Abstract
Cyclic guanosine monophosphate (cGMP) is a second messenger molecule that transduces nitric oxide (NO) and natriuretic peptide (NP) coupled signaling, stimulating phosphorylation changes by protein kinase G (PKG). Enhancing cGMP synthesis or blocking its degradation by phosphodiesterase type 5A (PDE5A) protects against cardiovascular disease1,2. However, cGMP stimulation alone is limited by counter-adaptions including PDE upregulation3. Furthermore, though PDE5A regulates NO-generated cGMP4,5, NO-signaling is often depressed by heart disease6. PDEs controlling NP-coupled cGMP remain uncertain. Here we show that cGMP-selective PDE9A7,8 is expressed in mammalian heart including humans, and is upregulated by hypertrophy and cardiac failure. PDE9A regulates NP rather than NO-stimulated cGMP in heart myocytes and muscle, and its genetic or selective pharmacological inhibition protects against pathological responses to neuro-hormones, and sustained pressure-overload stress. PDE9A inhibition reverses pre-established heart disease independent of NO-synthase (NOS) activity, whereas PDE5A inhibition requires active NOS. Transcription factor activation and phospho-proteome analyses of myocytes with each PDE selectively inhibited reveals substantial differential targeting, with phosphorylation changes from PDE5A inhibition being more sensitive to NOS activation. Thus, unlike PDE5A, PDE9A can regulate cGMP signaling independent of the NO-pathway, and its role in stress-induced heart disease suggests potential as a therapeutic target.
The PDE super-family contains eleven sub-genes conferring different cyclic nucleotide and tissue selectivity9. PDE5A was the first cyclic-GMP selective enzyme discovered, and plays a major role in erectile and pulmonary vasomotor control. PDE9A was cloned 20 years later7,8 and has the highest affinity and selectivity for cGMP with a Km for cGMP 1000-fold lower than for cAMP8. PDE5A and PDE9A share only 28% homology8 and PDE9A lacks N-terminus cGMP/PKG stimulatory regulatory domains present in PDE5A9. PDE9A is expressed primarily in the brain, gut, and kidney. To date, studies have focused on its role in cognitive function10,11 and while mRNA is detectable in the heart and other tissues7,8, its role remains largely unknown.
To test PDE9A involvement in the heart, gene and protein expression were assessed in myocardial tissue and isolated myocytes. Fig. 1a shows PDE9A immunostaining in rat neonatal cardiomyocytes (RNCMs) and adult mouse myocytes, with targeted gene deletion (siRNA or PDE9A−/− mice, Extended Data Fig. 1) as a negative control. Protein detection by immunoblot in neonatal myocytes is shown in Extended Data Fig. 2a. Basal gene expression is low but increases with agonist (e.g. phenylephrine (PE)) or mechanical (in vivo pressure-overload) stimulation (Fig. 1b). Increased PDE9A protein expression and cGMP-esterase activity is found in left ventricular (LV) myocardium from humans with heart failure and depressed function (Fig. 1c–e, Extended Data Fig. 2b, c and Extended Data Table 1). Protein expression also increases in human LV hypertrophy from aortic stenosis (pressure-overload), and most strikingly in heart failure and a preserved ejection fraction (HFPEF, Fig. 1f), a prevalent form of HF wherein contractile function appears normal despite symptoms12. Human PDE9A expression primarily localizes to myocytes based on co-localization with troponin-T (Fig. 1g–i) and in situ hybridization staining (Fig. 1j, k). Whereas Pde5a is expressed in fibroblasts9, Pde9a is essentially undetectable in isolated human fibroblasts (qPCR-threshold cycle= 39).
Figure 1. PDE9A expression in heart and myocytes increases with disease.
a, PDE9A in neonatal and adult cardiomyocytes; gene silencing as negative controls (scale: 20μm). b, PDE9A gene expression in RNCMs with phenylephrine (PE) or adult myocytes/heart (mouse) after TAC; *p<0.01-vs-Control. c, Immunoblot/immunostaining of PDE9A from human dilated cardiomyopathy (DCM) and non-failing (NF) myocardium (scale: 200μm). d, PDE9A expression and e, activity in NF and DCM, f, PDE9A protein expression in human myocardium for NF, HFPEF, and AS. *p<0.0001,†p<0.05-vs-NF;‡p<0.0001-vs-AS and DCM. g–i, Co-localization of cardiac troponin T (cTnT) and PDE9A in adult mouse myocytes. j–k, in situ hybridization of Pde9a in human NF and DCM myocardium. Summary values are mean±SEM and biological replicates indicated in the figure panels or legend. This holds for all subsequent figures.
PDE9A upregulation by heart disease suggested its inhibition might blunt pathological stress responses. To test this, RNCMs and adult myocytes were stimulated with phenylephrine or endothelin-1 (ET-1), increasing protein synthesis and hypertrophic fetal gene (Nppa, Nppb) expression. Co-incubation with a selective PDE9A antagonist (PF-04449613 [PF-9613], 5μM, Extended Data Fig. 3a for selectivity of PDE9A vs PDE5A), or Pde9a gene silencing/deletion (Fig. 2a, b; upper) reversed these changes. Cells lacking Pde9a were unaffected by PF-9613, confirming the drug’s selectivity (Fig. 2a). Similar results were obtained with PF-04447943, another PDE9A-inhibitor now used in human trials (NCT00930059, Extended Data Fig. 3b). Anti-hypertrophic effects of PDE9A inhibition required activation of PKG, as they were blocked DT3 (Fig. 2b; lower and Extended Data Fig. 3c).
Figure 2. PDE9A inhibition suppresses cardiac hypertrophy via NP-cGMP pathway.
a, Effect of PF-9613 or gene-silencing on hypertrophic gene-activation in RNCMs (n=12/group for all but Nppa-siRNA-PDE9A−/− n=8) and adult myocytes (n=6); #-p<0.05;$-p<0.01;§-p<0.01;*-p<0.001;. b, Upper: PDE9A-inhibition reduces agonist-stimulated protein synthesis; *-p<0.001-vs-baseline; #-p<0.01 vs PE/ET-1. Lower: PKG-inhibitor DT3 prevents anti-hypertrophic effect of PF-9613;*-p<0.001;#-p<0.05. c, d, PF-9613 or PDE9A-siRNA augments cGMP from ANP but not DEANO simulation in neonatal and adult myocytes; all groups p<0.01 vs baseline; *-p<0.01 vs ANP. e, FlincG-cGMP fluorescence in RNCM before and after ANP stimulation (color-coded for sampled intracellular location); right: time course normalized to baseline. f, Myocyte cGMP stimulated by ANP rises with PF-9613 in RNCMs but not cells with Pde9a silenced. g, PF-9613 does not alter DEANO stimulated cGMP whereas PDE5A inhibitor (SIL) does. h, ANP±PF-9613 effects are unchanged by ODQ, whereas DEANO±SIL are inhibited; *-p<0.01-versus-baseline; #-p<0.01-vs-ANP; $-p<0.05 vs DEANO. i, RNMCs exposed to L-NAME and PE±siRNA to PDE5A, PDE9A, or both; *p<0.05 vs baseline; #p<0.05-vs-PE-scrambled siRNA.
