Abstract
Background:
Disturbances in circadian rhythm-related physiological and behavioral processes are frequently observed in depressed patients and several clock genes have been identified as risk factors for the development of mood disorders. However, the particular involvement of the circadian system in the pathophysiology of depression and its molecular regulatory interface is incompletely understood.
Methods:
A naturalistic animal model of depression based upon exposure to chronic mild stress was used to induce anhedonic behavior in mice. Micro-punch dissection was used to isolate basolateral amygdala tissue from anhedonic mice followed by quantitative real-time PCR–based analysis of gene expression.
Results:
Here we demonstrate that chronic mild stress-induced anhedonic behavior is associated with disturbed diurnal oscillation of the expression of Clock, Cry2, Per1, Per3, Id2, Rev-erbα, Ror-β and Ror-γ in the mouse basolateral amygdala. Clock gene desynchronization was accompanied by disruption of the diurnal expressional pattern of vascular endothelial growth factor A expression in the basolateral amygdala of anhedonic mice, also reflected in alterations of circulating vascular endothelial growth factor A levels.
Conclusion:
We propose that aberrant control of diurnal rhythmicity related to depression may indeed directly result from the illness itself and establish an animal model for the further exploration of the molecular mechanisms mediating the involvement of the circadian system in the pathophysiology of mood disorders.
Keywords: chronic mild stress, depression, clock gene, amygdala, VEGF
Introduction
Mood disorder patients frequently present with disturbances in circadian rhythm-related physiological and behavioral processes, such as alterations in sleep/wake cycles, daily dependent patterns of body temperature and hormone release and diurnal mood changes (as reviewed in Bunney and Bunney, 2013). Conversely, disruptions of circadian rhythms, as resulting from shift work, have been reported to negatively affect mood and emotionality (Gordon et al., 1986; Costa, 1997; Choobineh et al., 2006). In experimental animals, light deprivation has been found to induce depression-like behavior together with cellular and molecular alterations related to mood disorders (Gonzalez and Aston-Jones, 2008). Correspondingly, bright light therapy has been proven to be effective as a nonpharmacological intervention for the treatment of specific forms of depression, mainly seasonal affective disorders (as reviewed in Golden et al., 2005).
In mammals, the endogenous molecular circadian rhythm is organized by autonomously regulated clock genes. These genes form a complex mechanism of positive and negative transcription-translation feedback loop which is orchestrated by the suprachiasmatic nucleus (SCN) of the hypothalamus as the master neuronal circadian pacemaker (as reviewed in Albrecht, 2012). Interestingly, the functional activity of the SCN is not only regulated by light but is also subject to modulation by emotion- related physiological states (Lamont et al., 2005). Clock genes have also been found to be expressed in other areas of the brain (Lamont et al., 2005; Yang et al., 2007; Jilg et al., 2010) as well as nonneuronal tissues (Storch et al., 2002).
With regard to the association between emotions and alterations thereof in mood disorders and circadian rhythms, there are reports on the expression of clock genes in brain areas forming part of the neural circuitry involved in depression. As such, in the amygdala, which is critically involved in the expression and modulation of emotions and associated to the pathophysiology of depression (as reviewed in Ressler and Mayberg, 2007), clock gene expression has been reported both in experimental animals and human samples (Ino, 2004; Lamont et al., 2005; Kitajima et al., 2006; Segall et al., 2009; Mozhui et al., 2010; Wyse and Coogan, 2010; Li et al., 2013). Specifically, a seminal study recently described a flattening in the circadian pattern of gene expression in human amygdala tissue derived from postmortem brains of major depressive disorder patients (Li et al., 2013).
We here aimed to build upon this work by investigating whether comparable alterations could be observed in a naturalistic animal model of the disorder which would allow to further investigate the molecular basis of the described association between circadian abnormalities and mood disorders (Li et al., 2013). To this end we used the chronic mild stress (CMS) model of depression to systematically evaluate clock gene rhythmicity and its modulation by induction of a depressive-like state in the mouse basolateral amygdala (BLA). Specifically, given the critical importance of growth factor support and deficiencies thereof in the pathophysiology of depression, we investigated whether the vascular endothelial growth factor (VEGF)—which has been recently identified as a direct target of a circadian regulatory gene (Jensen and Cao, 2013) and associated with stress-related disorders, including depressive disorders (Kahl et al., 2009; Elfving et al., 2010; Khan et al., 2014)—showed diurnally oscillating expression patterns in the mouse BLA and might be linked to stress-induced anhedonia.
Methods
Animals
C57BL/6N male mice (Charles-River Laboratories, Germany), aged 8 to 10 weeks at the onset of experiments, were used in all instances. All animals were naïve, that is, without any prior manipulation, at the onset of experiments. Mice were single-housed in a sound-attenuated room with constant temperature of approximately 21°C under a 12-h–light/–dark cycle with light on at 7:00 AM (defined as Zeitgeber Time [ZT] 0). The light intensity at the level of the animals’ cages was approximately 200 lux. Food and water were available ad libitum throughout the experiment, unless otherwise specified. All experiments were designed to reduce animal suffering and keep the number of animals used at the minimum level. Sample sizes were similar to those reported in previous studies (Griesauer et al., 2014; Khan et al., 2014). Animal experiments described in this study were approved by the national ethical committee on animal care and use (GZ-66.009/0215-II/10b/2009; Bundesministerium für Wissenschaft und Forschung) and carried out according to international laws and policies.
Behavioral Paradigms
CMS Procedure
Upon delivery of mice to the local animal facility, all animals were randomly (alternating within each cage) assigned to one of the two experimental groups (CMS and control, respectively), and behavioral assessments were carried out blind to the experimental conditions. The CMS procedure was based on the protocol described by Strekalova et al., 2004 and was carried out with minor modifications. Briefly, for 4 weeks mice were daily exposed to one of the following stressors: exposure to rat (15 hours), space restraint (in a plastic transparent tube with a 2.6-cm internal diameter; 2 hours) and tail suspension (6 minutes). Stressors were applied in the following order: exposure to rat (days 1–7), restraint (days 8–10), exposure to rat (days 11–17), tail suspension (days 18–22), restraint (days 23–25), and tail suspension (days 26–28). All procedures, except the rat exposure (overnight), were carried out during the light phase. Body weight measurements and fur state were monitored weekly. Control animals were handled only for several minutes every day.
Sucrose Preference Test
The Sucrose Preference Test (SPT) was carried out essentially as previously described (Khan et al., 2014). Briefly, during a 4-day training phase, mice were habituated to drink a 2% sucrose solution. The day before the SPT, mice were deprived of food and water for 18 hours. During the test, subjects were given a free choice between 2 bottles, one with the sucrose solution and the other with water. Mice were tested for 3 hours, starting at 9:00 am. To prevent possible effects of side preference in drinking behavior, the position of the bottles (right/left) was alternated between animals. Total liquid consumption was measured by weighing the bottles before and after the SPT. Sucrose preference was calculated as percentage of sucrose solution consumed relative to the total amount of liquid intake.
Tissue Dissection
Mice were sacrificed by cervical dislocation at 6 equally spaced time points during a 24-hour interval and brains were rapidly removed and frozen on dry ice. Samples of the BLA were isolated using a micro-punch procedure. Briefly, 3 brain coronal sections of 300 μm, starting at −0.94mm from bregma point (Paxinos and Franklin, 2001), were collected. Then, 3 bilateral samples of BLA were extracted with a blunted 0.69-mm–diameter sample cannula (Fine Science Tools, Germany), put into 700 μL Qiazol lysis buffer, vortexed and kept at −80°C until use.
