Abstract
T cells play a crucial role for viral clearance or persistence; however, the precise mechanisms that control their responses during viral infection remain incompletely understood. microRNAs (miR) have been implicated as key regulators controlling diverse biological processes through posttranscriptional repression. Here, we demonstrate that hepatitis C virus (HCV)-mediated decline of miR-181a expression impairs CD4+ T cell responses via over-expression of dual specific phosphatase 6 (DUSP6). Specifically, a significant decline of miR-181a expression along with over-expression of DUSP6 were observed in CD4+ T cells from chronically HCV-infected individuals compared to healthy subjects, and the levels of miR-181a loss were found to be negatively associated with the levels of DUSP6 over-expression in these cells. Importantly, reconstitution of miR-181a or blockade of DUSP6 expression in CD4+ T cells led to improved T cell responses including enhanced CD25 and CD69 expressions, increased IL-2 expression, and improved proliferation of CD4+ T cells derived from chronically HCV-infected individuals. Since a decline of miR-181a concomitant with DUSP6 over-expression are the signature markers for age-associated T cell senescence, these findings provide novel mechanistic insights into HCV-mediated premature T cell aging via miR-181a-regulated DUSP6 signaling, and reveal new targets for therapeutic rejuvenation of impaired T cell responses during chronic viral infection.
Keywords: CD4 T cells, DUSP6, hepatitis C virus, immune regulation, miR-181a
Introduction
Hepatitis C virus (HCV) is a blood-borne viral infection characterized by a high rate (over 80%) of chronicity. HCV has evolved multiple strategies to evade host immunity, thus becoming an excellent model to study the mechanisms of persistent viral infections [1–2]. While the use of direct antiviral agents (DAAs) has resulted in a significant improvement in the outcome of HCV treatment, this therapeutic cocktail is still in the development, costly, and likely to face new issues including viral mutation, relapse, and reinfection following therapy [3–4]. Additionally, the lack of a vaccine for this virus is a major hurdle to control this global infection. The failure to successfully manage this chronic viral infection and to develop an effective vaccine stems from our incomplete understanding of the host immunity to HCV that lead to viral persistence.
T cells play a crucial role for viral clearance or persistence; however, the precise mechanisms that control their responses during viral infection remain largely unknown. microRNAs (miRNAs or miR) are non-coding 12–23 nucleotide RNA molecules that regulate gene expression through post-transcriptional repression or target mRNA degradation [5]. Compelling evidence has suggested that some miRNAs are able to regulate HCV replication and its related liver diseases by directly interacting with the HCV genome or indirectly controlling virus-associated host pathways [6–9]. How HCV modulates the expression of miRNAs and host cellular proteins in infected individuals, particularly in T cells for the benefit of its survival and persistence in vivo, is less understood. A recent study has revealed that the signature marker for T cell aging - dual specific phosphatase 6 (DUSP6) – is regulated by miR-181a and raises the threshold for T cell activation after T cell receptor (TCR) stimulation [10]. DUSP6 is a cytoplasmic phosphatase that inactivates TCR signaling by dephosphorylating the phosphoserine and phosphotyrosine residues of extracellular signal-regulated kinase (ERK) to regulate T cell proliferation and differentiation. It remains unknown whether miR-181a or DUSP6 function as cellular mediators by which HCV shapes T cell responses to facilitate the establishment and maintenance of viral persistence.
In this study, we provide preliminary evidence demonstrating that miR-181a and DUSP6 are utilized by HCV to dampen T cell responses. We demonstrate that an HCV-mediated decline of miR-181a expression impairs CD4+ T cell responses via up-regulation of DUSP6 expression, suggesting that these molecules may serve as novel targets for restoration of impaired T cell functions during chronic viral infection.
Materials and Methods
Subjects
The study protocol was approved by a joint institutional review board at East Tennessee State University and James H. Quillen VA Medical Center (ETSU/VA IRB, Johnson City, TN). The study subjects comprised two populations: 1) 48 chronically HCV-infected patients, all virologically and serologically positive for HCV prior to antiviral treatment, with HCV genotype 1 (70%), type 2 or 3 (30%), and viral loads ranging from 12,300 ~ 500,000 IU/ml; 2) 18 healthy subjects (HS), the buffy coat while blood cells derived from Key Biologics, LLC., Memphis, TN. Written informed consent was obtained from all participants. The majority of subjects were male. The mean age of HCV-infected subjects was comparable to HS (P>0.05).
Cell isolation
Peripheral blood mononuclear cells (PBMCs) were isolated from the subjects by Ficoll-density centrifugation with lympho-H (Atlanta biological, Lawrenceville, GA) and then viably cryopreserved in freezing medium in liquid nitrogen. If indicated, CD4+ T cells were further purified from isolated PBMCs by negative selection with magnetic beads using a CD4+ T cell Isolation Kit (Cell purity > 95%, Miltenyi Biotec, Auburn, CA).
Co-culture of human primary CD4+ T cells with HCV+ or HCV− Huh7 hepatocytes
Huh7 cells stably transfected with or without HCV genomic RNA were co-cultured with purified healthy CD4+ T cells as described previously [12]. Following 72 h co-culture in the medium supplemented with 100 IU/ml rhIL-2 with or without 1 μg/ml anti-CD3/CD28 stimulation, CD4+ T cells were harvested and the expressions of DUSP6 and IL-2 were examined by flow cytometry. miRNA in CD4+ T cells were extracted, and miR-181a was quantified by real-time RT-PCR.
miScript miRNA array
Total cellular RNA in CD4+ T cells from 6 chronic HCV-infected patients and 6 HS was isolated and cleaned using the miRNeasy Mini kit (Qiagen) according to the manufacturer’s instructions. RNA quality and quantity were measured using a BioPhotometer spectrophotometer, and RNA integrity was determined using gel electrophoresis. The primers for the 384 miRNAs and PCR reagents were purchased from Qiagen. Data were analyzed using RT2 profiler PCR array data analysis software (http://www.sabiosciences.com/pcrarraydataanalysis.php). Fold-change values >2 were indicative of up-regulated miRNAs; fold-change values <0.5 and fold-regulation values <−2 were indicative of down-regulated miRNAs.
