Abstract
While being devoid of the ability to recognize ligands itself, the WW2 domain is believed to aid ligand binding to WW1 domain in the context of WW1-WW2 tandem module of WWOX tumor suppressor. In an effort to test the generality of this hypothesis, we have undertaken here a detailed biophysical analysis of the binding of WW domains of WWOX alone and in the context of WW1-WW2 tandem module to an array of putative PPXY ligands. Our data show that while the WW1 domain of WWOX binds to all ligands in a physiologically-relevant manner, the WW2 domain does not. Moreover, ligand binding to WW1 domain in the context of WW1-WW2 tandem module is two-to-three-fold stronger than when treated alone. We also provide evidence that the WW domains within the WW1-WW2 tandem module physically associate so as to adopt a fixed spatial orientation relative to each other. Of particular note is the observation that the physical association of WW2 domain with WW1 blocks access to ligand. Consequently, ligand binding to WW1 domain not only results in the displacement of WW2 lid but also disrupts the physical association of WW domains in the liganded conformation. Taken together, our study underscores a key role of allosteric communication in the ability of WW2 orphan domain to chaperone physiological action of WW1 domain within the context of the WW1-WW2 tandem module of WWOX.
Keywords: WW tandem module, WW-ligand thermodynamics, Binding-coupled dissociation, Allosteric communication, Equilibrium shift
INTRODUCTION
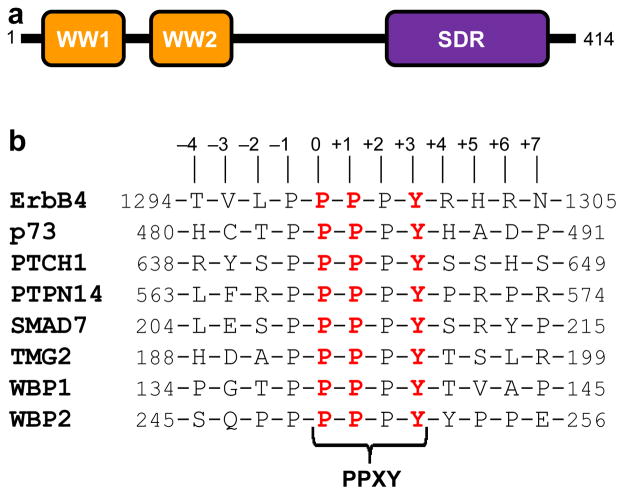
With a modular architecture comprised of a tandem copy of WW domains (designated WW1 and WW2) located N-terminal to the short-chain dehydrogenase/reductase (SDR) domain (Figure 1a), the WWOX tumor suppressor mediates a multitude of cellular activities including growth, proliferation, apoptosis and tumor suppression (1–3). In particular, aberrant expression of WWOX is believed to be linked to the progression of many forms of cancer, including those of breast and prostate (4–11). Notably, the ability of WWOX to drive cellular signaling is largely dependent upon the ability of its WW1 domain to recognize PPXY motifs located within cognate ligands such as WBP1/2 signaling adaptors (12, 13), ErbB4 receptor kinase (14), p73 tumor suppressor (15), and many others (16).
Figure 1.
Modular organization of human WWOX and putative ligands. (a) WWOX is comprised of a tandem copy of WW domains, designated WW1 and WW2, located N-terminal to the short-chain dehydrogenase/reductase (SDR) domain. (b) Amino acid sequence of 12-mer peptides, derived from various cognate ligands of WWOX and YAP, containing the PPXY motifs and flanking residues. Note that the numerals indicate the nomenclature used in this study to distinguish residues within and flanking the motifs relative to the first proline within the PPXY motif, which is arbitrarily assigned zero.
Additionally, these WWOX ligands are cellular targets of WW domains of YAP, a key regulator of Hippo signaling cascade involved in regulating the size of organs and in the suppression of tumors through inhibiting cellular proliferation and promoting apoptosis (17–21). In addition to WBP1/2 (22, 23), ErbB4 (24, 25) and p73 (26), YAP transcriptional regulator interacts with a wide variety of other cellular proteins such as TMG2 transmembrane protein (27), PTPN14 phosphatase (28), SMAD7 signaling adaptor (29) and PTCH1 transmembrane receptor (30). Importantly, it is believed that WWOX antagonizes the transactivation function of YAP by virtue of its ability to competitively bind to these proteins and plays a central role in the maintenance of cellular homeostasis (14, 18).
While WW1 domain of WWOX is critical to its ability to recognize putative PPXY ligands, no physiological ligands of the WW2 domain have hitherto been identified. Interestingly, we previously reported that the WW2 domain structurally stabilizes the WW1 domain within the context of WW1-WW2 tandem module and augments ligand binding to WWOX (13, 31). This earlier work prompted us to postulate that the WW2 domain is an orphan domain whose primary physiological role is to chaperone ligand binding to WW1 domain within WWOX. However, this hypothesis is only based on a limited set of ligands, namely WBP1/2 (13) and ErbB4 (31), thereby questioning the extent to which this proposed model might also hold true in the case of other ligands. In an effort to address this important question, we have undertaken here a detailed biophysical analysis of the binding of WW domains of WWOX alone and in the context of WW1-WW2 tandem module to an array of putative PPXY ligands—both known ligands of WWOX as well as other ligands of YAP that appear to be strong candidates for interacting with WWOX (Figure 1b). Our new study corroborates the notion that WW2 is indeed a bona fide orphan domain and additionally provides the physical basis underlying its ability to chaperone the WW1 domain within the context of the WW1-WW2 tandem module of WWOX.
MATERIALS and METHODS
Protein preparation
WW1 domain (residues 16–50), WW2 domain (residues 57–91) and WW1-WW2 tandem module (residues 16–91) of human WWOX were cloned into pET30 bacterial expression vectors with an N-terminal His-tag using Novagen LIC technology as described earlier (13). All recombinant proteins were subsequently expressed in Escherichia coli BL21*(DE3) bacterial strain (Invitrogen) and purified on a Ni-NTA affinity column using standard procedures as described previously (13). Further treatment on a Hiload Superdex 200 size-exclusion chromatography (SEC) column coupled in-line with GE Akta FPLC system led to purification of recombinant domains to apparent homogeneity as judged by SDS-PAGE analysis. Final yield was typically between 50–100mg protein of apparent homogeneity per liter of bacterial culture. Protein concentration was determined spectrophotometrically on the basis of extinction coefficients calculated for each recombinant construct using the online software ProtParam at ExPasy Server (32).
Peptide synthesis
12-mer peptides spanning PPXY motifs located within various cognate ligands of WWOX and YAP were commercially obtained from GenScript Corporation. The amino acid sequence of these peptides is shown in Figure 1b. The peptide concentrations were measured gravimetrically.
Isothermal titration calorimetry
Isothermal titration calorimetry (ITC) experiments were performed on a Microcal VP-ITC instrument. All measurements were repeated at least three times. Briefly, WW domains of WWOX alone or in the context of the WW1-WW2 tandem module were dialyzed in 50mM Sodium phosphate, 100mM NaCl, 1mM EDTA and 5mM β-mercaptoethanol at pH 7.0. All experiments were initiated by injecting 25 × 10μl aliquots of 2–4mM of each peptide from the syringe into the calorimetric cell containing 1.46ml of 50–100μM of WW domains of WWOX alone or in the context of WW1-WW2 tandem module at 25°C. The change in thermal power as a function of each injection was automatically recorded using the ORIGIN software and the raw data were further processed to yield binding isotherms of heat release per injection as a function of molar ratio of each peptide to WW domain construct. The heats of mixing and dilution were subtracted from the heat of binding per injection by carrying out a control experiment in which the same buffer in the calorimetric cell was titrated against each peptide in an identical manner. To determine the equilibrium dissociation constant (Kd) and the enthalpic change (ΔH) associated with binding, the ITC isotherms were iteratively fit to a one-site binding model by non-linear least squares regression analysis using the integrated ORIGIN software as described earlier (13, 33). Notably, all binding stoichiometries were fixed to unity while ΔH and Kd were allowed to float during the fitting procedure to improve the accuracy of thermodynamic parameters. The free energy change (ΔG) upon peptide binding was calculated from the relationship:
| (1) |
where R is the universal molar gas constant (1.99 cal/K/mol) and T is the absolute temperature.