RNA Purification and qRT-PCR
Total RNA from the BLA was isolated using a miRNA Micro kit (QUIAGEN, CA), amplified using T-Script reverse transcriptase (QuantiTect Whole Transcriptome, QUIAGEN, CA), and quantified by the Quant-iT PicoGreen dsDNA Reagent (Invitrogen CA). Then 2 μL of cDNA from each sample were subjected to quantitative real-time PCR (qRT-PCR) using Power SYBR Green PCR Master Mix (Applied Biosystems, CA) according to the recommendations of the manufacturer. Transcription levels of target genes were assayed in duplicates and normalized against the amount of β-actin mRNA (delta cycle threshold (dCT)). Primer sequences for mouse clock genes were as follows: mus_Clock, forward, 5’-GGCGTTGTTGATTGGACTAGG-3’, reverse, 5’-GAATGGAGTCTCCAACACCCA-3’; mus_Bmal1, forward, 5’-AA CCTTCCCGCAGCTAACAG-3’; reverse, 5’-AGTCCTCTTTGGGCC ACCTT-3’; mus_Cry1, forward, 5’-AGGAGGACAGATCCCAATG GA-3’; reverse, 5’-GCAACCTTCTGGATGCCTTCT-3’; mus_Cry2, forward, 5’-AGCTGATGTGTTCCCAAGGCT-3’; reverse, 5’-CATAATG GCTGCATCCCGTT-3’; mus_Per1, forward, 5’-CCAGATTGGTGGAG GTTACTGAGT-3’; reverse, 5’-GCGAGAGTCTTCTTGGAGCAGT AG-3’; mus_Per2, forward, 5’-AGAACGCGGATATGTTTGCTG-3’; reverse, 5’-ATCTAAGCCGCTGCACACACT-3’; mus_Per3, forward, 5’-CCGCCCCTACAGTCAGAAAG-3’; reverse, 5’-GCCCCACGTGCTT AAATCCT-3’; mus_Id2, forward 5’-AGGCATCTGAATTCCCTTCT GA-3’; reverse, 5’-AGTCCCCAAATGCCATTTATTTAG-3’; mus_Rev-erb α, forward, 5’-CCCTGGACTCCAATAACAACACA-3’; reverse, 5’-GCCATTGGAGCTGTCACTGTAG-3’; mus_Rev-erb β, forward, 5’-GGAACGGACCGTCACCTTT-3’; reverse, 5’-TCCCCTGCTCCCA TTGAGT-3’; ROR-α, forward, 5’-TTGCCAAACGCATTGATGG-3’; reverse, 5’-TTCTGAGAGTCAAAGGCACGG-3’; ROR-β, forward, 5’-ATGGCAGACCCACACCTACG-3’; reverse, 5’-TATCCGCTTGGCGA ACTCC-3’; ROR-γ, forward, 5’-CGAGATGCTGTCAAGTTTGGC-3’; reverse, 5’-TGTAAGTGTGTCTGCTCCGCG-3’; Dec1, forward, 5’-CC CGTCTCTGATGAATAAAGACCA-3’; reverse, 5’-GGACAGCATGCC GTAGAAGTGA-3’; Dec2, forward, 5’-ATGAATGCATTGCTCAGCTG AAAG-3’; reverse, 5’-GCTGCTGCTCAGTTAAGGCTGTTAG-3’; mus_ E4bp4, forward, 5’-AGAACCACGATAACCCATGAAAG-3’; reverse, 5’-GACTTCAGCCTCTCATCCATCAA-3’; mus_Npas2, forward, 5’-AC GCAGATGTTCGAGTGGAAA-3’; reverse, 5’-CGCCCATGTCAAGTGC ATT-3’; mus_Neuro D1, forward, 5’-CGAGTCATGAGTGCCCAGC TTA-3’; reverse, 5’-CCGGGAATAGTGAAACTGACGTG-3’. Primers used for amplification of mouse VEGFA, VEGFB, VEGFC, VEGFR1, VEGFR2, and VEGFR3 were previously described (Catteau et al., 2011).
Enzyme-Linked Immunosorbent Assay
Animals were deeply anaesthetized with an intraperitoneal injection of ketamine/xylazine (100/40mg/kg), and blood samples were collected by cardial puncture at 6 equally spaced time points as described above. Then 15 μL of heparin (Sigma, MO) wer immediately added to blood samples, and plasma was obtained by centrifugation for 5 minutes at 14000rpm at 4°C. VEGFA concentration levels were determined using a commercially available assay kit following the manufacturer’s instructions (R&D Systems, MN). All samples were analyzed in duplicates. Photometric quantification was performed on an enzyme-linked immunosorbent assay plate reader (Stat Fax-2100, Awareness Technologies, CT).
Immunohistochemistry
Mice were deeply anaesthetized with an intraperitoneal injection of ketamine/xylazine (100/40mg/kg) at ZT12 and perfused intracardially with 4% paraformaldehyde. Brains were removed, cryoprotected for 48 hours and cut into 30 μm–thick coronal sections at a section interval of 60 μm. Free-floating sections were incubated in 10mM citrate buffer (pH 6) for 30 minutes at 70°C for antigen retrieval. Sections were then blocked in Tris-buffered saline solution containing 0.25% Triton Tx-100, 2.5% bovine serum albumin, and 2% normal goat serum for 1 hour, prior to an overnight incubation at 4°C with a rabbit polyclonal antibody against CLOCK (1:500; PA1–520, Thermo Scientific, MA). Slices were then incubated for 2 hours with Alexa Fluor 488 goat anti-rabbit (1:1000, Invitrogen, MA) at room temperature in Tris-buffered saline solution containing 0.25% Triton, 2.5% bovine serum albumin, and 2% normal goat serum for 1 hour. Slices were incubated with red Fluoro Myelin (1:300; Molecular Probes, OR) for 20 minutes, then mounted and cover-slipped using Fluorogel with Tris mounting medium (Electron Microscopy Sciences, PA).
Image Analysis
Tissue sections containing the BLA were analyzed on a confocal microscope (Zeiss Axiovert 200, Zeiss, Germany). Immunofluorescent signals were sequentially captured using the 488 (green) and 543 (red) channels and processed using Zeiss LSM510 Image Browser. Eight images of the BLA (4 right and 4 left) were evaluated for each subject, starting at −0.94mm from bregma point (Paxinos and Franklin, 2001). NIH ImageJ software (National Institute of Health, MD) was used for image processing and densitometric analysis of positive cells. BLA borders were delineated according to the Fluoro Myelin staining and the mouse brain atlas (Paxinos and Franklin, 2001). Background staining was subtracted from each image. Profile of expression was determined by densitometric analysis of particles, represented by CLOCK immunopositive cells. For each particle, the integrated density (mean grey value×area, expressed in μm2) was measured. Immunoreactivity for each slice was obtained by the average of the integrated density within that slice.
Data Collection and Statistical Analysis
Cosinor analysis was performed to determine significant circadian patterns in mRNA expression. Data were analyzed using CircWave statistic software (V1.4) according to the equation Y=c+aSIN (i2π t/24)+bCOS (i2π t/24), where Y is delta cycle threshold, t is ZT, and a, b, and c were predicted. Further nonlinear regression analysis was used to determine peak time, trough, and amplitude of the curve, using the center of gravity parameter. Comparisons of mRNA expression levels were made using a 2-way analysis of variance (ANOVA), with treatment and ZT as factors. For VEGF protein studies, the relative expression of VEGF was analyzed by 2-way ANOVA, with treatment and ZT as factors. Post-hoc pairwise comparisons, with Bonferroni correction for multiple comparisons were conducted where appropriate. For statistical analysis of CLOCK protein expression, data were tested for normality using the Kolmogorov–Smirnov test followed by an unpaired 2-tailed Student’s t test. Reductions in sucrose preference and body weight during the experimental period were analyzed by 1-way ANOVA followed by posthoc pairwise comparisons, with Bonferroni correction for multiple comparisons. All data were analyzed using SPSS (SPSS 18.0, IBM, NY) statistical software with the alpha level set at 0.05 at all instances.
Results
Diurnal Pattern of Clock Gene Expression in the Mouse BLA Is Disrupted in Anhedonic Mice
To first assess the presence and potential cyclic rhythmicity of the expression of 18 clock genes constituting major elements of the endogenous molecular circadian machinery in the BLA, brain samples of control mice were collected every 4 hours for a 24-hour period (7:00 am defined as ZT 0). qRT-PCR analysis indicated the expression of all the clock genes examined in the mouse BLA (listed in Table 1a). Subsequent cosinor analysis revealed a statistically significant rhythmic pattern of expression for the following clock genes: Clock (P=.004) peaking in the morning (ZT4); Bmal1 (P=.005) peaking at night (ZT16); Cry2 (P=.008) peaking in the late night (ZT21); Per1 (P=.04) and Per3 (P=.0005) peaking in the morning (ZT3 and ZT4); Id2 (P=.005) peaking at night (ZT18); Rev-erbα (P=.03) peaking in the morning (ZT3); Ror-β (P=.007) peaking in the morning (ZT4); and Ror-γ (P=.03) peaking in the late night (ZT21) (Table 1b and1c).