Real-time PCR quantification of microRNA and mRNA
miRNA were isolated from CD4+ T cells using RNAzol®RT (Molecular Research Center, Inc. Cincinnati, OH). cDNA was generated by TaqMan miRNA Reverse Transcription kit (Life Technologies, Grand Island, NY). Real-time PCR was conducted using a CFX96™Real-time PCR Detection System (Bio-Rad Laboratories, Inc. Hercules, CA). miR-181a levels, normalized to the U6 small nucleolar RNA (snRU6), were quantified with the relative quantification method (2−ΔΔCt) [10]. Cellular total mRNA was isolated from CD4+ T cells using the RNeasy Mini Kit (Qiagen Inc, Valencia, CA). cDNA was generated with the RT2 First Strand Kit (Qiagen). DUSP6 transcripts were quantified by RT2 SYBR Green qPCR Master mix and RT2 qPCR Primer Assay (Qiagen), normalized to the GAPDH transcripts, and expressed as relative quantification method (2−ΔΔCt).
Flow cytometry
The following antibodies were used for immunostaining: FITC-DUSP6 (Bioss, Inc. Wobum, MA), APC-CD4/PE-IL-2 (eBioscience, San Diego, CA), PE-CD69/Percp-CD25 (Biolengend, San Diego, CA), and Alexa Fluor 488-Phospho-Akt (ser473) (193H12; Cell Signaling, Danvers, MA). Procedures for intracellular and cytokine staining were performed essentially as described previously [12–15]. The isotype-matched controls (eBioscience) were used to determine the level of background staining; Fluorescence minus one (FMO) strategy was used to determine background levels of staining and adjust multicolor compensation for cell gating. The cells were analyzed on an Accuri™ C6 flow cytometer (BD, Franklin Lakes, NJ) using FlowJo software (Tree Star, Inc., Ashland, OR).
Transfection of human primary CD4+ T cells with miR-181a precursors
Purified CD4+ T cells from HCV-infected individuals were transfected with 50 pmol of miR-181a precursors or negative control (Life technologies, Grand Island, NY) using the Human T Lymphocyte Nucleofector Kit and the Nucleofector I Device (Lonza, Allendale, NJ) according to the manufacturer’s instructions.
DUSP6 blockade
Purified CD4+ T cells from HCV patients were incubated with different concentrations of the DUSP6 inhibitor, BCI (EMD Millipore, Billerica, MA) at 37°C for 2 h. Cells were washed three times and then stimulated with anti-CD3/CD28 antibodies (1 μg/ml). After 20 h incubation and 4 h brefeldin, the cells were harvested for analysis of CD69, CD25, and IL-2 expression by Accuri™ C6 flow cytometer; DUSP6 expression by real-time PCR.
Proliferation assays
PBMCs or CD4+ T cells isolated from chronic HCV patients were labeled with CFSE (10 μM, Invitrogen) for 15 min at 37°C per manufacture’s instruction, washed with complete medium, and cultured (1 × 106) in a 12-well plate in the presence anti-CD3/CD28 (1 μg/ml) and rhIL-2 (100 U/ml, R&D System). After incubation for 5~7 d, the cells were immunostained with PE-CD4 and analyzed by Accuri™ C6 flow cytometer and FlowJo software.
Statistical analysis
Study results were expressed as the mean ± standard deviation (SD). Comparison between two groups was performed by multiple comparisons testing/least significant difference or Tukey’s procedure depending on the ANOVA F test Prism software (version 4; GraphPad Software) by a nonparametric Mann–Whitney U test. A pair-wise t test was used to compare the significance of changes in DUSP6 inhibition or miR-181a precursor transfection experiments. Correlations between DUSP6 expression on CD4+ T cells and miR-181a expression were analyzed using a Pearson Correlation program. Values of p <0.05, p<0.01, and p<0.001 were considered significant or very significant, respectively.
Results
miRNA expression profiling in CD4+ T cells from HCV-infected patients versus HS
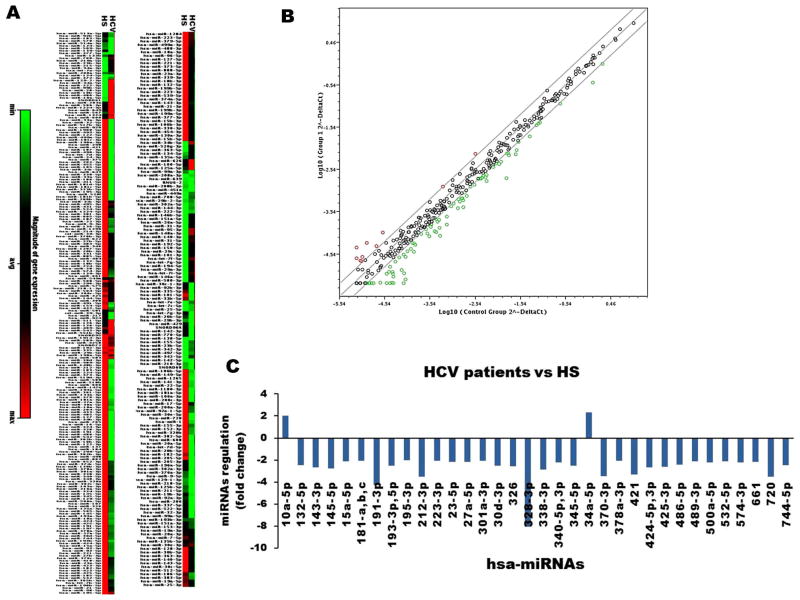
It is well-established that persistent viruses such as HCV and HIV can lead to T cell exhaustion and senescence by encoding or altering the expression and function of miRNAs that target cellular and viral mRNAs to regulate virus replication and cell functions [16–21]. In order to identify human miRNAs involved in regulating T cell responses during HCV infection, we compared miRNA arrays in CD4+ T cells from chronically HCV-infected individuals versus HS. A total of 384 well-known miRNAs were measured in this array. Differentially expressed miRNAs from HCV versus HS are shown as heat maps in Fig. 1A and a scatter plot of the log base 10 for the hybridization intensity of each miRNA in the two groups in Fig 1B. Expression of 336 miRNAs was found to be unchanged between HCV and HS. A total of 38 miRNAs were identified differentially expressed in CD4+ T cells from HCV compared to the HS; 2 miRNAs were up-regulated, and 36 miRNAs were down-regulated (Fig. 1C). We were particularly interested in miR-181a, b, and c, whose expressions were found consistently low in HCV compared to HS in this array, and these miRs had been reported by others to be relevant to age-associated T cell senescence [10].
