Abstract
Long-term mammalian spermatogenesis requires proper development of spermatogonial stem cells (SSCs) that replenish the testis with germ cell progenitors during adult life. TAF4b is a gonadal-enriched component of the general transcription factor complex, TFIID, which is required for the maintenance of spermatogenesis in the mouse. Successful germ cell transplantation assays into adult TAF4b-deficient host testes suggested that TAF4b performs an essential germ cell autonomous function in SSC establishment and/or maintenance. To elucidate the SSC function of TAF4b, we characterized the initial gonocyte pool and rounds of spermatogenic differentiation in the context of the Taf4b-deficient mouse testis. Here we demonstrate a significant reduction in the late embryonic gonocyte pool and a deficient expansion of this pool soon after birth. Resulting from this reduction of germ cell progenitors is a developmental delay in meiosis initiation, as compared to age-matched controls. While GFRα1+ spermatogonia are appropriately present as Asingle and Apaired in wild type testes, TAF4b-deficient testes display an increased proportion of long and clustered chains of GFRα1+ cells. In the absence of TAF4b, seminiferous tubules in the adult testis either lack germ cells altogether or are found to have missing generations of spermatogenic progenitor cells. Together these data indicate that TAF4b-deficient spermatogenic progenitor cells display a tendency for differentiation at the expense of self-renewal and a renewing pool of SSCs fail to establish during the critical window of SSC development.
Keywords: TAF4b, Spermatogonial Stem Cells, Self-Renewal, Spermatogenesis, Meiosis
Introduction
Throughout most of an adult male lifespan, unipotent stem cells in the testis called spermatogonial stem cells (SSCs) support long-term spermatogenesis. These SSCs undergo both self-renewing and differentiating divisions, allowing for the production of millions of sperm each day. In mammals, the process of germ cell development is complex and highly coordinated. Following specification in the proximal epiblast, primordial germ cells (PGCs) migrate from the hindgut through the dorsal mesentery, proliferate and colonize the nascent somatic gonad between embryonic days (E)8.5 and E13.5 [1-4]. Once inside the male gonad around E13.5, PGCs transition to undifferentiated gonocytes that become enveloped in testicular cords with the somatic support cell precursors. In mice, gonocytes proliferate until about E16 and then become mitotically quiescent until postnatal day (P)4 when they migrate to the basement membrane and resume proliferation, to produce differentiating spermatogonia, as well as a population of SSCs that will support long-term spermatogenesis [5]. As with all stem cell populations, there is a delicate balance between self-renewal and differentiation of SSCs that is highly regulated and required for long-term spermatogenic differentiation. The signals and molecular mechanisms governing the decision of SSCs to renew or differentiate remain enigmatic. In the adult testes, SSCs represent only 0.03% of all germ cells and are difficult to distinguish from closely related and positioned differentiating spermatogonia [6]. However, over the past two decades, several experimental advances in stable SSC culturing and transplantation paradigms have allowed us to explore many aspects of SSC biology [7-12]. These advances facilitated the discovery and characterization of several genes important for SSC function, which include transcription factors (Bcl6b, Lhx1, Etv5, Id4 and Plzf), translational regulators (Nanos2), extracellular signaling factors (GDNF, GFRα1, and Ret), intracellular signaling factors (PI3K/AKT, SFK) and more recently, microRNAs (miR-21)[3, 12-21]. Together, morphological and molecular analyses identified Asingle (As) spermatogonia as a population of undifferentiated germ cells that contain SSCs. Recent identification of the transcription factors Pax7 and Id4 marking two independent As subpopulations, each containing robust SSC properties, underscores the potential cell type and lineage heterogeneity that exists within As spermatogonia [21, 22]. During spermatogenesis, As spermatogonia give rise to 2 Apaired (Apr) and 4-16 Aaligned (Aal) spermatogonial chains through cell division and incomplete cytokinesis [23-27]. Aal spermatogonia synchronously differentiate into the first generation of differentiating type spermatogonia that progress through differentiating spermatogonial divisions, followed by meiotic and post-meiotic differentiation to produce mature sperm. This cyclical process includes 12 (I-XII) subsequent morphologically distinct seminiferous epithelial stages [28]. Glial cell line-derived neurotrophic factor (GDNF) signaling from Sertoli cells promotes proliferation of undifferentiated spermatogonia and is also required for SSC maintenance [29-32]. A subset of SSC-containing undifferentiated As and Apr spermatogonia express the GPI-anchored cell surface GDNF receptor, GFRα1 [26, 33, 34]. GFRα1 has proven a valuable marker for observation of As/Apr/Aal dynamics during spermatogenesis, as well as their isolation and characterization [35, 36].
Transcriptional regulation plays a central role in the precise control of animal growth, development and fertility. RNA polymerase II (PolII)-mediated transcription initiation depends upon a complex assembly of general transcription factor complexes on core promoter elements [37, 38]. TFIID contains the TATA-box binding protein (TBP) along with 14 TBP-associated factors (TAFs) and is required for core promoter recognition and activator-dependent PolII recruitment [39-44]. Over the past 15 years, several studies have identified TBP-like and TAF-like paralogs that confer unique transcriptional regulation in a variety of cell types, most notably in germ cells [45, 46]. Therefore, germ cell-specific gene expression programs require not only specialized transcriptional activators and repressors, but also alternate forms of the general transcription machinery [46, 47].
TAF4b is a gonadal-enriched TFIID subunit required for both male and female fertility in the mouse [48-52]. Male Taf4b-deficient mice are subfertile, exhibiting extensive germ cell loss and impaired expression of several SSC-specific genes by 3 months of age [49]. Furthermore, long-term cultured mouse SSCs and human SSCs display elevated expression of TAF4b [53, 54]. While our previous study indicates that TAF4b is required for long-term spermatogenesis, it is unknown whether TAF4b is required for the establishment and/or maintenance of SSCs. Therefore, to better understand TAF4b function in male germ cell development and fertility, we have characterized the establishment of gonocytes during late embryonic development and the initial rounds of spermatogenic differentiation in the context of the Taf4b-deficient testis. Surprisingly, TAF4b is already required for gonocyte development during embryogenesis. While TAF4b-deficient gonocytes can ultimately differentiate into healthy sperm, they are unable to maintain a self-renewing SSC pool. Together these data uncover a novel role of TAF4b in embryonic spermatogonia development and highlight the role of TAF4b in promoting the establishment of the initial SSC pool.
Materials and Methods
Animals
Wild type C57BL/6 and Taf4b −/− mice were housed and maintained in the Brown University Animal Care Facility in accordance with the NIH Guide for the Care and Use of Laboratory Animals. The Taf4b −/− mice have a C57BL/6 genetic background and were generated as described previously [48]. All offspring were genotyped using genomic DNA from tail-snips and PCR analysis of the Taf4b gene locus targeted by homologous recombination.