Table 1.
Characterization of Clock Gene Expression in the Mouse BLA. (a) Comparison of cosinor analysis of the cyclic expression of 18 clock genes in basolateral amygdala (BLA) tissue between control (total of n = 49) and anhedonic (total of n = 30) mice). P values <.05 denote a statistically significant cosinor analysis fitted curve, indicating diurnally rhythmic gene expression. (b) Peak time, trough and amplitude of clock genes exhibiting diurnally rhythmic gene expression in the mouse BLA. (c) Comparison of BLA and suprachiasmatic nucleus (SCN) expression pattern of clock genes.
| 1a | |||
|---|---|---|---|
| Cosinor Analysis P Value | |||
| Gene | Control | Anhedonic | |
| Clock | .004 | .63 | |
| Bmal1 | .005 | .01 | |
| Cry1 | .07 | .97 | |
| Cry2 | .008 | .52 | |
| Per1 | .04 | .22 | |
| Per2 | .82 | .53 | |
| Per3 | .0005 | .92 | |
| Id2 | .005 | .06 | |
| Rev-erbα | .03 | .93 | |
| Rev-erbβ | .63 | .94 | |
| Ror-α | .3 | .51 | |
| Ror-β | .007 | .19 | |
| Ror-γ | .03 | .38 | |
| Dec1 | .22 | .31 | |
| Dec2 | .12 | .75 | |
| E4bp4 | .13 | .09 | |
| Npas2 | .34 | .35 | |
| NeuroD1 | .2 | .84 | |
| 1b | |||
|---|---|---|---|
| Gene | Peak (ZT) | Trough (ZT) | Amplitude |
| Clock | 4 | 16 | 1.42 |
| Bmal1 | 16/23 (anhedonic) | 4/11 (anhedonic) | 0.67/0.42 (anhedonic) |
| Cry2 | 21 | 9 | 0.48 |
| Per1 | 3 | 15 | 1.32 |
| Per3 | 4 | 16 | 1.81 |
| Id2 | 18 | 6 | 0.34 |
| Rev-erbα | 3 | 15 | 1.04 |
| Ror-β | 4 | 16 | 0.85 |
| Ror-γ | 21 | 9 | 1.47 |
| Abbreviation: ZT, Zeitgeber Time. | |||
| 1c | |||
|---|---|---|---|
| Peak (ZT) mRNA expression | |||
| Gene | BLA | SCN | |
| Clock | 4 | Constitutive | (Zheng et al., 1999; Shearman et al., 2000) |
| Bmal1 | 16 | 16–18 | (Shearman et al., 2000) |
| Cry2 | 21 | —/15 | controversial (Kume et al., 1999; Mendoza et al., 2005) |
| Per1 | 3 | 4 | (Shearman et al., 1997; Zheng et al., 1999) |
| Per3 | 4 | 6–9 | (Zylka et al., 1998; Shearman et al., 2000) |
| Id2 | 18 | 18 | (Ueda et al., 2002) |
| Rev-erbα | 3 | 4 | (Onishi et al., 2002) |
| Ror-β | 4 | 4 | (Sumi et al., 2002) |
| Ror-γ | 21 | — | not expressed in SCN (Ueda et al., 2002) |
Abbreviation: ZT, Zeitgeber Time.
To investigate whether depression-like behavior may affect diurnal patterns of clock gene expression, mice were exposed to a CMS paradigm to induce a depressive-like state. Sucrose intake was evaluated before and after the CMS procedure (SPT1 and SPT2). The mean percentage change (14.51%) in sucrose preference between SPT1 and SPT2 in control animals, which were handled only for the same period of time, was used as the criterion to separate responders (susceptible) from nonresponders (resilient).
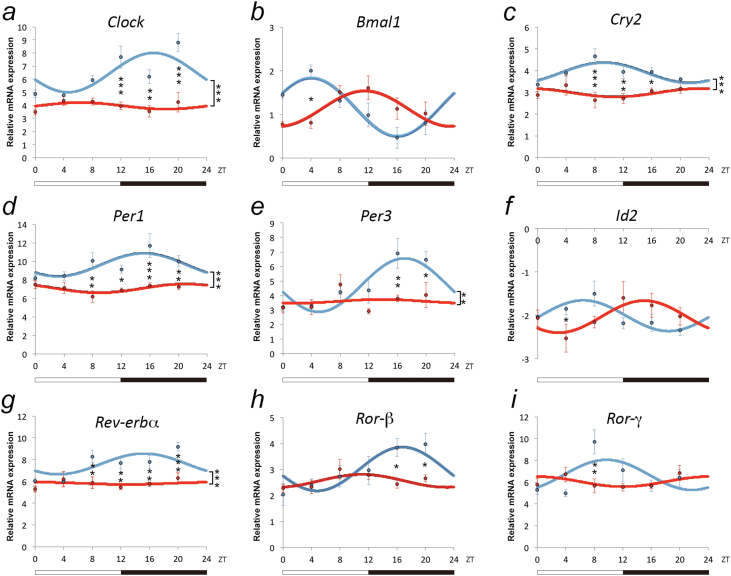
A total of 46.27% of mice responded to the CMS procedure by a significant reduction in sucrose preference (P=.001) (Figure 1a), which was paralleled by a significant reduction in the percentage of body weight gain (P=.001) during the experimental period (Figure 1b) and were termed “anhedonic.” Change in sucrose preference and percentage of body weight gain was comparable between the remaining “nonanhedonic” (53.73% of all animals) and handled-only “control” mice. Cosinor analysis of clock gene expression in the BLA of anhedonic mice revealed a disruption in the diurnal expression pattern of all clock genes analyzed (Table 1a). For all clock genes (with the exception of Bmal1, where the induction of a depressive-like state resulted in a shift in the rhythmic expression pattern as indicated by an alteration in the peak time [from ZT16 to ZT23] compared with control mice [Table 1b]), the diurnal rhythmicity in clock gene expression was abolished in anhedonic mice (Table 1a; Figure 2; Supplementary Table S1).
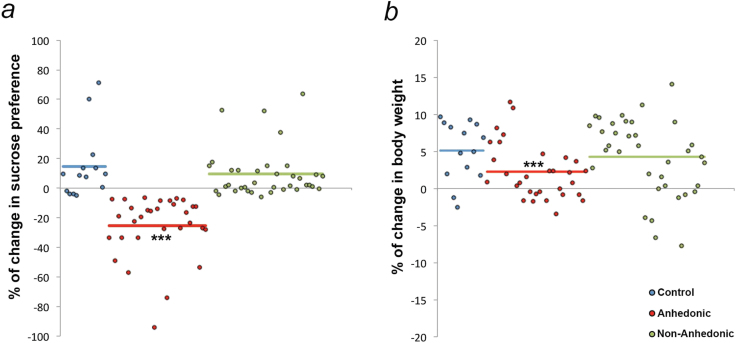
Figure 1.
Chronic mild stress (CMS) induces anhedonic behavior and reduction in body weight gain in a subset of mice. After 28 days of CMS, a subset of mice (46.37%) displayed (a) a decrease in sucrose preference compared with the baseline evaluation before the start of the CMS; and (b) a reduction in body weight gain over the experimental period and were termed “anhedonic.” Sucrose preference and percentage of body weight gain were comparable with control animals in the remaining animals (53.72%) (nonanhedonic). ***P<.001 denotes comparison by posthoc tests between anhedonic and control and nonanhedonic mice, respectively.
Figure 2.