Circular dichroism
Far-UV circular dichroism (CD) measurements were conducted on a Jasco J-815 spectropolarimeter thermostatically controlled at 25°C. Briefly, 12-mer peptides spanning PPXY motifs located within various cognate ligands of WWOX and YAP were dialyzed in 10mM Sodium phosphate at pH 7.0 and experiments were conducted on 100μM sample of each peptide. Data were collected using a quartz cuvette with a 2-mm pathlength in the 185–255 nm wavelength range and with a slit bandwidth of 2nm at a scan rate of 10 nm/min. All data were normalized against reference spectra to remove the contribution of buffer. Each data set represents an average of four scans acquired at 0.1 nm intervals. Data were converted to mean ellipticity, [θ], as a function of wavelength (λ) of electromagnetic radiation using the following equation:
| (2) |
where Δε is the observed ellipticity in mdeg, c is the peptide concentration in μM, and l is the cuvette pathlength in cm.
Molecular modeling
Structural models of WW1-WW2 tandem module of WWOX alone (unliganded) and in complex with p73 peptide (liganded) containing the PPXY motif were built using the MODELLER software based on homology modeling (34). Briefly, the structural model of unliganded WW1-WW2 tandem module of WWOX was obtained using the NMR structure of WW1-WW2 tandem module of FBP21 pre-mRNA splicing factor as a template (PDBID 2JXW). It should be noted here that the WW domains within the WW1-WW2 tandem module of FBP21 are tethered together via a short flexible interdomain linker, which is similar to that separating the WW domains of WWOX. To obtain the liganded structure, the 12-mer p73 peptide was docked onto the WW1 domain within the unliganded structure of WW1-WW2 tandem module of WWOX in a 1:1 stoichiometry using the NMR structure of the homologous WW domain of YAP bound to a peptide containing the PPXY motif as a template (PDBID 1JMQ). In each case, a total of 100 atomic models were calculated and the structure with the lowest energy, as judged by the MODELLER Objective Function, was selected for further analysis. The atomic models were rendered using RIBBONS (35). The quality of the structural models was compared with template structures using DOPE, a built-in statistical potential in MODELLER similar to ERRAT, and the B-factor plots. The B-factors for both the model and template structures were calculated in MODELLER for consistency. It should be noted here that the WW domains of WWOX share greater than 50% strong sequence similarity at the amino acid level with their counterparts in both the FBP21 and YAP. This implies that the structural model of the WW1-WW2 tandem module of WWOX presented here can be relied upon with a high degree of confidence bar the physical association, or lack thereof, of constituent WW domains within the modular assembly. This latter structural feature was further interrogated using molecular dynamics (vide infra).
Molecular dynamics
Molecular dynamics (MD) simulations were performed with the GROMACS software (36) using the integrated AMBER99SB-ILDN force field (37, 38). Briefly, the structural models of WW1-WW2 tandem module of WWOX alone (unliganded) and in complex with p73 peptide (liganded) containing the PPXY motif were each centered in a cubic box and explicitly hydrated with a water layer that extended 10Å (box size) from the protein surface along each orthogonal direction using the extended simple point charge (SPC/E) water model (39, 40). The ionic strength of solution was set to 100mM with NaCl and the hydrated structures were energy-minimized with the steepest descent algorithm prior to equilibration under the NPT ensemble conditions, wherein the number of atoms (N), pressure (P) and temperature (T) within the system were kept constant. The Particle-Mesh Ewald (PME) method (41) was employed to compute long-range electrostatic interactions with a spherical cut-off of 10Å and a grid space of 1.6Å with a fourth order interpolation. The Linear Constraint Solver (LINCS) algorithm was used to restrain bond lengths (42). All MD simulations were performed at 300K under periodic boundary conditions (PBC), so as to mimic the bulk solvent effect, using the standard “md” leap-frog integrator to solve Newton’s equations of motion with a time step of 2fs. For the final MD production runs, data were collected every ns over a time scale of 4μs. All MD simulations were performed on a Linux workstation using parallel processors at the High Performance Computing (HPC) facility within the Center for Computational Science (CCS) of the University of Miami. Structural snapshots taken at 200-ns time intervals during the second half of simulation (2–4μs) were superimposed using MOLMOL (43) and rendered with RIBBONS (35).
RESULTS and DISCUSSION
WW1 but not WW2 domain of WWOX binds to putative ligands
To test the generality of the hypothesis that the WW2 domain of WWOX is an orphan domain, we conducted ITC analysis on the binding of WW domains alone and in the context of WW1-WW2 tandem module to an array of putative PPXY ligands (Figure 1a). Representative ITC isotherms obtained for such measurements are shown in Figure 2, while detailed thermodynamic parameters are presented in Tables 1 and 2. Our data show that while the WW1 domain of WWOX binds to all ligands in a physiologically-relevant manner, the WW2 domain does not (Table 1). Notably, ligand binding to WW1 domain in the context of WW1-WW2 tandem module is two-to-three-fold stronger than when treated alone (Table 2). These observations are in an excellent agreement with our earlier studies indicating that the WW2 domain not only structurally stabilizes the WW1 domain within the context of WW1-WW2 tandem module but also augments ligand binding to WWOX (13, 31). On the basis of these extensive data, we postulate that the WW2 is indeed a bona fide orphan domain and that its sole physiological function appears to lie in its ability to act as a chaperone and facilitate ligand binding to WW1 domain. It is noteworthy that such synergistic action observed here between the WW domains of WWOX appears to be a hallmark of WW tandem domains in general (44–50).
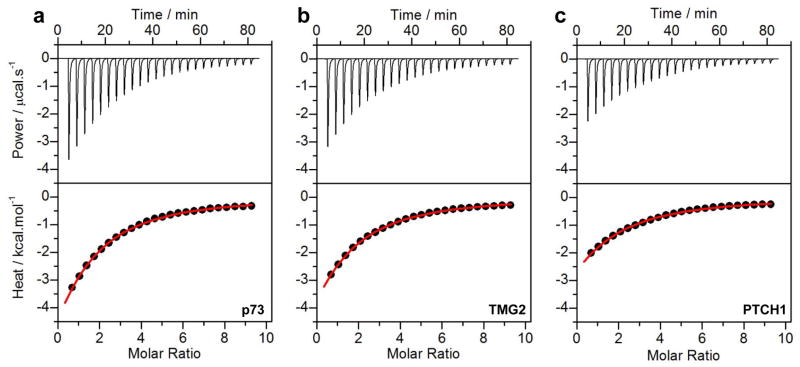
Figure 2.
Representative ITC isotherms for the binding of isolated WW1 domain of WWOX to p73 (a), TMG2 (b) and PTCH1 (c) peptides. The upper panels show the raw ITC data expressed as change in thermal power with respect to time over the period of titration. In the lower panels, change in molar heat is expressed as a function of molar ratio of corresponding peptide to WW1 domain of WWOX. The solid red lines in the lower panels show the fit of data to a one-site binding model using the integrated ORIGIN software as described earlier (13, 33).
Table 1.
Thermodynamic parameters for the binding of WW1 domain of WWOX to PPXY-containing peptides derived from various putative ligands
| Ligand | Sequence | Kd / μM | ΔH / kcal.mol−1 | TΔS / kcal.mol−1 | ΔG / kcal.mol−1 |
|---|---|---|---|---|---|
| ErbB4 |
|
179 ± 10 | −7.85 ± 0.80 | −2.73 ± 0.83 | −5.12 ± 0.03 |
| p73 |
|
107 ± 9 | −16.65 ± 0.78 | −11.23 ± 0.82 | −5.42 ± 0.05 |
| PTCH1 |
|
278 ± 19 | −10.17 ± 0.62 | −5.31 ± 0.57 | −4.86 ± 0.04 |
| PTPN14 |
|
763 ± 62 | −5.96 ± 0.72 | −1.70 ± 0.77 | −4.26 ± 0.04 |
| SMAD7 |
|
272 ± 29 | −22.60 ± 0.99 | −17.73 ± 1.05 | −4.87 ± 0.06 |
| TMG2 |
|
202 ± 23 | −14.15 ± 0.49 | −9.10 ± 0.43 | −5.05 ± 0.07 |
| WBP1 |
|
368 ± 44 | −17.55 ± 0.78 | −12.86 ± 0.85 | −4.69 ± 0.07 |
| WBP2 |
|
133 ± 18 | −15.15 ± 0.49 | −9.85 ± 0.42 | −5.30 ± 0.08 |
The consensus residues within the PPXY motif of each peptide are colored red for clarity. All parameters were obtained from ITC measurements at 25°C and pH 7. Binding stoichiometries were fixed to unity to improve the accuracy of thermodynamic parameters. Errors were calculated from at least three independent measurements to one standard deviation.
Table 2.
