Abstract
Background
A growing collection of retrospective studies have suggested that TP53 mutations and/or CDKN2A deletions have prognostic significance in Ewing sarcoma. We sought to evaluate these variables in patients with localized disease treated prospectively on a single Children’s Oncology Group protocol.
Procedure
Of the 568 patients enrolled on Children’s Oncology Group protocol AEWS0031 (NCT00006734), 112 had tumor specimens of sufficient quality and quantity to allow for analysis of TP53 mutations status by DNA sequencing, and CDKN2A deletion by dual color fluorescent in situ hybridization.
Results
Eight of 93 cases (8.6%) were found to have TP53 point mutations and 12 of 107 cases (11.2%) demonstrated homozygous CDKN2A deletion. Two cases were found have an alteration in both genes. There was no significant difference in event-free survival of patients with TP53 mutations and/or CDKN2A deletions compared to patients with normal TP53/CDKN2A gene status, as demonstrated by log rank test (p = 0.58).
Conclusions
Although previous retrospective studies suggest their significance, TP53 mutation and/or CDKN2A deletion are not reliable prognostic biomarkers in localized Ewing sarcoma.
Keywords: Ewing sarcoma, TP53, CDKN2A, Prognosis, Biomarker, Outcomes
INTRODUCTION
Ewing sarcoma is the second most common primary malignancy of bone in the pediatric population.[1] Prior to the development of systemic chemotherapy, 90% of patients with Ewing sarcoma died of distant metastases.[2,3] With current therapeutic protocols the 5-year survival rate for patients with localized disease is roughly 70%.[4] Approximately 25% of patients have metastatic disease at presentation, portending a poorer prognosis, with a 24-month event-free survival (EFS) of 35%.[5]
Current chemotherapeutic agents are generally non-specific cytotoxins associated with significant short- and long-term side effects. Further advances in therapy are likely to include increased dose-intensity and newly introduced agents, both of which may impart significant risk to the patient. Therefore, it is increasingly important to identify patients that are at a high risk for treatment failure with current protocols in order to justify the potential danger associated with escalated treatment regimens. Better understanding of the pathogenesis of Ewing sarcoma has created interest in the identification of prognostic molecular biomarkers to define this high-risk patient population.
Ewing sarcomas are defined by characteristic somatic chromosomal translocations that usually fuse the EWSR1 gene (encoding the EWS protein) to an ETS family transcription factor gene. In approximately 85% of cases the translocation (11;22)(q24;q12) occurs, resulting in the EWS/FLI fusion protein.[6-9] This fusion protein is necessary, but not sufficient for the oncologic phenotype of Ewing sarcoma cells.[10-15] EWS/FLI induction of oncogenesis is host cell line dependent, suggesting that additional cell-specific mutations are required.[16-19] Specifically, inhibition of the p53 or p16INK4a pathways appears to facilitate this process.[18,19]
The p53 protein (encoded by the TP53 gene) is responsible for cellular growth arrest and apoptosis in response to cell stress signals, such as DNA damage and oncogenic mutations.[20-23] The p53 pathway is negatively regulated by MDM2, which itself is sequestered and inhibited by p14ARF.[24,25] The gene locus CDKN2A encodes the p14ARF and p16INK4a proteins from overlapping transcripts.[26] The p16INK4a protein helps to prevent cell cycle progression by inhibiting cyclin-dependent kinase-mediated phosphorylation of the RB protein.[27,28] Therefore, deletion of the CDKN2A locus usually results in the loss of p14ARF and p16INK4a, thus eliminating the growth inhibition provided by both the p53 and RB pathways.[26,29]
Alterations in p53 and p16INK4a proteins have been identified in approximately 10% and 20% of Ewing sarcoma cases, respectively.[30] Over the past twenty years a growing series of retrospective studies have suggested that alterations in the p53 and p16INK4a pathways have prognostic significance in Ewing sarcoma.[31-36] However, other retrospective series have failed to demonstrate a correlation between these genetic alterations and clinical outcome.[37-39]
Given the importance of identifying molecular biomarkers able to distinguish high-risk patient populations and the conflicting data from the prior studies, we designed a prospective study to clarify the relationship between TP53 and/or CDKN2A status and clinical outcomes in patients with localized Ewing sarcoma. The current study focuses on a well-defined group of patients with localized disease treated in a prospective fashion on a single Children’s Oncology Group (COG) study. The genetic analysis techniques employed by this study could be readily translated into the clinical setting if TP53 and/or CDKN2A proved to be clinically-valuable biomarkers.
METHODS
Clinical Specimens and Treatment
All patients were enrolled on COG protocol AEWS0031 (NCT00006734).[4] Inclusion criteria were patients with Ewing sarcoma (diagnosed by pathologic review at enrolling institutions and centrally, but no requirement for molecular confirmation) who had localized disease without overt evidence of metastasis. Protocol treatment consisted of alternating cycles of vincristine/doxorubicin/cyclophosphamide and ifosfamide/etoposide for 12 weeks, at which time local control (surgery and/or radiation therapy) was performed, followed by continuation of chemotherapy until a total of 14 cycles were received. The protocol was designed to evaluate the effect of interval time compression between administered chemotherapy cycles and patients were randomly assigned to receive either standard timing (with chemotherapy cycles administered every three weeks) or interval-compressed timing (with chemotherapy cycles administered every two weeks).