Modulation of circadian patterns of clock genes expression in the basolateral amygdala (BLA) of anhedonic mice. Data represent relative mRNA expression (dCT) of (a) Clock, (b) Bmal1, (c) Cry2, (d) Per1, (e) Per3, (f) Id2, (g) Rev-erbα, (h) Ror-β, and (i) Ror-γ determined by quantitative real-time PCR (qRT-PCR) of BLA tissue of control (blue) and anhedonic mice (red); total of n=49 for control and n=30 for anhedonic mice. Data are reported as dCT values (inversely proportional to the amount of gene expression) and displayed as mean values ± SEM. Posthoc pair-wise comparisons with Bonferroni correction were carried out when a statistically significant interaction between treatment and Zeitgeber Time (ZT) was detected to determine differences between groups at individual time points. *P<.05, **P<.01, ***P<.001, denote differences revealed by posthoc tests.
Changes in BLA CLOCK Protein Expression Induced by Chronic Stress Parallel Alterations at the mRNA Level
To exemplarily demonstrate that the observed changes of clock gene expression in anhedonic mice were paralleled by respective alterations of protein levels, BLA protein expression of CLOCK was compared between anhedonic and control mice at ZT12 (Figure 3a). ZT12 was chosen as representative time point, since 2-way ANOVA analysis followed by Bonferroni posthoc tests of mRNA expression levels had indicated significant differences between control and anhedonic mice specifically during the night phase, at ZT12 (P=.2E-4); ZT16 (P=.001); ZT20 (P=1.56E-07), with higher Clock expression in the BLA of anhedonic mice (significant main effect of treatment (F (1,61)=50.72, P=1.43E-09), significant main effect of time points (F (5,61)=4.61, P=.001) and significant interaction between treatment and time points [F (5,61) = 3.60, P=.006]).
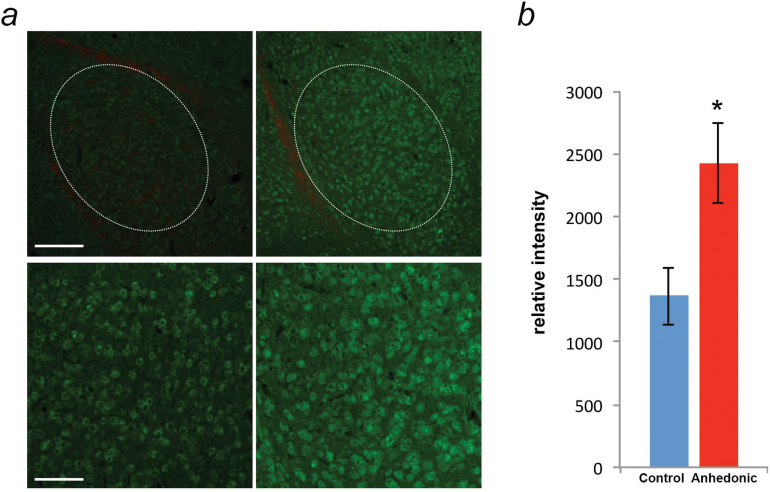
Figure 3.
Alterations of CLOCK protein expression in the basolateral amygdala (BLA) of anhedonic mice. (a) Representative immunofluorescence images of CLOCK protein expression in the BLA of control (left) and anhedonic (right) mice during late day-dusk (Zeitgeber Time [ZT]12). The 30-μm amygdala coronal sections were stained with CLOCK antibody (green) and red Fluoro Myelin (red). Dotted white lines indicate the areas analyzed in each tissue section. Scale bar: upper 200 μm, lower 100 μm. (b) Quantification of relative intensities of CLOCK protein expression in the BLA of control (n=6; blue) and anhedonic (n=7; red) mice at ZT12. Data are displayed as mean ± SEM, * P<.05.
Thus, for protein expression analysis, the CMS procedure was carried out in a parallel group of mice, and handled mice again served as controls. CLOCK was found to be expressed in the BLA of both control and anhedonic mice. Analysis of the relative intensities of CLOCK protein expression revealed a significant difference between the 2 groups (t(11)=−2.61; P=.02), with higher relative intensity in the anhedonic group (Figure 3b), thus paralleling the results at the mRNA level.
Diurnal Pattern of VEGFA Expression Is Abolished in the BLA of Anhedonic Mice
Searching for the molecular link between alterations in clock gene expression and the observed depressive-like state of anhedonic mice, we focused on the analysis of VEGF, previously described to be both directly regulated by circadian genes (Koyanagi et al., 2003) and related to the pathophysiology of depression (Kahl et al., 2009; Elfving et al., 2010; Khan et al., 2014). Thus, mRNA levels of VEGFA, VEGFB, VEGFC, and their receptors VEGFR1, VEGFR2, and VEGFR3 were analyzed by qRT-PCR at the same 6 time points used for clock gene expression evaluation in the BLA of control mice. Cosinor analysis revealed a rhythmic diurnal pattern of expression for VEGFA (P=.02) peaking in the early morning (ZT1), VEGFB (P=.007) peaking at night (ZT18), and VEGFC (P=.005) peaking at dawn (ZT23) (Table 2a-b). None of the VEGFRs analyzed was found to exhibit diurnal expression patterns (Table 2a). To investigate the effect of a depressive-like state on the rhythmicity of VEGF expression, VEGF gene expression was subsequently examined in the BLA of anhedonic mice. Cosinor analysis provided evidence that the rhythmic expression of VEGFA was completely abolished in anhedonic mice (P=.25) (Figure 4a). Although the diurnal oscillation of VEGFB (P=.009) and VEGFC (P=.01) was preserved in anhedonic mice (Figure 4b-c), a slight shift in peak time expression was observed (Table 2b; Supplementary Table S2).
Table 2.
Characterization of VEGF and VEGFR Expression in the Mouse BLA. (a) Comparison of cosinor analysis of the cyclic expression of vascular endothelial growth factor VEGFA, VEGFB, and VEGFC in basolateral amygdala (BLA) tissue (total of n = 52 for control and n = 26 for anhedonic mice). P values < .05 denote a statistically significant cosinor analysis fitted curve, indicating diurnally rhythmic gene expression. (b) Peak time, trough, and amplitude of VEGFs exhibiting diurnally rhythmic gene expression in the mouse BLA. 2a
| Gene | Cosinor Analysis P Value | |
|---|---|---|
| Control | Anhedonic | |
| VEGFA | .02 | .25 |
| VEGFB | .007 | .009 |
| VEGFC | .005 | .01 |
| VEGFR1 | .94 | |
| VEGFR2 | .39 | |
| VEGFR3 | .43 | |
| 2b | |||
|---|---|---|---|
| Gene | Peak (ZT) | Trough (ZT) | Amplitude |
| VEGFA | 1 | 13 | 1.77 |
| VEGFB | 18/2 (anhedonic) | 6/14 (anhedonic) | 0.55 |
| VEGFC | 23/1 (anhedonic) | 11/13 (anhedonic) | 2.36 |
Abbreviation: ZT, Zeitgeber Time.
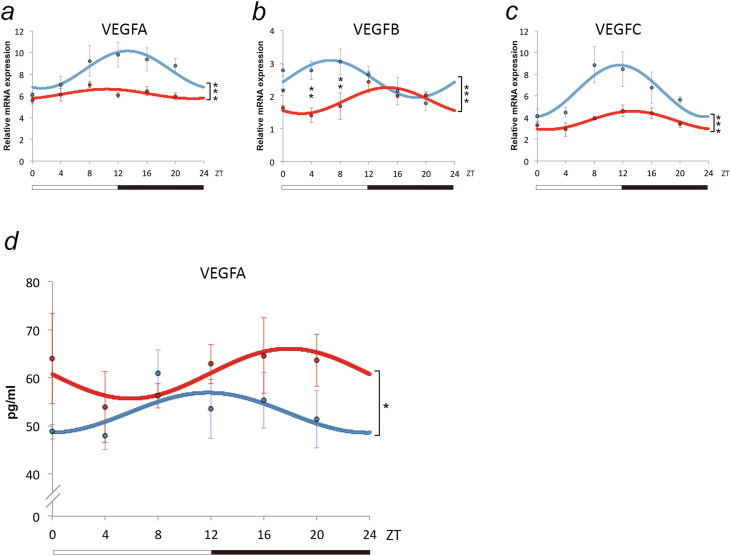
Figure 4.