Both PDE5A and PDE9A regulate cGMP-PKG activity; therefore we tested if this regulation is redundant or targets different cGMP pools. Gene silencing of PDE9A in RNCMs had no effect on cGMP augmentation from an NO-donor (DEANO, 1μM) but enhanced cGMP (Fig. 2c) and PKG activity (Extended Data Fig. 4a) triggered by natriuretic peptide (ANP, 1μM). Adult myocytes exposed to PF-9613 also augmented cGMP only with ANP-stimulation (Fig. 2d; PDE9A−/− cells were negative controls). We also measured intracellular cGMP generation in adult myocytes expressing the cGMP-fluorescent sensor, FlincG13 (Fig. 2e). ANP-stimulated cGMP increased more after PF-9613 in control cells, but not those lacking PDE9A (Fig. 2f). By contrast, PF-9613 did not alter cGMP stimulated by DEANO, whereas the latter increased with PDE5A-inhibition (sildenafil (SIL), 1μM, Fig. 2g). Neither SIL nor PDE5A-siRNA altered ANP-stimulated cGMP (Extended Data Fig. 4b, c). Pre-incubation with soluble guanylate cyclase (sGC) inhibitor ODQ (10μM, 1H-[1,2, 4]oxadiazolo[4,3,-a]quinoxalin-1-one) had no effect on ANP or ANP+PF-9613 stimulated cGMP, but fully blocked the rise from DEANO or DEANO+SIL (Fig. 2h). Lastly, RNCMs with gene-silencing of PDE5A, PDE9A, or both were incubated with the NOS-inhibitor L-NAME (1 mM). PE-stimulated hypertrophic genes were suppressed only if PDE9A was silenced (Fig. 2i). Thus, opposite to PDE5A, PDE9A hydrolyzes cGMP coupled to NP but not NO-stimulation.
We hypothesized that disparate cGMP-targeting related to intracellular localization and thus PDE-compartmentation. Confocal immunohistochemistry confirmed this, revealing PDE5A but not PDE9A co-localization with α-actinin at the z-disk, and PDE9A but not PDE5A co-localization with t-tubular membranes (sarcoplasmic reticulum ATPase-2a) (Extended Data Fig. 5). Interestingly, NP receptor-type A also displays a striation-pattern in myocytes14.
To test if PDE9A contributes to maladaptive hypertrophy and heart remodeling in vivo, PDE9A−/− mice were exposed to sustained pressure-overload. Their hearts developed less dilation and dysfunction, had reduced heart and lung weights, interstitial fibrosis, and myocyte hypertrophy versus TAC-littermate controls (Fig. 3a–d). Myocardial cGMP increased more in PDE9A−/− hearts (Fig. 3e), and this was accompanied by suppression of pathological genes including connective tissue growth factor, fibronectin, and transient receptor potential canonical channel type-6 (Fig. 3f). The latter signals upstream of calcineurin-NFAT, transduces myofibroblast transformation and hypertrophy15, and is directly inhibited by PKG16,17. Cyclic GMP can also suppress cAMP by activating PDE218 to blunt hypertrophy and fibrosis; however cAMP rose similarly in both groups after TAC (Extended Data Fig. 6).
Figure 3. Pressure-overload induced cardiac pathobiology is suppressed in PDE9A−/− mice.
a, Echocardiography of littermate-control and PDE9A−/− left-ventricle subjected to TAC. b, Summary data for fractional shortening (FS), LV end-systolic dimension (LV-ESD), and LV mass; *-p<0.01-vs-TAC-WT. c, Fibrosis and myocyte enlargement in TAC control-TAC versus PDE9A−/−-TAC; *p<0.001 vs Sham-WT; #-p<0.001 vs TAC-WT. d, Heart and lung weight normalized to tibia length (TL); *-p<0.001 vs sham-WT; #-p<0.001;§-p<0.05-vs-TAC-WT. e, Myocardial cGMP after TAC; *-p<0.05 vs sham-control; #-p<0.05 vs TAC-WT. f, g, Nppa, β-myosin heavy chain (Myh7), connective tissue growth factor (Ctgf); fibronectin (Fn1); collagen type-1a (Col1a2); metallo-proteinase-2, (Mmp2); and transient receptor potential canonical–6, (Trpc6) expression; all normalized to GAPDH; *-p<0.001 vs sham-WT;#-p<0.001;§-p<0.05 vs WT/TAC.
The clinically relevant question is whether inhibition of PDE9A reverses pre-established hypertrophy/dysfunction, and if this is differentiable from the protection afforded by PDE5A inhibition? To address this, C57BL/6J mice were subjected to a severe-TAC to rapidly induce chamber dilation, dysfunction, and hypertrophy, and on day-8, mice divided into three groups: PF-9613, SIL, or vehicle, and treated for 4 additional weeks while maintaining TAC. Oral PF-9613 itself had no effect on blood pressure or heart function (Extended Data Fig. 7). To test if cGMP-targeting by PDE9A differed from PDE5A in vivo, the experiment was repeated adding the NOS inhibitor L-NAME to the drinking water.
All groups developed marked chamber dysfunction, dilation, and hypertrophy after 1-week TAC (pre-treatment; Fig. 4a, b). In L-NAME(−) mice, inhibiting either PDE reversed changes to near sham-control levels, lowered post-mortem LV mass, lung weight, and abnormal molecular signatures (Extended Data Fig. 8). However, in L-NAME(+) mice, only PDE9A inhibition was effective (Fig. 4a, b). Blocking either PDE increased myocardial cGMP in L-NAME(−)-TAC but only PDE9A-inhibition did so in L-NAME(+)-TAC mice (Fig. 4c). L-NAME prevented PKG activation by SIL but not PF-9613 (Fig. 4d). In vitro PKG activity in PDE9A treated myocardium was itself little altered. This parallels reported data with NP stimulation, but differing from increases seen with PDE5A inhibition5.