Fig. 1. miRNA expression array in CD4+ T cells from chronically HCV-infected individuals versus HS.
A) Heat maps showing differentially expressed miRNAs in CD4+ T cells. Each column represents results from samples of 6 HCV-infected individuals and 6 HS, respectively. Each row corresponds to a single miRNA probe. Colors indicate the level of miRNA expression (red, high level; green, low level). B) Scatter plot of the log base 10 for the intensity of each miRNA in the two groups (x-axis, HS shown as control group; y-axis, HCV-infected group). The middle line indicates a fold-change (2−ΔCt) of 1. The top and the bottom lines indicate the desired 2 fold-change in miRNA expression threshold. The red points above the top line represent upregulated miRNAs. The green points below the bottom line represent downregulated miRNAs. C) Differentially upregulated and downregulated miRNAs from HCV-infected individuals compared to those from HS.
HCV infection down-regulates the expression of miR-181a in CD4+ T cells
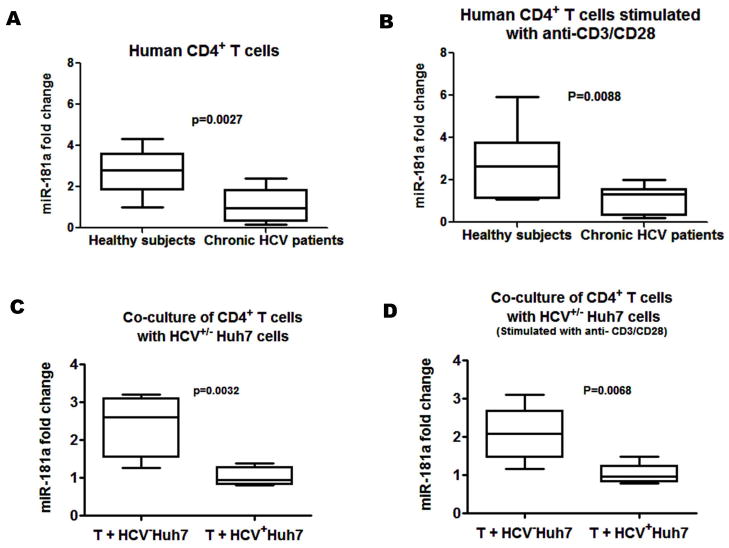
A recent study has shown that a decline in miR-181a expression with age impairs TCR sensitivity [10]. To determine whether HCV infection caused the decline in miR-181a expression, we validated the miR-181a expression by real-time RT-PCR in CD4+ T cells from 20 HCV-infected patients and 14 HS. As shown in Fig. 2A and 2B, the levels of miR-181a in both unstimulated (A) and TCR-stimulated (B) CD4+ T cells, in the absence or presence of anti-CD3/CD28 stimulation ex vivo, were significantly decreased from HCV-infected patients compared to HS.
Fig. 2. miR-181a expression in CD4+ T cells from HCV-infected individuals versus HS.
A and B) CD4+ T cells were purified from PBMCs isolated from 20 HCV-infected individuals and 14 HS using magnetic beads. miRNA were isolated from CD4+ T cells in the presence and absence of anti-CD3/CD28 antibody stimulation ex vivo. miR-181a expression was measured by real-time PCR by specific Taqman assays, normalized to snRU6 internal control, and quantified with the 2−ΔΔCt relative quantification method. Medians, twenty-fifth and seventy-fifth percentiles as boxes and tenth and ninetieth percentiles as whiskers are shown for the miR-181a fold changes of HCV-infected individuals compared to the HS. C and D) Purified CD4+ T cells from 6 HS were incubated with HCV+/− Huh7 hepatocytes at a ratio 4:1 in vitro for 72 h with or without 1 μg/ml anti-CD3/CD28 stimulation. miR-181a expression in CD4+ T cells were examined by real-time PCR and analyzed for the fold change of those co-cultured with HCV− Huh7 compared to the one co-cultured with HCV+ Huh7.
To further assess the role of HCV in modulation of miR-181a expression, we examined the miR-181a expression in purified healthy CD4+ T cells co-cultured with HCV+/− Huh7 hepatocytes in vitro for 72 h, with or without anti-CD3/CD28 stimulation. Again, a more than 2-fold decline in miR-181a expression was observed in CD4+ T cells co-cultured with HCV+ Huh7 cells compared with those co-cultured with HCV− Huh7 cells (Fig. 2C and 2D). These findings suggest that HCV induces a decline in miR-181a expression that may influence target gene expression to facilitate viral hijacking of critical host pathways associated with T cell dysfunction.