Thermodynamic parameters for the binding of WW1-WW2 tandem module of WWOX to PPXY-containing peptides derived from various putative ligands
| Ligand | Sequence | Kd / μM | ΔH / kcal.mol−1 | TΔS / kcal.mol−1 | ΔG / kcal.mol−1 |
|---|---|---|---|---|---|
| ErbB4 |
|
75 ± 8 | −5.58 ± 0.33 | +0.06 ± 0.39 | −5.64 ± 0.06 |
| p73 |
|
60 ± 6 | −7.33 ± 0.26 | −1.55 ± 0.33 | −5.78 ± 0.07 |
| PTCH1 |
|
79 ± 8 | −5.86 ± 0.54 | −0.25 ± 0.60 | −5.61 ± 0.06 |
| PTPN14 |
|
214 ± 30 | −4.69 ± 0.42 | +0.33 ± 0.51 | −5.02 ± 0.08 |
| SMAD7 |
|
53 ± 8 | −12.10 ± 0.57 | −6.25 ± 0.65 | −5.85 ± 0.09 |
| TMG2 |
|
84 ± 9 | −9.28 ± 0.41 | −3.71 ± 0.48 | −5.57 ± 0.07 |
| WBP1 |
|
141 ± 12 | −10.46 ± 0.76 | −5.20 ± 0.81 | −5.26 ± 0.05 |
| WBP2 |
|
42 ± 7 | −7.51 ± 0.68 | −1.53 ± 0.78 | −5.98 ± 0.10 |
The consensus residues within the PPXY motif of each peptide are colored red for clarity. All parameters were obtained from ITC measurements at 25°C and pH 7. Binding stoichiometries were fixed to unity to improve the accuracy of thermodynamic parameters. Errors were calculated from at least three independent measurements to one standard deviation.
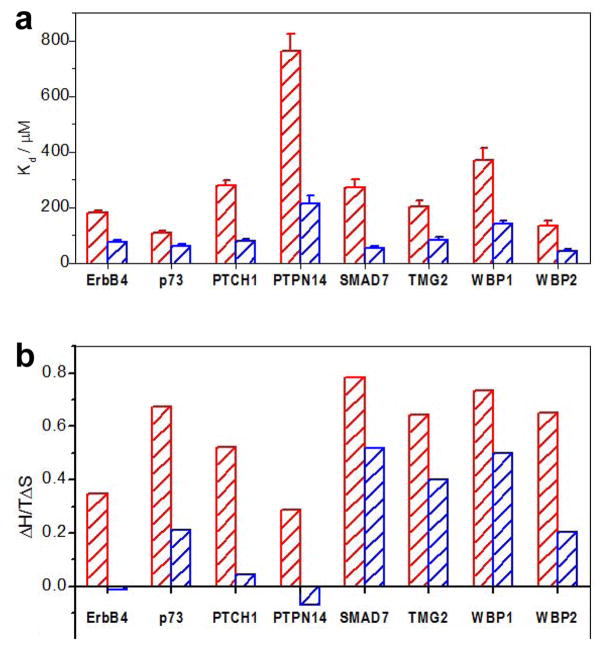
Importantly, ligand binding to WW1 domain in the context of WW1-WW2 tandem module of WWOX is accompanied by entropic change that is more favorable relative to that observed for ligand binding to WW1 domain alone (Figure 3). This finding strongly argues that the enhancement in ligand binding to WW1-WW2 tandem module likely results from a diminished change in the loss of the disorder of the system, and/or alternatively, from an increase in the disorder upon complex formation compared to ligand binding to WW1 domain alone. Interestingly, the first possibility can be easily rationalized in light of the knowledge that the WW1 domain alone is partially unstructured and undergoes folding upon ligand binding, while ligand binding in the context of WW1-WW2 tandem module occurs to a fully folded WW1 domain (13), thereby reducing the entropic cost. Nevertheless, the possibility that ligand binding to WW1-WW2 tandem module may also be accompanied by an increase in the disorder cannot be ruled out. Thus, for example, it is conceivable that the WW2 orphan domain physically associates with the WW1 domain in the context of WW1-WW2 tandem module so as to structurally stabilize it. However, ligand binding to WW1 domain results in the dissociation of WW2 domain, presumably due to a combination of structural and thermodynamic factors. In particular, dissociation of WW2 domain from WW1 domain may be necessary to easily accommodate the incoming ligand due to steric reasons, while at the same time being entropically favorable due to an increase in the disorder of the system.
Figure 3.
Comparison of thermodynamic properties for the binding of WW1 domain alone (red) and in the context of WW1-WW2 tandem module (blue) of WWOX to various peptides. (a) Comparison of equilibrium dissociation constant (Kd) associated with binding. Error bars were calculated from at least three independent measurements to one standard deviation. (b) Comparison of TΔS/ΔH ratio, where ΔH and TΔS are the enthalpic and entropic changes associated with binding, respectively.
Differential binding of various ligands to WW1 domain of WWOX does not correlate with their structure
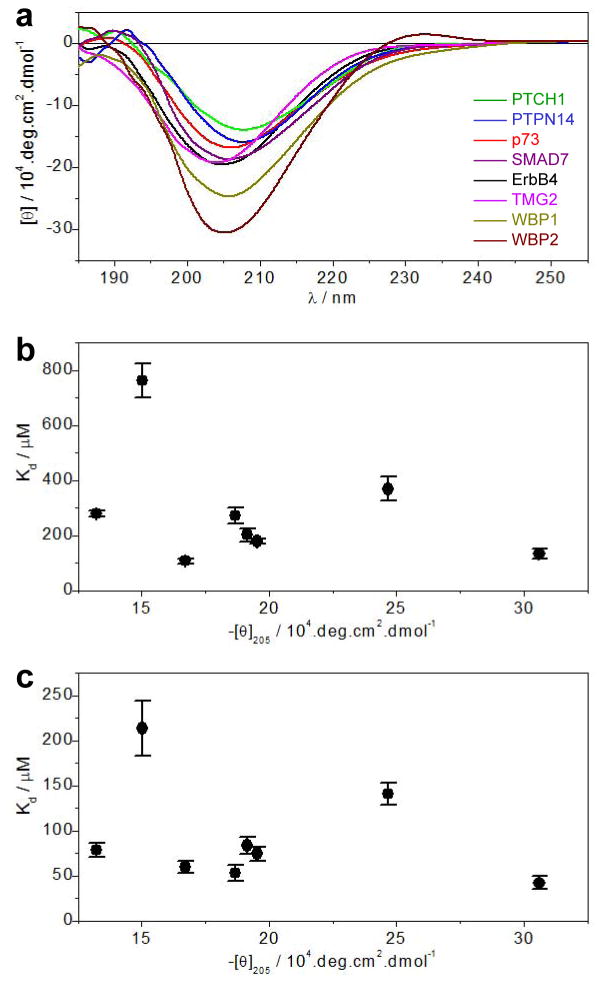
Our data presented above show that while WW1 domain of WWOX binds to all putative ligands, it does so with differential affinities both alone and in the context of WW1-WW2 tandem module (Tables 1 and 2). One likely explanation lies in the fact that this is due to the differences in residues located within and flanking the consensus PPXY motif of various ligands (Figure 1b). Indeed, our earlier studies have shown that the requirement of such accessory residues seems to be important for binding affinity though they may not always have a critical role (13, 31). However, it is also worth noting that the PPXY-containing peptides adopt polyproline type II (PPII) helical conformation upon binding to WW domains (51–54). Accordingly, peptide ligands that harbor a PPII-helix in solution in isolation should be expected to have an entropic advantage, which could correlate with higher binding affinity to cognate WW partners. In order to test whether such conformational advantage could also account for the differential binding of various PPXY motifs to WW1 domain of WWOX observed here, we conducted far-UV CD analysis on the corresponding peptides (Figure 4a).
Figure 4.
Comparison of secondary structure as probed by mean ellipticity at 205nm ([θ]205) and equilibrium dissociation constant (Kd) associated with the binding of various peptides to WW1 domain of WWOX. (a) Far-UV CD spectra of various peptides as indicated. Note that the mean ellipticity, [θ], was calculated using Eq [2]. (b) Dependence of Kd on [θ]205 for the binding of various peptides to WW1 domain alone. (c) Dependence of Kd on [θ]205 for the binding of various peptides to WW1 domain in the context of WW1-WW2 tandem module.
Our data show that the far-UV CD spectra of all peptides are predominantly characterized by the presence of a negative band centered around 205nm with varying intensity. While this spectral signature is well-documented for proline-rich peptides harboring random coil conformation, PPII-helical peptides are also characterized by the presence of a sharp positive band around 225nm (55, 56). The absence of a well-defined 225-nm band is thus indicative of the fact that all cognate peptides of WW1 domain of WWOX analyzed here possess random coil conformation in solution and thus must undergo a conformational change to PPII-helix upon binding. Importantly, the differential intensity of the 205-nm band further suggests that the extent of random coil within these peptides substantially varies with the WBP2 peptide apparently harboring the largest random coil content while PTCH1 occupies the other end of the spectrum. It is also noteworthy that there appears to be no correlation between the mean ellipticity at 205nm ([θ]205), which is used here as a measure of the extent of random coil content, and the equilibrium dissociation constant (Kd) observed for the binding of various peptides to WW1 domain of WWOX (Figures 4a and 4b). This implies that the molecular origin of the differential binding of various peptides most likely resides in their sequence rather than structure.