Modulation of circadian patterns of vascular endothelial growth factor (VEGF) gene expression in amygdala and plasma of anhedonic mice. Data are reported as dCT values (inversely proportional to the amount of gene expression) and displayed as mean values ± SEM. Patterns of (a) VEGFA, (b) VEGFB, and (c) VEGFC determined by quantitative real-time PCR (qRT-PCR) analysis of BLA tissue of control (blue) and anhedonic mice (red); total of n=52 for control and n=26 for anhedonic mice. Two-way ANOVA analysis revealed a significant effect of treatment (ie, anhedonic vs control) for the expression of all individual VEGF genes analyzed (VEGFA, F (1,68)=16.75, P=.0001; VEGFB, F (1,71)=13.80, P=.0004; VEGFC, F (1,65)=16.33, P=.0001). Posthoc pairwise comparisons with Bonferroni correction have been carried out when a statistically significant interaction between treatment and Zeitgeber Time (ZT) was detected to determine differences between groups at individual time points. *P<.05, **P<.01, denote differences revealed by posthoc tests. (d) Concentration of VEGF at 6 time points in plasma of control mice (n=23; blue) and anhedonic mice (n=24; red), with 3 to 4 animals per time point. VEGF levels (pg/mL) were determined by enzyme-linked immunosorbent assay. Two-way analysis of variance (ANOVA) revealed a significant main effect of treatment (ie, anhedonic vs control) (F (1,35)=5.43, P<.05). Data are displayed as mean ± SEM, *P<.05.
Depressive-Like States Are Reflected in Alterations of Plasma VEGFA Levels
To test whether modulation of VEGFA expression in the BLA of anhedonic mice was paralleled by respective changes in peripheral concentrations of VEGFA, VEGFA plasma levels were analyzed at the same 6 time points in control and anhedonic mice. No diurnal rhythmicity of plasma VEGFA levels was observed (P=.33) (Figure 4d). Two-way ANOVA analysis, however, revealed a significant main effect of treatment on VEGFA concentration (F (1,35)=5.43, P=.02). The enhancement of plasma VEGFA concentration in anhedonic mice (Figure 4d) is thus paralleling the observed changes in the BLA. No significant effect of time point (F (5,35)=0.59, P=.71) or interaction between treatment and time points (F (5,35)=0.65, P=.67) was observed.
Discussion
Despite the tight association between alterations in circadian rhythms and mood disorders, a systematic investigation of the endogenous molecular circadian machinery in the amygdala, a central component of the neural circuitry involved in emotional regulation and its modulation by a depressive-like state in experimental animals has not been carried out so far.
Using CMS-induced anhedonia as a naturalistic animal model of depression, we propose that aberrant control of diurnal rhythmicity related to affective disorders may indeed directly result from the illness itself and extend from the regulation of clock genes to other molecular key players relevant to the pathophysiology of depression. This conclusion is based upon: (1) the disruption of the diurnal expressional oscillation of Clock, Cry2, Per1, Per3, Id2, Rev-erbα, Ror-β, and Ror-γ in BLA tissue of anhedonic mice; (2) an anhedonia-induced modulation of the cyclic pattern of VEGFA, VEGFB, and VEGFC expression resulting in augmented mRNA levels with a complete abolishment of VEGFA diurnal amygdala expressional oscillations; and (3) a corresponding enhancement of peripheral VEGFA concentration in anhedonic vs control mice.
Amygdala Clock Gene Expression
Reports on the presence of individual clock genes in the amygdala are available (Ino, 2004; Lamont et al., 2005; Kitajima et al., 2006; Segall et al., 2009; Mozhui et al., 2010; Wyse and Coogan, 2010). The present study for the first time simultaneously analyzed 18 canonical clock genes and showed that all of them are detectable at the mRNA level in the mouse BLA, with 9 of them showing clear diurnal rhythmicity. Of those, there are parallels to reported peak times even in other species, for example, Per1 peaks at ZT3 both in the present study in mice and in humans (Li et al., 2013). For other genes, circadian oscillations were observed in the current analysis that had not been detected by others, for example Bmal1. Vice versa, in the present study, was not observed circadian oscillation of Per2, as described in previous studies (Lamont et al., 2005; Li et al., 2013). This may relate to differences in mRNA vs protein expression, species specificity, or technical aspect of the individual detection methods applied.
To our knowledge, this is the first description of Ror-α, part of an additional regulatory loop that activates Bmal1 transcription in the SCN (Sato et al., 2004) and Ror-γ gene expression in the amygdala. The present study showed also for the first time the expression of Cry2 and Dec1, two important negative regulatory components of the core clock machinery (Vitaterna et al., 1999; Honma et al., 2002) and E4bp4, a basic leucine zipper transcription factor controlling the circadian oscillation of expression of its target genes (Mitsui et al., 2001), in the amygdala. Detection of all core clock genes examined suggests that control by the circadian system may importantly contribute to the regulation of BLA function, thereby contributing to the modulation of emotional states. The functional relevance of diurnally oscillating clock gene expression in the BLA is further validated by the fact that, as in the SCN (Shearman et al., 1997; Shearman et al., 2000), BLA Per1 expression is also at an ~180° antiphase angle to the Bmal1 expression. The overall temporal phasing of most clock genes observed in the BLA was synchronous with previous reports of the expression of the respective genes in the mouse SCN (Shearman et al., 1997, 2000; Zylka et al., 1998; Zheng et al., 1999; Onishi et al., 2002; Sumi et al., 2002; Ueda et al., 2002) (Table 1c). As in other brain regions (Jilg et al., 2010) and in peripheral tissues (Storch et al., 2002), the expression of Clock showed a clear diurnal rhythmicity in the BLA while being constitutively expressed in the SCN (Zheng et al., 1999; Shearman et al., 2000). Similarly, mRNA levels of Ror-γ, which is not expressed in the SCN (Ueda et al., 2002), robustly cycle in the BLA, as previously described in extra-neural tissues (Kang et al., 2007). Currently, is not clear why some extra-SCN cerebral clocks oscillate in phase, others in antiphase to clock gene expression in the SCN (Girotti et al., 2009). However, given the tight coupling of temporal phasing of the major core components of the BLA molecular clock to the SCN, a direct regulatory control of the master clock on the circadian machinery of the BLA can be hypothesized, as also suggested by previous findings (Lamont et al., 2005). In principle, the SCN could be acting on the BLA via neural or humoral routes. The paraventricular nucleus of the hypothalamus is thus far the only identified brain region directly relaying synaptic signals from the SCN to the amygdala (Peng and Bentivoglio, 2004), but the expression of several clock genes in the paraventricular nucleus has been demonstrated to be in antiphase with the corresponding gene products in the SCN (Girotti et al., 2009), the neural pathway is most likely not mediating the impact of the SCN on BLA cyclic activity. With regard to the humoral factors, a likely candidate potentially synchronizing the transcriptional regulation of BLA clock genes to SCN rhythms is Gastrin-releasing peptide (GRP). GRP, which is known to communicate photic signals within the SCN hereby regulating circadian behavior and clock gene expression (Aida et al., 2002), is also highly expressed in the amygdala where, through binding to its receptor GRPR, it is involved in the regulation of emotional behaviors (Monje et al., 2011; Martel et al., 2012).
Alternatively, it has to be considered that non-SCN mediated rhythms may also be involved in the regulation of amygdala clock gene expression, as direct photic inputs to the amygdala have been described (Hattar et al., 2006; Morin and Studholme, 2014).
This observation proposes SCN-independent modulatory effects of light exposure on molecular events in the amygdala and related physiological and behavior outputs as intriguing novel aspects that should be further addressed in future studies.
We next set out to address the question whether the diurnal oscillation of BLA clock genes was disrupted in a naturalistic animal model of depression. Using the CMS paradigm, we observed that, in agreement with previous reports, only a subset of animals displays a reduction in sucrose preferences after exposure to CMS (“anhedonic”), an effect described as partial depressive-like outcome in chronic stress models of depression in the literature (Strekalova et al., 2004, 2006; Bergstrom et al., 2007; Krishnan et al., 2007; Li et al., 2010; Vialou et al., 2010). Anhedonic mice, but not nonanhedonic or control animals, also presented with a significant decrease in percent body weight change during the experimental period, as previously described by others (Strekalova et al., 2004, 2006; Strekalova et al., 2008; Strekalova and Steinbusch, 2010).