Quantitative RT-PCR (qPCR)
RNA was purified from detunicated whole testes by Trizol extraction (Invitrogen, Carlsbad, CA). Template cDNA was generated using the iScript cDNA Synthesis Kit (Bio-Rad, Hercules, CA). Primer sets were designed to amplify 100-175 base pair-sized products (Table 1). The ABI 7900H Real-Time PCR system (Applied Biosystems) and the Power SYBR® Green qPCR Master Mix with ROX™ (Invitrogen, Carlsbad, CA) were used for qPCR data acquisition. The qPCR reactions were incubated at 50°C for 2 minutes, 95°C for 10 minutes followed by 40 cycles of 95°C for 15 seconds and 60°C for seconds and a dissociation curve analysis. Data from each gene were normalized to 18S rRNA levels and represented as a fold change relative to RNA levels in the indicated embryonic or postnatal testes for each respective qPCR figure panel. Each qPCR reaction was performed in triplicate and averaged. Error bars indicate the standard deviation resulting from experimental and normalized triplicate qPCR reactions.
Table 1.
| Forward | Reverse | |
|---|---|---|
| 18S rRNA | 5’-CCG CGG TTC TAT TTT GTT GG-3’ | 5’-GGC GCT CCC TCT TAA TCA TG-3’ |
| Taf4b | 5’-GAT GTT ACT AAA GGC AGC CAA GAG T-3’ | 5’-CTG CTC TGG ATC TTC TTT ATT GGA G-3’ |
Immunofluorescence
Testes were dissected from various embryonic and postnatal stage mice, embedded in Tissue-Tek® O.C.T. Compound (Sakura Finetek, Torrance, CA) and frozen in liquid nitrogen. Serial sections were cut at 8 μM thickness onto glass slides with a Leica cryostat and stored at °80°C until use. Prior to incubation, slides were fixed in 4% paraformaldehyde for 20 minutes and permeabilized in 0.5% Triton X-100 for 20 minutes. They were then incubated in blocking buffer (PBS with 3% goat serum, 1% BSA and 0.5% Tween-20), incubated in primary antibody for 1 hour, washed 3X in PBS-0.5%Tween-20 (PBST), incubated in secondary antibody for 1 hour, washed 3X in PBST and mounted in DAPI-containing Vectashield® Mounting Medium (Vector Laboratories, Burlingame, CA). Primary antibodies used were affinity purified rabbit anti-TAF4b (1:200;[48]), anti-TRA98 (1:500; B-Bridge, Cupertino, CA), anti-γH2Ax (1:500; EMD Millipore, Billerica, MA), anti-GFRα1 (1:200; R&D Systems, Minneapolis, Minnesota), anti-PLZF mouse mAb (2A9)(1:200; EMD Millipore, Billerica, MA), anti-cKit rabbit (1:100; Cell Signaling, Danvers, MA) and anti-Cleaved Caspase-3 rabbit (1:100; Cell Signaling, Danvers, MA). For immunofluorescence, the secondary antibodies used were Alexa Fluor® 488-conjugated anti-rat IgG (H+L)(1:500; Invitrogen, Carlsbad, CA), Alexa Fluor® 594-conjugated anti-rabbit IgG (H+L)(1:500; Invitrogen, Carlsbad, CA), Alexa Fluor® 488-conjugated anti-mouse IgG (H+L)(1:500; Invitrogen, Carlsbad, CA). Images were acquired with a Zeiss Axio Imager M1 and a Zeiss LSM 710 Confocal Laser Scanning Microscope (Carl Zeiss Inc., Thornwood, NY).
Immunohistochemistry and Histology
Testes were dissected from various embryonic and postnatal stage mice, fixed overnight in 1:10 formalin, dehydrated through serial washes of 70%, 95% and 100% ethanol followed by a final wash with xylene and allowed to perfuse in paraffin overnight before embedding. Serial sections were cut to 5 μm thickness onto glass slides and oven dried overnight. Slides for staining were incubated in xylene, hydrated in serial washes of 100%, 95% and 70% ethanol, treated for antigen retrieval using 1% Antigen Unmasking Solution (Vector Laboratories, Burlingame, CA) in a steamer for 20 minutes, permeabilized with 0.1% Triton and 0.1% sodium citrate buffer and blocked for endogenous peroxidase activity using Bloxall Solution (Vector Laboratories, Burlingame, CA) each for 5 minutes. They were then blocked with Vectastain R.T.U. 2.5% normal horse or goat serum (Vector Laboratories, Burlingame, CA), incubated in primary antibody as previously described overnight at 4°C and in biotinylated anti-rabbit secondary antibody (1:500; Vector Laboratories, Burlingame, CA) for 1 hour with 3X PBS wash after each incubation. To develop the staining, slides were incubated with R.T.U. Vectastain® Elite® ABC System for 45 minutes, NovaRED substrate kit for 2 minutes and counterstained with methyl green for 10 minutes (Vector Laboratories, Burlingame, CA). Brown University’s Molecular Pathology Core performed PAS staining for histology sections. Images were obtained using the Scanscope CS Aperio (Leica Microsystems Inc., Buffalo Grove, IL).
Results
TAF4b expression during testis development
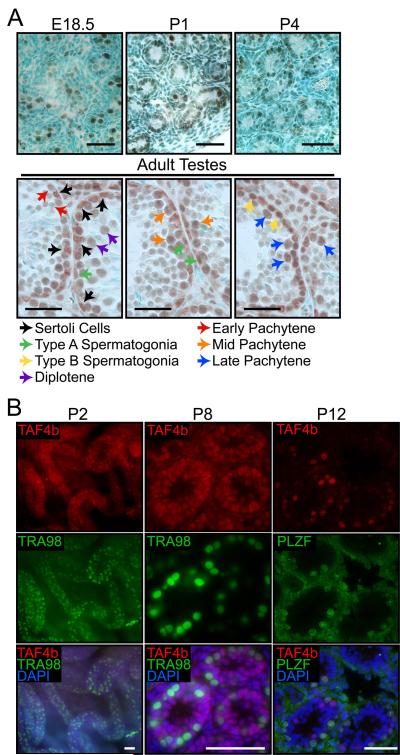
To better understand the cell-autonomous functions of TAF4b, we determined Taf4b mRNA expression and protein enrichment during embryonic and neonatal testes development in wild type mice. Previous protein localization analyses found TAF4b to be highly enriched in neonatal gonocytes and to be present in adult spermatogonia and spermatids, whereas TAF4b was not detected in meiotic spermatocytes or somatic cells [49]. We extended this analysis to embryonic day (E) 18.5 testes, as well as postnatal day (P) 1, P2, P3, P4, P8 and P12 testes. Consistent with previous results, TAF4b is enriched in early postnatal gonocytes. In addition, it is also expressed in embryonic gonocytes at E18.5 (Figure 1A). TAF4b colocalizes with the germ cell marker TRA98 in P2 seminiferous tubule whole mounts and P8 testes sections (Figure 1B)[55]. In P12 testes, TAF4b is readily detected in PLZF+ undifferentiated spermatogonia, as well as PLZF- differentiating spermatogonia (Figure 1B). In adult testes, we found TAF4b protein in nuclei of type A and B spermatogonia, as well as of pachytene and diplotene spermatocytes in various stages of the epithelial cycle (Figure 1A). Furthermore, TAF4b is detected in the somatic Sertoli cell nuclei as well (Figure 1A; black arrows). Quantitative RT-PCR (qPCR) analysis of Taf4b mRNA shows an increase in transcript abundance between P7 and P19, coincident with the onset of meiosis (Supplemental Figure 1). This increase also coincides with the expanded Taf4b expression in somatic and differentiating germ cells. The presence of TAF4b protein in late embryonic gonocytes suggests it might play a much earlier role in embryonic and neonatal spermatogonial pool establishment than previously known. The detection of TAF4b protein in Sertoli cell nuclei is also intriguing. Germ cell transplantation experiments, using Taf4b −/− recipient testes, support a nonessential role for TAF4b in Sertoli cells regarding spermatogenesis [49]. However, Sertoli cell-specific TAF4b may function in proper testes physiology beyond germ cell colonization and subsequent initial rounds of spermatogenesis. Together, these data suggest that TAF4b may have several novel and cell type-specific functions during germ cell development and spermatogenesis.