DUSP6 is over-expressed in CD4+ T cells with HCV infection
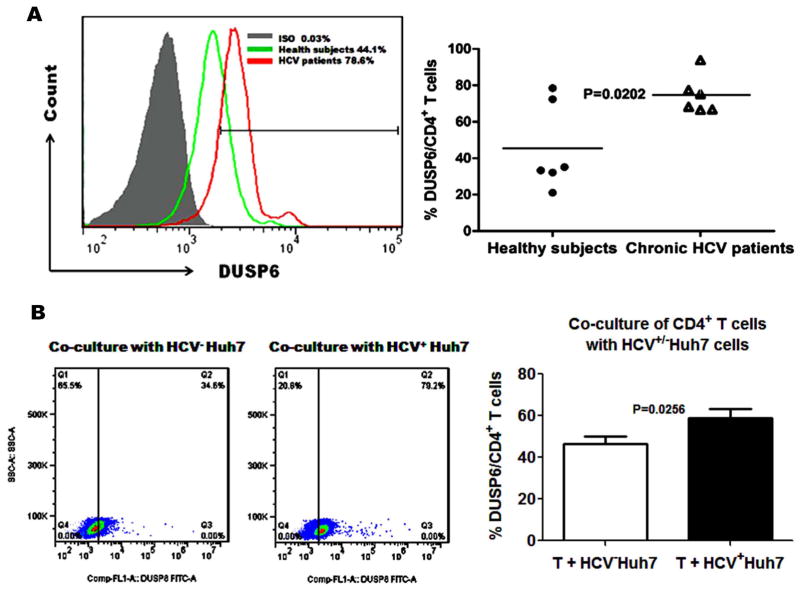
We have previously demonstrated that HCV core, the first protein to be expressed and circulating in the blood of infected individuals, impairs human T cell response by inhibiting the phosphorylation of TCR-induced ERK and mitogen-activated ERK kinase (MEK) [20]. One major feedback loop that controls the activation of the ERK pathway and attenuates TCR signaling involves DUSP6, a cytoplasmic phosphatase with substrate specificity for phosphorylated ERK. Increased DUSP6 protein expression during T cell senescence has been implicated in the reduced TCR sensitivity with aging [10]. To study the role of DUSP6 in HCV-induced ERK inhibition and CD4+ T cell suppression during HCV infection, we examined the expression of DUSP6 in CD4+ T cells from HCV-infected patients versus HS. As shown in the representative histogram and summary data in Fig. 3A, DUSP6 was over-expressed in anti-CD3/CD28-stimulated CD4+ T cells from HCV-infected patients compared to age-matched HS as determined by flow cytometry analysis. Again, we examined DUSP6 expression in purified healthy CD4+ T cells co-cultured with HCV+/− Huh7 hepatocytes in vitro for 72 h with anti-CD3/CD28 stimulation. As shown in the representative dot plots and summary data in Fig. 3B, the DUSP6+ cell frequencies and mean fluorescence intensity in CD4+ T cells co-cultured with HCV+ Huh7 cells were significantly increased compared to those co-cultured with HCV− Huh7 cells. These results indicate that HCV infection, while inhibiting miR-181a expression, induces DUSP6 over-expression in CD4+ T cells.
Fig. 3. HCV-induced DUSP6 expression.
A) PBMCs isolated from 6 HCV patients and 6 age-comparable HS were stimulated with anti-CD3/CD28 for 24 h ex vivo. Cells were immunostained with FITC-conjugated anti-human DUSP6 antibody and PE-conjugated anti-CD4 antibody and analyzed by flow cytometry. Representative histogram of DUSP6 expression by CD4+ T cells in HCV patients and HS versus isotype-IgG control is shown in the left panel. Percentages of DUSP6-expressing CD4+ T cells in HCV patients and HS are shown in the right panel. Each symbol represents an individual subject, and the horizontal lines represent median values. B) CD4+ T cells were purified from PBMCs isolated from 6 HS using magnetic beads. Purified CD4+ T cells were co-cultured with HCV+/− Huh7 hepatocytes at a ratio 4:1 in vitro for 72 h with 1 μg/ml of anti-CD3/CD28 stimulation and immunostained with FITC-conjugated anti-human DUSP6 followed by flow cytometric analysis. Representative dot plots of DUSP6 expression in CD4+ T cells from co-cultured with HCV+ Huh7 and HCV− Huh7 are shown in the left panel. Percentages of DUSP6-expressing CD4+ T cells in those co-cultured with HCV+ Huh7 and HCV− Huh7 are shown in the right panel.
miR-181a controls T cell responses through regulating DUSP6 expression in HCV infection
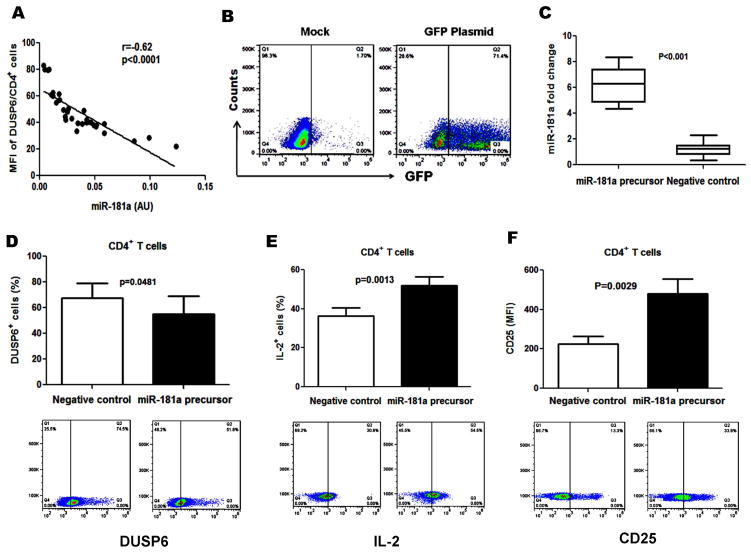
Previous work has suggested that DUSP6 is one of the phosphatases controlled by miR-181a [6, 10, 22]. To determine the relationship between DUSP6 and miR-181a expression in CD4+ T cells with HCV infection, we analyzed miR-181a levels and DUSP6 protein expression in CD4+ T cells from 20 HCV-infected patients and 6 HS co-cultured with HCV+ Huh7 cells. As shown in Fig. 4A, miR-181a expression negatively correlates with the DUSP6 expression in CD4+ T cells in the setting of HCV infection (r = −0.62, P<0.0001).
Fig. 4. miR-181a regulates DUSP6 expression and CD4+ T cell function.
A) Negative association between DUSP6 and miR-181a expression in CD4+ T cells. miR-181a expression levels (2−ΔCt) plotted against DUSP6 protein expression in CD4+ T cells from 20 HCV patients and 6 HS co-cultured with HCV+ Huh7 cells. Pearson correlation was used to analyze their relationship (r=−0.62, p<0.0001). B) Transfection efficiency using Amaxa Nucleofector System to transfect GFP and control vectors to CD4+ T cells from HCV-infected individuals and analyzed by flow cytometry. C) miR-181a expressions were measured by real-time PCR in CD4+ T cells transfected with miR-181a precursor and miRNA precursor negative control in 8 HCV-infected individuals. D, E, F) DUSP6, IL-2, and CD25 expressions were analyzed by flow cytometry in CD4+ T cells transfected by miR-181a precursor or negative control.