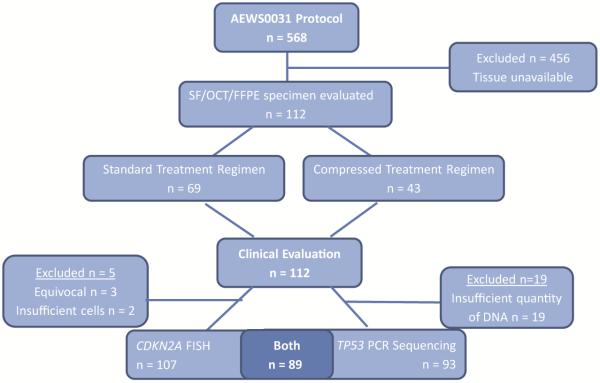
Of the 568 patients enrolled on COG protocol AEWS0031[4], 112 patient specimens were included in the current analysis under COG protocol AEWS08B1 (NCT00898053). Inclusion criteria for the current analysis were: 1) the availability of banked specimens from patients enrolled on COG protocol AEWS0031, 2) sufficient quality and quantity of genetic material for analysis, and 3) identifiable specimen source for correlation with clinical outcome (Figure 1). This biological-correlative analysis was approved as COG protocol AEWS08B1 and received all requisite IRB approvals.
Figure 1.
CONSORT diagram demonstrating patient specimen inclusion. SF= snap frozen, OCT= optimal cutting temperature, FFPE= formalin-fixed paraffin-embedded.
CDKN2A Analysis by FISH
Dual color fluorescent in situ hybridization (FISH) was used to assess for deletion of the CDKN2A gene locus. Commercially available CDKN2A locus-specific and chromosome 9 centromere-specific probes were utilized (LSI p16INK4a [9p21] Spectrum Orange/CEP 9 Spectrum Green Dual Color Probe Set, Vysis, Des Plaines, IL). Cytologic touch preparations were performed on 112 specimens comprising snap frozen, optimal cutting temperature (OCT) compound-embedded, or formalin-fixed paraffin-embedded (FFPE) tissue preservations. Of these 112 specimens, 107 produced interpretable results. Five specimens were excluded from analysis due to insufficient cells (two) and equivocal FISH results (three). Positive and negative controls were established with a Malignant Fibrous Histiocytoma sample with a del(9)(p12p24) karyotype, and with a Ewing sarcoma sample demonstrating a +9 karyotype, respectively.
Hybridization signals were assessed independently by two trained individuals in 100-200 interphase nuclei with strong and well-delineated signals. An interphase cell specimen was determined to be abnormal if the copy number for the LSI p16INK4a and/or CEP 9 probe signals were greater than two signals per probe in more than 10% of the cells evaluated, or less than two signals per probe in 20% of the cells evaluated (more than two standard deviations above the average false-positive rate). Cytologic touch preparations of pathologically unremarkable skeletal muscle served as normal controls.
TP53 Analysis by PCR Sequencing
Genomic DNA extraction was performed with EPICENTRE Biotechnologies MasterPure Complete DNA and RNA Purification kit. Extracted DNA was evaluated by agrose gel electrophoresis and spectrophotometry. Of the 112 specimens processed, 98 yielded a sufficient amount of genomic DNA, defined to be greater than 0.25 μg.
PCR amplification of TP53 exons 5 through 8 was performed with the following paired primer sequences:
Exon 5:
Forward: 5'GTAAAACGACGGCCAGTACTTTCAACTCTGTCTCCTTCCTCTTC 3'
Reverse: 5' CAGGAAACAGCTATGACCAGCCCTGTCGTCTCTCCAG 3'
Exon 6:
Forward: 5' GTAAAACGACGGCCAGTCACTGATTGCTCTTAGGTCT 3'
Reverse: 5' CAGGAAACAGCTATGACAGTTGCAAACCAGACCTCAGG 3'
Exon 7:
Forward: 5’ CTGCTTGCCACAGGTCTC 3’
Nested: 5' GTAAAACGACGGCCAGTTCATCTTGGGCCTGTGTTATCTC 3'
Reverse: 5’-TGGATGGGTAGTAGTATGGAAC-3’
Nested: 5' AGGAAACAGCTATGACGTGCAGGGTGGCAAGTGG 3'
Exon 8:
Forward: 5' GTAAAACGACGGCCAGTGACCTGATTTCCTTACTGCC 3'
Reverse: 5' CAGGAAACAGCTATGACCCACCGCTTCTTGTCCTGCT 3’
Amplicons were purified and sequenced. Forward and reserve exon sequencing provided internal confirmation of the mutations, with positive mutations being identified in both directions. Controls for normal and mutant TP53 were normal peripheral blood lymphocyte DNA and known TP53 mutation-containing cell line DNA, respectively. Titration experiments demonstrated that as little as 10% mutated DNA could be detected.
Whole Genome Amplification
Genomic DNA (20 ng) from each of the 59 snap frozen and 39 OCT embedded specimens with a sufficient amount of DNA remaining following the PCR-sequencing studies were subjected to the QIAGEN’s REPLI-g (Venlo, NL) service for whole genomic amplification (WGA). The WGA resulted in 100-152 μg of DNA from each starting material. The WGA DNA was subjected to PCR and sequencing for TP53 mutations as described above.
Data Analysis
The primary study endpoint was event-free survival, defined as the time from study entry until disease progression, death without progression of disease, occurrence of a second malignant neoplasm or last follow-up, whichever occurred first. If a patient was alive and without malignancy, the patient was considered censored for analysis; in all other cases, an event was considered to have occurred.