Figure 4. Chronic PDE9A-inhibition reverses pre-established hypertrophy/dysfunction in NOS-independent manner.
a, M-mode echocardiograms from mice ±L-NAME exposed to 5-wk TAC±PDE5 or PDE9-inhibition starting 1-week after TAC. b, PF-9613 and SIL reversed cardiac dysfunction from TAC in L-NAME(−), but only PF-9613 was effective in L-NAME(+) mice; *-p<0.01;#-p<0.05-vs-TAC. c, cyclic-GMP levels and d, In vitro PKG activity measured in the same experiments. e, Transcription factor activation in RNCMs stimulated by PE±gene-silencing of PDE5A or PDE9A; (Symbols for panels c–e; *p<0.005,#-p<0.01,†p<0.001,$-p<0.05. f, Differential phosphorylation of target proteins following PDE9A versus PDE5A inhibition. L-NAME co-treatment reversed PDE5A-selective changes (red) far more than PDE9A-selective changes.
To further explore differential signaling from each PDE inhibitor, RNCMs were infected with luciferase reporter plasmids to assess activation of transcription factors known to regulate cardiac growth/hypertrophy and survival. NFAT, MEF2, and CREB all increased with PE stimulation. NFAT declined with PDE5 or -9 inhibition, consistent with results for TRPC6 (Fig. 4e and Extended Data Fig. 8) and studies showing both NP and NO-stimulation blocks this pathway16,17. MEF2 only declined with PDE5A-I while both GATA4 and CREB rose only with PDE9A-I (Fig. 4e). The latter two are linked to NP/cGMP signaling, enhanced survival, and adaptive myocardial stress responses19,20.
Lastly, we performed unbiased proteomic analysis to detect serine/threonine phosphorylation increases in myocyte proteins altered by PDE9A-I or PDE5A-I, with or without concomitant L-NAME. The majority (85%) of phosphorylated amino acid residues were modified by one or the other PDE inhibitor and the rest by both (Fig. 4f and Supplementary Table 1). Of these residues, L-NAME reduced 21% of those specific to PDE5A-I versus 5.2% to PDE9A-I (red symbols, p<0.02), supporting targeting of PDE9A to non-NO dependent cGMP (Supplementary Table 2).
The efficacy of inhibiting a cGMP-PDE to counter myocardial responses to pathological stress requires having sufficient cGMP synthesized and PDE expressed, and pathogenic signaling suppressible by PKG. Though prior work indicated PDE5A might fulfill these criteria. However, its preferred targeting to NOS-dependent cGMP is a potential limitation, as this pool is often depressed in cardiovascular disease. We have now identified PDE9A-I as an alternative, that unlike PDE5A-I, remains effective even when NOS-cGMP synthesis is suppressed. This non-redundant function is consistent with intracellular compartmentation21. The revelation that PDE9A serves as a NP-cGMP targeted PDE is important as this source of cGMP often rises in heart disease whereas the NOS-derived pool declines. As with PDE5A, there are multiple downstream targets stemming from PDE9A regulation that collectively impact myocardial biology and disease. The current data identifies transcriptional controllers and protein substrates, setting the stage for future work.
The observation of heightened PDE9A expression in human heart failure, particularly HFPEF is exciting. Morbidity and mortality from HFPEF is high and with still no effective therapies this remains a major unmet medical need worldwide12. While there is considerable enthusiasm for cGMP-PKG targeted treatment for this disease22, recent data from a multi-center clinical trial employing PDE5A-I was disappointing23. Among potential reasons are a lack of PDE5A upregulation and low myocardial cGMP attributed to depressed NO-signaling24. Our results suggest PDE9A-I is an attractive alternative. The recent success of a combined angiotensin-receptor blocker and neprilysin inhibitor25 (the latter blunting NP-proteolysis) that is being tested in HFPEF (NCT01920711), advances in synthetic NP therapies26, offer opportunities for combined treatment. PDE9A inhibitors appear well tolerated in humans and are being studied for neurocognitive disease (c.f. clinicaltrials.gov/NCT00930059). The current results support exploring these agents as new avenues for treatment of the heart, and potentially other organs in which PDE9A and the NP-signaling system play a role.
METHODS
Human Myocardial Tissue
Procurement of human myocardial tissue was performed under protocols approved by Institutional Review Boards at the University of Pennsylvania (Philadelphia, PA), Johns Hopkins University (Baltimore, MD), and VU University Medical Center, (Amsterdam, The Netherlands) and its coordinated affiliated centers24 and consent for biopsy procedures or use of explanted tissues prospectively obtained in all cases. Explant dilated-non-ischemic failing human hearts were procured at the time of orthotopic heart transplantation at the Hospital of University of Pennsylvania. Non-failing hearts were obtained at the time organ donation from cadaveric donors. In all cases, hearts were arrested in situ using ice-cold cardioplegia solution, transported on wet ice, and flash frozen in liquid nitrogen within four hours of explantation. All samples were full-thickness biopsies obtained from the free wall of the left ventricle. HFPEF patients were referred for cardiac catheterization and LV endomyocardial biopsy because of clinical suspicion of restrictive cardiomyopathy. LV biopsies were procured using femoral artery access and a long bioptome. Diagnostic criteria and clinical characteristics of the HFPEF and AS patients have been previously reported24. Control samples for these studies were obtained from explanted unused donor hearts.
PDE9A Knockout Mouse
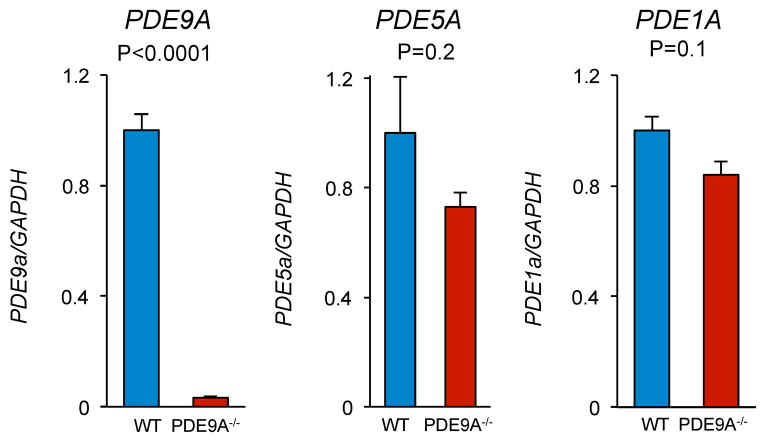
All protocols involving animals followed U.S. National Institute of Health guidelines and were approved by the animal and care use committee of the Johns Hopkins Medical Institutions. PDE9A global knock-out (PDE9A−/−) mice were developed by Pfizer Incorporated. The model replaced exon-12 in the catalytic domain of PDE9A with a LacZ-neomycin cassette (see Extended Data Fig. 1). Mice did not express any splice variants as they all share this sequence. PDE9A−/− mice were born in normal Mendelian ratios, and had no evident physiological or behavioral abnormalities (Supplementary Table 3). Expression of alternative cGMP-targeting PDEs, PDE1A and PDE5A were not significantly different in PDE9A−/− versus littermate control hearts (Extended Data Fig. 9).