Structural insights into the physical basis underlying the ability of WW2 domain to chaperone and aid ligand binding to WW1 domain of WWOX
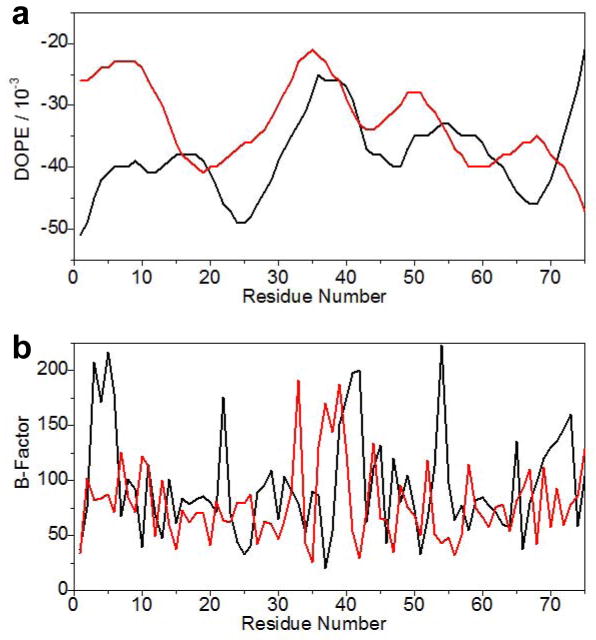
In order to shed light on the physical basis underlying the ability of WW2 domain to chaperone and aid ligand binding to WW1 domain, we modeled the structure of WW1-WW2 tandem module of WWOX using the NMR structure of homologous WW1-WW2 tandem module of FBP21 pre-mRNA splicing factor as a template (45). Additionally, the p73 peptide was docked into the binding groove of WW1 domain in the context of WW1-WW2 tandem module using the NMR structure of the homologous WW domain of YAP bound to a peptide containing the PPXY motif as a template (52). As shown in Figure 5, our structural analysis reveals that the WW1-WW2 tandem module adopts a dumbbell-like conformation with the WW domains tethered together with what appears to be a highly flexible linker. We add that the DOPE and B-factor plots of the structural model of WW1-WW2 tandem module of WWOX are comparable to the structure of WW1-WW2 tandem module of FBP21 used as a template (Figures 6a and 6b), thereby further validating our structural analysis though it does not support or argue against the physical association of constituent WW domains within the modular assembly.
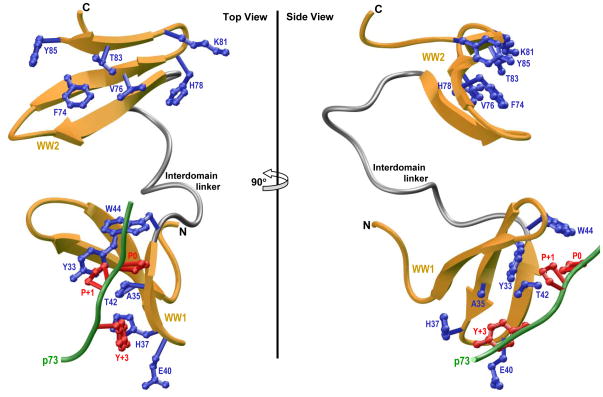
Figure 5.
Ribbon representation of the structural model of WW1-WW2 tandem module of WWOX in complex with the p73 peptide containing the PPXY motif bound to the WW1 domain. Two alternative orientations related by a 90°-rotation about the vertical axis are depicted for the inquisitive eye. In each case, the WW domains are shown in yellow with the interdomain linker depicted in gray, and the bound peptide is colored green. The sidechain moieties of residues (blue) within the WW1 domain engaged in intermolecular contacts with the consensus residues (red) within the PPXY motif of p73 peptide are shown. For comparison, the sidechain moieties of structurally-equivalent residues (blue) within the putative ligand binding groove of WW2 domain are also shown.
Figure 6.
Comparison of the quality of the structural model of WW1-WW2 tandem module of WWOX (red) and the solution structure of WW1-WW2 tandem module of FBP21 (black) as assessed by DOPE (a) and B-factor (b) plots. Note that both the DOPE potential and B-factor are dimensionless parameters. The residue number is arbitrarily assigned so as to directly compare the equivalently-aligned residues in both the model (WWOX) and the template (FBP21).
Notably, the p73 peptide roughly adopts the PPII-helical conformation and binds within the hydrophobic groove on the concave face of the triple-stranded β-sheet fold of WW1 domain in a canonical manner (51–54) (Figure 5). Moreover, the p73 peptide is largely stabilized by intermolecular contacts between sidechain moieties of consensus residues P0, P+1 and Y+3 located within the PPXY motif of p73 peptide and several highly conserved residues lining the hydrophobic groove within the WW1 domain. Thus, the pyrrolidine moiety of P0, the first proline within the PPXY motif, stacks against the indole sidechain of W44 in WW1 domain. The sidechain moieties of Y33/T42 residues within the WW1 domain sandwich the pyrrolidine ring of P+1 within the PPXY motif. Finally, the phenyl moiety of Y+3, the terminal tyrosine within the PPXY motif, buries deep into the hydrophobic groove and is escorted by sidechain atoms of the A35/H37/E40 triad in WW1 domain. It is noteworthy here that the apparent lack of ligand binding to WW2 domain largely resides in the lack of conservation of W44 of WW1 domain in the structurally-equivalent position, occupied by Y85, within the WW2 domain (13, 31).
Of particular significance is the observation that the WW domains within the WW1-WW2 tandem module of WWOX apparently do not engage in interdomain contacts (Figure 5). Such lack of physical association between the WW domains is indeed also noted for the WW1-WW2 tandem module FBP21 (45). Importantly, the rather flexible nature of the interdomain linker also implies that the WW domains within the WW1-WW2 tandem module are unlikely to adopt a fixed spatial orientation and thus should be expected to move freely with respect to each other. In light of such apparent lack of spatial orientation and interdomain interaction, the physical basis of how WW2 domain chaperones and augments ligand binding to WW1 domain within the WW1-WW2 tandem module of WWOX appears to be somewhat mysterious. While it is conceivable that our structural model has failed to capture the true picture of how WW domains are bundled together within the WW1-WW2 tandem module, the possibility that factors other than structure, such as protein dynamics and/or cooperativity, may hold clues to addressing this phenomenon cannot be excluded. For example, ligand binding to WW1 domain could trigger a conformational change within the WW1-WW2 tandem module so as to enhance its degrees of freedom and the resulting entropic gain could in turn lower its free energy.
Ligand binding augments conformational dynamics within the WW1-WW2 tandem module of WWOX
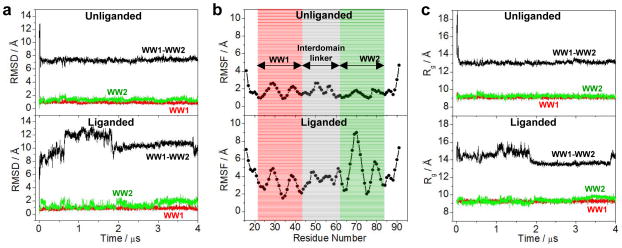
To investigate the extent to which WW domains may physically associate with each other and to probe the effect of ligand binding on protein dynamics, we conducted MD simulations on the structural model of WW1-WW2 tandem module of WWOX alone (unliganded) as well as in complex with p73 peptide (liganded) over a μs-timescale. It should be noted here that the starting conformation of unliganded WW1-WW2 tandem module was assumed to be identical to that of the liganded protein for the purpose of MD simulations. As shown in Figure 7, our MD analysis reveals that the unliganded and liganded proteins exhibit highly distinct conformational profiles. Thus, the unliganded protein initially undergoes a rapid “burst” phase over the first 100ns reaching a root mean square deviation (RMSD) for the backbone atoms in excess of 12Å. This is rapidly followed by structural equilibration of the unliganded protein with an overall RMSD of around 7Å after about 200ns during the course of the simulation (Figure 7a). This is evidence that the WW domains within the unliganded WW1-WW2 tandem module undergo a conformational transition from the initial state and attain a new stable conformation in which they either become physically associated and/or adopt a fixed spatial orientation relative to each other. In remarkable contrast, the liganded protein experiences a slow-and-steady rise in the RMSD reaching a maximum of close to 14Å and then slowly reaches some degree of structural equilibrium after 2μs during the course of simulation (Figure 7a). This implies that while the WW domains within the liganded WW1-WW2 tandem module may also attain a fixed spatial orientation, their relative stability is likely to be very low compared to the unliganded state. It is noteworthy that the dynamics of the constituent WW domains within both the unliganded and liganded proteins are comparable with an RMSD of around 1Å in sharp contrast to those of WW1-WW2 tandem module. This salient observation argues that while WW1-WW2 tandem module is likely to possess a rather high degree of structural plasticity, the triple-stranded β-sheet fold of constituent WW domains nonetheless retains remarkable structural rigidity and stability.