The study population was segregated into two subpopulations according to the presence (yes v. no) of a TP53 gene mutation and/or CDKN2A allele deletion. The prognostic significance of a TP53 mutation and/or CDKN2A deletion was assessed using a two-sided log rank test. EFS as a function of time since study enrollment was estimated using the method of Kaplan and Meier.[40] Equality of risk for EFS-event across patient groups was assessed using the log rank statistic.[40] A p-value of 0.05 or less was considered indicative of a significant relationship between gene mutation/deletion status and clinical outcome. Confidence intervals were constructed using a relative risk regression model[40] with the characteristic of interest as the only variable in the model.
The secondary goals for the study were to estimate the incidence of TP53 mutations and CDKN2A deletion in tumor samples derived from the population of patients with localized Ewing sarcoma. To do this, the proportion of specimens determined to have a genetic alteration were used to estimate of the mutation/deletion fraction.
In determining whether the evaluated population of 112 patients was representative of the entire protocol population (n = 568) and the prognostic significance of TP53/CDKN2A alterations, subgroup analysis of standard and interval-compressed protocols was not performed in order to maintain statistically significant populations. Therefore, the presented analysis includes patients treated with both the standard and interval-compressed treatment protocols considered as one population.[4]
Chi-squared and Fisher’s exact test were used as appropriate to identify significant demographic and/or outcome variances between the evaluated study group versus the protocol enrolled group at large, and between patients with versus without TP53/CDKN2A alterations.
RESULTS
Population characteristics
The 112 specimens evaluated were a subset of the 568 patients enrolled in COG protocol AEWS0031. The demographic data and EFS of the evaluated patient subset was statistically equivalent to the larger population, with the exception of patient age (Table I), and is thus generally representative of the whole patient population with localized Ewing sarcoma treated on protocol AEWS0031.
Table 1.
COG AEWS0031 patient demographics
|
Not Evaluated
N=456 |
Evaluated
N=112 |
P-value | |||
|---|---|---|---|---|---|
| Demographics | N | % | N | % | |
| Age | 0.04 | ||||
| <=9 | 124 | 27% | 38 | 34% | |
| 10-17 | 271 | 59% | 68 | 61% | |
| 18<= | 61 | 13% | 6 | 5% | |
| Sex | 0.8 | ||||
| Male | 246 | 56% | 62 | 55% | |
| Female | 210 | 46% | 50 | 45% | |
| Race | 0.9* | ||||
| Caucasian | 401 | 88% | 101 | 90% | |
| African American | 12 | 3% | 2 | 2% | |
| Other | 19 | 4% | 3 | 3% | |
| Missing | 24 | 5% | 6 | 5% | |
| Primary Tumor Site | 0.4 | ||||
| Appendicular | 151 | 33% | 44 | 39% | |
| Thoracic | 73 | 16% | 16 | 14% | |
| Pelvic | 70 | 15% | 20 | 18% | |
| Other Axial | 66 | 14% | 9 | 8% | |
| Extraosseous | 96 | 21% | 23 | 21% | |
Fischer's Exact Test. A comparison between patients evaluated and not evaluated in the current study (AEWS08B1).
TP53 mutations and CDKN2A deletions
Ninety-three of 112 patient tumor specimens (83%) yielded genomic DNA that was of sufficient quantity and quality to be evaluated for TP53 mutation. PCR amplification and sequencing of exons 5 through 8 identified a point mutation in 8 of 93 specimens (8.6%). This mutation rate is consistent with previously published reports that establish the Ewing sarcoma tumoral mutation rate from 4.2% to 20%.[41,42] Seven of eight TP53 alterations occurred within a region commonly affected by oncogenic mutations [43]. Mutations were identified in each examined exon.
Dual color FISH identified homozygous CDKN2A deletions in 12 of 107 analyzable specimens, demonstrating a CDKN2A loss incidence of 11.2%. Again, this incidence is consistent with previous reports in Ewing sarcoma tumor specimens of 7% to 21%.[33,41] Note that in keeping with prior studies, only homozygous CDKN2A loss was considered significant, as cells with heterozygous CDKN2A loss would be expected to have a normal phenotype.
Eighty-nine (79.5%) of the initial 112 specimens were analyzable for both TP53 mutation and CDKN2A deletion. Of those 89 specimens, two (2.24%) had an alteration in both genes, while 17 specimens (19.1%) had an alteration in either TP53 or CDKN2A (Table II).
Table II.
CDKN2A and/or TP53 alteration rate
| CDKN2A | TP53 | Either/Both* | |
|---|---|---|---|
| Alterations | 12 | 8 | 17 |
| Normal | 95 | 85 | 72 |
|
| |||
| Percentage | 11.20% | 8.60% | 19.10% |
two patients had both p16 loss and p53 mutation
Whole Genome Amplification Analysis
Direct Sanger sequencing of the 98 WGA DNA specimens demonstrated 100% concordance with genomic DNA sequencing. The eight specimens with mutations identified by sequencing of genomic DNA exons 5 through 8 were also identified in the WGA DNA samples.