Figure 1. Germ and somatic cell TAF4B protein enrichment during testes development.
(A) Immunohistochemistry analysis of TAF4b protein localization during late embryonic, early postnatal and adult testes sections. Corresponding colored arrows indicate examples of TAF4b protein enrichment in different somatic and germ cell types. Scale bars = 50 μm. (B) P2 testes whole mount and P8 testes section immunofluorescence show colocalization of TAF4b (red) with TRA98 (green) in gonocytes, as well as TAF4b enrichment in TRA98-negative somatic cells. P12 testes sections show TAF4b protein enrichment (red) in both PLZF-positive (green) and PLZF-negative spermatogonia. DAPI (blue). Scale bars = 50 μm.
Taf4b-deficient testes display a reduced embryonic gonocyte pool and a deficient neonatal germ cell expansion
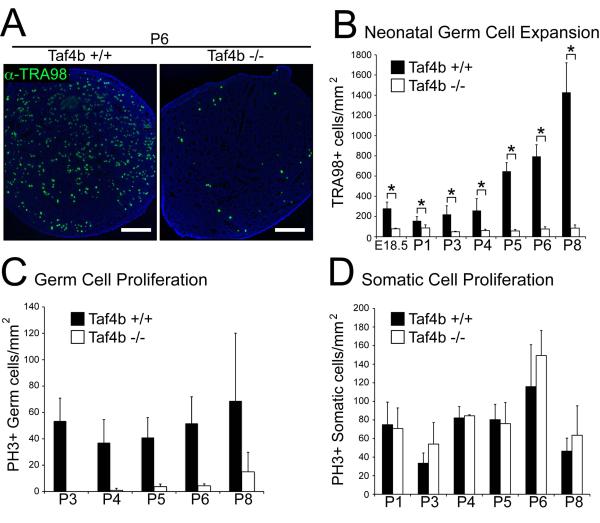
Germ cell deficiencies were previously reported in P2 Taf4b −/− males [49]. However, in view of the strong TAF4b expression in embryonic gonocytes (Figure 1B), Taf4b −/− animals may exhibit germ cell defects at a much earlier time point. To test this possibility, we assayed germ and somatic cell numbers and proliferation, using immunostaining with the germ cell marker TRA98 and phospho-histone H3 Ser10 as a marker of mitosis, during late embryonic and early postnatal testes development in litter matched wild type and Taf4b −/− animals. Strikingly, Taf4b-deficient testes have 3.6-fold fewer germ cells at E18.5 compared to wild type (Figure 2B). Moreover, while TRA98-staining in P3-P8 testes illustrates the normal neonatal germ cell expansion in wild type testes, Taf4b −/− testes fail to expand their germ cell population (Figures 2A, 2B, and Supplementary Figure 2).
Figure 2. Reduced Embryonic testes germ cell number and delayed neonatal gonocyte proliferation in Taf4b −/− testes.
(A) Identification of germ cell numbers in wild type and Taf4b-deficient testes using TRA98 immunofluorescence (green) at indicated embryonic (E) and postnatal (P) developmental time points. DAPI (blue). Scale bars = 200 μm. (B) Germ cell quantification normalized to total cross section area. Comparative Taf4b +/+ and −/− germ cell differences are statistically significant at all time points. * (p<0.05). Phosphorylated histone H3 (PH3) and TRA98 immunofluorescence distinguished proliferating germ and somatic cells. The number of germ cells (C) and somatic cells (D) positive for PH3 were normalized to total cross-section area in both Taf4b +/+ and −/− testes. N≥3 for each data point. ** (p<0.001), ns = not significant.
One of the most prominent differences between the wt and Taf4b−/− testes is the virtual absence of gonocyte proliferation at P3 and P4 in the mutant testes. This cell proliferation defect is unique to germ cells in Taf4b −/− testes, as somatic cell proliferation is unaffected (Figure 2D). Despite fewer germ cells in E18.5 Taf4b −/− testes, when tubules do contain germ cells they have nearly the same germ cell density as in the wild type (Supplemental Figure 2). However, this germ cell density similarity is lost by P8, following neonatal germ cell expansion. Although P8 Taf4b −/− testes have fewer germ cells, a significantly higher percentage of germ cells are actively proliferating compared to wild type (Figure 2C and Supplemental Figure 3). This may be due to the appearance of non-mitotic spermatocytes, diluting the percentage of mitotic germ cells in wt testes. These results suggest that Taf4b −/− germ cells are capable of proliferation, but the initiation of spermatogenesis, with its concomitant postnatal increase in proliferative activity, is delayed in Taf4b−/− testes.
Altered GFRα1+ spermatogonial chain dynamics during Taf4b −/− testes development
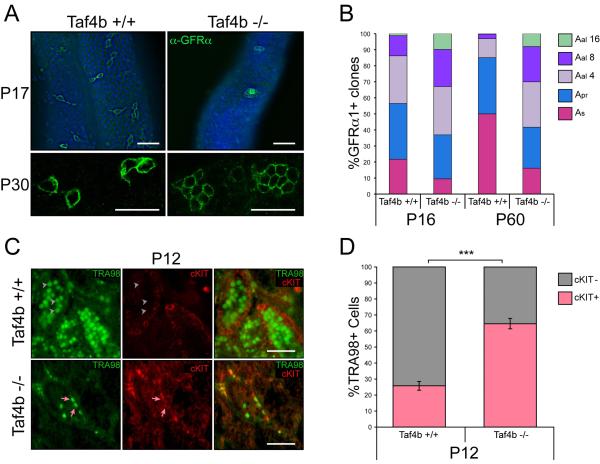
Taf4b −/− testes have reduced germ cells numbers because of the low number of gonocytes they start with and their delayed spermatogenesis. To learn more about these germ cells, we studied GFRα1 expression in spermatogonia in mutant testes. GFRα1 immunofluorescence in P16 and P60 testes reveals fewer GFRα1+ cells in Taf4b −/− animals compared to wild type mice. Furthermore, there is a conspicuous disruption in their cellular organization within the seminiferous tubules. In the wild type testis, GFRα1+ spermatogonia were predominantly in As and Apr undifferentiated spermatogonia. However, most GFRα1+ Taf4b −/− spermatogonia were in Aal-like spherical cell aggregates (Figure 3A, Supplemental Movies 1 and 2). Quantitative analysis of GFRα1+ spermatogonia revealed an altered distribution in spermatogonial chain lengths between Taf4b −/− and wild type animals (Figure 3A and 3B). These results are strikingly similar to those observed in GFRα1+ spermatogonia following germ cell transplantation and may reflect a shared compensatory mechanism used by testes to cope with a severely limited spermatogonial population [35]. Remarkably, relatively few As spermatogonia are present at P16 in Taf4b −/− mice, while one would expect this number to be sufficiently high to enable recovery from the few gonocytes and spermatogonia present at the start of spermatogenesis. This indicates a problem in the regulation of the balance between self-renewal and differentiation of the SSCs. To directly test whether Taf4b −/− germ cells exhibit a higher propensity for differentiation, we assayed differentiating germ cell numbers in litter matched P12 wild type and Taf4b −/− testes using immunostaining with TRA98 (germ cells) and c-KIT (differentiating spermatogonia) (Figure 3C). Remarkably, while 25.8% of the wild type TRA98+ germ cells were committed to differentiation, we observe a significantly higher proportion of Taf4b −/− germ cells, at 64.6%, are committed to differentiation (Figure 3D).