Transverse Aortic Constriction (TAC) and Chronic Drug Studies
Pressure overload was performed by surgical placement of suture around the transverse aorta sized to a 27G needle, as described1. For chronic drug treatment studies, size, age, and sex (male) matched C57BL/6J mice (Jackson Labs) were randomized to receive vehicle, PF-9613 (30 mg/kg 2x/day by oral gavage), or sildenafil (200mg/kg/day with Bioserv soft diet). The mean free plasma concentration of PF-9613 was 77 nM, (peak of 1.5 μM at 30 minutes, t1/2 = 1.2 hours) within the selective range, while sildenafil yielded 30 nM1 also in the selective range. Mice subjected to TAC or sham operation. Mice were sacrificed at 5 weeks for tissue analysis. This protocol was then repeated in mice administered L-NAME (1mg/ml in drinking water) initiated one week prior to TAC, and continued for the full 5-week TAC. Tissue histology and echocardiography followed reported methods27. For these studies, in vivo analysis and post-sacrifice myocardial analysis were performed blinded as to experimental group. For in vivo PDE9A−/−analysis included all animals studied. For the drug-intervention study, which was designed to test reversal of heart disease established after 1-week of TAC, animals dying in the first week (before drug assignment) or who failed to develop disease after TAC (likely related to inadequate constriction) were excluded from analysis. Chronic PF-9613 had no effect on cardiac function or systemic pressures (e.g. no evidence of arterial vasodilation) in shams (Extended Data Fig. 7). There was also no systemic vasodilation from drug treatment in TAC hearts (e.g. systemic resistance 11.5±1 vehicle, 10.6±2 mmHg/mL•min PF-9613, p>0.5).
PDE9A inhibitors: PF-04449613 and PF-04447943 and siRNA vectors
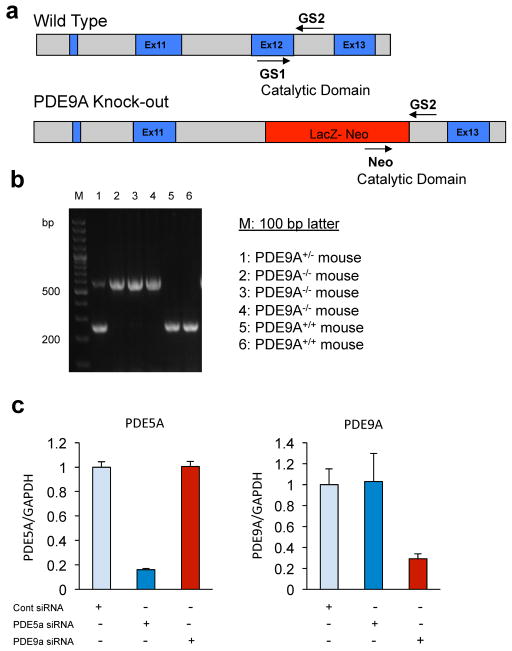
The structure and pharmacology of PF-04449613 and PF-04447943 have been reported10,28. We performed a dose-ranging study for PDE9A and PDE5A selectivity using a fluorescent polarization assay1 (Molecular Devices) (Extended Data Fig. 3a). This identified 5μM PF-04449613 inhibited 70% of PDE9A without altering PDE5A activity, whereas 1 μM sildenafil blocked PDE5A>70% with no effect on PDE9A. These concentrations were used in the subsequent cell culture studies. For gene silencing, we used targeted siRNA (PDE9A; L-098890-02, PDE5A; L-093210-02, control; D-001810-10) on-target siRNA smart pool, Dharmacon) or scrambled controls, achieving 70–80% knockdown (Extended Data Fig. 1c).
Isolated Myocyte Hypertrophy and Immunohistochemistry
Cell isolation methods for both neonatal and adult myocytes have been reported27. For adult cell studies, the isolation process itself produces variance in the absolute agonist response observed. Thus, quantitative response comparisons are best considered within a given cell type (e.g. control or PDE9a−/−). RNCMs were isolated from 1 to 2-day-old Sprague–Dawley rats and cultured for 2 days prior to study, while adult mouse myocytes were studied in primary culture for no more than 24 hours. For immunohistochemistry, myocytes were plated on laminin-coated imaging dishes, fixed with 50% methanol and 50% acetone, permeabilized with 0.1% saponin in PBS, and blocked in 10% BSA in PBS. Cells were then incubated overnight with primary antibodies at 4°C (rabbit anti-PDE9A (using human peptide), gift of Dr. Laurinda Jaffe, Univ. of CT, Farmington, CT; 1:100 or 1:50 dilution for wild-type and knock-out, respectively; rabbit anti-PDE5 (1:250, Cell Signaling); mouse anti-SERCA2 (1:250, Abcam); mouse anti-α actinin (1:500, Sigma)); then with secondary antibodies for 1h at room temperature (Alexa Fluor 488 or Alexa Fluor 546–conjugated; Invitrogen); then imaged on an inverted epifluorescent microscope with argon-krypton laser confocal scanning (Zeiss, UltraView; Perkin Elmer Life Science Inc.).
Human Heart Immunostaining and in situ hybridization
Sectioned human heart tissues were dehydrated with PBS in buffer containing 10mM sodium citrate, 0.05% Tween 20 at pH 6.0. Sections were blocked for 1h at room temperature in PBS with 0.1% bovine serum albumin and 0.1% Tween-20) followed by incubation with PDE9A (1:200) or cardiac TnT antibody (1:100, Thermo).
In situ hybridization with digoxigenin (DIG)-labeled PDE9A probe was performed as described previously29. We generated an anti-sense RNA probe to human PDE9A using partial sequence of human PDE9A, flanked with forward (5′-TGT CCT AGA GAA ACG CGT GG-3′) and reverse (5′-GGT GAC AGG GTT GAT GCT GA-3′) primers, amplified by PCR, and ligated into pGEM-T-easy vector (Promega). Anti-sense RNA probe labeled with DIG was prepared using the RNA labeling kit (Roche).
