mayol L. (2011). Targeting g-quadruplex structure in the human c-kit promoter with short PNA sequences. Bioconjug Chem 22:654–663 [DOI] [PubMed] [Google Scholar]
- 11.Zhang X, Ishihara T. and Corey DR. (2000). Strand invasion by mixed base PNAs and a PNA-peptide chimera. Nucleic Acids Res 28:3332–3338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Onyshchenko MI, Gaynutdinov TI, Englund EA, Appella DH, Neumann RD. and Panyutin IG. (2009). Stabilization of g-quadruplex in the bcl2 promoter region in double-stranded DNA by invading short pnas. Nucleic Acids Res 37:7570–7580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Englund EA, Gupta P, Micklitsch CM, Onyshchenko MI, Remeeva E, Neumann RD, Panyutin IG. and Appella DH. (2014). Ppg peptide nucleic acids that promote DNA guanine quadruplexes. Chembiochem 15:1887–1890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ishizuka T, Yang J, Komiyama M. and Xu Y. (2012). G-rich sequence-specific recognition and scission of human genome by pna/DNA hybrid g-quadruplex formation. Angew Chem Int Ed Engl 51:7198–7202 [DOI] [PubMed] [Google Scholar]
- 15.Klobutcher LA, Swanton MT, Donini P. and Prescott DM. (1981). All gene-sized DNA molecules in four species of hypotrichs have the same terminal sequence and an unusual 3′ terminus. Proc Natl Acad Sci (USA) 78:3015–3019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Englund EA. and Appella DH. (2007). Gamma-substituted peptide nucleic acids constructed from l-lysine are a versatile scaffold for multifunctional display. Angew Chem Int Ed Engl 46:1414–1418 [DOI] [PubMed] [Google Scholar]
- 17.He Y, Neumann RD. and Panyutin IG. (2004). Intramolecular quadruplex conformation of human telomeric DNA assessed with 125i-radioprobing. Nucleic Acids Res 32:5359–5367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sambrook J, Fritsch EF. and Maniatis T, eds. Molecular cloning, 2nd ed. (1989) Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 19.Maxam AM. and Gilbert W. (1980). Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol 65:499–560 [DOI] [PubMed] [Google Scholar]
- 20.Datta B, Schmitt C. and Armitage BA. (2003). Formation of a pna2-dna2 hybrid quadruplex. J Am Chem Soc 125:4111–4118 [DOI] [PubMed] [Google Scholar]
- 21.Belousov ES, Afonina IA, Kutyavin IV, Gall AA, Reed MW, Gamper HB, Wydro RM. and Meyer RB. (1998). Triplex targeting of a native gene in permeabilized intact cells: Covalent modification of the gene for the chemokine receptor ccr5. Nucleic Acids Res 26:1324–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gehring K, Leroy JL. and Gueron M. (1993). A tetrameric DNA structure with protonated cytosine. Cytosine base pairs. Nature 363:561–565 [DOI] [PubMed] [Google Scholar]
- 23.Bentin T. and Nielsen PE. (1996). Enhanced peptide nucleic acid binding to supercoiled DNA: Possible implications for DNA “breathing” dynamics. Biochemistry 35:8863–8869 [DOI] [PubMed] [Google Scholar]
- 24.Kuhn H, Demidov VV, Nielsen PE. and Frank-Kamenetskii MD. (1999). An experimental study of mechanism and specificity of peptide nucleic acid (PNA) binding to duplex DNA. J Mol Biol 286:1337–1345 [DOI] [PubMed] [Google Scholar]
- 25.Demidov VV. and Frank-Kamenetskii MD. (2004). Two sides of the coin: affinity and specificity of nucleic acid interactions. Trends Biochem Sci 29:62–71 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.