Survival Analysis
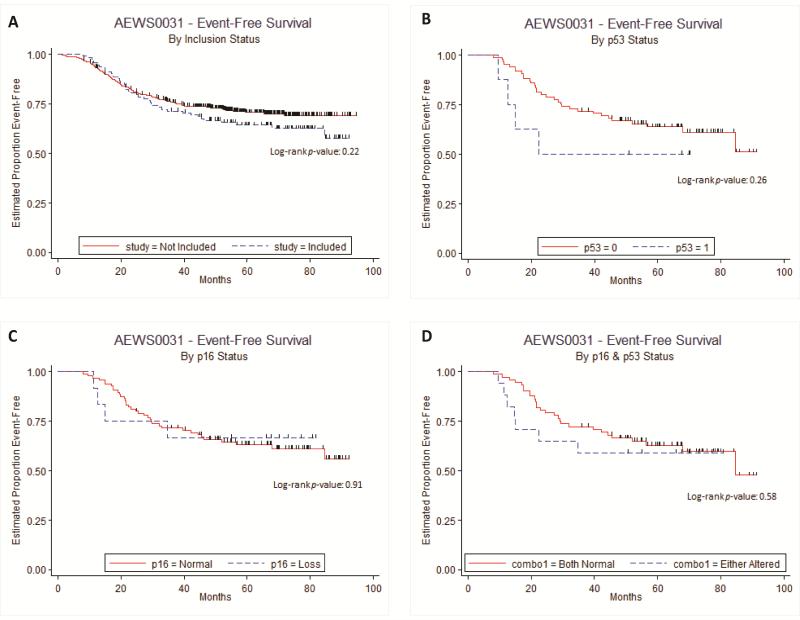
EFS analysis was performed on all of the patients enrolled in COG protocol AEWS0031. Demographic data of the evaluated (n = 112) and not evaluated (n = 456) patients demonstrated similarity between the two groups, with the exception of age (p = 0.04; Table I). Although the evaluated patient population was younger, there was no difference in EFS compared to excluded patient population, demonstrated by a relative hazard ratio of 1.248 [95% CI: 0.8779 to 1.7742] (Table III). Since younger age at time of diagnosis has been shown to be of positive prognostic significance,[44] it does not appear that the age discrepancy between the two groups significantly influenced the clinical outcomes. Kaplan-Meier EFS comparisons between the included and excluded patient populations also failed to demonstrate a significant difference between the two groups (p = 0.22; Figure 2A). Therefore, the evaluated population appears to be representative of the total patient population treated on protocol AEWS0031.
Table III.
Relative Hazard Ratios for inclusion in the current study and gene alteration
| Relative Hazard Ratio | 95% Confidence Interval | Median follow-up* | Maximum follow-up* | |
|---|---|---|---|---|
|
| ||||
| Evaluated Status | ||||
| Not Evaluated | 1.0000 | 4.93 | 7.86 | |
| Evaluated | 1.2480 | 0.8779 to 1.7742 | 5.78 | 7.67 |
| CDKN2A Status | ||||
| Normal | 1.0000 | 5.62 | 7.67 | |
| Deleted | 0.9397 | 0.3337 to 2.6459 | 5.95 | 6.79 |
| TP53 Status | ||||
| Normal | 1.0000 | 5.50 | 7.60 | |
| Mutated | 1.8288 | 0.6450 to 5.1856 | 5.71 | 5.83 |
| CDKN2A&TP53 Status | ||||
| Both normal | 1.0000 | 5.44 | 7.60 | |
| Either/Both altered | 1.2638 | 0.5499 to 2.9046 | 5.82 | 6.71 |
Among patients without an event. Relative hazard ratio with 95% confidence intervals and follow-up durations for (1) AEWS0031 patients evaluated and not evaluated in the current study (AEWS08B1), and (2) patients with and without CDKN2A and/or TP53 alterations.
Figure 2.
A. Kaplan-Meier EFS curve of evaluated (included) v. not evaluated (excluded) patients from protocol AEWS0031. B. Kaplan-Meier EFS curve of patients with v. without TP53 mutation. C. Kaplan-Meier EFS curve of patients with v. without CDKN2A deletion. D. Kaplan-Meier EFS curve of patients with v. without TP53 mutation and/or CDKN2A deletion
Comparison in patient outcomes between those whose tumors harbored mutations in TP53 versus those who did not demonstrated a statistically similar EFS for the two groups (p = 0.26; Figure 2B). TP53 mutation was found to have an insignificant relative hazard ratio of 1.8288 [95% CI: 0.645 to 5.1856] compared to the wild-type gene (Table III). It should be noted that in this study a trend was observed, that patients with TP53 mutations represented an older population compared to patients without TP53 mutations (p = 0.67, Table IV). Additionally, patients with TP53 mutations were racially diverse (75% Caucasian) compared to patients without TP53 mutation (95% Caucasian, p = 0.014, Table IV). However, given the small number of patients with p53 mutation (n = 8) the racial difference is accounted for by two patients, one who self-identified as Asian and another as African-American. Non-Caucasian race and older age at diagnosis portends a worse prognosis than Caucasian and younger patients,[44] thus any difference between TP53 mutant and non-mutant populations should have been accentuated by the age and race differences between these groups; despite this, no difference was observed.
Table IV.