Figure 7.
Conformational dynamics as probed through MD analysis conducted on the structural model of WW1-WW2 tandem module of WWOX alone (unliganded) and in complex with p73 peptide containing the PPXY motif bound to the WW1 domain (liganded). (a) RMSD of backbone atoms (N, Cα and C) within each simulated structure relative to the initial modeled structure of unliganded (top panel) and liganded (bottom panel) WW1-WW2 tandem module as a function of simulation time. (b) RMSF of backbone atoms (N, Cα and C) averaged over the entire course of corresponding MD trajectory of unliganded (top panel) and liganded (bottom panel) WW1-WW2 tandem module as a function of residue number. Note that the red and green vertical rectangular boxes respectively demarcate the core residue boundaries (excluding the terminal loops) of WW1 and WW2 domains, while the gray vertical rectangular box denotes the residues spanning the interdomain linker. (c) Radius of gyration (Rg) of each simulated structure relative to the initial modeled structure of unliganded (top panel) and liganded (bottom panel) WW1-WW2 tandem module as a function of simulation time. Note that in (a) and (c), the overall physical parameter for each WW1-WW2 tandem module spanning residues 16–91 (black) is deconvoluted into the corresponding core regions (excluding the terminal loops) of constituent WW1 domain spanning residues 22–43 (red) and WW2 domain spanning residues 63–84 (green).
In order to shed light on the contribution of individual residues to the distinct dynamic behaviors observed for unliganded and liganded WW1-WW2 tandem module, we next analyzed the root mean square fluctuation (RMSF) of backbone atoms over the course of MD simulations (Figure 7b). Strikingly, our RMSF analysis reveals that not only the constituent WW domains but also the interdomain linker attain a relatively stable conformation in the unliganded protein. Additionally, the major source of structural instability within the unliganded protein—as reflected by a rather high RMSD of 7Å at structural equilibrium (Figure 1a)—arises from the relatively high mobility of residues spanning the N- and C-terminal loops compared to the rest of the protein. Surprisingly, while the pronounced flexibility of the interdomain linker is also a contributor, the apparent lack of structural equilibrium observed in the liganded protein appears to be largely due to the high mobility of residues located not only within the N- and C-terminal loops but also within the intradomain loops of WW2 domain. This suggests that the WW2 domain adopts a much more stable structure within the unliganded conformation compared to liganded protein—implying that the WW domains are likely to be physically associated within the unliganded protein. This notion is further supported when we compare the dependence of radius of gyration (Rg) of unliganded protein versus liganded conformation as a function of simulation time (Figure 7c). Thus, while Rg rapidly reaches a stable value of 13Å after the initial burst phase in the unliganded protein, it seems to hover between a high of 16Å and a low of 13Å for the liganded protein across the entire course of 2-μs simulation. In other words, the Rg comparison shows that while the unliganded protein attains a well-defined conformation, the liganded protein continues to sample various distinct conformations.
Ligand binding to WW1 domain is coupled to the dissociation of WW2 domain within the WW1-WW2 tandem module of WWOX
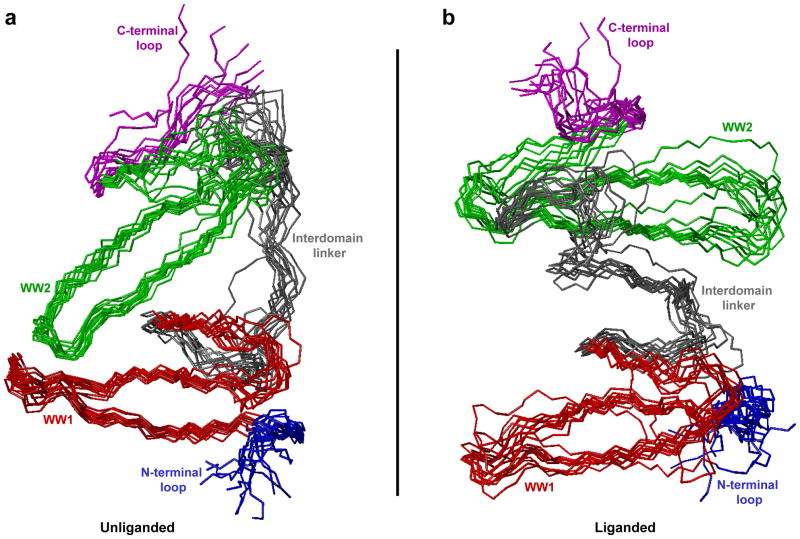
To understand the physical basis underlying the association of WW domains within the unliganded WW1-WW2 tandem module of WWOX and the apparent lack thereof in the liganded protein, we superimposed structural snapshots of corresponding conformations observed at various time intervals during the second half of MD simulations (Figure 8). Consistent with our dynamics analysis presented above (Figure 7), the structural superimposition of various simulated conformations suggests that the unliganded protein indeed adopts a well-defined conformation so as to allow the WW domains to physically associate with each other and attain a highly stable and fixed spatial orientation relative to each other. In contrast, while the WW domains also appear to attain some level of fixed spatial orientation relative to each other over the second half of simulation within the liganded state, this appears to be largely due to the structural ordering of the interdomain linker in lieu of direct physical association between the WW domains. Equally importantly, the rather pronounced dynamics behavior observed in the liganded state suggests that such spatial orientation is likely to be relatively unstable over the course of longer time scales.
Figure 8.
Superimposition of simulated structures as derived from MD analysis conducted on the structural model of WW1-WW2 tandem module of WWOX alone (unliganded) and in complex with p73 peptide containing the PPXY motif bound to the WW1 domain (liganded). Note that the superimposed structures for the unliganded (a) and liganded (b) WW1-WW2 tandem module were obtained at 200-ns time intervals over the second half of simulation (2–4μs). All 11 structures were superimposed with respect to the backbone atoms (N, Cα and C) of the core regions of WW1 (residues 22–43) and WW2 (residues 63–84) domains. In each case, the constituent WW1 (residues 22–43) and WW2 (residues 63–84) domains are respectively colored red and green, while the N-terminal loop (residues 16–21), interdomain linker (residues 44–62) and C-terminal loop (residues 85–91) are respectively shown in blue, gray and magenta. In (b), the p73 peptide is not shown for clarity.
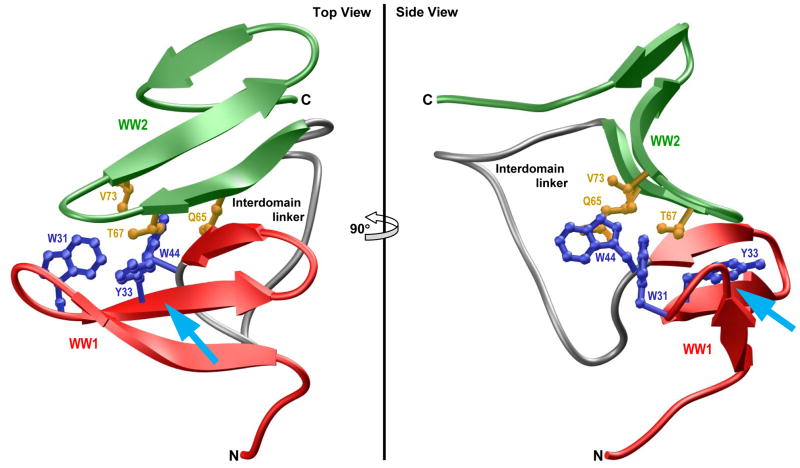
To further understand the atomic basis of the physical association between WW domains within the unliganded WW1-WW2 tandem module and how ligand binding may disrupt such association, we next analyzed the structure of unliganded protein observed at the end of 2-μs simulation (Figure 9). Interestingly, the physical association between the WW domains in the unliganded protein is driven by the docking of the convex side of WW2 domain onto the concave face of WW1 domain. These domain-domain interactions are primarily mediated via a two-prong mechanism: firstly, the indole moiety of W44 within the WW1 domain is sandwiched by the sidechain atoms of Q65 and V73 located within WW2 domain; secondly, the sidechain group of T67 within WW2 domain is sandwiched between aromatic rings of W31 and Y33 located within WW1 domain. Such van der Waals contacts are further buttressed by hydrogen bonding between the Nε1 atom of W44 within WW1 domain and Oε1 atom of Q65 within WW2 domain. Of particular note is the observation that the juxtaposition of WW2 domain on the concave face of WW1 domain partially blocks the ligand binding groove of the latter—the WW2 domain essentially acts like a “lid” that needs to be displaced in order to provide access to the incoming ligand. Accordingly, ligand binding must somehow result in the dissociation of WW2 domain so as to allow the WW1 domain to fully accommodate the ligand. Importantly, the free energy required to overcome such kinetic barrier must somehow be recouped. As noted above, the entropic gain resulting from the dissociation of WW2 domain should render such binding-coupled dissociation thermodynamically feasible. Consistent with this notion, our data presented above indeed suggest that favorable entropic factors underscore the ability of WW2 domain to aid ligand binding to WW1-WW2 tandem module of WWOX (Figure 3).