Circadian analysis of clock gene expression revealed that, with the exception of Bmal1, whose peak was shifted, the rhythmic expression of all clock genes that were diurnally expressed in BLA of control mice was abolished in CMS-induced anhedonic mice. However, rhythm desynchronization among the individual subjects may also contribute to the apparent loss of clock gene oscillation in the anhedonic group, which is indeed analyzed as a “population rhythm.”
The CMS paradigm employed herein does not allow for a concomitant analysis of circadian wheel running activity; thus, an independent study will be required to examine whether disrupted amygdala clock gene oscillation is also paralleled by disturbed behavioral rhythms. However, a previous study showed that CMS caused disturbances of the diurnal and circadian rhythm of wheel-running locomotor activity in rats (Gorka et al., 1996). Along these lines, the physiological rhythmic pattern of the core body temperature has been found to be disrupted in rats exposed to a CMS paradigm (Ushijima et al., 2006). Considering that it has been previously shown that CMS alters hepatic clock gene expression without affecting the respective rhythms in the SCN (Takahashi et al., 2013), it can be speculated that circulating factors, such as corticosterone, may directly confer CMS-induced alterations in amygdala clock gene oscillation. This hypothesis gains further support from a previous finding demonstrating an altered pattern of circadian corticosterone secretion in chronically stressed mice (Dalm et al., 2012), as rhythms in corticosterone have been identified as modulators of clock gene expression (Lamont et al., 2005; Segall et al., 2006).
A large body of clinical data has proved a correlation between clock gene functions and mood disorders, including bipolar disorder (Benedetti et al., 2003), seasonal affective disorder (Partonen et al., 2007), and major depression (Li et al., 2013). A seminal study recently for the first time identified disruption of diurnal oscillatory patterns of gene expression, including elements of the molecular circadian machinery, in several brain regions using postmortem material of depressed patients (Li et al., 2013). Although in rats the effect of chronic stress on individual clock genes in the hippocampus has been assessed (Jiang et al., 2013), this is the first comprehensive analysis of the diurnal expression of all core elements of the molecular clock in the mouse amygdala and its disruption in a validated animal model of depression. Whereas the nature of the design of the present study does not allow inferring a direct causal relationship between the alteration of clock gene rhythmicity and the depressive-like state, it further corroborates evidence for the pathophysiological relevance of several clock genes, including Clock, Bmal1, Cry2, Per1, and Per3, which have been previously strongly associated with mood disorders based upon genetic studies in the human population (Benedetti et al., 2003; Carpen et al., 2006; Artioli et al., 2007; Partonen et al., 2007; Lavebratt et al., 2010) and relevant animal models (Griesauer et al., 2014).
VEGFA Cyclic Pattern of Expression and Disruption in Anhedonic Mice
Searching for the downstream effectors potentially mediating the effects of altered clock gene rhythmicity on depressive-like behavior, we turned towards the analysis of growth factors previously implicated in mood disorders, specifically VEGF (Kahl et al., 2009; Elfving et al., 2010; Khan et al., 2014). The VEGF-signaling pathway was a likely candidate considering previously published evidence for a direct regulatory impact of elements of the molecular clock machinery on VEGF expression (Koyanagi et al., 2003).
We here firstly describe the diurnal oscillation of VEGF expression in the central nervous system by examining the cyclic pattern of expression of VEGFA, VEGFB and VEGFC in the mouse BLA. Interestingly, though all 3 genes exhibited a clear diurnal rhythmicity, mRNA levels of VEGFA and VEGFC peaked in the light phase, while VEGFB expression was higher in the dark phase of the cycle in control mice. In agreement with our findings, rhythmic expression of VEGFA (isoform 120 and 164) was previously observed in implanted tumor cells (Koyanagi et al., 2003), with a peak in the light phase. Strikingly, the diurnal oscillation of VEGFA was completely abolished and a shift in the peak time expression of VEGFB and VEGFC was recorded in the BLA of anhedonic mice. Considering the observed alterations in clock gene expression in anhedonic mice, these findings suggest that correct functioning of the molecular clock is required for the proper diurnal oscillation of VEGF in the amygdala. Moreover, in view of the available evidence for a link between altered VEGF levels and depression (Kahl et al., 2009; Elfving et al., 2010; Khan et al., 2014), it can be hypothesized that disruption of the cyclic nature of VEGF expression, potentially resulting from aberrant rhythmicity of the endogenous molecular clock, presents a pathophysiological correlate of altered emotional states in mood disorders.
Interestingly, although no diurnal pattern of plasma VEGFA levels was detected, augmented peripheral VEGFA concentrations paralleled the general trend for increased levels of the 3 individual members of the VEGF family of growth factors in the BLA of anhedonic mice. These results support previous findings of increased plasma VEGF concentrations in depressed patients (Kahl et al., 2009), which, however, had not been observed in other studies (Dome et al., 2009; Ventriglia et al., 2009).
To conclude, this study establishes an animal model for the impact of prodepressogenic environmental stimuli on diurnal oscillation of elements of the endogenous clock as a valid tool to further explore the molecular mechanisms mediating the involvement of the circadian system in the pathophysiology of mood disorders. Moreover, the results presented herein lay the ground for future investigations aiming at the identification of alternative drug targets and novel biomarkers for depression and invite consideration of the relevance of amygdala circadian rhythmicity in therapeutic interventions for the disorder.
Supplementary materials
For supplementary material accompanying this paper, visit http://www.ijnp.oxfordjournals.org/
Statement of Interest
None.
Author Contributions
D.D.P. designed the study and wrote the manuscript. G.S. carried out behavioral, molecular, and biochemical analysis and wrote the manuscript. W.F.D., K.T., and S.S. carried out gene expression analyses.
Acknowledgments
Daniela D. Pollak is supported by the Austrian Science Fund (FWF): stand-alone project P22424 and member of special research network (SFB 35): F3516-B20.