Characteristics of patients with and without CDKN2A and/or TP53 alterations
|
CDKN2A Deleted N=12 |
CDKN2A Normal N = 95 |
P-value |
TP53 Mutation N=8 |
TP53 Normal N=85 |
P-value | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Demographics | N | % | N | % | N | % | N | % | ||
| Age | 0.22 | 0.067 | ||||||||
| <=9 | 3 | 25% | 33 | 35% | 0 | 0% | 31 | 36% | ||
| 10-17 | 7 | 58% | 58 | 61% | 7 | 88% | 50 | 59% | ||
| 18<= | 2 | 17% | 4 | 4% | 1 | 13% | 4 | 5% | ||
| Sex | 0.76 | 0.72 | ||||||||
| Male | 6 | 50% | 53 | 56% | 4 | 50% | 50 | 59% | ||
| Female | 6 | 50% | 42 | 44% | 4 | 50% | 35 | 41% | ||
| Race | 0.75 | 0.013 | ||||||||
| Caucasian | 11 | 92% | 85 | 89% | 6 | 75% | 81 | 95% | ||
| African American | 0 | 0% | 2 | 2% | 1 | 13% | 0 | 0% | ||
| Asian | 0 | 0% | 1 | 1% | 1 | 13% | 0 | 0% | ||
| Other | 0 | 0% | 2 | 2% | 0 | 0% | 2 | 2% | ||
| Missing | 1 | 8% | 5 | 5% | 0 | 0% | 2 | 2% | ||
| Primary Tumor Site | 0.2 | 0.4 | ||||||||
| Appendicular | 4 | 33% | 38 | 40% | 2 | 25% | 31 | 36% | ||
| Thoracic | 0 | 0% | 15 | 16% | 1 | 13% | 13 | 15% | ||
| Pelvic | 4 | 33% | 15 | 16% | 2 | 25% | 1 | 1% | ||
| Other Axial | 2 | 17% | 6 | 6% | 0 | 0% | 9 | 11% | ||
| Extraosseous | 2 | 17% | 21 | 22% | 3 | 38% | 15 | 18% | ||
Comparison in patient outcomes between those whose tumors contained CDKN2A deletion and those who did not, also demonstrated a statistically similar EFS (p = 0.91; Figure 2C) and an insignificant relative hazard ratio (0.9397 [95% CI: 0.3337 to 2.6459], Table III) between the two groups. Patients with and without CDKN2A deletions were demographically equivalent (Table IV). Thus, age, sex, race, and primary tumor location would not have prevented our ability to detect a difference in outcome, if one had existed.
To determine if alteration in either TP53 and/or CDKN2A correlated with EFS in patients with localized Ewing sarcoma, we considered the evaluable population for either alteration in toto. Kaplan-Meier comparisons again showed no statistically-significant difference in EFS between the groups (p = 0.58; Figure 2D).
DISCUSSION
This study identified an incidence of TP53 mutation (8.4%) and CDKN2A deletion (11.2%) in tumor specimens from patients with Ewing sarcoma that was consistent with rates identified in previous series.[30,32-36,42] However, in contrast to those retrospective studies, we did not find a correlation between either of these genetic alterations and clinical outcome. EFS rates were equivalent between patients whose tumors did and did not have alternations in either TP53 and/or CDKN2A.
Our study differs from the previous series in multiple ways, each of which could help explain our discrepant findings. The current study consists of prospective specimen collection, obtained from patients on a standardized treatment protocol. While our study population was larger than most of the prior reports, it was confined to patients with localized disease. And lastly, the methodologies utilized to assess gene status could be promptly translated into clinical use.
The current study’s prospective patient enrollment and specimen collection imparts a significantly higher level of evidence than the proceeding small retrospective studies that lacked “prospectively dictated therapy, follow-up, specimen selection, or statistical analysis.”[45] In addition to the inherent bias of retrospective reviews, prior studies’ inclusion of patients treated on a variety of therapeutic protocols over a range of treatment eras introduced a significant variable when evaluating clinical outcomes.
The 112 evaluated specimens are a subpopulation of the 568 patients enrolled in COG protocol AEWS0031. With the exception of age at enrollment, the demographic and EFS data of the evaluated subpopulation is equivalent to that of the entire protocol enrolled patient population. Due to the congruity between these groups, the results from the 112 evaluated specimens are believed to be representative of the whole protocol population. Regardless, the 112 specimens in the current study is a larger population than any prior which has shown prognostic significance of TP53/CDKN2A alterations.[30-36,46] The largest retrospective review was able to assess 107 specimens for TP53 mutations and/or CDKN2A deletions; consistent with our findings, their multivariate analysis failed to demonstrate an association between these genetic alterations and progression free or disease specific survival.[37]
The presence of metastatic disease upon presentation is a well-recognized negative prognostic marker. Other clinical features—tumor size, location, response to neoadjuvent therapy and patients age at presentation—have a weaker correlation with negative outcomes leaving few, if any, prognostic markers available for patients with localized disease.[47] By excluding patients with metastatic disease we eliminated this confounding variable and focused on the population for which prognostication is most difficult.
PCR sequencing analysis of TP53 exons 5 through 8 produced an accurate and clinically replicable assay. This region of p53 contains 90% of the identified oncogenic mutations and the DNA binding domain.[43] Using this strategy we identified a TP53 mutation rate of 8.6%. While this rate is well within the reported range of 3% to 19%,[41,42] it does fall below the mutation rate reported in the largest retrospective series of 15%.[37] However in that review mutation status was assessed by immunohistochemistry (IHC), which has been shown to be less specific than DNA sequencing, as demonstrated in a series from Memorial Sloan Kettering Cancer Center, where two of 10 Ewing sarcoma samples with positive IHC staining for p53 were ultimately determined to be negative for TP53 mutations by GeneChip analysis.[30]
Our analysis of the CDKN2A locus was performed with a commercially available CDKN2A specific-probe that is larger than the probes used in previously published reports. While this may have caused false negative results if small deletion segments were missed, our CDKN2A homozygous deletion rate of 11.2% was consistent with previously published reports of 7% to 21%.[33,41] The utilization of a commercially available CDKN2A probe established the parameters of clinically applicable assay.