Figure 9.
Ribbon representation of the simulated structure of unliganded WW1-WW2 tandem module of WWOX obtained after 2μs of simulation time. Two alternative orientations related by a 90°-rotation about the vertical axis are depicted for closer inspection. The WW1 and WW2 domains are respectively colored red and green, while the interdomain linker is depicted in gray. Additionally, the sidechain moieties of residues within the WW1 and WW2 domains engaged in interdomain contacts are shown in blue and yellow, respectively. The cyan arrow indicates the direction of the incoming ligand as it would dock into the hydrophobic groove located on the concave face of WW1 domain, the same face that partially engages in interdomain contacts with the WW2 domain, in a competitive manner.
CONCLUSIONS
The WWOX tumor suppressor mediates a multitude of cellular activities including growth, proliferation, apoptosis and tumor suppression (1–3). While its mechanism of action remains largely elusive, it is well-established that the ability of WWOX to mediate protein-protein interactions in cellular signaling is largely mediated by the WW1 domain (12–16). Importantly, we postulated that the WW2 is an orphan domain that not only structurally stabilizes the WW1 domain within the context of WW1-WW2 tandem module but it also augments ligand binding to WWOX (13, 31). Our data reported herein not only further validate this hypothesis but also provide the physical basis of the action of WW2 domain. Notably, on the basis of MD simulations, our new study demonstrates that the WW2 domain physically associates with WW1 domain in the context of WW1-WW2 tandem module in the absence of ligand and that such association serves two purposes: firstly, it enables the WW2 domain to chaperone and promote the folding of WW1 domain to a triple-stranded β-sheet fold, a pre-requisite for subsequent ligand binding; secondly, the association of WW domains occurs in a manner such that it creates a kinetic barrier for ligand binding to WW1 domain—the WW2 domain essentially acts like a lid that is subsequently displaced to provide access to the incoming ligand. Remarkably, while ligand binding disrupts physical association of WW domains within the WW1-WW2 tandem module of WWOX, they nonetheless reorient to attain some level of spatial orientation in the liganded state though this does appear to be somewhat transient on a μs-timescale. We note that while our MD simulations reported herein were conducted using AMBER99SB-ILDN force field (37, 38), their reproducibility was further confirmed with GROMOS96-53A6 (57), CHARMM-27 (58, 59), and OPLS-AA (60, 61) force fields. The fact that all major MD force fields employed here reach a consensus on the conformational space available to the WW1-WW2 tandem module both in the unliganded and liganded state further solidifies the novel findings of our study.
In the language of statistical mechanics, the WW1-WW2 tandem module of WWOX most probably exists in an equilibrium exchange between a closed state and an open state (Figure 10). In the closed state, the physical association between the WW domains prevents ligand binding. In the open state, the WW domains are no longer physically associated but may still be somewhat spatially oriented so as to allow unhindered access to the ligand. In the absence of ligand, the equilibrium between the open and the closed state predominantly lies in favor of the latter. Subsequent ligand binding to WW1 domain within the open state shifts the equilibrium in its direction by virtue of the ability of WW1-WW2 tandem module to undergo a conformational switch so as to trigger the dissociation of WW2 domain. Consequently, the resulting gain in the conformational entropy of WW1-WW2 tandem module not only facilitates but also augments ligand binding. It is noteworthy that the ability of WW domains within the WW1-WW2 tandem module of WWOX to reorient themselves upon ligand binding to attain some level of fixed orientation appears to be somewhat similar to that observed for the tandem WW domains of Prp40 yeast splicing factor (47). In Prp40, the interdomain linker adopts α-helical conformation, thereby imparting a relatively stable and fixed spatial orientation upon tandem WW domains. However, in a manner akin to the liganded WW1-WW2 tandem module of WWOX, the WW domains of Prp40 do not physically associate but rather act as independent rigid bodies yet bounded together. This distinguishing feature of tandem WW domains of Prp40 is necessary in order to allow their ligand binding grooves to point outwards, thereby enabling them to bind to distinct ligands and bridge precisely between target sites within the splicing machinery.
Figure 10.
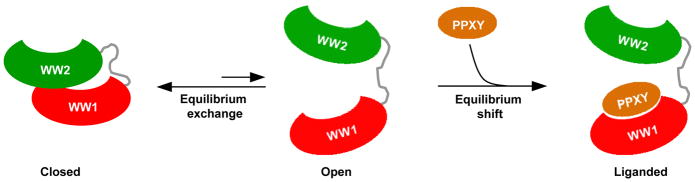
A model for ligand binding to WW1-WW2 tandem module of WWOX. In the unliganded state, the WW1-WW2 tandem module is proposed to exist in an equilibrium exchange between a closed state and an open state. In the closed state, the physical association between the WW domains prevents ligand binding. In the open state, the WW domains are no longer physically associated but may still be somewhat spatially oriented so as to allow unhindered access to the ligand. In the absence of PPXY ligand, the equilibrium between the open and the closed state predominantly lies in favor of the latter. Subsequent binding of PPXY ligand to WW1 domain within the open state shifts the equilibrium in its direction by virtue of the ability of WW1-WW2 tandem module to undergo a conformational switch so as to displace the WW2 lid.
In short, our study provokes the notion that the WW domains within the WW1-WW2 tandem module of WWOX are physically associated into a single supramodular unit with the WW2 domain acting as a lid over the ligand binding groove of WW1 domain. Ligand binding to WW1 domain displaces the WW2 domain in an allosteric manner and the resulting gain of entropy aids ligand binding to WW1 domain. Our findings thus underscore the importance of allosteric communication in the molecular operation of tandem protein modules.
Acknowledgments
This work was supported by the National Institutes of Health Grant R01-GM083897 and funds from the Sylvester Comprehensive Cancer Center (to AF), and by Breast Cancer Coalition grants (RFA #50709 & RFA #60707) from the Department of Health of Pennsylvania (to MS). CBM is a recipient of a postdoctoral fellowship from the National Institutes of Health (Award# T32-CA119929).