References
- Aida R, Moriya T, Araki M, Akiyama M, Wada K, Wada E, Shibata S. (2002). Gastrin-releasing peptide mediates photic entrainable signals to dorsal subsets of suprachiasmatic nucleus via induction of Period gene in mice. Mol Pharmacol 61:26–34. [DOI] [PubMed] [Google Scholar]
- Albrecht U. (2012). Timing to perfection: the biology of central and peripheral circadian clocks. Neuron 74:246–260. [DOI] [PubMed] [Google Scholar]
- Artioli P, Lorenzi C, Pirovano A, Serretti A, Benedetti F, Catalano M, Smeraldi E. (2007). How do genes exert their role? Period 3 gene variants and possible influences on mood disorder phenotypes. Eur Neuropsychopharmacol 17:587–594. [DOI] [PubMed] [Google Scholar]
- Benedetti F, Serretti A, Colombo C, Barbini B, Lorenzi C, Campori E, Smeraldi E. (2003). Influence of CLOCK gene polymorphism on circadian mood fluctuation and illness recurrence in bipolar depression. Am J Med Genet B Neuropsychiatr Genet 123B:23–26. [DOI] [PubMed] [Google Scholar]
- Bergstrom A, Jayatissa MN, Thykjaer T, Wiborg O. (2007). Molecular pathways associated with stress resilience and drug resistance in the chronic mild stress rat model of depression: a gene expression study. J Mol Neurosci 33:201–215. [DOI] [PubMed] [Google Scholar]
- Bunney BG, Bunney WE. (2013). Mechanisms of rapid antidepressant effects of sleep deprivation therapy: clock genes and circadian rhythms. Biol Psychiatry 73:1164–1171. [DOI] [PubMed] [Google Scholar]
- Carpen JD, von Schantz M, Smits M, Skene DJ, Archer SN. (2006). A silent polymorphism in the PER1 gene associates with extreme diurnal preference in humans. J Hum Genet 51:1122–1125. [DOI] [PubMed] [Google Scholar]
- Catteau J, Gernet JI, Marret S, Legros H, Gressens P, Leroux P, Laudenbach V. (2011). Effects of antenatal uteroplacental hypoperfusion on neonatal microvascularisation and excitotoxin sensitivity in mice. Pediatr Res 70:229–235. [DOI] [PubMed] [Google Scholar]
- Choobineh A, Rajaeefard A, Neghab M. (2006). Problems related to shiftwork for health care workers at Shiraz University of Medical Sciences. East Mediterr Health J 12:340–346. [PubMed] [Google Scholar]
- Costa G. (1997). The problem: shiftwork. Chronobiol Int 14:89–98. [DOI] [PubMed] [Google Scholar]
- Dalm S, de Kloet ER, Oitzl MS. (2012). Post-training reward partially restores chronic stress induced effects in mice. PLoS One 7:e39033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dome P, Teleki Z, Rihmer Z, Peter L, Dobos J, Kenessey I, Tovari J, Timar J, Paku S, Kovacs G, Dome B. (2009). Circulating endothelial progenitor cells and depression: a possible novel link between heart and soul. Mol Psychiatry 14:523–531. [DOI] [PubMed] [Google Scholar]
- Elfving B, Plougmann PH, Wegener G. (2010). Differential brain, but not serum VEGF levels in a genetic rat model of depression. Neuroscience letters 474:13–16. [DOI] [PubMed] [Google Scholar]
- Girotti M, Weinberg MS, Spencer RL. (2009). Diurnal expression of functional and clock-related genes throughout the rat HPA axis: system-wide shifts in response to a restricted feeding schedule. Am J Physiol Endocrinol Metab 296:E888–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golden RN, Gaynes BN, Ekstrom RD, Hamer RM, Jacobsen FM, Suppes T, Wisner KL, Nemeroff CB. (2005). The efficacy of light therapy in the treatment of mood disorders: a review and meta-analysis of the evidence. Am J Psychiatry 162:656–662. [DOI] [PubMed] [Google Scholar]
- Gonzalez MM, Aston-Jones G. (2008). Light deprivation damages monoamine neurons and produces a depressive behavioral phenotype in rats. Proc Natl Acad Sci U S A 105:4898–4903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon NP, Cleary PD, Parker CE, Czeisler CA. (1986). The prevalence and health impact of shiftwork. Am J Public Health 76:1225–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorka Z, Moryl E, Papp M. (1996). Effect of chronic mild stress on circadian rhythms in the locomotor activity in rats. Pharmacol Biochem Behav 54:229–234. [DOI] [PubMed] [Google Scholar]
- Griesauer I, Diao W, Ronovsky M, Elbau I, Sartori S, Singewald N, Pollak DD. (2014). Circadian abnormalities in a mouse model of high trait anxiety and depression. Ann Med. 46:148–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattar S, Kumar M, Park A, Tong P, Tung J, Yau KW, Berson DM. (2006). Central projections of melanopsin-expressing retinal ganglion cells in the mouse. J Comp Neurol 497:326–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honma S, Kawamoto T, Takagi Y, Fujimoto K, Sato F, Noshiro M, Kato Y, Honma K. (2002). Dec1 and Dec2 are regulators of the mammalian molecular clock. Nature 419:841–844. [DOI] [PubMed] [Google Scholar]
- Ino H. (2004). Immunohistochemical characterization of the orphan nuclear receptor ROR alpha in the mouse nervous system. J Histochem Cytochem 52:311–323. [DOI] [PubMed] [Google Scholar]
- Jensen LD, Cao Y. (2013). Clock controls angiogenesis. Cell Cycle 12:405–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang WG, Li SX, Liu JF, Sun Y, Zhou SJ, Zhu WL, Shi J, Lu L. (2013). Hippocampal CLOCK protein participates in the persistence of depressive-like behavior induced by chronic unpredictable stress. Psychopharmacology (Berl) 227:79–92. [DOI] [PubMed] [Google Scholar]
- Jilg A, Lesny S, Peruzki N, Schwegler H, Selbach O, Dehghani F, Stehle JH. (2010). Temporal dynamics of mouse hippocampal clock gene expression support memory processing. Hippocampus 20:377–388. [DOI] [PubMed] [Google Scholar]
- Kahl KG, Bens S, Ziegler K, Rudolf S, Kordon A, Dibbelt L, Schweiger U. (2009). Angiogenic factors in patients with current major depressive disorder comorbid with borderline personality disorder. Psychoneuroendocrinology 34:353–357. [DOI] [PubMed] [Google Scholar]
- Kang HS, Angers M, Beak JY, Wu X, Gimble JM, Wada T, Xie W, Collins JB, Grissom SF, Jetten AM. (2007). Gene expression profiling reveals a regulatory role for ROR alpha and ROR gamma in phase I and phase II metabolism. Physiol Genomics 31:281–294. [DOI] [PubMed] [Google Scholar]
- Khan D, Fernando P, Cicvaric A, Berger A, Pollak A, Monje FJ, Pollak DD. (2014). Long-term effects of maternal immune activation on depression-like behavior in the mouse. Transl Psychiatry 4:e363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajima K, Takahashi R, Yokota Y. (2006). Localization of Id2 mRNA in the adult mouse brain. Brain Res 1073–1074:93–102. [DOI] [PubMed] [Google Scholar]
- Koyanagi S, Kuramoto Y, Nakagawa H, Aramaki H, Ohdo S, Soeda S, Shimeno H. (2003). A molecular mechanism regulating circadian expression of vascular endothelial growth factor in tumor cells. Cancer Res 63:7277–7283. [PubMed] [Google Scholar]
- Krishnan V, et al. (2007). Molecular adaptations underlying susceptibility and resistance to social defeat in brain reward regions. Cell 131:391–404. [DOI] [PubMed] [Google Scholar]
- Kume K, Zylka MJ, Sriram S, Shearman LP, Weaver DR, Jin X, Maywood ES, Hastings MH, Reppert SM. (1999). mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell 98:193–205. [DOI] [PubMed] [Google Scholar]
- Lamont EW, Robinson B, Stewart J, Amir S. (2005). The central and basolateral nuclei of the amygdala exhibit opposite diurnal rhythms of expression of the clock protein Period2. Proc Natl Acad Sci U S A 102:4180–4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavebratt C, Sjoholm LK, Soronen P, Paunio T, Vawter MP, Bunney WE, Adolfsson R, Forsell Y, Wu JC, Kelsoe JR, Partonen T, Schalling M. (2010). CRY2 is associated with depression. PLoS One 5:e9407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JZ, Bunney BG, Meng F, Hagenauer MH, Walsh DM, Vawter MP, Evans SJ, Choudary PV, Cartagena P, Barchas JD, Schatzberg AF, Jones EG, Myers RM, Watson SJ, Jr., Akil H, Bunney WE. (2013). Circadian patterns of gene expression in the human brain and disruption in major depressive disorder. Proc Natl Acad Sci U S A 110:9950–9955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Zheng X, Liang J, Peng Y. (2010). Coexistence of anhedonia and anxiety-independent increased novelty-seeking behavior in the chronic mild stress model of depression. Behav Processes 83:331–339. [DOI] [PubMed] [Google Scholar]