As in this investigation, for large scale genomic studies to be performed in rare diseases, organized longitudinal tumor banking is essential.[48] While COG’s Biopathology Center is an invaluable resource of tumor specimens accumulated over many years, the quantity of each sample is finite, necessitating a judicious evaluation of study merit prior to specimen utilization. Careful utilization of this limited and valuable resource is of critical importance. Although the evaluation of TP53 and CKDN2A alterations in patients with localized Ewing sarcoma is a meaningful clinical question, we recognize the value of other genomic questions and the scarcity of tumor specimen tissue. Therefore, following PCR analysis, WGA was performed in order to replenish the Ewing sarcoma sample DNA. By report from the commercial vendor (QIAGEN) and confirmed by our experience, WGA produced an accurate amplification of genomic DNA. With this technology the valuable resource of tumor specimen DNA can be restored and maintained for use in future projects.
A useful biomarker, as described by Hodgson et al., must demonstrate a clear prognostic relationship, and must be evaluable on easily obtained specimens, in an efficient and cost effective manner with currently available technology.[49] The prior studies that demonstrated a correlation between TP53/CDKN2A alterations and clinical outcomes, promoted these genetic alterations as prognostic biomarkers.29-35, 46 While strong associations were identified within their study population, our contrary findings demonstrate that TP53/CDKN2A alterations are not universally applicable biomarkers and therefore have no prognostic significance in localized Ewing sarcoma.
Identification of characteristics with which to differentiate the presenting patient population continues to be elusive in localized Ewing sarcoma. Despite early optimism, TP53/CDKN2A alterations prove to be unreliable biomarkers. This finding serves as a reminder that a potential biomarker’s prognostic value is limited to the study population in which it was initially identified and its utility should not be extrapolated without validation by large prospective trials.
ACKNOWLEDGEMENTS
Support for this study was provided by the Nicholas Currey Fund and the Thrasher Research Foundation. Dr. Lessnick also acknowledges support from the NIH/NCI via R01 CA140394 and from the Huntsman Cancer Institute via P30CA042014. Dr. Womer received support from the Daniel P. Sullivan Fund.
Footnotes
Conflict of Interest Statement: The authors have no conflicts of interest to report in relation to this manuscript.
REFERENCES
- 1.Pizzo PA, Poplack DG. Principles and practice of pediatric oncology. Wolters Kluwer/Lippincott Williams & Wilkins Health; Philadelphia: 2011. [Google Scholar]
- 2.Wang CC, Schulz MD. Ewing's sarcoma; a study of fifty cases treated at the Massachusetts General Hospital, 1930-1952 inclusive. N Engl J Med. 1953;248(14):571–576. doi: 10.1056/NEJM195304022481401. [DOI] [PubMed] [Google Scholar]
- 3.Dahlin DC, Coventry MB, Scanlon PW. Ewing's sarcoma. A critical analysis of 165 cases. The Journal of bone and joint surgery American volume. 1961;43-A:185–192. [PubMed] [Google Scholar]
- 4.Womer RB, West DC, Krailo MD, et al. Randomized controlled trial of interval-compressed chemotherapy for the treatment of localized Ewing sarcoma: a report from the Children's Oncology Group. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2012;30(33):4148–4154. doi: 10.1200/JCO.2011.41.5703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Felgenhauer JL, Nieder ML, Krailo MD, et al. A pilot study of low-dose anti-angiogenic chemotherapy in combination with standard multiagent chemotherapy for patients with newly diagnosed metastatic Ewing sarcoma family of tumors: A Children's Oncology Group (COG) Phase II study NCT00061893. Pediatric blood & cancer. 2013;60(3):409–414. doi: 10.1002/pbc.24328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Turc-Carel C, Philip I, Berger MP, et al. Chromosome study of Ewing's sarcoma (ES) cell lines. Consistency of a reciprocal translocation t(11;22)(q24;q12) Cancer Genet Cytogenet. 1984;12(1):1–19. doi: 10.1016/0165-4608(84)90002-5. [DOI] [PubMed] [Google Scholar]
- 7.Turc-Carel C, Aurias A, Mugneret F, et al. Chromosomes in Ewing's sarcoma. I. An evaluation of 85 cases of remarkable consistency of t(11;22)(q24;q12). Cancer Genet Cytogenet. 1988;32(2):229–238. doi: 10.1016/0165-4608(88)90285-3. [DOI] [PubMed] [Google Scholar]
- 8.Whang-Peng J, Triche TJ, Knutsen T, et al. Chromosome translocation in peripheral neuroepithelioma. N Engl J Med. 1984;311(9):584–585. doi: 10.1056/NEJM198408303110907. [DOI] [PubMed] [Google Scholar]
