Abstract
A major goal of our laboratory is to understand the molecular mechanisms that underlie the development and functions of diverse macrophage phenotypes in health and disease. Recent studies using genetic and genomic approaches suggest a relatively simple model of collaborative and hierarchical interactions between lineage-determining and signal-dependent transcription factors that enable selection and activation of transcriptional enhancers that specify macrophage identity and function. In addition, we have found that it is possible to use natural genetic variation as a powerful tool for advancing our understanding of how the macrophage deciphers the information encoded by the genome in order to attain specific phenotypes in a context-dependent manner. Here, I will describe our recent efforts to extend genetic and genomic approaches to investigate the roles of distinct tissue environments in determining the phenotypes of different resident populations of macrophages.
Preface
It is a great honor to give this year’s Russell Ross Memorial Lectureship in Vascular Biology. Dr. Ross made many seminal discoveries in this field, trained a generation of leading investigators, and was one of the major proponents of the concept that atherosclerosis is an inflammatory disease. I initially met Dr. Ross at a Gordon Research Conference on Atherosclerosis that was the very first scientific meeting I attended. My graduate work at that time was focused on lipoprotein metabolism and the idea that atherosclerosis was a lipid disease. The opportunity to discuss my work with Dr. Ross and to get his perspective at this meeting and during subsequent encounters had a major influence on my ultimate scientific directions. A longstanding interest of my laboratory has been to integrate the inflammation and lipid theories of atherosclerosis by studying the intersection of lipid metabolism and macrophage gene expression. In this lecture, I will present recent studies that attempt to advance our understanding of how tissue environment drives the selection and function of enhancers that control tissue-specific macrophage identities.
Introduction
Macrophages reside in essentially all tissues of the body and play key roles as sentinels of infection and injury 1-3. In addition, each population of macrophages within a tissue takes on specialized functions that are tuned to the developmental and functional requirements of that tissue. For example, microglia, representing the main population of macrophages within the nervous system, play roles in phagocytosis of apoptotic neurons and synaptic pruning. In the spleen, macrophages phagocytose senescent red blood cells and participate in iron recycling. Even within a single tissue, macrophages can exhibit heterogeneous phenotypes. Distinct populations of macrophages resident in the peritoneal cavity can be distinguished based on morphological criteria and different levels of MHC class II expression 4, 5. While macrophage heterogeneity is normally tuned to support normal tissue homeostasis, the ability of these cells to acquire distinct phenotypes in response to their environments can also result in pathogenic consequences. This scenario is exemplified by a diversity of macrophage phenotypes within atherosclerotic lesions as defined by variation in lipid accumulation and distinct surface markers 6. While most macrophages within the artery wall are thought to promote lesion development, some may be protective.
These observations raise the question of how distinct populations of macrophages are established and the extent to which different tissue environments play instructive roles with respect to their phenotypes. The recent development of genomic approaches that are based on the ability to sequence millions of short DNA fragments has revolutionized the approach to this type of question. It is now possible to globally quantify the broad spectrum of RNAs that are produced by a cell or tissue (mRNAs, miRNAs, etc.) by converting these RNAs to libraries of DNA copies that can be deeply sequenced (referred to as RNA-Seq) 7. In addition, it is also possible to globally define the genomic locations of specific histone modifications and transcription factors of interest using chromatin immunoprecipitation linked to deep sequencing (referred to as ChIP-Seq) 8. In this method, cells are treated with a crosslinking agent to covalently link transcription factors and histones to DNA. The DNA is then sheared into small fragments and subjected to immunoprecipitation with antibodies to the histone modification or transcription factor of interest. The crosslinks are subsequently reversed and the purified DNA fragments are subjected to deep sequencing. The sequenced ‘tags’ are then mapped to the genome. Tag accumulations at specific regions of the genome indicate that the marked histone or transcription factor of interest was present, with the overall pattern providing a genome wide histogram of their locations. By combining ChIP-Seq and RNA-Seq approaches, it has been possible to investigate mechanism by which transcription factors drive cell-specific patterns of gene expression on a global scale.