To further evaluate the relationship between miR-181a and DUSP6 expression, we transfected CD4+ T cells from HCV-infected individuals with miR-181a precursor and miRNA precursor negative control, followed by determination of miR-181a expression with real-time PCR as well as DUSP6, CD25 and IL-2 protein expression by flow cytometry. Since the transfection efficiency in primary T cells is a major challenge in studying the role of miRNAs in gene regulation, we employed the Amaxa Nucleofector System to transfect GFP and control vectors, and achieved a transfection efficiency of up to 70% in human CD4+ T cells (Fig. 4B). When we transfected with a miR-181a precursor, miR-181a expression in human CD4+ T cells was increased more than 6 times compared to the negative control (Fig. 4C). Meanwhile, DUSP6+ T cell frequencies in CD4+ T cells transfected with miR-181a precursor were significantly decreased compared with those transfected with miRNA precursor negative control (Fig. 4D). In parallel, TCR activation-induced expression of IL-2 (Fig. 4E) and CD25 (Fig. 4F) in CD4+ T cells were significantly increased with transfection of miR-181a precursor compared to negative control. These data indicate that reconstitution of miR-181a restores the impaired T cell responses with HCV infection, likely via the DUSP6 pathway.
miR-181a regulates DUSP6 gene expression at the translational level
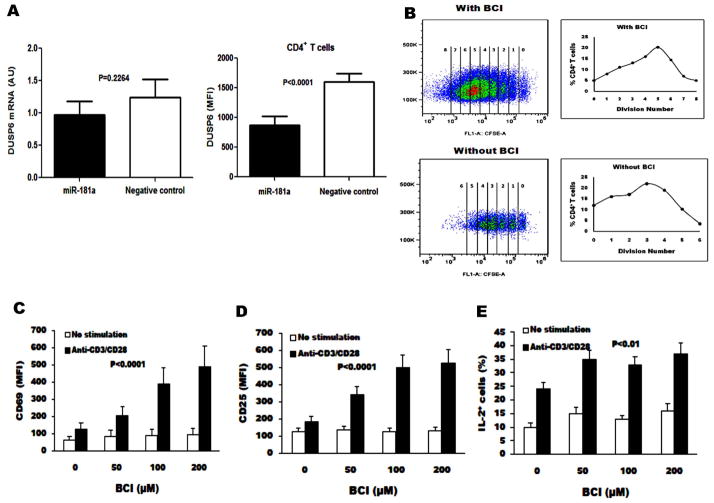
The regulation of miRNAs occurs post-transcriptionally through binding to messenger RNA (mRNA), primarily at the 3′-untranslated region of the mRNA [23–24]. To determine whether the inhibitory effect of miRNA-181a on DUSP6 protein expression is due to mRNA degradation or inhibition of mRNA expression, we measured the levels of DUSP6 mRNA by real-time PCR and DUSP6 protein expression by flow cytometry in CD4+ T cells from HCV-infected individuals following transfection of miR-181a precursor or negative control. As shown in Fig. 5A, the levels of DUSP6 mRNA in CD4+ T cells transfected with miR-181a precursor were reduced slightly compared with those transfected with negative control, but not to a statistically significant degree (left panel). However, DUSP6 protein expression, shown as mean fluorescent intensity (MFI), in CD4+ T cells transfected with miR-181a precursor was significantly reduced when compared with those transfected with negative control (P<0.0001) (right panel). These data indicate that miR-181a down-regulates DUSP6 expression in CD4+ T cells from HCV infected individuals, primarily by inhibiting target mRNA translation but not mRNA degradation.
Fig. 5. miR-181a regulates DUSP6 gene expression at the translational level and DUSP6 inhibition augments T cell activation.
A) The levels of DUSP6 mRNA in CD4+ T cells from HCV-infected individuals with transfection of miR-181a precursor and negative control were analyzed by real-time PCR (left panel), and DUSP6 protein expression by flow cytometry. DUSP6 expression was shown as mean fluorescent intensity (MFI) in CD4+ T cells transfected with miR-181a precursor compared with those transfected with negative control (right panel). B) Purified CD4+ T cells from 6 HCV-infected individuals were labeled with CFSE, stimulated with anti-CD3/CD28 in the presence of IL-2 and (E)-2-benzylidene-3-(cyclohexylamino)-2, 3-dihydro-1H-inden-1-one (BCI) ex vivo for 5 d, and followed by flow cytometry analysis. Representative dot plots of cell division as CFSE-dilution in the gated CD4+ T cells from BCI treated and non-treated are shown here. C–E) Purified CD4+ T cells from HCV-infected individuals with anti-CD3/CD28 antibodies in the presence of increasing concentrations of BCI. The MFI of CD69, CD25, and IL-2 expressions were analyzed by flow cytometry and shown as mean±SE from 6 HCV-infected individuals.
DUSP6 inhibition augments T cell activation and proliferation
Given that increased expression of DUSP6 is the major consequence of miR-181a loss in CD4+ T cells from HCV-infected individual CD4+ T cells, we rationalized that inhibition of DUSP6 might be a reasonable approach to restore CD4+ T cell responses. (E)-2-benzylidene-3-(cyclohexylamino)-2, 3-dihydro-1H-inden-1-one (BCI) has been shown to be an inhibitor of DUSP6 activity by specifically binding to an allosteric site but not the catalytic phosphatase domain [10, 25]. We stimulated purified CD4+ T cells from HCV-infected individuals with anti-CD3/CD28 antibodies in the presence of increasing concentrations of BCI. TCR stimulation-induced proliferation of CD4+ T cells was measured by CFSE dilution. We also quantified the MFI of CD69 and CD25 as markers of T cell activation and measured activation-induced IL-2 expression. As shown in Fig. 5B, inhibition of DUSP6 signaling with BCI significantly enhanced the proliferative capacity of CD4+ T cells in response to TCR stimulation compared to those without BCI inhibition. In addition, the inhibition of DUSP6 by BCI enhanced expression of CD69 (Fig. 5C) and CD25 (Fig. 5D) on CD4+ T cells (P < 0.0001) in a dose dependent manner. BCI treatment also increased IL-2 production in CD4+ T cells (Fig. 5E). These results indicate that miR-181a-regulated DUSP6 negatively controls CD4+ T cell activation and proliferation, supporting the notion that miR-181a and DUSP6 are potential targets for improving CD4+ T cell responses in HCV infection.