Molecular Analyses
PDE and PKG activity, cAMP and cGMP levels, were performed as previously described1. Quantitative PCR was used to assess RNA expression using standard procedures. PCR primers were: TaqMan primers used for Col1a2 (Mm00483888_m1), Ctgf (Mm01192933_g1), Fn1 (Mm01256744_m1), Gapdh (Mm99999915_g1), Myh7 (Mm00600555_m1), Nppa (Mm01255747_g1), Nppb (Mm01255770_g1), Mmp2 (Mm00439498_m1), PDE9a (Mm00501049_m1), PDE1a (Mn00450244_m1), PDE5a (Mn00463177_m1), Trpc6 (Mm01176083_m1); For SYBR primers used for rat ANP (Forward 5′-ATACAGTGCGGTGTCCAACACAGA-3′, Reverse 5′-TGACCTCATCTTCTACCGGCATCT-3′), rat BNP (Forward 5′-ATGCAGAAGCTGCTGGAGCTGATA-3′, Reverse 5′-CTTCTGCCCAAAGCAGCTTGAACT-3′), and rat GAPDH (Forward 5′-GACATGCCGCCTGGAGAAAC-3′, Reverse 5′-AGCCCAGGATGCCCTTTAGT-3′), rat PDE5A (Forward 5′-AACTCGTGGCAGCCGAATTCTTTG-3′, Reverse 5′-TGTTCATTAGATCAGCGGGCTCCA-3′), rat PDE9A (Forward 5′-TACGGAAGCCCACCTTTGATGTCT-3′, Reverse 5′-TTGGGTTGATGCTGAAGTCCCTGA-3′). All PCR samples were run in duplicate and normalized to GAPDH. Specificity of the SYBR green assays was confirmed by dissociation curve analysis.
Protein electrophoresis and immunoblot assays followed standard procedures, using lysis buffer (Cell Signaling) with protease inhibitor PMSF (1 mM), and run on NuPAGE 4 to 12% gel (Invitrogen). Membranes were probed for human anti-PDE9A (1:1,000), mouse anti-GAPDH or human anti-actin (1:10,000, Cell Signaling). Quantitation used fluorescence detection (Odyssey, Licor, and Odyssey Application Software 3.0). For human LV biopsy tissue analysis, samples were applied at 25 μg (dry weight), and probed with primary antibody (Millipore, ABN32, dilution 1:1000), and visualized using secondary horseradish peroxidase-labeled, goat-anti-rabbit/mouse antibody (dilution 1:1000; DakoCytomation) and enhanced chemiluminescence (ECL Western blotting detection, Amersham Biosciences). Signals were normalized to actin (dilution 1:1000; clone KJ43A; Sigma) stained on the same blots. Leucine incorporation assays were performed as described16
Real-time myocyte cGMP Analysis
RNCMs plated on gelatin-coated 35 mm glass bottom culture dishes (MatTek) were incubated overnight, and at 50% confluence, exposed to adenovirus expressing FlincG13 (Courtesy of W. Dostmann, Univ. of Vermont, Burlington, VT) at an MOI-10, for 24–48 h at 37°C, 5% CO2 until a 90% transfection efficiency was achieved. Since the absolute level of expressed FlincG varied among cells, each cell was used as their own control, with stimulation protocols compared within a given cell by paired analysis. Cell imaging was performed in imaging buffer (Hank’s balanced salt solution (HBSS), Mediatech, Inc.) by using a 3i spinning disk confocal system on a ZEISS microscope with SlideBook 5.0 software (Intelligent Imaging Innovations, Inc.), a 63x oil dipping objective (N.A. 1.0), and iXon ENCCD DVB camera with 1 second acquisitions and 20-ms exposure time, exciting with a solid state laser at 488 nm and collecting the emission at 510 nm at 37°C. cGMP responses are investigated upon local application (20 to 50 ml) and subsequent diffusion of DEANO (Calbiochem), ANP (Sigma), ODQ (Sigma), and specific PDE inhibitors (Pfizer). Data is analyzed by using the Slidebook software.
Histology
Myocardium was fixed with 10% formaldehyde, paraffin embedded and sectioned into 4 μm slices. Masson’s trichrome staining was used to visualize collagen. Quantification of fibrosis content was performed in 4–6 regions of each heart. Wheat germ agglutinin staining of mouse heart sections: Slides were deparaffinized, rehydrated, and subjected to citrate-based heat-mediated antigen retrieval. Slides were incubated with 5 μg/ml Alexa Fluor 647-conjugated wheat germ agglutinin (Invitrogen) overnight at 4 °C and mounted using Prolong Gold mounting medium (Invitrogen)27. Myocyte cross sectional area was measured using an automated algorithm with NIH Image J 1.47i software. Image acquisition was performed on a Zeiss LSM510-META laser scanning microscope.
Luciferase Reporter Assay
RNCMs were transfected with NFAT-luc, CREB1-luc, GATA4-luc, TK-luc (Promega) or MEF2-luc (Addgene), and using X-fect reagent (Clontech) according to manufacturer’s protocol. After 24 hours transfection, cells were stimulated with 25 μM of PE for 6 hours, lysates harvested using passive lysis buffer (Promega) and luciferase activity determined using the Dual Luciferase assay kit (Promega) and Veritas 96 well microplate luminometer (Turner Biosystems) following manufacturer’s protocol.
Phosphoproteomic Analysis
Samples (n=4/group) were lysed in 8M Urea, 0.5% SDS with brief sonication and protein concentration determination by the BCA method. For each sample, 200 μg of total protein was digested with trypsin/Lys-C protease mixture (Promega) according to previously published methods30, samples desalted on 10 mg Oasis HLB cartridges (Waters) and eluted in 300 μL of 80% acetonitrile, 5% trifluoroacetic acid, 1 M glyocolic acid and enriched by titanium dioxide (TiO2). Enriched peptides were desalted as above but eluted in 200 μL of 80% ACN, 0.1% formic acid and dried under vacuum. Dried peptides were re-suspended in 20 μL of 0.1% FA for LC-MS/MS analysis.
For each sample, 4 μL was injected in duplicate onto an EASY-nLC 1000 (mobile phase A was 0.1% FA in water and mobile phase B was 0.1 % FA in ACN) connected to a Q-Exactive Plus (Thermo) equipped with a nano-electrospray ion source. All raw MS/MS data was searched using the Sorcerer 2TM-SEQUEST® algorithm (Sage-N Research) using default peak extraction parameters. Post-search analysis was performed using Scaffold 4 (Proteome Software, Inc.) with protein and peptide probability thresholds set to 95% and 90%, respectively, and one peptide required for identification, and these the spectra were manually validated. Phosphosite localization was determined using Scaffold PTM version 2.1.3 and phosphosites with probabilities less than 90% were ignored. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD001585.
Statistics and Reproducibility
For all analyses, sample size is reported in the figure legends or figures themselves. Most of in vitro studies were done with 2–3 sets of independent experiments. Comparisons of multiple groups were performed using either 1-way or 2-way ANOVA (if appropriate). If normality or equal variance tests failed, then a Kruskal Wallis test was used. Two-group anlaysis used either Student’s t-test or non-parametric Mann-Whitney test. Post-hoc multiple comparisons testing used either a Tukey or Dunns test. Analysis of time-dependent changes were performed by analysis of covariance with repeated measures. Formal power analysis was not prospectively performed though for variables where variance was known, we could estimate sample size based on an anticipated mean effect.