- 9.Delattre O, Zucman J, Plougastel B, et al. Gene fusion with an ETS DNA-binding domain caused by chromosome translocation in human tumours. Nature. 1992;359(6391):162–165. doi: 10.1038/359162a0. [DOI] [PubMed] [Google Scholar]
- 10.Kinsey M, Smith R, Lessnick SL. NR0B1 is required for the oncogenic phenotype mediated by EWS/FLI in Ewing's sarcoma. Mol Cancer Res. 2006;4(11):851–859. doi: 10.1158/1541-7786.MCR-06-0090. [DOI] [PubMed] [Google Scholar]
- 11.Smith R, Owen LA, Trem DJ, et al. Expression profiling of EWS/FLI identifies NKX2.2 as a critical target gene in Ewing's sarcoma. Cancer Cell. 2006;9(5):405–416. doi: 10.1016/j.ccr.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 12.Ouchida M, Ohno T, Fujimura Y, et al. Loss of tumorigenicity of Ewing's sarcoma cells expressing antisense RNA to EWS-fusion transcripts. Oncogene. 1995;11(6):1049–1054. [PubMed] [Google Scholar]
- 13.Kovar H, Aryee DN, Jug G, et al. EWS/FLI-1 antagonists induce growth inhibition of Ewing tumor cells in vitro. Cell Growth Differ. 1996;7(4):429–437. [PubMed] [Google Scholar]
- 14.Tanaka K, Iwakuma T, Harimaya K, et al. EWS-Fli1 antisense oligodeoxynucleotide inhibits proliferation of human Ewing's sarcoma and primitive neuroectodermal tumor cells. The Journal of clinical investigation. 1997;99(2):239–247. doi: 10.1172/JCI119152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Toretsky JA, Connell Y, Neckers L, et al. Inhibition of EWS-FLI-1 fusion protein with antisense oligodeoxynucleotides. J Neurooncol. 1997;31(1-2):9–16. doi: 10.1023/a:1005716926800. [DOI] [PubMed] [Google Scholar]
- 16.Thompson AD, Teitell MA, Arvand A, et al. Divergent Ewing's sarcoma EWS/ETS fusions confer a common tumorigenic phenotype on NIH3T3 cells. Oncogene. 1999;18(40):5506–5513. doi: 10.1038/sj.onc.1202928. [DOI] [PubMed] [Google Scholar]
- 17.May WA, Gishizky ML, Lessnick SL, et al. Ewing sarcoma 11;22 translocation produces a chimeric transcription factor that requires the DNA-binding domain encoded by FLI1 for transformation. Proc Natl Acad Sci U S A. 1993;90(12):5752–5756. doi: 10.1073/pnas.90.12.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lessnick SL, Dacwag CS, Golub TR. The Ewing's sarcoma oncoprotein EWS/FLI induces a p53-dependent growth arrest in primary human fibroblasts. Cancer Cell. 2002;1(4):393–401. doi: 10.1016/s1535-6108(02)00056-9. [DOI] [PubMed] [Google Scholar]
- 19.Deneen B, Denny CT. Loss of p16 pathways stabilizes EWS/FLI1 expression and complements EWS/FLI1 mediated transformation. Oncogene. 2001;20(46):6731–6741. doi: 10.1038/sj.onc.1204875. [DOI] [PubMed] [Google Scholar]
- 20.Crawford LV, Pim DC, Gurney EG, et al. Detection of a common feature in several human tumor cell lines--a 53,000-dalton protein. Proc Natl Acad Sci U S A. 1981;78(1):41–45. doi: 10.1073/pnas.78.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oliner JD, Kinzler KW, Meltzer PS, et al. Amplification of a gene encoding a p53-associated protein in human sarcomas. Nature. 1992;358(6381):80–83. doi: 10.1038/358080a0. [DOI] [PubMed] [Google Scholar]
- 22.Lin AW, Barradas M, Stone JC, et al. Premature senescence involving p53 and p16 is activated in response to constitutive MEK/MAPK mitogenic signaling. Genes Dev. 1998;12(19):3008–3019. doi: 10.1101/gad.12.19.3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Serrano M, Lin AW, McCurrach ME, et al. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 1997;88(5):593–602. doi: 10.1016/s0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- 24.Stott FJ, Bates S, James MC, et al. The alternative product from the human CDKN2A locus, p14(ARF), participates in a regulatory feedback loop with p53 and MDM2. EMBO J. 1998;17(17):5001–5014. doi: 10.1093/emboj/17.17.5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Momand J, Zambetti GP, Olson DC, et al. The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell. 1992;69(7):1237–1245. doi: 10.1016/0092-8674(92)90644-r. [DOI] [PubMed] [Google Scholar]
- 26.Quelle DE, Zindy F, Ashmun RA, et al. Alternative reading frames of the INK4a tumor suppressor gene encode two unrelated proteins capable of inducing cell cycle arrest. Cell. 1995;83(6):993–1000. doi: 10.1016/0092-8674(95)90214-7. [DOI] [PubMed] [Google Scholar]
- 27.Serrano M, Hannon GJ, Beach D. A new regulatory motif in cell-cycle control causing specific inhibition of cyclin D/CDK4. Nature. 1993;366(6456):704–707. doi: 10.1038/366704a0. [DOI] [PubMed] [Google Scholar]
- 28.Weinberg RA. The retinoblastoma protein and cell cycle control. Cell. 1995;81(3):323–330. doi: 10.1016/0092-8674(95)90385-2. [DOI] [PubMed] [Google Scholar]
- 29.Chin L, Pomerantz J, DePinho RA. The INK4a/ARF tumor suppressor: one gene--two products--two pathways. Trends Biochem Sci. 1998;23(8):291–296. doi: 10.1016/s0968-0004(98)01236-5. [DOI] [PubMed] [Google Scholar]