Discussion
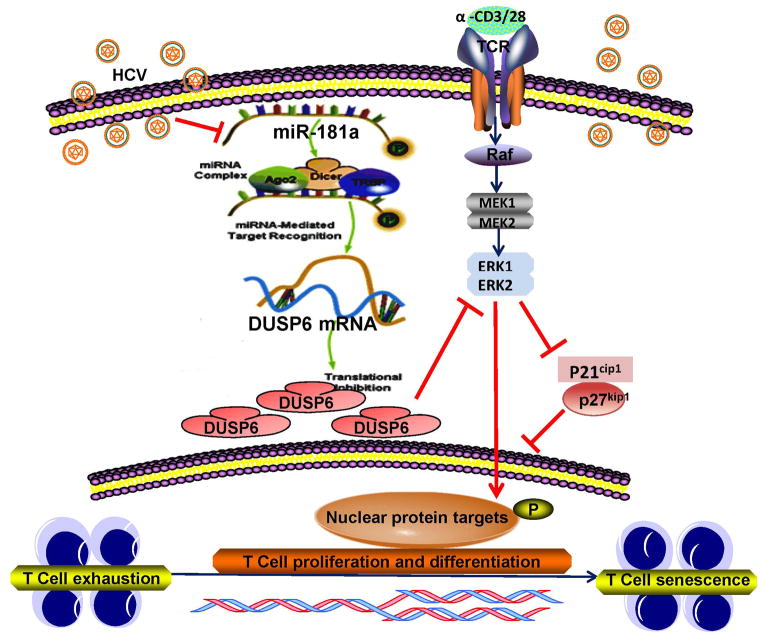
HCV infection is a world-wide infectious disease that can lead to chronic hepatitis, liver cirrhosis, and hepatocellular carcinoma. After decades of studying this immunomodulatory virus, it has become evident that HCV-mediated T cell dysfunction plays a major role in viral persistence as well as disease progression [35–39]. We have previously reported that CD4+ T cells play a pivotal role in the host immune responses to pathogenic infection and vaccination [11–12, 18–20, 35–39]. In this study, we further explored the mechanisms of CD4+ T cell dysfunction in chronic HCV infection. Our studies demonstrate that HCV-induced, miR-181a- mediated DUSP6 over-expressions in CD4+ T cells recalibrate the TCR sensitivity threshold, thus controlling the signaling strength of CD4+ T cells from HCV-infected individuals, which evidenced by a more profound dysfunction of CD4+ T cell activation, proliferation and IL-2 secretion when compared to HS. Importantly, reconstitution of miR-181a or inhibition of DUSP6 leads to a significant improvement of CD4+ T cell activation, proliferation, and IL-2 production in HCV-infected individuals. Thus, manipulation of miR-181a and/or DUSP6 expression may represent a key intervention to strengthen and broaden CD4+ T cell responses in HCV-infected individuals. Based on this study, our previous reports [11–12, 18–20, 35–39], and other investigators’ work [10], we propose a model depicted in Fig. 6, to illustrate the role and pathways involved in the HCV-induced, miR-181a-mediated DUSP6 over-expression in CD4+ T cells to control T cell responses in the setting of chronic viral infection.
Fig. 6. Schematic model for premature T cell aging by HCV-induced, miR-181a-mediated DUSP6 regulatory signaling pathways.
HCV-induced decline of miR-181a expression facilitates DUSP6 over-expression in CD4+ T cells. This, in turn, may negatively affect the TCR-induced signaling pathways, such as ERK/MAPK phosphorylation and then alters cell cycle regulators p21cip1/p27kip1 and cyclins and cyclin-dependent kinases (CDKs) activities. Therefore, reconstitution of miR-181a and/or inhibition of DUSP6 may provide a novel approach to improve T cell responses in virally infected individuals.
Recent studies suggest that miRNA-mediated gene regulation may represent a fundamental layer of post-transcriptional genetic programs in metazoan genomes [27–29]. Many miRNAs are differentially regulated in hematopoietic lineages, and some have been shown to play roles in controlling the development of immune cells [6, 29–32]. However, the role of miRNAs in the adaptive immune response to viral infection is largely unknown. Our findings demonstrate that HCV infection aberrantly regulates a significant number of miRNAs in human CD4+ T cells, and miR-181a is developmentally regulated to refine T cell function through DUSP signaling. Since a decline of miR-181a, along with DUSP6 over-expression, are hallmarks of age-associated T cell senescence [10, 22, 29], our findings in this study provide novel mechanistic insights for the HCV-mediated premature T cell aging via miR-181a-regulated DUSP6 signaling. This provides new insight into the role of HCV-induced changes in human miRNA expression and their significance in T cell immune senescence.
Though multiple miRNAs may attribute to the regulation of a single target gene, and one particular miRNA could regulate multi-targets [27, 33, 34], our results demonstrate that a single miR-181a can very efficiently regulate the expression of DUSP6, and a single molecule - DUSP6 - can impair T cell receptor sensitivity significantly. Our data show that miR-181a slightly reduces the mRNA levels of DUSP6, but significantly down-regulates the DUSP6 protein expression, supporting the notion that miR-181a down-regulates DUSP6 expression by inhibiting mRNA translation rather than mRNA degradation. Nevertheless, we do not exclude the possibility that other TCR signaling components or other miRNAs may also contribute to changes in T cell response. Indeed, by prediction of miRNA targets through Target Scan Human (TSH) bioinformatics analysis, other tyrosine phosphatases, such as SHP-2, PTPN2, and DUSP5, were also found to be potential miR-181a targets that deserve further investigations.
We propose that HCV may employ two critical cell regulatory mechanisms, cell exhaustion and cell senescence (Fig. 6), through up-regulation of different inhibitory pathways, such as PD-1/Tim-3, KLRG1/p16ink4a, and miR181a/DUSP6, to dampen the functions of immune cells to respond appropriately to vaccines during chronic infection [35]. Although there has been substantial progress in identifying the mechanisms that regulate their processes separately, it is unclear how these processes interrelate, and whether blocking pathways that maintain either the exhausted or the senescent state, or both, can boost vaccine responses, especially in virally-infected individuals. A recent study demonstrated that a young HIV-infected patient with less than 4 years duration of infection have early immune exhaustion leading to premature aging and senescence comparable to the elderly, suggesting virus induces premature immune senescence associated with high rates of immune exhaustion following short-term infection [40]. We have demonstrated that HCV-infected individuals exhibit higher expression of both exhaustion and senescence markers, including PD-1, SOCS-1, Tim-3, and KLRG-1, in APC or helper T cells, which is associated with an impaired cellular function that is more profound in HCV-infected, HBV vaccine non-responders when compared to responders [12, 14, 18–19]. Given the fact that the most common reason for vaccine failure in chronic viral infection is limited T cell proliferative potential, a better understanding of this mechanism by which virus usurps host signaling machinery to modulate immune responses may open a new avenue to enhance vaccine efficacy and immunotherapy.