Figure 3. Altered GFRα1-positive spermatogonia chain length in Taf4b −/− testes.
(A) GFRα1 immunofluorescence (green) and DAPI (blue) of wild type and Taf4b −/− whole-mount P17 testes and confocal imaging of whole-mount P30 wild type and Taf4b −/− testes. Scale bars = 50 μm. (B) Quantification of GFRα1+ spermatogonial chain length in P16 and P60 testes from both wild type and Taf4b −/− mice. Each time point represents 250-400 chains from a minimum of 3 animals. (C) Immunofluorescence analysis of P12 testes sections show nuclear TRA98 (green) staining in germ cells and cell surface cKIT (red) staining in differentiating germ cells. Grey arrowheads indicate TRA98+/cKIT- cells more prevalent in Taf4b −/− testes and pink arrows indicate TRA98+/cKIT+ cells more prevalent in wild type testes. Scale bars = 50 μm. (D) Quantification of TRA98+ germ cells exhibiting or lacking cKIT staining in both wild type (n=142 seminiferous tubules) and Taf4b −/− (n=255 tubules) testes sections. Data are represented as mean and error bars indicate +/− SEM. *** (p<0.0001).
TAF4b is required for timely formation of spermatocytes and meiotic entry
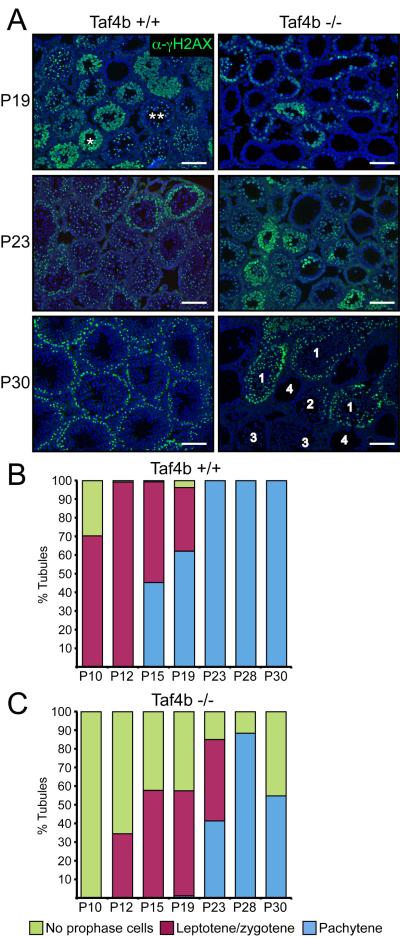
During juvenile testes development and maturation, spermatogenic initiation occurs in a coordinated fashion. To test whether Taf4b-deficiency affects the timely formation of spermatocytes and their entry into meiosis, we quantified meiotic initiation and progression by way of staining for γH2AX in wild type and Taf4b −/− testes at several time points between P10 and P30. During meiosis, the histone variant H2AX is rapidly phosphorylated at serine 139 (γH2AX) and accumulates at DNA double-strand breaks. The pattern of γH2AX localization is dynamic through meiosis I prophase and can distinguish between the earlier leptotene/zygotene and pachytene stages. While leptotene/zygotene spermatocytes exhibit nuclear foci of γH2AX staining, pachytene spermatocytes have XY body-specific γH2AX localization [56, 57].
Wild type testes have progressed to the leptotene/zygotene spermatocytes stage or beyond, in 70% of their tubules at P10 and nearly all tubules at P12. By P15, 45% of the tubules progressed to the pachytene spermatocytes stage or beyond and this number increased to 100% by P23 (Figure 4A and 4B). Taf4b −/− testes contain no prophase spermatocytes at P10 and only 35% of their tubules have leptotene/zygotene stage spermatocytes at P12. We first observe tubules with pachytene spermatocytes at P19 in Taf4b −/− testes and this number peaks at 90% by P28, while the remaining tubules contain no cells in meiotic prophase (Figure 4A and 4C). By P30, we observe various degrees of germ cell depletion in Taf4b −/− testes. We characterized phenomenon in 4 categories: normal (1), partial absence of spermatocytes (2), having spermatids undergoing spermiogenesis but lacking spermatocytes (3) or completely devoid of meiotic spermatocytes and spermatids (4) (Figure 4A). Empty tubules have either lost all their germ cells due to sloughing or were part of the 10% observed with no cells in meiotic prophase. An important distinction between wild type and Taf4b −/− testes at P30 is the appearance of category 3 degenerate tubules in Taf4b −/− testes, which contain advanced post-meiotic cells, such as round spermatids, but lack spermatocytes in meiotic prophase. This missing generation of germ cells suggests a paucity of differentiating spermatogonia, either caused by a too low number of SSCs or a problem in the differentiation of spermatogonia.
Figure 4. Delayed meiosis initiation and progression in Taf4b −/− testes.
(A) γH2AX immunofluorescence (green) of P19, P23 and P30 testis sections from wild type and Taf4b −/− mice. P30 Taf4b −/− seminiferous tubule phenotypic categories: (1) normal, (2) partial absence of spermatocytes, (3) having spermatids undergoing spermiogenesis but lacking spermatocytes, (4) completely devoid of meiotic spermatocytes and spermatids. DAPI (blue) * Denotes leptotene/zygotene stage spermatocytes with diffuse γH2AX staining, ** denotes pachytene stage spermatocytes with XY-body γH2AX foci. Scale bars = 100 μm. Quantitative analysis of γH2AX staining and meiotic progression during the first wave of spermatogenesis in wild type (B) and Taf4b −/− (C) testes. Tubules containing multiple meiotic cell types are categorized by furthest stage. Between 100 and 200 tubules were counted for each data point.
The data over the period of P10 to P30 show that Taf4b−/− tubules have a delay in the production of spermatocytes. This delay likely reflects the earlier defect in spermatogonial population development and expansion. Although a vast majority of Taf4b −/− tubules are able to initiate meiosis and progress through prophase, they lag developmentally by approximately 4-5 days compared to wild type. Seminiferous tubule diameter measurements between wild type and Taf4b −/− littermates show slight but statistically significant differences and are in agreement with the observed developmental lag (Supplemental Figure 4). Taf4b −/− testes at least partially recover from this delay and ultimately produce mature gametes. However, germ cell exhaustion ensues shortly thereafter. Together, these results suggest that while Taf4b is not required for entry into meiotic prophase or progression, it is essential for punctual meiotic initiation during the initial rounds of spermatogenesis and maintenance of the progenitor cells required for subsequent rounds of spermatogenesis.