Extended Data
Extended Data Figure 1. Development of PDE9A Knockout (PDE9A−/−), and specificity of PDE5A or PDE9A siRNA.
a, PDE9A knockout (PDE9A−/−) mice were developed by replacing of Exon-12 region with Lac-Z-neomycin in the catalytic domain of c-terminal in PDE9a gene. The genotyping was performed using specific primers designed between Exon-11 and -13 including neomycin as following; GS1 (5′-cacagatgatgtacagtatggtctgg-3′), GS2 (5′-tgcagtcatcaggaccaagatgtcc-3′) and Neo (5′-gacgagttcttctgaggggatcgatc-3′). b, The typical genotyping pattern of PDE9A−/− mice was shown on 2% agarose gel (250 bp for WT and 500 bp for PDE9A−/− mice). c, Selective gene silencing using siRNA targeting PDE5A or PDE9A. PCR confirms specificity and substantial gene knockdown achieved in cell culture (n=6/group).
Extended Data Figure 2. Expression of PDE9A protein in RNCM, mouse brain, and human heart.
a, Immunoblot for PDE9A in neonatal cardiomyocytes transfected with either scrambled control siRNA (Sc) or siRNA to PDE9A (siR) - confirming suppression of protein expression by the siRNA. A control gel for comparison was derived from brain tissue using PDE9A−/− mice and littermate controls. The band identified at ~60 to 65 kDa was similar in both tissues. PDE9A bands are usually identified between 55–70 KDa depends on the splice variants expressed in a given tissue and species. b, Control immunohistochemistry showing PDE9A detected by antibody can be largely quenched (inactivated) selectivity by preincubation with recombinant ligand (scale bar: 50 μm). c, Immunostaining of PDE9A from all 10 control and DCM patients; (scale bar: 200 μm). There was consistent enhanced staining in DCM patients versus controls. All images were obtained at the identical level of laser illumination and have not been altered in any way.
Extended Data Figure 3. Selectivity of PDE9A inhibitors.
a, The dose-responses of selective PDE9A inhibitor (PF-9613) on recombinant PDE9A or PDE5A. Data performed in triplicate at each point. A dose of 5 μM of PF-9613 inhibited PDE9A effectively, but had negligible impact on PDE5A. By contrast, the PDE5A inhibitor sildenafil inhibited PDE5A by 80% at a dose of 1 μM, commonly used for cells and tissue, but had no impact on PDE9A at this dose. These doses were therefore used in our cell-based studies. b, Confirmation that an alternative PDE9A inhibitor (PF-04447943) currently being tested in humans shows similar anti-hypertrophic effects as PF-9613 in cardiac myocytes (n=4/group); *p<0.001 vs baseline; #p<0.01 vs PE. c, Inhibition of PKG activity with DT3 reverses the suppression of PE-stimulated BNP gene expression by PF-9613. This is a companion panel to Fig. 2b (lower) (n=6 for basal (no drugs, n=8 other groups); *p<0.001 vs baseline; #p<0.01 vs PE..
Extended Data Figure 4. PKG activity or cGMP measurement of ANP with inhibitors in RNCM.
a, PF-9613 significantly increases PKG activity assessed by in vitro assay upon stimulation with ANP (n=4/group); *p<0.01 vs other groups. b, PF-9613 augmentation of ANP-stimulated cGMP is not altered due to gene silencing of PDE5A. (n=5) This differs from the complete suppression of cGMP modulation by PF-9613 in myocytes with PDE9A genetically silenced (c.f. Fig 1g). c, SIL does not enhance cGMP stimulated by ANP. This contrasts to its augmentation of NO-donor derived cGMP (c.f. Fig. 1h). (n=4); *P < 0.01 vs basal state, #P < 0.01 vs ANP.
Extended Data Figure 5. Confocal immunostaining of cardiomyocyte PDE9A and PDE5A.
a, PDE9A does not colocalize with α-actinin at the z-band, where as PDE5A does. b, PDE9A does not co-localize with α-actinin in rat neonatal myocytes. c) PDE9A co-localizes with T-tubular membranes as defined by antibody staining against the sarcoplasmic reticular ATPase-2 (SERCA2a). This differed from the localization of PDE5A (scale bar: 20 μm).
Extended Data Figure 6. Myocardial cAMP levels in controls (sham-WT) and PDE9A−/− mice before and after trans-aortic constriction (TAC).
The cAMP levels were increased in TAC-WT, but they were not affected by modulation of PDE9A expression (n=Sham-WT (4), TAC-WT (5), Sham-PDE9A−/− (6), TAC-PDE9A−/− (10)).
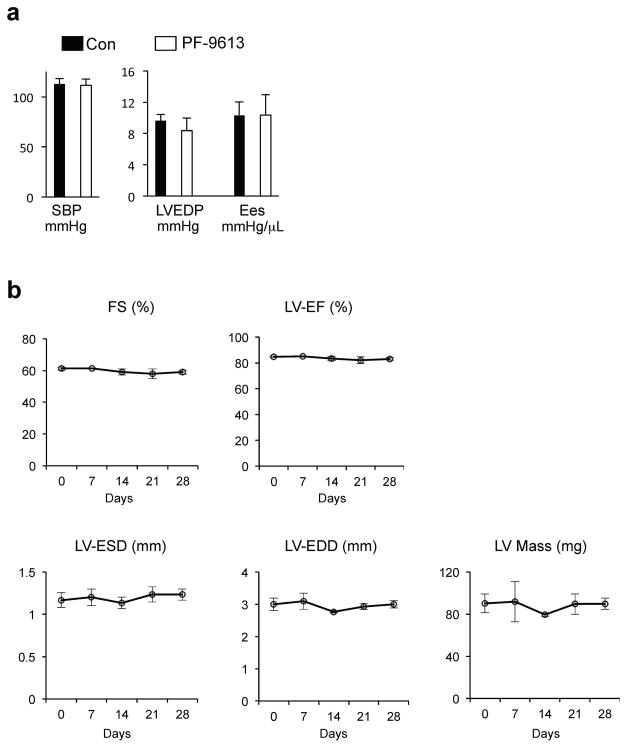
Extended Data Figure 7. Effect of PF-9613 on blood pressure and cardiac function in mouse.
a, Acute administration of PF-9613 by gavage was studied to assess effects on cardiac pressures, and contractility (end-systolic elastance; Ees). Over a 1 hour observation period (peak plasma concentrations found after 30 minutes) there was no change in any of these parameters. (n=3) b, Chronic treatment of sham control mice with PF-9613 for 3-weeks (n=3) revealed no effect on cardiac function, mass, or volumes.
Abbreviations: FS% - fractional shortening, LV-ESD and LV-EDD - end-systolic and end-diastolic left ventricular cross sectional dimension; EF% - ejection fraction.