In summary, this study delineates the mechanisms of viral infection-induced immune senescence or cell aging by measuring T cell responses to TCR stimulation. Our findings suggest that a decline of miR-181a expression leads to DUSP6 over-expression and CD4+ T cell dysfunction during HCV infection. Reconstitution of miR-181a and/or inhibition of DUSP6 can restore the impaired functions of CD4+ T cells from chronically HCV-infected individuals. Further characterization of miR-181a and DUSP6 expression and functional changes in CD4+ T cells will not only help us to better understand the pathogenesis of chronic HCV infection but may reveal a potential therapeutic target for managing this chronic viral disease.
Acknowledgments
This work was supported by an NIH NIDDK grant to ZQY/JPM (R01DK093526) and an NIH NIAID grant to ZQY/JPM (R01AI114748). Mr. Jeddidiah Griffin is supported by an NIH NIAID grant to JPM/ZQY (R15AI072750). Ms. Yun Zhou, a joint PhD student, was supported in part by the China Scholarship Council (CSC 201306590012). Dr. Ying RS, a visiting scholar, was supported by a viral hepatitis research grant from Guangzhou Municipal Health Bureau, China. Dr. Shi L is a visiting scholar, partially supported by the Guanghua Foundation of Xian Jiaotong University, China. Dr. Cheng YQ is a visiting scholar, partially supported by Beijing 302 Hospital, Beijing, China. We are greatly appreciated the support from Dr. T. Wakita, Department of Virology II, NIH of Japan, for transferring HCV JFH-1 strain through a MTA; and Dr. T.J. Liang, Liver Section, NIH/NIDDK, for sending Huh-7 cells. This publication is the result of work supported with resources and the use of facilities at the James H. Quillen Veterans Affairs Medical Center.
Footnotes
The contents in this publication do not represent the views of the Department of Veterans Affairs or the United States Government.
References
- 1.Park SH, Rehermann B. Immune responses to HCV and other hepatitis viruses. Immunity. 2014;40:13–24. doi: 10.1016/j.immuni.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rosen HR. Emerging concepts in immunity to hepatitis C virus infection. J Clin Invest. 2013;123:4121–4130. doi: 10.1172/JCI67714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Au JS, Pockros PJ. Novel therapeutic approaches for hepatitis C. Clin Pharmacol Ther. 2014;95:78–88. doi: 10.1038/clpt.2013.206. [DOI] [PubMed] [Google Scholar]
- 4.Manns MP1, von Hahn T. Novel therapies for hepatitis C - one pill fits all? Nat Rev Drug Discov. 2013;12:595–610. doi: 10.1038/nrd4050. [DOI] [PubMed] [Google Scholar]
- 5.Chen K, Rajewsky N. The evolution of gene regulation by transcription factors and microRNAs. Nat Rev Genet. 2007;8:93–103. doi: 10.1038/nrg1990. [DOI] [PubMed] [Google Scholar]
- 6.Lee CH, Kim JH, Lee SW. The role of microRNAs in hepatitis C virus replication and related liver diseases. J Microbiol. 2014;52:445–451. doi: 10.1007/s12275-014-4267-x. [DOI] [PubMed] [Google Scholar]
- 7.Zhang Y, Wei W, Cheng N, Wang K, Li B, et al. Hepatitis C virus-induced up-regulation of microRNA-155 promotes hepatocarcinogenesis by activating Wnt signaling. Hepatology. 2012;56:1631–1640. doi: 10.1002/hep.25849. [DOI] [PubMed] [Google Scholar]
- 8.Mukherjee A, Shrivastava S, Bhanja Chowdhury J, Ray R, Ray RB. Transcriptional suppression of miR-181c by hepatitis C virus enhances homeobox A1 expression. J Virol. 2014;88:7929–7940. doi: 10.1128/JVI.00787-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Estrabaud E, Lapalus M, Broët P, Appourchaux K, De Muynck S, et al. Reduction of microRNA 122 expression in IFNL3 CT/TT carriers and during progression of fibrosis in patients with chronic hepatitis C. J Virol. 2014;88:6394–6402. doi: 10.1128/JVI.00016-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li G, Yu M, Lee WW, Tsang M, Krishnan E, et al. Decline in miR-181a expression with age impairs T cell receptor sensitivity by increasing DUSP6 activity. Nat Med. 2012;18:1518–1524. doi: 10.1038/nm.2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li G, Li K, Lea AS, Li NL, Abdulla NE, et al. In situ hybridization for the detection of hepatitis C virus RNA in human liver tissue. J Viral Hepat. 2013;20:183–192. doi: 10.1111/j.1365-2893.2012.01642.x. [DOI] [PubMed] [Google Scholar]
- 12.Shi L, Wang JM, Ren JP, Cheng YQ, Ying RS, et al. KLRG1 impairs CD4+ T cell responses via p16ink4a and p27kip1 pathways: role in hepatitis B vaccine failure in individuals with hepatitis C virus infection. J Immunol. 2014;192:649–657. doi: 10.4049/jimmunol.1302069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Y, Ma CJ, Ni L, Zhang CL, Wu XY, et al. Cross-talk between programmed death-1 and suppressor of cytokine signaling-1 in inhibition of IL-12 production by monocytes/macrophages in hepatitis C virus infection. J Immunol. 2011;186:3093–3103. doi: 10.4049/jimmunol.1002006. [DOI] [PubMed] [Google Scholar]
- 14.Wang JM, Shi L, Ma CJ, Ji XJ, Ying RS, et al. Differential regulation of IL-12/IL-23 by Tim-3 drives Th17 cell development during HCV infection. J Virol. 2013;87(8):4372–4383. doi: 10.1128/JVI.03376-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ma CJ, Ren JP, Li GY, Wu XY, Brockstedt DG, et al. Enhanced virus-specific CD8+ T cell responses by Listeria monocytogenes-infected dendritic cells in the context of Tim-3 blockade. PLoS One. 2014;9(1):e87821. doi: 10.1371/journal.pone.0087821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Skalsky RL, Cullen BR. Viruses, microRNAs, and host interactions. Annu Rev Microbiol. 2010;64:123–141. doi: 10.1146/annurev.micro.112408.134243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Singaravelu R, Russell RS, Tyrrell DL, Pezacki JP. Hepatitis C virus and microRNAs: mired in a host of possibilities. 2014;7:1–10. doi: 10.1016/j.coviro.2014.03.004. [DOI] [PubMed] [Google Scholar]