Taf4b is required for spermatogonial progenitor maintenance and renewal
Taf4b −/− testes undergo an age-dependent premature decline in germ cells and fertility [49]. To directly test whether adult Taf4b −/− testes lack or display impaired SSC renewal, we examined germ cell differentiation in P60 testes by detailed histological analysis. All tubules in wild type P60 testes contain a variety of spermatogenic cell types (Figure 5B). The tubules in Taf4b −/− P60 testes, however, are remarkably heterogeneous. While many tubules appear normal, several show one or more missing generations of germ cells up to a complete Sertoli cell-only phenotype lacking all germ cells (Figure 5A and 5C). We also observed tubules lacking spermatogonia and spermatocytes, but containing round and elongated spermatids (Figure 5A and 5D). Tubules in mature testes missing all types of spermatogonia suggest they have also lost their ability to maintain an undifferentiated spermatogonial progenitor pool, which includes SSCs. One source for this observed germ cell loss may be enhanced apoptosis in Taf4b −/− testes. To test this possibility, we assayed the apoptosis in litter matched P21 wt and Taf4b −/− germ cells by immunostaining with TRA98 (germ cells) and cleaved Caspase 3 protein (apoptotic cells). We observed no significant increase in cleaved Caspase 3-stained germ cells in Taf4b −/− testes, suggesting apoptosis is not a major mechanism for germ cell loss in Taf4b-deficient mice (Supplementary Table 2). Together, these data indicate that TAF4b is required for the proper embryonic gonocyte development and suggest that TAF4b is required for establishing the proper balance between SSC renewal and differentiation. In the absence of TAF4b, this balance is disrupted and the testis displays an age-dependent depletion of germ cells progressive increase in Sertoli cell-only tubules [49](data not shown).
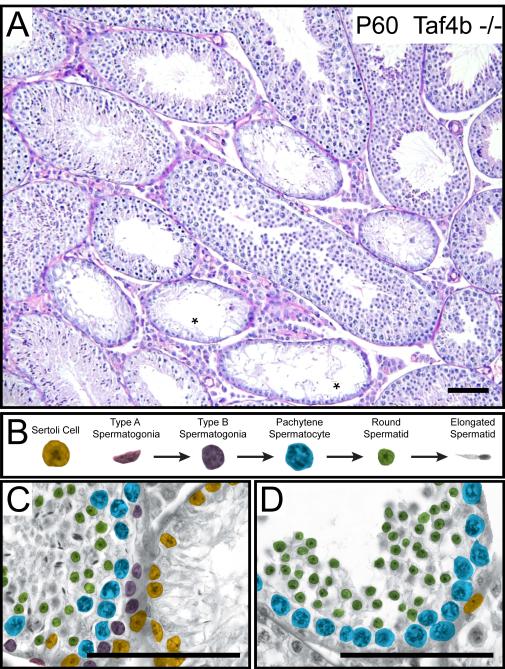
Figure 5. Taf4b −/− testes have missing generations of progenitor spermatogonia.
(A) Hemoxylin and eosin stained P60 Taf4b −/− testis section, * indicate seminiferous tubules devoid of germ cells. (B) Somatic and germ cell types present in the P60 Taf4b −/− tubule. Cell types are distinguished by their respective pseudocoloring. (C) Left tubule – Complete complement of germ cells present. Right tubule – Sertoli-cell only phenotype. (D) A tubule missing progenitor spermatogonia and elongated spermatids but containing differentiated pachytene spermatocyte and round spermatid cell types. Scale bars = 100 μm.
Discussion
Understanding how stem cells balance self-renewal with differentiation is paramount in understanding the regulatory mechanisms governing their behavior and exploring their therapeutic potential in regenerative medicine. In the mammalian testis, unipotent spermatogonial stem cells (SSCs) are required for long-term sperm production and male fertility. Loss of function genetic analyses in mice have revealed several factors required for SSC maintenance such as Nanos2, Bcl6b, PLZF, Foxo1, GFRα1, Ret, Etv5, Rb, and Id4 [16, 19, 21, 30, 33, 58-62]. Our results indicate a requirement for Taf4b in SSC maintenance as well. TAF4b-deficient male mice are initially fertile, but they exhibit a progressive germ cell loss leading to complete infertility. Taf4b-deficiency does not completely block a particular stage in germ cell development or spermatogenic differentiation. The initial fertility followed by complete infertility in Taf4b −/− mice is consistent with aberrant SSC development and is a characteristic phenotype for SSC maintenance defects [63]. Furthermore, wild type spermatogonia can repopulate adult Taf4b −/− testes, also suggesting a germ cell-autonomous requirement for TAF4b in SSCs [49]. However, the pathways in which TAF4b functions in germ cell development and SSC maintenance are unclear.
Here we report an unexpected pattern of TAF4b expression, localization and function during gonocyte development and the first wave of spermatogenesis. Taf4b −/− mice have significantly fewer gonocytes during late embryogenesis, which subsequently fail to timely produce differentiating type spermatogonia at the start of spermatogenesis. These mice exhibit a significant delay in the formation of spermatocytes and subsequent meiotic initiation. A potential cause for the delay in the appearance of spermatocytes is the reduced numbers of neonatal gonocytes in Taf4b −/− mice. Indeed, mice lacking a functional Bmp8b gene have a similar neonatal phenotype with germ cell proliferation defects and delayed occurrence of meiotic initiation [64]. Mice may require a minimum number of gonocytes in the neonatal testis before they form differentiating type spermatogonia and Taf4b −/− testes may not have sufficient gonocyte numbers for this process to appropriately occur in a timely fashion.
The insufficient number of gonocytes in neonatal Taf4b −/− testes may alter SSC balance and drive differentiation at the expense of self-renewal, whereby a renewing pool of SSCs fail to establish during the critical window of SSC development. Alternatively, TAF4b may have additional unique functions in SSCs that promote their establishment and/or maintenance regardless of the neonatal gonocyte pool size. The latter possibility seems to be the more likely one. In general, stem cells will increase their rate of self-renewal in response to a shortage in differentiated cell numbers. A recent study by Hara and coworkers suggests that GFRα1+ chain fragmentation is important in SSC renewal [65]. The many long and clustered GFRα1+ chains we observe in the TAF4b-deficient testes do not seem to contribute to normal SSC numbers unless TAF4b is involved in SSC chain fragmentation regulation. However, a recent study by Chakraborty et al., failed to find a relation between chains of Aal spermatogonia and SSC renewal [66]. We observed TAF4b expression in embryonic gonocytes, as well as in spermatocytes and Sertoli cells. While TAF4b may not be required in all these cell types for the initial rounds of spermatogenesis, it may have cell type-specific functions that are masked by the SSC renewal phenotype of Taf4b −/− mice.
The present results support a functional requirement for TAF4b in gonocyte development and proliferation, as well as in a proper establishment of the SSC population required for long-term spermatogenesis in the mouse (Figure 6). Indeed, TAF4b may play a similar role in human spermatogenesis. A recent genetic analysis supports a functional requirement for TAF4b in male reproductive health as well. Ayhan and colleagues identified a nonsense mutation in the human Taf4b gene. Four brothers that were homozygous for this mutation had reduced sperm levels, three being azoospermic and one oligospermic [67]. These findings are compatible with defective SSCs and the progressive loss of germ cells in these individuals. Further studies in our mouse model will test whether PGC proliferation or migration is impaired in Taf4b −/− mice, which genes are regulated by TAF4b in embryonic gonocytes to ensure proper establishment and proliferation, and which cofactors function with TAF4b in gene regulation. Understanding how TAF4b regulates gene expression programs necessary for SSC development may help us to better address fertility defects in men and may uncover common general transcription factor-based regulatory principles for self-renewal in other important stem cell populations.