ABBREVIATIONS
- CD
Circular dichroism
- DOPE
Discrete optimized protein energy
- ErbB4
Erythroblastic (Erb) leukemia viral oncogene homolog B4
- FBP21
Formin-binding protein 21
- ITC
Isothermal titration calorimetry
- LIC
Ligation-independent cloning
- MD
Molecular dynamics
- MM
Molecular modeling
- p73
Tumor protein 73
- PPII
Polyproline type II (helix)
- PTCH1
Protein patched homolog 1
- PTPN14
Protein tyrosine phosphatase (non-receptor type) 14
- SEC
Size-exclusion chromatography
- SMAD7
Mothers against decapentaplegic homolog 7
- TMG2
Transmembrane gamma-carboxyglutamic acid protein 2
- YAP
YES-associated protein
- WBP1/2
WW domain-binding proteins 1 and 2
- WWOX
WW domain-containing oxidoreductase
References
- 1.Bednarek AK, Laflin KJ, Daniel RL, Liao Q, Hawkins KA, Aldaz CM. WWOX, a novel WW domain-containing protein mapping to human chromosome 16q23.3–24.1, a region frequently affected in breast cancer. Cancer Res. 2000;60:2140–2145. [PubMed] [Google Scholar]
- 2.Bednarek AK, Keck-Waggoner CL, Daniel RL, Laflin KJ, Bergsagel PL, Kiguchi K, Brenner AJ, Aldaz CM. WWOX, the FRA16D gene, behaves as a suppressor of tumor growth. Cancer Res. 2001;61:8068–8073. [PubMed] [Google Scholar]
- 3.Hezova R, Ehrmann J, Kolar Z. WWOX, a new potential tumor suppressor gene. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub. 2007;151:11–15. doi: 10.5507/bp.2007.002. [DOI] [PubMed] [Google Scholar]
- 4.Nunez MI, Ludes-Meyers J, Abba MC, Kil H, Abbey NW, Page RE, Sahin A, Klein-Szanto AJ, Aldaz CM. Frequent loss of WWOX expression in breast cancer: correlation with estrogen receptor status. Breast Cancer Res Treat. 2005;89:99–105. doi: 10.1007/s10549-004-1474-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aqeilan RI, Kuroki T, Pekarsky Y, Albagha O, Trapasso F, Baffa R, Huebner K, Edmonds P, Croce CM. Loss of WWOX expression in gastric carcinoma. Clin Cancer Res. 2004;10:3053–3058. doi: 10.1158/1078-0432.ccr-03-0594. [DOI] [PubMed] [Google Scholar]
- 6.Aqeilan RI, Croce CM. WWOX in biological control and tumorigenesis. J Cell Physiol. 2007;212:307–310. doi: 10.1002/jcp.21099. [DOI] [PubMed] [Google Scholar]
- 7.Aqeilan RI, Hagan JP, Aqeilan HA, Pichiorri F, Fong LY, Croce CM. Inactivation of the Wwox gene accelerates forestomach tumor progression in vivo. Cancer Res. 2007;67:5606–5610. doi: 10.1158/0008-5472.CAN-07-1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pluciennik E, Kusinska R, Potemski P, Kubiak R, Kordek R, Bednarek AK. WWOX--the FRA16D cancer gene: expression correlation with breast cancer progression and prognosis. Eur J Surg Oncol. 2006;32:153–157. doi: 10.1016/j.ejso.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 9.Lewandowska U, Zelazowski M, Seta K, Byczewska M, Pluciennik E, Bednarek AK. WWOX, the tumour suppressor gene affected in multiple cancers. J Physiol Pharmacol. 2009;60(Suppl 1):47–56. [PubMed] [Google Scholar]
- 10.Zelazowski MJ, Pluciennik E, Pasz-Walczak G, Potemski P, Kordek R, Bednarek AK. WWOX expression in colorectal cancer--a real-time quantitative RT-PCR study. Tumour Biol. 2011;32:551–560. doi: 10.1007/s13277-010-0150-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sudol M, Hunter T. NeW wrinkles for an old domain. Cell. 2000;103:1001–1004. doi: 10.1016/s0092-8674(00)00203-8. [DOI] [PubMed] [Google Scholar]
- 12.Ludes-Meyers JH, Kil H, Bednarek AK, Drake J, Bedford MT, Aldaz CM. WWOX binds the specific proline-rich ligand PPXY: identification of candidate interacting proteins. Oncogene. 2004;23:5049–5055. doi: 10.1038/sj.onc.1207680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McDonald CB, Buffa L, Bar-Mag T, Salah Z, Bhat V, Mikles DC, Deegan BJ, Seldeen KL, Malhotra A, Sudol M, Aqeilan RI, Nawaz Z, Farooq A. Biophysical basis of the binding of WWOX tumor suppressor to WBP1 and WBP2 adaptors. J Mol Biol. 2012;422:58–74. doi: 10.1016/j.jmb.2012.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aqeilan RI, Donati V, Palamarchuk A, Trapasso F, Kaou M, Pekarsky Y, Sudol M, Croce CM. WW domain-containing proteins, WWOX and YAP, compete for interaction with ErbB-4 and modulate its transcriptional function. Cancer Res. 2005;65:6764–6772. doi: 10.1158/0008-5472.CAN-05-1150. [DOI] [PubMed] [Google Scholar]
- 15.Aqeilan RI, Pekarsky Y, Herrero JJ, Palamarchuk A, Letofsky J, Druck T, Trapasso F, Han SY, Melino G, Huebner K, Croce CM. Functional association between Wwox tumor suppressor protein and p73, a p53 homolog. Proc Natl Acad Sci U S A. 2004;101:4401–4406. doi: 10.1073/pnas.0400805101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Del Mare S, Salah Z, Aqeilan RI. WWOX: its genomics, partners, and functions. J Cell Biochem. 2009;108:737–745. doi: 10.1002/jcb.22298. [DOI] [PubMed] [Google Scholar]
- 17.Zhao B, Wei X, Li W, Udan RS, Yang Q, Kim J, Xie J, Ikenoue T, Yu J, Li L, Zheng P, Ye K, Chinnaiyan A, Halder G, Lai ZC, Guan KL. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 2007;21:2747–2761. doi: 10.1101/gad.1602907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bertini E, Oka T, Sudol M, Strano S, Blandino G. YAP: at the crossroad between transformation and tumor suppression. Cell Cycle. 2009;8:49–57. doi: 10.4161/cc.8.1.7259. [DOI] [PubMed] [Google Scholar]
- 19.Sudol M. Newcomers to the WW Domain-Mediated Network of the Hippo Tumor Suppressor Pathway. Genes Cancer. 2010;1:1115–1118. doi: 10.1177/1947601911401911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sudol M, Harvey KF. Modularity in the Hippo signaling pathway. Trends Biochem Sci. 2010;35:627–633. doi: 10.1016/j.tibs.2010.05.010. [DOI] [PubMed] [Google Scholar]
- 21.Salah Z, Aqeilan RI. WW domain interactions regulate the Hippo tumor suppressor pathway. Cell Death Dis. 2011;2:e172. doi: 10.1038/cddis.2011.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen HI, Sudol M. The WW domain of Yes-associated protein binds a proline-rich ligand that differs from the consensus established for Src homology 3-binding modules. Proc Natl Acad Sci U S A. 1995;92:7819–7823. doi: 10.1073/pnas.92.17.7819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen HI, Einbond A, Kwak SJ, Linn H, Koepf E, Peterson S, Kelly JW, Sudol M. Characterization of the WW domain of human yes-associated protein and its polyproline-containing ligands. J Biol Chem. 1997;272:17070–17077. doi: 10.1074/jbc.272.27.17070. [DOI] [PubMed] [Google Scholar]
- 24.Komuro A, Nagai M, Navin NE, Sudol M. WW domain-containing protein YAP associates with ErbB-4 and acts as a co-transcriptional activator for the carboxyl-terminal fragment of ErbB-4 that translocates to the nucleus. J Biol Chem. 2003;278:33334–33341. doi: 10.1074/jbc.M305597200. [DOI] [PubMed] [Google Scholar]
- 25.Omerovic J, Puggioni EM, Napoletano S, Visco V, Fraioli R, Frati L, Gulino A, Alimandi M. Ligand-regulated association of ErbB-4 to the transcriptional co-activator YAP65 controls transcription at the nuclear level. Exp Cell Res. 2004;294:469–479. doi: 10.1016/j.yexcr.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 26.Levy D, Adamovich Y, Reuven N, Shaul Y. Yap1 phosphorylation by c-Abl is a critical step in selective activation of proapoptotic genes in response to DNA damage. Mol Cell. 2008;29:350–361. doi: 10.1016/j.molcel.2007.12.022. [DOI] [PubMed] [Google Scholar]
- 27.Kulman JD, Harris JE, Xie L, Davie EW. Proline-rich Gla protein 2 is a cell-surface vitamin K-dependent protein that binds to the transcriptional coactivator Yes-associated protein. Proc Natl Acad Sci U S A. 2007;104:8767–8772. doi: 10.1073/pnas.0703195104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu X, Yang N, Figel SA, Wilson KE, Morrison CD, Gelman IH, Zhang J. PTPN14 interacts with and negatively regulates the oncogenic function of YAP. Oncogene. 2013;32:1266–1273. doi: 10.1038/onc.2012.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ferrigno O, Lallemand F, Verrecchia F, L’Hoste S, Camonis J, Atfi A, Mauviel A. Yes-associated protein (YAP65) interacts with Smad7 and potentiates its inhibitory activity against TGF-beta/Smad signaling. Oncogene. 2002;21:4879–4884. doi: 10.1038/sj.onc.1205623. [DOI] [PubMed] [Google Scholar]