- Martel G, Hevi C, Wong A, Zushida K, Uchida S, Shumyatsky GP. (2012). Murine GRPR and stathmin control in opposite directions both cued fear extinction and neural activities of the amygdala and prefrontal cortex. PLoS One 7:e30942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza J, Graff C, Dardente H, Pevet P, Challet E. (2005). Feeding cues alter clock gene oscillations and photic responses in the suprachiasmatic nuclei of mice exposed to a light/dark cycle. J Neurosci 25:1514–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsui S, Yamaguchi S, Matsuo T, Ishida Y, Okamura H. (2001). Antagonistic role of E4BP4 and PAR proteins in the circadian oscillatory mechanism. Genes Dev 15:995–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monje FJ, Kim EJ, Cabatic M, Lubec G, Herkner KR, Pollak DD. (2011). A role for glucocorticoid-signaling in depression-like behavior of gastrin-releasing peptide receptor knock-out mice. Ann Med 43:389–402. [DOI] [PubMed] [Google Scholar]
- Morin LP, Studholme KM. (2014). Light pulse duration differentially regulates mouse locomotor suppression and phase shifts. J Biol Rhythms 29:346–354. [DOI] [PubMed] [Google Scholar]
- Mozhui K, Karlsson RM, Kash TL, Ihne J, Norcross M, Patel S, Farrell MR, Hill EE, Graybeal C, Martin KP, Camp M, Fitzgerald PJ, Ciobanu DC, Sprengel R, Mishina M, Wellman CL, Winder DG, Williams RW, Holmes A. (2010). Strain differences in stress responsivity are associated with divergent amygdala gene expression and glutamate-mediated neuronal excitability. J Neurosci 30:5357–5367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onishi H, Yamaguchi S, Yagita K, Ishida Y, Dong X, Kimura H, Jing Z, Ohara H, Okamura H. (2002). Rev-erbalpha gene expression in the mouse brain with special emphasis on its circadian profiles in the suprachiasmatic nucleus. J Neurosci Res 68:551–557. [DOI] [PubMed] [Google Scholar]
- Partonen T, Treutlein J, Alpman A, Frank J, Johansson C, Depner M, Aron L, Rietschel M, Wellek S, Soronen P, Paunio T, Koch A, Chen P, Lathrop M, Adolfsson R, Persson ML, Kasper S, Schalling M, Peltonen L, Schumann G. (2007). Three circadian clock genes Per2, Arntl, and Npas2 contribute to winter depression. Ann Med 39:229–238. [DOI] [PubMed] [Google Scholar]
- Paxinos G, Franklin K. (2001). The mouse brain in stereotaxic coordinates, 2nd Edition. USA: Elsevier Academic Press. [Google Scholar]
- Peng ZC, Bentivoglio M. (2004). The thalamic paraventricular nucleus relays information from the suprachiasmatic nucleus to the amygdala: a combined anterograde and retrograde tracing study in the rat at the light and electron microscopic levels. J Neurocytol 33:101–116. [DOI] [PubMed] [Google Scholar]
- Ressler KJ, Mayberg HS. (2007). Targeting abnormal neural circuits in mood and anxiety disorders: from the laboratory to the clinic. Nat Neurosci 10:1116–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato TK, Panda S, Miraglia LJ, Reyes TM, Rudic RD, McNamara P, Naik KA, FitzGerald GA, Kay SA, Hogenesch JB. (2004). A functional genomics strategy reveals Rora as a component of the mammalian circadian clock. Neuron 43:527–537. [DOI] [PubMed] [Google Scholar]
- Segall LA, Milet A, Tronche F, Amir S. (2009). Brain glucocorticoid receptors are necessary for the rhythmic expression of the clock protein, PERIOD2, in the central extended amygdala in mice. Neurosci Lett 457:58–60. [DOI] [PubMed] [Google Scholar]
- Segall LA, Perrin JS, Walker CD, Stewart J, Amir S. (2006). Glucocorticoid rhythms control the rhythm of expression of the clock protein, Period2, in oval nucleus of the bed nucleus of the stria terminalis and central nucleus of the amygdala in rats. Neuroscience 140:753–757. [DOI] [PubMed] [Google Scholar]
- Shearman LP, Zylka MJ, Weaver DR, Kolakowski LF, Jr., Reppert SM. (1997). Two period homologs: circadian expression and photic regulation in the suprachiasmatic nuclei. Neuron 19:1261–1269. [DOI] [PubMed] [Google Scholar]
- Shearman LP, Sriram S, Weaver DR, Maywood ES, Chaves I, Zheng B, Kume K, Lee CC, van der Horst GT, Hastings MH, Reppert SM. (2000). Interacting molecular loops in the mammalian circadian clock. Science 288:1013–1019. [DOI] [PubMed] [Google Scholar]
- Storch KF, Lipan O, Leykin I, Viswanathan N, Davis FC, Wong WH, Weitz CJ. (2002). Extensive and divergent circadian gene expression in liver and heart. Nature 417:78–83. [DOI] [PubMed] [Google Scholar]
- Strekalova T, Steinbusch HW. (2010). Measuring behavior in mice with chronic stress depression paradigm. Prog Neuropsychopharmacol Biol Psychiatry 34:348–361. [DOI] [PubMed] [Google Scholar]
- Strekalova T, Spanagel R, Bartsch D, Henn FA, Gass P. (2004). Stress-induced anhedonia in mice is associated with deficits in forced swimming and exploration. Neuropsychopharmacology 29:2007–2017. [DOI] [PubMed] [Google Scholar]
- Strekalova T, Gorenkova N, Schunk E, Dolgov O, Bartsch D. (2006). Selective effects of citalopram in a mouse model of stress-induced anhedonia with a control for chronic stress. Behav Pharmacol 17:271–287. [DOI] [PubMed] [Google Scholar]
- Strekalova T, van Miegem V, Redkozubova O, Dolgov O, Larde G, Beznosko B, Vankin G, Bachurin S. (2008). Sucrose test method: Facts, artifacts and application in anhedonia models with young and old C57BL/6 mice. Int J Nuropsychopharm 11(Suppl 1):128. [Google Scholar]
- Sumi Y, Yagita K, Yamaguchi S, Ishida Y, Kuroda Y, Okamura H. (2002). Rhythmic expression of ROR beta mRNA in the mice suprachiasmatic nucleus. Neuroscience Lett 320:13–16. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamada T, Tsukita S, Kaneko K, Shirai Y, Munakata Y, Ishigaki Y, Imai J, Uno K, Hasegawa Y, Sawada S, Oka Y, Katagiri H. (2013). Chronic mild stress alters circadian expressions of molecular clock genes in the liver. Am J Physiol Endocrinol Metab 304:E301–309. [DOI] [PubMed] [Google Scholar]
- Ueda HR, Chen W, Adachi A, Wakamatsu H, Hayashi S, Takasugi T, Nagano M, Nakahama K, Suzuki Y, Sugano S, Iino M, Shigeyoshi Y, Hashimoto S. (2002). A transcription factor response element for gene expression during circadian night. Nature 418:534–539. [DOI] [PubMed] [Google Scholar]
- Ushijima K, Morikawa T, To H, Higuchi S, Ohdo S. (2006). Chronobiological disturbances with hyperthermia and hypercortisolism induced by chronic mild stress in rats. Behav Brain Res 173:326–330. [DOI] [PubMed] [Google Scholar]
- Ventriglia M, Zanardini R, Pedrini L, Placentino A, Nielsen MG, Gennarelli M, Bocchio-Chiavetto L. (2009). VEGF serum levels in depressed patients during SSRI antidepressant treatment. Prog Neuropsychopharmacol Biol Psychiatry 33:146–149. [DOI] [PubMed] [Google Scholar]
- Vialou V, et al. (2010). DeltaFosB in brain reward circuits mediates resilience to stress and antidepressant responses. Nat Neurosci 13:745–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitaterna MH, Selby CP, Todo T, Niwa H, Thompson C, Fruechte EM, Hitomi K, Thresher RJ, Ishikawa T, Miyazaki J, Takahashi JS, Sancar A. (1999). Differential regulation of mammalian period genes and circadian rhythmicity by cryptochromes 1 and 2. Proc Natl Acad Sci U S A 96:12114–12119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyse CA, Coogan AN. (2010). Impact of aging on diurnal expression patterns of CLOCK and BMAL1 in the mouse brain. Brain Res 1337:21–31. [DOI] [PubMed] [Google Scholar]
- Yang S, Wang K, Valladares O, Hannenhalli S, Bucan M. (2007). Genome-wide expression profiling and bioinformatics analysis of diurnally regulated genes in the mouse prefrontal cortex. Genome Biol 8:R247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng B, Larkin DW, Albrecht U, Sun ZS, Sage M, Eichele G, Lee CC, Bradley A. (1999). The mPer2 gene encodes a functional component of the mammalian circadian clock. Nature 400:169–173. [DOI] [PubMed] [Google Scholar]
- Zylka MJ, Shearman LP, Weaver DR, Reppert SM. (1998). Three period homologs in mammals: differential light responses in the suprachiasmatic circadian clock and oscillating transcripts outside of brain. Neuron 20:1103–1110. [DOI] [PubMed] [Google Scholar]