- 30.Huang HY, Illei PB, Zhao Z, et al. Ewing sarcomas with p53 mutation or p16/p14ARF homozygous deletion: a highly lethal subset associated with poor chemoresponse. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2005;23(3):548–558. doi: 10.1200/JCO.2005.02.081. [DOI] [PubMed] [Google Scholar]
- 31.Amir G, Issakov J, Meller I, et al. Expression of p53 gene product and cell proliferation marker Ki-67 in Ewing's sarcoma: correlation with clinical outcome. Hum Pathol. 2002;33(2):170–174. doi: 10.1053/hupa.2002.31475. [DOI] [PubMed] [Google Scholar]
- 32.Mangham DC, Cannon A, Li XQ, et al. p53 overexpression in Ewing's sarcoma/primitive neuroectodermal tumour is an uncommon event. Clin Mol Pathol. 1995;48(2):M79–82. doi: 10.1136/mp.48.2.m79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lopez-Guerrero JA, Pellin A, Noguera R, et al. Molecular analysis of the 9p21 locus and p53 genes in Ewing family tumors. Lab Invest. 2001;81(6):803–814. doi: 10.1038/labinvest.3780290. [DOI] [PubMed] [Google Scholar]
- 34.Wei G, Antonescu CR, de Alava E, et al. Prognostic impact of INK4A deletion in Ewing sarcoma. Cancer. 2000;89(4):793–799. doi: 10.1002/1097-0142(20000815)89:4<793::aid-cncr11>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 35.Abudu A, Mangham DC, Reynolds GM, et al. Overexpression of p53 protein in primary Ewing's sarcoma of bone: relationship to tumour stage, response and prognosis. British journal of cancer. 1999;79(7-8):1185–1189. doi: 10.1038/sj.bjc.6690190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.de Alava E, Antonescu CR, Panizo A, et al. Prognostic impact of P53 status in Ewing sarcoma. Cancer. 2000;89(4):783–792. [PubMed] [Google Scholar]
- 37.Lopez-Guerrero JA, Machado I, Scotlandi K, et al. Clinicopathological significance of cell cycle regulation markers in a large series of genetically confirmed Ewing's sarcoma family of tumors. Int J Cancer. 2011;128(5):1139–1150. doi: 10.1002/ijc.25424. [DOI] [PubMed] [Google Scholar]
- 38.Park YK, Chi SG, Kim YW, et al. Mutational alteration of the p16CDKN2a tumor suppressor gene is infrequent in Ewing's sarcoma. Oncol Rep. 1999;6(6):1261–1266. doi: 10.3892/or.6.6.1261. [DOI] [PubMed] [Google Scholar]
- 39.Brownhill SC, Taylor C, Burchill SA. Chromosome 9p21 gene copy number and prognostic significance of p16 in ESFT. British journal of cancer. 2007;96(12):1914–1923. doi: 10.1038/sj.bjc.6603819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kalbfleisch JD, Prentice RL. The statistical analysis of failure time data. xiii. J. Wiley; Hoboken, N.J.: 2002. p. 439. [Google Scholar]
- 41.Patino-Garcia A, Sierrasesumaga L. Analysis of the p16INK4 and TP53 tumor suppressor genes in bone sarcoma pediatric patients. Cancer Genet Cytogenet. 1997;98(1):50–55. doi: 10.1016/s0165-4608(96)00397-4. [DOI] [PubMed] [Google Scholar]
- 42.Tsuchiya T, Sekine K, Hinohara S, et al. Analysis of the p16INK4, p14ARF, p15, TP53, and MDM2 genes and their prognostic implications in osteosarcoma and Ewing sarcoma. Cancer Genet Cytogenet. 2000;120(2):91–98. doi: 10.1016/s0165-4608(99)00255-1. [DOI] [PubMed] [Google Scholar]
- 43.Hernandez-Boussard T, Rodriguez-Tome P, Montesano R, et al. IARC p53 mutation database: a relational database to compile and analyze p53 mutations in human tumors and cell lines. International Agency for Research on Cancer. Hum Mutat. 1999;14(1):1–8. doi: 10.1002/(SICI)1098-1004(1999)14:1<1::AID-HUMU1>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 44.Lee J, Hoang BH, Ziogas A, et al. Analysis of prognostic factors in Ewing sarcoma using a population-based cancer registry. Cancer. 2010;116(8):1964–1973. doi: 10.1002/cncr.24937. [DOI] [PubMed] [Google Scholar]
- 45.Hayes DF, Bast RC, Desch CE, et al. Tumor marker utility grading system: a framework to evaluate clinical utility of tumor markers. Journal of the National Cancer Institute. 1996;88(20):1456–1466. doi: 10.1093/jnci/88.20.1456. [DOI] [PubMed] [Google Scholar]
- 46.Yang Y, Zhen T, Zhang F, et al. p53 and hepatoma-derived growth factor expression and their clinicopathological association with Ewing family tumour. J Clin Pathol. 2013 doi: 10.1136/jclinpath-2013-201705. [DOI] [PubMed] [Google Scholar]
- 47.Shukla N, Schiffman J, Reed D, et al. Biomarkers in Ewing Sarcoma: The Promise and Challenge of Personalized Medicine. A Report from the Children's Oncology Group. Frontiers in oncology. 2013;3:141. doi: 10.3389/fonc.2013.00141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Borinstein SC, Beeler N, Block JJ, et al. A Decade in Banking Ewing Sarcoma: A Report from the Children's Oncology Group. Frontiers in oncology. 2013;3:57. doi: 10.3389/fonc.2013.00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hodgson DR, Whittaker RD, Herath A, et al. Biomarkers in oncology drug development. Mol Oncol. 2009;3(1):24–32. doi: 10.1016/j.molonc.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]