- 30.Linn H, Ermekova KS, Rentschler S, Sparks AB, Kay BK, Sudol M. Using molecular repertoires to identify high-affinity peptide ligands of the WW domain of human and mouse YAP. Biol Chem. 1997;378:531–537. doi: 10.1515/bchm.1997.378.6.531. [DOI] [PubMed] [Google Scholar]
- 31.Schuchardt BJ, Bhat V, Mikles DC, McDonald CB, Sudol M, Farooq A. Molecular origin of the binding of WWOX tumor suppressor to ErbB4 receptor tyrosine kinase. Biochemistry. 2013;52:9223–9236. doi: 10.1021/bi400987k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A. Protein Identification and Analysis Tools on the ExPASy Server. In: Walker JM, editor. The Proteomics Protocols Handbook. Humana Press; Totowa, New Jersey, USA: 2005. pp. 571–607. [Google Scholar]
- 33.Wiseman T, Williston S, Brandts JF, Lin LN. Rapid measurement of binding constants and heats of binding using a new titration calorimeter. Anal Biochem. 1989;179:131–137. doi: 10.1016/0003-2697(89)90213-3. [DOI] [PubMed] [Google Scholar]
- 34.Marti-Renom MA, Stuart AC, Fiser A, Sanchez R, Melo F, Sali A. Comparative Protein Structure Modeling of Genes and Genomes. Annu Rev Biophys Biomol Struct. 2000;29:291–325. doi: 10.1146/annurev.biophys.29.1.291. [DOI] [PubMed] [Google Scholar]
- 35.Carson M. Ribbons 2.0. J Appl Crystallogr. 1991;24:958–961. [Google Scholar]
- 36.Van Der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJ. GROMACS: fast, flexible, and free. J Comput Chem. 2005;26:1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 37.Lindorff-Larsen K, Piana S, Palmo K, Maragakis P, Klepeis JL, Dror RO, Shaw DE. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins. 2010;78:1950–1958. doi: 10.1002/prot.22711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hornak V, Abel R, Okur A, Strockbine B, Roitberg A, Simmerling C. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins. 2006;65:712–725. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Toukan K, Rahman A. Molecular-dynamics study of atomic motions in water. Physical Review B. 1985;31:2643–2648. doi: 10.1103/physrevb.31.2643. [DOI] [PubMed] [Google Scholar]
- 40.Berendsen HJC, Grigera JR, Straatsma TP. The Missing Term in Effective Pair Potentials. J Phys Chem. 1987;91:6269–6271. [Google Scholar]
- 41.Darden TA, York D, Pedersen L. Particle mesh Ewald: An N.log(N) method for Ewald sums in large systems. J Chem Phys. 1993;98:10089–10092. [Google Scholar]
- 42.Hess B, Bekker H, Berendsen HJC, Fraaije JGEM. LINCS: A linear constraint solver for molecular simulations. J Comput Chem. 1997;18:1463–1472. [Google Scholar]
- 43.Koradi R, Billeter M, Wuthrich K. MOLMOL: a program for display and analysis of macromolecular structures. J Mol Graph. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 44.Sudol M, Recinos CC, Abraczinskas J, Humbert J, Farooq A. WW or WoW: the WW domains in a union of bliss. IUBMB Life. 2005;57:773–778. doi: 10.1080/15216540500389039. [DOI] [PubMed] [Google Scholar]
- 45.Huang X, Beullens M, Zhang J, Zhou Y, Nicolaescu E, Lesage B, Hu Q, Wu J, Bollen M, Shi Y. Structure and function of the two tandem WW domains of the pre-mRNA splicing factor FBP21 (formin-binding protein 21) J Biol Chem. 2009;284:25375–25387. doi: 10.1074/jbc.M109.024828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fedoroff OY, Townson SA, Golovanov AP, Baron M, Avis JM. The Structure and Dynamics of Tandem WW Domains in a Negative Regulator of Notch Signaling, Suppressor of Deltex. J Biol Chem. 2004;279:34991–35000. doi: 10.1074/jbc.M404987200. [DOI] [PubMed] [Google Scholar]
- 47.Wiesner S, Stier G, Sattler M, Macias MJ. Solution Structure and Ligad Recognition of the WW Domain of the Yeast Splicing Factor Prp40. J Mol Biol. 2002;324:807–822. doi: 10.1016/s0022-2836(02)01145-2. [DOI] [PubMed] [Google Scholar]
- 48.Kanelis V, Farrow NA, Kay LE, Rotin D, Forman-Kay JD. NMR studies of tandem WW domains of Nedd4 in complex with a PY motif-containing region of the epithelial sodium channel. Biochem Cell Biol. 1998;76:341–350. doi: 10.1139/bcb-76-2-3-341. [DOI] [PubMed] [Google Scholar]
- 49.Chong PA, Lin H, Wrana JL, Forman-Kay JD. Coupling of tandem Smad ubiquitination regulatory factor (Smurf) WW domains modulates target specificity. Proc Natl Acad Sci U S A. 2010;107:18404–18409. doi: 10.1073/pnas.1003023107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Webb C, Upadhyay A, Giuntini F, Eggleston I, Furutani-Seiki M, Ishima R, Bagby S. Structural features and ligand binding properties of tandem WW domains from YAP and TAZ, nuclear effectors of the Hippo pathway. Biochemistry. 2011;50:3300–3309. doi: 10.1021/bi2001888. [DOI] [PubMed] [Google Scholar]
- 51.Macias MJ, Hyvonen M, Baraldi E, Schultz J, Sudol M, Saraste M, Oschkinat H. Structure of the WW domain of a kinase-associated protein complexed with a proline-rich peptide. Nature. 1996;382:646–649. doi: 10.1038/382646a0. [DOI] [PubMed] [Google Scholar]
- 52.Pires JR, Taha-Nejad F, Toepert F, Ast T, Hoffmuller U, Schneider-Mergener J, Kuhne R, Macias MJ, Oschkinat H. Solution structures of the YAP65 WW domain and the variant L30 K in complex with the peptides GTPPPPYTVG, N-(n-octyl)-GPPPY and PLPPY and the application of peptide libraries reveal a minimal binding epitope. J Mol Biol. 2001;314:1147–1156. doi: 10.1006/jmbi.2000.5199. [DOI] [PubMed] [Google Scholar]
- 53.Huang X, Poy F, Zhang R, Joachimiak A, Sudol M, Eck MJ. Structure of a WW domain containing fragment of dystrophin in complex with beta-dystroglycan. Nat Struct Biol. 2000;7:634–638. doi: 10.1038/77923. [DOI] [PubMed] [Google Scholar]
- 54.Kanelis V, Rotin D, Forman-Kay JD. Solution structure of a Nedd4 WW domain-ENaC peptide complex. Nat Struct Biol. 2001;8:407–412. doi: 10.1038/87562. [DOI] [PubMed] [Google Scholar]
- 55.Woody RW. Circular dichroism spectrum of peptides in the poly(Pro)II conformation. J Am Chem Soc. 2009;131:8234–8245. doi: 10.1021/ja901218m. [DOI] [PubMed] [Google Scholar]
- 56.Rabanal F, Ludevid MD, Pons M, Giralt E. CD of proline-rich polypeptides: application to the study of the repetitive domain of maize glutelin-2. Biopolymers. 1993;33:1019–1028. doi: 10.1002/bip.360330704. [DOI] [PubMed] [Google Scholar]
- 57.Oostenbrink C, Villa A, Mark AE, van Gunsteren WF. A biomolecular force field based on the free enthalpy of hydration and solvation: the GROMOS force-field parameter sets 53A5 and 53A6. J Comp Chem. 2004;25:1656–1676. doi: 10.1002/jcc.20090. [DOI] [PubMed] [Google Scholar]
- 58.Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M. CHARMM: A program for macromolecular energy, minimization, and dynamics calculations. J Comp Chem. 1983;4:187–217. [Google Scholar]
- 59.Brooks BR, Brooks CLr, Mackerell ADJ, Nilsson L, Petrella RJ, Roux B, Won Y, Archontis G, Bartels C, Boresch S, Caflisch A, Caves L, Cui Q, Dinner AR, Feig M, Fischer S, Gao J, Hodoscek M, Im W, Kuczera K, Lazaridis T, Ma J, Ovchinnikov V, Paci E, Pastor RW, Post CB, JZP, Schaefer M, Tidor B, Venable RM, Woodcock HL, Wu X, Yang W, York DM, Karplus M. CHARMM: The biomolecular simulation program. J Comp Chem. 2009;30:1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jorgensen WL, Tirado-Rives J. The OPLS Force Field for Proteins: Energy Minimizations for Crystals of Cyclic Peptides and Crambin. J Am Chem Soc. 1988;110:1657–1666. doi: 10.1021/ja00214a001. [DOI] [PubMed] [Google Scholar]
- 61.Kaminski GA, Friesner RA, Tirado-Rives J, Jorgensen WL. Evaluation and Reparametrization of the OPLS-AA Force Field for Proteins via Comparison with Accurate Quantum Chemical Calculations on Peptides. J Phys Chem B. 2001;105:6474–6487. [Google Scholar]