Extended Data Figure 8. Effect of chronic PDE9A inhibition on LV mass, lung weight, and alteration of TAC-responsive genes.
a, Post-mortem analysis of heart mass and lung weight (both normalized to tibia length) from mice subjected to 5-weeks of pressure overload (TAC), and co-treated with either vehicle control, PDE9A inhibitor, or PDE5A inhibitor. A sham operated control group is also shown (n= sham (6), TAC (9), TAC+PF9613 (9), TAC+SIL (5)). b, Molecular analysis of TAC-responsive (increased expression) genes, including showing similar reductions from either PDE inhibitor in some (e.g. Trpc6), a disparity with significant or borderline greater efficacy from PDE9A inhibition on others (e.g. Ctgf, Nppa, p<0.02 and p<0.1, respectively between PDE5A and PDE9A-inhibitor response), and substantial disparities in others (e.g. Fn1, p<0.001 between PDE5A and PDE9A-inhibition). (n= sham (5), TAC (5), TAC+PF9613 (6), TAC+SIL (5)); * p<0.01 vs Sham; † p<0.001 vs Sham ; # p≤0.05, ‡p<0.01, §p<0.001 vs TAC.
Extended Data Figure 9. Gene expression of cGMP-hydrolyzing PDEs in PDE9A−/− and littermate controls.
(n=10/group)
Extended Data Table 1.
Clinical characteristics of dilated heart failure and donor control patients.
| Non-Failing Controls | Dilated HF | p | |
|---|---|---|---|
| Heart Weight/Body Weight (g/kg) | 4.9 ± 0.6 | 7.1 ± 1.0 | <0.0001 |
| Ejection Fraction (%] | 53.1 ± 2.0 | 14.3 ± 4.2 | <0.0001 |
| Biventricular Pacing [CRT] | 0 | 70% | 0.003 |
| History of VT/VF | 0 | 60% | 0.01 |
| History of Diabetes | 30% | 30% | NS |
| Drugs: | |||
| Amiodarone | 0% | 60% | 0.01 |
| Beta-blockers | 0% | 90% | <0.001 |
| ACE-inhibitors | 40% | 20% | NS |
| Nitrates | 0% | 30% | NS |
VT – ventricular tachycardia; VF – ventricular fibrillation; CRT – cardiac resynchronization therapy
Supplementary Material
Supplementary Table 1: Proteomic analysis of protein amino acid residues with increased phosphorylation in response to inhibition of PDE5A, PDE9A, or both.
Supplementary Table 2: Proteomic analysis of amino acid residues where increased phosphorylation from PDE5A or PDE9A inhibition was reduced by co-incubation with L-NAME.
Supplementary Table 3: Mice lacking PDE9A displayed similar heart function and mass as in littlemate controls.
Acknowledgments
We thank students Robert D. Wardlow and Xueying Hu for their assistance with some of the assays and studies. This research was supported by: The National Institute of Health (HL-119012, HL-089297, HL-07227), Fondation Leducq TransAtlantic Network of Excellence, The Peter Belfer Foundation, and Abraham and Virginia Weiss Professorship (D.A.K.); HL-093432 (E.T.), American Heart Association (D.I.L), Max Kade Fellowship of the Austrian Academy of Sciences (P.P.R). Procurement of human heart tissue was enabled by grants from the National Institutes of Health (HL089847 and HL105993) to Dr. Kenneth B. Margulies. Nazha Hamdani and Walter J. Paulus were supported by the European Commission FP7 project 2010 Health (MEDIA; 261409). Ronald Holewinski and Jennifer E. Van Eyk were supported by The Johns Hopkins Innovation Proteomics Center in Heart Failure (NHLBI-HV-10-05 (2) and HHSN268201000032C). We thank Pfizer and in particular Christopher Schmidt and Robin Kleiman for providing the PDE9A−/− mouse and PF-04449613, and Laurinda Jaffe at the University of Connecticut Health Center for providing PDE9A antibody.
Footnotes
Supplementary information is linked to the online version of the paper at www.nature.com/nature
Author Contributions
D.I.L. and D.A.K conceived and directed the project, designed experiments and prepared the manuscript. D.I.L. conducted most of the experiments and analyzed the data. G.Z. helped with in vivo experiments. T.S., S.J., T.D., M.Z., and P.P.R. contributed molecular biology experiments. G.C. and C.K. contributed immunostaining and in situ hybridization analyses. N.H. and W.P. performed experiments for HFPEF and AS human samples. M.J.R and R.H. performed the experiments for phospho-proteomics and J.A.K contributed to the data analysis. D.B. performed echocardiography and drug treatment. K.B.M. coordinated the human sample and data collection at the U. of Penn, and W.R.D., J.E.V., and E.T. helped with data interpretation and presentation.
The authors declare no competing financial interests.
Readers are welcome to comment on the online version of this article at www.nature.com/nature.
This file contains Supplementary Tables 1–3.