- 18.Wang JM, Ma CJ, Li GY, Wu XY, Thayer P, et al. Tim-3 alters the balance of IL-12/IL-23 and drives TH17 cells: Role in hepatitis B vaccine failure during hepatitis C infection. Vaccine. 2013;31:2238–2245. doi: 10.1016/j.vaccine.2013.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moorman JP, Zhang CL, Ni L, Ma CJ, Zhang Y, et al. Impaired hepatitis B vaccine responses during chronic hepatitis C infection: involvement of the PD-1 pathway in regulating CD4+ T cell responses. Vaccine. 2011;29:3169–3176. doi: 10.1016/j.vaccine.2011.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yao ZQ, Nguyen DT, Hiotellis AI, Hahn YS. Hepatitis C virus core protein inhibits human T lymphocyte responses by a complement-dependent regulatory pathway. J Immunol. 2001;167(9):5264–5272. doi: 10.4049/jimmunol.167.9.5264. [DOI] [PubMed] [Google Scholar]
- 21.Beld M, Penning M, McMorrow M, Gorgels J, van den Hoek A, Goudsmit J. Different hepatitis C virus (HCV) RNA load profiles following seroconversion among injecting drug users without correlation with HCV genotype and serum alanine aminotransferase levels. J Clin Microbiol. 1998;36:872–877. doi: 10.1128/jcm.36.4.872-877.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li QJ, Chau J, Ebert PJ, Sylvester G, Min H, et al. miR-181a is an intrinsic modulator of T cell sensitivity and selection. Cell. 2007;129:147–161. doi: 10.1016/j.cell.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 23.Cannell IG, Kong YW, Bushell M. How do microRNAs regulate gene expression? Biochem Soc Trans. 2008;36:1224–1231. doi: 10.1042/BST0361224. [DOI] [PubMed] [Google Scholar]
- 24.Jackson RJ, Standart N. How do microRNAs regulate gene expression? Sci STKE. 2007;367:re 1. doi: 10.1126/stke.3672007re1. [DOI] [PubMed] [Google Scholar]
- 25.Molina G, Vogt A, Bakan A, Dai W, Queiroz de Oliveira P, et al. Zebrafish chemical screening reveals an inhibitor of Dusp6 that expands cardiac cell lineages. Nat Chem Biol. 2009;5:680–687. doi: 10.1038/nchembio.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ciccarelli C, Marampon F, Scoglio A, Mauro A, Giacinti C, et al. P21 WAF1 expression induced by MEK/ERK pathway activation or inhibition correlates with growth arrest, myogenic differentiation and onco-phenotype reversal in rhabdomyosarcoma cells. Mol Cancer. 2005;4:41. doi: 10.1186/1476-4598-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ambros V. microRNA pathways in flies and worms: growth, death, fat, stress, and timing. Cell. 2003;113:673–676. doi: 10.1016/s0092-8674(03)00428-8. [DOI] [PubMed] [Google Scholar]
- 28.Bartel DP. microRNA genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 29.Papapetrou EP1, Kovalovsky D, Beloeil L, Sant’angelo D, Sadelain M. Harnessing endogenous miR-181a to segregate transgenic antigen receptor expression in developing versus post-thymic T cells in murine hematopoietic chimeras. J Clin Invest. 2009;119:157–168. doi: 10.1172/JCI37216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bartel DP, Chen CZ. Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs. Nat Rev Genet. 2004;5:396–400. doi: 10.1038/nrg1328. [DOI] [PubMed] [Google Scholar]
- 31.Chen CZ, Li L, Lodish HF, Bartel DP. microRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 32.Fazi F, Rosa A, Fatica A, Gelmetti V, De Marchis ML, et al. A minicircuitry comprised of microRNA-223 and transcription factors NFI-A and C/EBPalpha regulates human granulopoiesis. Cell. 2005;123:819–831. doi: 10.1016/j.cell.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 33.Krek A, Grün D, Poy MN, Wolf R, Rosenberg L, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 34.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 35.Yao ZQ, Moorman JP. Immune exhaustion and immune senescence – two distinct pathways for HBV vaccine failure during HCV and/or HIV infection (Review) Arch Immunol Ther Exp. 2013;61:193–201. doi: 10.1007/s00005-013-0219-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moorman JP, Wang JM, Zhang Y, Ji XJ, Ma CJ, et al. Tim-3 pathway controls regulatory and effector T cell balance during hepatitis C virus infection. J Immunol. 2012;189:755–766. doi: 10.4049/jimmunol.1200162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yao ZQ, Ni L, Zhang Y, Ma CJ, Zhang CL, et al. Differential regulation of T and B lymphocytes by PD-1 and SOCS-1 signaling in hepatitis C virus-associated non-Hodgkin’s lymphoma. Immunol Invest. 2011;40:243–264. doi: 10.3109/08820139.2010.534218. [DOI] [PubMed] [Google Scholar]
- 38.Ni L, Ma CJ, Zhang Y, Nandakumar S, Zhang CL, et al. PD-1 modulates regulatory T cells and suppresses T-cell responses in HCV-associated lymphoma. Immunol Cell Biol. 2011;89:535–539. doi: 10.1038/icb.2010.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Frazier AD, Zhang CL, Ni L, Ma CJ, Zhang Y, et al. Programmed death-1 affects suppressor of cytokine signaling-1 expression in T cells during hepatitis C infection. Viral Immunol. 2010;23:487–495. doi: 10.1089/vim.2010.0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ferrando-Martínez S, Ruiz-Mateos E, Romero-Sánchez MC, Muñoz-Fernández MÁ, Viciana P, et al. HIV Infection-Related Premature Immunosenescence: High Rates of Immune exhaustion after short time infection. Curr HIV Res. 2011;9:289–294. doi: 10.2174/157016211797636008. [DOI] [PubMed] [Google Scholar]