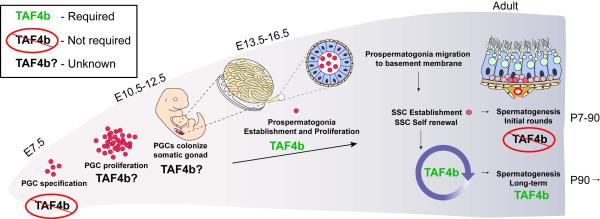
Figure 6. TAF4b function in germ cell development and spermatogenesis.
A schematic representation highlighting the major events in germ cell development from PGC specification to spermatogenesis. The requirement for TAF4b during each step is indicated.
Conclusion
Several lines of evidence suggest that the gonadal-enriched general transciption factor variant TAF4b is essential for SSC establishment and maintenance. Our results support an earlier requirement for TAF4b in embryonic and neonatal spermatogonial pool establishment than previously thought. We find testes lacking Taf4b have fewer gonocytes during late embryogenesis, deficient neonatal gonocyte expansion and several SSC-related abnormalities during the initial the initial waves of spermatogenesis. Our results indicate that TAF4b plays a critical role in the delicate balance between SSC self-renewal and differentiation and is required to sustain long-term and SSC-mediated fertility.
Supplementary Material
Supplemental Figure 1
Quantitative RT-PCR (qPCR) measured relative Taf4b mRNA levels in the indicated testes. All values were normalized against 18s rRNA and represent RNA levels as the fold difference relative to the respective RNA levels at postnatal day 1 (P1).
Supplemental Figure 2
Average number of germ cells (TRA98+) per tubule at E18.5 and P8 in wild type and Taf4b-deficient testes.
Supplemental Figure 3
The percentages of proliferating (PH3+) germ cells were measured in Taf4b +/+ and −/− testes at the indicated time points.
Supplemental Figure 4 - Delayed tubule growth in TAF4b-deficient testes
Sections of TAF4b −/− and their WT littermates were used to determine average diameter of testis tubules at indicated ages. Circular tubules were measured from the outside of the basement membrane through their widest point. Between 50 and 100 tubule diameters per data point were measured. Data are represented as mean +/− SEM.
Supplemental Movies 1 and 2 - Gfrα1+ clustered cells in TAF4b-deficient testis tubules span multiple layers
Movie of whole mount immunostaining of Gfrα1 was performed on P17 testis tubules from WT (1) and TAF4b −/− (2) littermates. Z-stack images were taken through 170 μm and spanning the outer layers of WT and TAF4b −/− tubules.
Table 2.
| Tubule Number | TRA98+ Cell Number | Cleaved Caspase 3+/ TRA98+ Cell Number |
%TRA98+ Cells with Cleaved Caspase 3 |
|
|---|---|---|---|---|
| Taf4b +/+ | 110 | 5363 | 27 | 0.5034496 |
| Taf4b −/− | 171 | 6319 | 11 | 0.1740782 |
p<0.0303
Acknowledgments
We thank K. Grive, J. Wardell and C. Brown for insightful comments on the manuscript. This work was supported by a New Scholar Award in Aging from the Ellison Medical Foundation to R.N.F. and a postdoctoral fellowship from the NICHD of the National Institutes of Health to E.A.G. (1F32HD077986).
References
- 1.Hayashi K, de Sousa Lopes SM, Surani MA. Germ cell specification in mice. Science. 2007;316:394–396. doi: 10.1126/science.1137545. [DOI] [PubMed] [Google Scholar]
- 2.Ewen KA, Koopman P. Mouse germ cell development: from specification to sex determination. Mol Cell Endocrinol. 2010;323:76–93. doi: 10.1016/j.mce.2009.12.013. [DOI] [PubMed] [Google Scholar]
- 3.Yoshida S. Stem cells in mammalian spermatogenesis. Dev Growth Differ. 2010;52:311–317. doi: 10.1111/j.1440-169X.2010.01174.x. [DOI] [PubMed] [Google Scholar]
- 4.Bendel-Stenzel M, Anderson R, Heasman J, et al. The origin and migration of primordial germ cells in the mouse. Semin Cell Dev Biol. 1998;9:393–400. doi: 10.1006/scdb.1998.0204. [DOI] [PubMed] [Google Scholar]
- 5.Phillips BT, Gassei K, Orwig KE. Spermatogonial stem cell regulation and spermatogenesis. Philos Trans R Soc Lond B Biol Sci. 2010;365:1663–1678. doi: 10.1098/rstb.2010.0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tegelenbosch RA, de Rooij DG. A quantitative study of spermatogonial multiplication and stem cell renewal in the C3H/101 F1 hybrid mouse. Mutat Res. 1993;290:193–200. doi: 10.1016/0027-5107(93)90159-d. [DOI] [PubMed] [Google Scholar]
- 7.Brinster RL, Avarbock MR. Germline transmission of donor haplotype following spermatogonial transplantation. Proc Natl Acad Sci U S A. 1994;91:11303–11307. doi: 10.1073/pnas.91.24.11303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brinster RL, Zimmermann JW. Spermatogenesis following male germ-cell transplantation. Proc Natl Acad Sci U S A. 1994;91:11298–11302. doi: 10.1073/pnas.91.24.11298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nagano M, Avarbock MR, Leonida EB, et al. Culture of mouse spermatogonial stem cells. Tissue Cell. 1998;30:389–397. doi: 10.1016/s0040-8166(98)80053-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kanatsu-Shinohara M, Ogonuki N, Inoue K, et al. Long-term proliferation in culture and germline transmission of mouse male germline stem cells. Biol Reprod. 2003;69:612–616. doi: 10.1095/biolreprod.103.017012. [DOI] [PubMed] [Google Scholar]
- 11.Brinster RL. Germline stem cell transplantation and transgenesis. Science. 2002;296:2174–2176. doi: 10.1126/science.1071607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kubota H, Avarbock MR, Brinster RL. Growth factors essential for self-renewal and expansion of mouse spermatogonial stem cells. Proc Natl Acad Sci U S A. 2004;101:16489–16494. doi: 10.1073/pnas.0407063101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ryu BY, Kubota H, Avarbock MR, et al. Conservation of spermatogonial stem cell self-renewal signaling between mouse and rat. Proc Natl Acad Sci U S A. 2005;102:14302–14307. doi: 10.1073/pnas.0506970102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sariola H, Saarma M. Novel functions and signalling pathways for GDNF. J Cell Sci. 2003;116:3855–3862. doi: 10.1242/jcs.00786. [DOI] [PubMed] [Google Scholar]
- 15.Oatley JM, Avarbock MR, Brinster RL. Glial cell line-derived neurotrophic factor regulation of genes essential for self-renewal of mouse spermatogonial stem cells is dependent on Src family kinase signaling. J Biol Chem. 2007;282:25842–25851. doi: 10.1074/jbc.M703474200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oatley JM, Avarbock MR, Telaranta AI, et al. Identifying genes important for spermatogonial stem cell self-renewal and survival. Proc Natl Acad Sci U S A. 2006;103:9524–9529. doi: 10.1073/pnas.0603332103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee J, Kanatsu-Shinohara M, Inoue K, et al. Akt mediates self-renewal division of mouse spermatogonial stem cells. Development. 2007;134:1853–1859. doi: 10.1242/dev.003004. [DOI] [PubMed] [Google Scholar]