References
- 1.Takimoto E, et al. Chronic inhibition of cyclic GMP phosphodiesterase 5A prevents and reverses cardiac hypertrophy. Nat Med. 2005;11:214–222. doi: 10.1038/nm1175. [DOI] [PubMed] [Google Scholar]
- 2.Kukreja RC, Salloum FN, Das A. Cyclic guanosine monophosphate signaling and phosphodiesterase-5 inhibitors in cardioprotection. J Am Coll Cardiol. 2012;59:1921–1927. doi: 10.1016/j.jacc.2011.09.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mullershausen F, Russwurm M, Koesling D, Friebe A. In vivo reconstitution of the negative feedback in nitric oxide/cGMP signaling: role of phosphodiesterase type 5 phosphorylation. Mol Biol Cell. 2004;15:4023–4030. doi: 10.1091/mbc.E03-12-0890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Castro LR, Verde I, Cooper DM, Fischmeister R. Cyclic guanosine monophosphate compartmentation in rat cardiac myocytes. Circulation. 2006;113:2221–2228. doi: 10.1161/CIRCULATIONAHA.105.599241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Takimoto E, et al. Compartmentalization of cardiac beta-adrenergic inotropy modulation by phosphodiesterase type 5. Circulation. 2007;115:2159–2167. doi: 10.1161/CIRCULATIONAHA.106.643536. [DOI] [PubMed] [Google Scholar]
- 6.Carnicer R, Crabtree MJ, Sivakumaran V, Casadei B, Kass DA. Nitric oxide synthases in heart failure. Antioxid Redox Signal. 2013;18:1078–1099. doi: 10.1089/ars.2012.4824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Soderling SH, Bayuga SJ, Beavo JA. Identification and characterization of a novel family of cyclic nucleotide phosphodiesterases. J Biol Chem. 1998;273:15553–15558. doi: 10.1074/jbc.273.25.15553. [DOI] [PubMed] [Google Scholar]
- 8.Fisher DA, Smith JF, Pillar JS, St Denis SH, Cheng JB. Isolation and characterization of PDE9A, a novel human cGMP-specific phosphodiesterase. J Biol Chem. 1998;273:15559–15564. doi: 10.1074/jbc.273.25.15559. [DOI] [PubMed] [Google Scholar]
- 9.Conti M, Beavo J. Biochemistry and Physiology of Cyclic Nucleotide Phosphodiesterases: Essential Components in Cyclic Nucleotide Signaling. Annu Rev Biochem. 2007;76:481–511. doi: 10.1146/annurev.biochem.76.060305.150444. [DOI] [PubMed] [Google Scholar]
- 10.Kleiman RJ, et al. Phosphodiesterase 9A regulates central cGMP and modulates responses to cholinergic and monoaminergic perturbation in vivo. J Pharmacol Exp Ther. 2012;341:396–409. doi: 10.1124/jpet.111.191353. [DOI] [PubMed] [Google Scholar]
- 11.Heckman PR, Wouters C, Prickaerts J. Phosphodiesterase inhibitors as a target for cognition enhancement in aging and Alzheimer’s disease: a translational overview. Current pharmaceutical design. 2014;21:317–331. doi: 10.2174/1381612820666140826114601. [DOI] [PubMed] [Google Scholar]
- 12.Sharma K, Kass DA. Heart failure with preserved ejection fraction: mechanisms, clinical features, and therapies. Circ Res. 2014;115:79–96. doi: 10.1161/CIRCRESAHA.115.302922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nausch LW, Ledoux J, Bonev AD, Nelson MT, Dostmann WR. Differential patterning of cGMP in vascular smooth muscle cells revealed by single GFP-linked biosensors. Proc Natl Acad Sci USA. 2008;105:365–370. doi: 10.1073/pnas.0710387105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Singh G, Kuc RE, Maguire JJ, Fidock M, Davenport AP. Novel snake venom ligand dendroaspis natriuretic peptide is selective for natriuretic peptide receptor-A in human heart: downregulation of natriuretic peptide receptor-A in heart failure. Circ Res. 2006;99:183–190. doi: 10.1161/01.RES.0000232322.06633.d3. [DOI] [PubMed] [Google Scholar]
- 15.Davis J, Burr AR, Davis GF, Birnbaumer L, Molkentin JD. A TRPC6-Dependent Pathway for Myofibroblast Transdifferentiation and Wound Healing In Vivo. Developmental cell. 2012;23:705–715. doi: 10.1016/j.devcel.2012.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koitabashi N, et al. Cyclic GMP/PKG-dependent inhibition of TRPC6 channel activity and expression negatively regulates cardiomyocyte NFAT activation Novel mechanism of cardiac stress modulation by PDE5 inhibition. J Mol Cell Cardiol. 2010;48:713–724. doi: 10.1016/j.yjmcc.2009.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kinoshita H, et al. Inhibition of TRPC6 channel activity contributes to the antihypertrophic effects of natriuretic peptides-guanylyl cyclase-A signaling in the heart. Circ Res. 2010;106:1849–1860. doi: 10.1161/CIRCRESAHA.109.208314. [DOI] [PubMed] [Google Scholar]
- 18.Stangherlin A, et al. cGMP signals modulate cAMP levels in a compartment-specific manner to regulate catecholamine-dependent signaling in cardiac myocytes. Circ Res. 2011;108:929–939. doi: 10.1161/CIRCRESAHA.110.230698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pilz RB, Broderick KE. Role of cyclic GMP in gene regulation. Front Biosci. 2005;10:1239–1268. doi: 10.2741/1616. [DOI] [PubMed] [Google Scholar]
- 20.Oka T, et al. Cardiac-specific deletion of Gata4 reveals its requirement for hypertrophy, compensation, and myocyte viability. Circ Res. 2006;98:837–845. doi: 10.1161/01.RES.0000215985.18538.c4. [DOI] [PubMed] [Google Scholar]
- 21.Maurice DH, et al. Advances in targeting cyclic nucleotide phosphodiesterases. Nature reviews. Drug discovery. 2014;13:290–314. doi: 10.1038/nrd4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Greene SJ, et al. The cGMP signaling pathway as a therapeutic target in heart failure with preserved ejection fraction. Journal of the American Heart Association. 2013;2:e000536. doi: 10.1161/JAHA.113.000536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Redfield MM, et al. Effect of phosphodiesterase-5 inhibition on exercise capacity and clinical status in heart failure with preserved ejection fraction: a randomized clinical trial. JAMA. 2013;309:1268–1277. doi: 10.1001/jama.2013.2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Heerebeek L, et al. Low myocardial protein kinase G activity in heart failure with preserved ejection fraction. Circulation. 2012;126:830–839. doi: 10.1161/CIRCULATIONAHA.111.076075. [DOI] [PubMed] [Google Scholar]
- 25.McMurray JJ, et al. Angiotensin-Neprilysin Inhibition versus Enalapril in Heart Failure N Engl J Med2014 [DOI] [PubMed] [Google Scholar]
- 26.Zakeri R, Burnett JC. Designer natriuretic peptides: a vision for the future of heart failure therapeutics. Can J Physiol Pharmacol. 2011;89:593–601. doi: 10.1139/y11-048. [DOI] [PubMed] [Google Scholar]
- 27.Seo K, et al. Combined TRPC3 and TRPC6 blockade by selective small-molecule or genetic deletion inhibits pathological cardiac hypertrophy. Proc Natl Acad Sci U S A. 2014;111:1551–1556. doi: 10.1073/pnas.1308963111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Verhoest PR, et al. Design and discovery of 6-[(3S,4S)-4-methyl-1-(pyrimidin-2-ylmethyl)pyrrolidin-3-yl]-1-(tetrahydro-2H-pyr an-4-yl)-1,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one (PF-04447943), a selective brain penetrant PDE9A inhibitor for the treatment of cognitive disorders. Journal of medicinal chemistry. 2012;55:9045–9054. doi: 10.1021/jm3007799. [DOI] [PubMed] [Google Scholar]
- 29.Zhou B, et al. Epicardial progenitors contribute to the cardiomyocyte lineage in the developing heart. Nature. 2008;454:109–113. doi: 10.1038/nature07060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kirk JA, et al. Cardiac resynchronization sensitizes the sarcomere to calcium by reactivating GSK-3beta. J Clin Invest. 2014;124:129–138. doi: 10.1172/JCI69253. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1: Proteomic analysis of protein amino acid residues with increased phosphorylation in response to inhibition of PDE5A, PDE9A, or both.
Supplementary Table 2: Proteomic analysis of amino acid residues where increased phosphorylation from PDE5A or PDE9A inhibition was reduced by co-incubation with L-NAME.
Supplementary Table 3: Mice lacking PDE9A displayed similar heart function and mass as in littlemate controls.