- 18.Braydich-Stolle L, Kostereva N, Dym M, et al. Role of Src family kinases and N-Myc in spermatogonial stem cell proliferation. Dev Biol. 2007;304:34–45. doi: 10.1016/j.ydbio.2006.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tyagi G, Carnes K, Morrow C, et al. Loss of Etv5 decreases proliferation and RET levels in neonatal mouse testicular germ cells and causes an abnormal first wave of spermatogenesis. Biol Reprod. 2009;81:258–266. doi: 10.1095/biolreprod.108.075200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Niu Z, Goodyear SM, Rao S, et al. MicroRNA-21 regulates the self-renewal of mouse spermatogonial stem cells. Proc Natl Acad Sci U S A. 2011;108:12740–12745. doi: 10.1073/pnas.1109987108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chan F, Oatley MJ, Kaucher AV, et al. Functional and molecular features of the Id4+ germline stem cell population in mouse testes. Genes Dev. 2014;28:1351–1362. doi: 10.1101/gad.240465.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aloisio GM, Nakada Y, Saatcioglu HD, et al. PAX7 expression defines germline stem cells in the adult testis. J Clin Invest. 2014;124:3929–3944. doi: 10.1172/JCI75943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huckins C. The spermatogonial stem cell population in adult rats. I. Their morphology, proliferation and maturation. Anat Rec. 1971;169:533–557. doi: 10.1002/ar.1091690306. [DOI] [PubMed] [Google Scholar]
- 24.Oakberg EF. Spermatogonial stem-cell renewal in the mouse. Anat Rec. 1971;169:515–531. doi: 10.1002/ar.1091690305. [DOI] [PubMed] [Google Scholar]
- 25.de Rooij DG. Spermatogonial stem cell renewal in the mouse. I. Normal situation. Cell Tissue Kinet. 1973;6:281–287. doi: 10.1111/j.1365-2184.1973.tb01617.x. [DOI] [PubMed] [Google Scholar]
- 26.Nakagawa T, Sharma M, Nabeshima Y, et al. Functional hierarchy and reversibility within the murine spermatogenic stem cell compartment. Science. 2010;328:62–67. doi: 10.1126/science.1182868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de Rooij DG, Griswold MD. Questions about spermatogonia posed and answered since 2000. J Androl. 2012;33:1085–1095. doi: 10.2164/jandrol.112.016832. [DOI] [PubMed] [Google Scholar]
- 28.Ahmed EA, de Rooij DG. Staging of mouse seminiferous tubule cross-sections. Methods Mol Biol. 2009;558:263–277. doi: 10.1007/978-1-60761-103-5_16. [DOI] [PubMed] [Google Scholar]
- 29.Meng X, Lindahl M, Hyvonen ME, et al. Regulation of cell fate decision of undifferentiated spermatogonia by GDNF. Science. 2000;287:1489–1493. doi: 10.1126/science.287.5457.1489. [DOI] [PubMed] [Google Scholar]
- 30.Naughton CK, Jain S, Strickland AM, et al. Glial cell-line derived neurotrophic factor-mediated RET signaling regulates spermatogonial stem cell fate. Biol Reprod. 2006;74:314–321. doi: 10.1095/biolreprod.105.047365. [DOI] [PubMed] [Google Scholar]
- 31.Hofmann MC. Gdnf signaling pathways within the mammalian spermatogonial stem cell niche. Mol Cell Endocrinol. 2008;288:95–103. doi: 10.1016/j.mce.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oatley JM, Brinster RL. Regulation of spermatogonial stem cell self-renewal in mammals. Annu Rev Cell Dev Biol. 2008;24:263–286. doi: 10.1146/annurev.cellbio.24.110707.175355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sada A, Suzuki A, Suzuki H, et al. The RNA-binding protein NANOS2 is required to maintain murine spermatogonial stem cells. Science. 2009;325:1394–1398. doi: 10.1126/science.1172645. [DOI] [PubMed] [Google Scholar]
- 34.Suzuki H, Sada A, Yoshida S, et al. The heterogeneity of spermatogonia is revealed by their topology and expression of marker proteins including the germ cell-specific proteins Nanos2 and Nanos3. Dev Biol. 2009;336:222–231. doi: 10.1016/j.ydbio.2009.10.002. [DOI] [PubMed] [Google Scholar]
- 35.Nagai R, Shinomura M, Kishi K, et al. Dynamics of GFRalpha1-positive spermatogonia at the early stages of colonization in the recipient testes of W/Wnu male mice. Dev Dyn. 2012;241:1374–1384. doi: 10.1002/dvdy.23824. [DOI] [PubMed] [Google Scholar]
- 36.Buageaw A, Sukhwani M, Ben-Yehudah A, et al. GDNF family receptor alpha1 phenotype of spermatogonial stem cells in immature mouse testes. Biol Reprod. 2005;73:1011–1016. doi: 10.1095/biolreprod.105.043810. [DOI] [PubMed] [Google Scholar]
- 37.Taatjes DJ, Marr MT, Tjian R. Regulatory diversity among metazoan co-activator complexes. Nat Rev Mol Cell Biol. 2004;5:403–410. doi: 10.1038/nrm1369. [DOI] [PubMed] [Google Scholar]
- 38.Thomas MC, Chiang CM. The general transcription machinery and general cofactors. Crit Rev Biochem Mol Biol. 2006;41:105–178. doi: 10.1080/10409230600648736. [DOI] [PubMed] [Google Scholar]
- 39.Pugh BF, Tjian R. Mechanism of transcriptional activation by Sp1: evidence for coactivators. Cell. 1990;61:1187–1197. doi: 10.1016/0092-8674(90)90683-6. [DOI] [PubMed] [Google Scholar]
- 40.Tanese N, Pugh BF, Tjian R. Coactivators for a proline-rich activator purified from the multisubunit human TFIID complex. Genes Dev. 1991;5:2212–2224. doi: 10.1101/gad.5.12a.2212. [DOI] [PubMed] [Google Scholar]
- 41.Dynlacht BD, Hoey T, Tjian R. Isolation of coactivators associated with the TATA-binding protein that mediate transcriptional activation. Cell. 1991;66:563–576. doi: 10.1016/0092-8674(81)90019-2. [DOI] [PubMed] [Google Scholar]
- 42.Moqtaderi Z, Yale JD, Struhl K, et al. Yeast homologues of higher eukaryotic TFIID subunits. Proc Natl Acad Sci U S A. 1996;93:14654–14658. doi: 10.1073/pnas.93.25.14654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Verrijzer CP, Tjian R. TAFs mediate transcriptional activation and promoter selectivity. Trends Biochem Sci. 1996;21:338–342. [PubMed] [Google Scholar]
- 44.Albright SR, Tjian R. TAFs revisited: more data reveal new twists and confirm old ideas. Gene. 2000;242:1–13. doi: 10.1016/s0378-1119(99)00495-3. [DOI] [PubMed] [Google Scholar]
- 45.Ohler U, Wassarman DA. Promoting developmental transcription. Development. 2010;137:15–26. doi: 10.1242/dev.035493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Freiman RN. Specific variants of general transcription factors regulate germ cell development in diverse organisms. Biochim Biophys Acta. 2009;1789:161–166. doi: 10.1016/j.bbagrm.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Goodrich JA, Tjian R. Unexpected roles for core promoter recognition factors in cell-type-specific transcription and gene regulation. Nat Rev Genet. 2010;11:549–558. doi: 10.1038/nrg2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Freiman RN, Albright SR, Zheng S, et al. Requirement of tissue-selective TBP-associated factor TAFII105 in ovarian development. Science. 2001;293:2084–2087. doi: 10.1126/science.1061935. [DOI] [PubMed] [Google Scholar]
- 49.Falender AE, Freiman RN, Geles KG, et al. Maintenance of spermatogenesis requires TAF4b, a gonad-specific subunit of TFIID. Genes Dev. 2005;19:794–803. doi: 10.1101/gad.1290105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Falender AE, Shimada M, Lo YK, et al. TAF4b, a TBP associated factor, is required for oocyte development and function. Dev Biol. 2005;288:405–419. doi: 10.1016/j.ydbio.2005.09.038. [DOI] [PubMed] [Google Scholar]
- 51.Lovasco LA, Seymour KA, Zafra K, et al. Accelerated ovarian aging in the absence of the transcription regulator TAF4B in mice. Biol Reprod. 2010;82:23–34. doi: 10.1095/biolreprod.109.077495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Voronina E, Lovasco LA, Gyuris A, et al. Ovarian granulosa cell survival and proliferation requires the gonad-selective TFIID subunit TAF4b. Dev Biol. 2007;303:715–726. doi: 10.1016/j.ydbio.2006.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kanatsu-Shinohara M, Ogonuki N, Iwano T, et al. Genetic and epigenetic properties of mouse male germline stem cells during long-term culture. Development. 2005;132:4155–4163. doi: 10.1242/dev.02004. [DOI] [PubMed] [Google Scholar]
- 54.Wu X, Schmidt JA, Avarbock MR, et al. Prepubertal human spermatogonia and mouse gonocytes share conserved gene expression of germline stem cell regulatory molecules. Proc Natl Acad Sci U S A. 2009;106:21672–21677. doi: 10.1073/pnas.0912432106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tanaka H, Pereira LA, Nozaki M, et al. A germ cell-specific nuclear antigen recognized by a monoclonal antibody raised against mouse testicular germ cells. Int J Androl. 1997;20:361–366. doi: 10.1046/j.1365-2605.1998.00080.x. [DOI] [PubMed] [Google Scholar]
- 56.Hunter N, Borner GV, Lichten M, et al. Gamma-H2AX illuminates meiosis. Nat Genet. 2001;27:236–238. doi: 10.1038/85781. [DOI] [PubMed] [Google Scholar]
- 57.Fernandez-Capetillo O, Mahadevaiah SK, Celeste A, et al. H2AX is required for chromatin remodeling and inactivation of sex chromosomes in male mouse meiosis. Dev Cell. 2003;4:497–508. doi: 10.1016/s1534-5807(03)00093-5. [DOI] [PubMed] [Google Scholar]
- 58.Costoya JA, Hobbs RM, Barna M, et al. Essential role of Plzf in maintenance of spermatogonial stem cells. Nat Genet. 2004;36:653–659. doi: 10.1038/ng1367. [DOI] [PubMed] [Google Scholar]
- 59.Buaas FW, Kirsh AL, Sharma M, et al. Plzf is required in adult male germ cells for stem cell self-renewal. Nat Genet. 2004;36:647–652. doi: 10.1038/ng1366. [DOI] [PubMed] [Google Scholar]
- 60.Goertz MJ, Wu Z, Gallardo TD, et al. Foxo1 is required in mouse spermatogonial stem cells for their maintenance and the initiation of spermatogenesis. J Clin Invest. 2011;121:3456–3466. doi: 10.1172/JCI57984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hu YC, de Rooij DG, Page DC. Tumor suppressor gene Rb is required for self-renewal of spermatogonial stem cells in mice. Proc Natl Acad Sci U S A. 2013;110:12685–12690. doi: 10.1073/pnas.1311548110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Oatley MJ, Kaucher AV, Racicot KE, et al. Inhibitor of DNA binding 4 is expressed selectively by single spermatogonia in the male germline and regulates the self-renewal of spermatogonial stem cells in mice. Biol Reprod. 2011;85:347–356. doi: 10.1095/biolreprod.111.091330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Song HW, Wilkinson MF. Transcriptional control of spermatogonial maintenance and differentiation. Semin Cell Dev Biol. 2014 doi: 10.1016/j.semcdb.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhao GQ, Hogan BL. Evidence that mouse Bmp8a (Op2) and Bmp8b are duplicated genes that play a role in spermatogenesis and placental development. Mech Dev. 1996;57:159–168. doi: 10.1016/0925-4773(96)00543-6. [DOI] [PubMed] [Google Scholar]
- 65.Hara K, Nakagawa T, Enomoto H, et al. Mouse Spermatogenic Stem Cells Continually Interconvert between Equipotent Singly Isolated and Syncytial States. Cell Stem Cell. 2014;14:658–672. doi: 10.1016/j.stem.2014.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chakraborty P, Buaas FW, Sharma M, et al. LIN28A marks the spermatogonial progenitor population and regulates its cyclic expansion. Stem Cells. 2014;32:860–873. doi: 10.1002/stem.1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ayhan O, Balkan M, Guven A, et al. Truncating mutations in TAF4B and ZMYND15 causing recessive azoospermia. J Med Genet. 2014;51:239–244. doi: 10.1136/jmedgenet-2013-102102. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1
Quantitative RT-PCR (qPCR) measured relative Taf4b mRNA levels in the indicated testes. All values were normalized against 18s rRNA and represent RNA levels as the fold difference relative to the respective RNA levels at postnatal day 1 (P1).
Supplemental Figure 2
Average number of germ cells (TRA98+) per tubule at E18.5 and P8 in wild type and Taf4b-deficient testes.
Supplemental Figure 3
The percentages of proliferating (PH3+) germ cells were measured in Taf4b +/+ and −/− testes at the indicated time points.
Supplemental Figure 4 - Delayed tubule growth in TAF4b-deficient testes
Sections of TAF4b −/− and their WT littermates were used to determine average diameter of testis tubules at indicated ages. Circular tubules were measured from the outside of the basement membrane through their widest point. Between 50 and 100 tubule diameters per data point were measured. Data are represented as mean +/− SEM.
Supplemental Movies 1 and 2 - Gfrα1+ clustered cells in TAF4b-deficient testis tubules span multiple layers
Movie of whole mount immunostaining of Gfrα1 was performed on P17 testis tubules from WT (1) and TAF4b −/− (2) littermates. Z-stack images were taken through 170 μm and spanning the outer layers of WT and TAF4b −/− tubules.