Environment is a major determinant of resident macrophage gene expression
As a starting point for investigating the influence of environment on macrophage gene expression, we performed RNA Seq analysis of three populations of resident macrophages; microglia (MG), large peritoneal macrophages (LPMs) and small peritoneal macrophages (SPMs) 4, 5. LPMS and SPMs share many features of macrophages, including expression of the CSF1 receptor, F4/80 and MerTK, but can be distinguished by low (LPM) or intermediate (SPMs) expression of MHC II. These resident macrophage populations were chosen for two reasons. First, they permitted an analysis of macrophages residing in different environments (e.g., LPMs and MG) and different macrophages in the same environment (LPMs and SPMs). Second, we were successful in developing methods of isolation that provided sufficient cells for genome-wide analysis and also preserved their in vivo gene expression and histone modification profiles.
RNA-Seq analysis indicated striking differences between LPMs and MG, with nearly 7000 mRNAs exhibiting significant differences in expression. Taking a very stringent threshold of a >16-fold difference, more than 500 mRNAs were preferentially expressed in MG, while more than 600 mRNAs were preferentially expressed in LPMs. In contrast, LPMs and SPMs exhibited a much more similar pattern of expression, with approximately 800 mRNAs exhibiting significant differences. 108 genes were expressed at >16-fold higher levels in SPMs, while only 5 mRNAs were expressed at >16-fold higher levels in LPMs. We compared these results with RNA-Seq data obtained for thioglycollate-elicited macrophages (TGEMs) and bone marrow derived macrophages (BMDMs), which are two widely used macrophage model systems. Clustering analysis indicated that SPMs and LPMs were closely related and TGEMs and BMDMs were closely related. In addition, these four cell types were much more similar to each other than they were to MG. We found that each type of macrophage was also distinguished by a unique gene expression pattern that was consistent with previous findings 9. For example, Cx3cr1 was much more highly expressed in MG than any of the other four macrophage populations, while peritoneal macrophages preferentially expressed Gata6. SPMs expressed much higher levels of the Ciita mRNA that encodes a transcription factor necessary for expression of MHC class II gene expression 10. Overall, the much greater differences in gene expression between LPMs and MG in comparison to LPMs and SPMs suggest an important role of environment in determining subset specific patterns of gene expression.
To directly examine the influence of environment on MG and LPM gene expression, we placed each cell type into culture for 7 days in the presence of M-CSF or IL-34, factors that induce signaling through the CSF1 receptor and maintain macrophage survival 3, 11-14. Using RNA-Seq to measure mRNA levels, we observed striking changes in gene expression in both cell types in comparison to the patterns observed in vivo. A large fraction of the genes exhibiting preferential expression in LPMs vs MG were significantly down-regulated in LPMs in culture. Similarly, a large fraction of the genes exhibiting preferential expression in MG vs LPMs were significantly down-regulated in MG in culture 9. Overall, each cell type preferentially lost expression of genes that represented the molecular signature of that cell type in vivo, indicating that the identities of MG and LPMs require constant environmental input.
Environment activates common and subset-specific enhancers
These findings raise the general question of how different macrophage identities are established and maintained. Gene expression is regulated at many levels, but gene transcription represents an essential and in many cases dominant point of control. Protein coding genes are transcribed from promoters, which represent genomic regions that recruit basal transcription factors and RNA polymerase II (Pol II). Physiologic levels of gene expression and responses to internal and external signals require the actions of additional sequence-specific transcription factors that recruit nucleosome remodeling complexes, histone modifying proteins and other factors to regulate Pol II activity. Such factors can bind in close proximity to promoters to influence gene expression. However, there is substantial evidence that additional genetic elements referred to as enhancers play major roles in determining cell-specific patterns of gene expression 15-17. Initially identified more than 30 years ago, enhancer elements can be located at various distances from promoters, typically between 1 and 50 kilobases away, and rarely as far as 500 kilobases distant. They can also be positioned upstream, downstream or within the genes they regulate. Like promoters, enhancers are occupied by sequence specific transcription factors that recruit nucleosome remodeling factors and histone modifying proteins. To a much greater extent than promoters, enhancers are occupied by lineage determining transcription factors that are required for the development of specific cell types. They also exhibit a distinct histone modification signature in which histone H3 lysine 4 exhibits more monomethylation (H3K4me1) or dimethylation (H3K4me2) than trimethylation (H3K4me3), which is a mark of promoters (Figure 1A, B). Remarkably, annotation of the human genome for histone modifications and other features of enhancers in dozens of different cell types suggest the existence of ~1 million enhancer elements, many more such elements than genes 18. These studies further suggest that each cell type selects a subset of ~30-50 thousand enhancers from this vast set of enhancers that in turn serve as the genetic regulatory elements specifying that cell’s identity and regulatory potential.
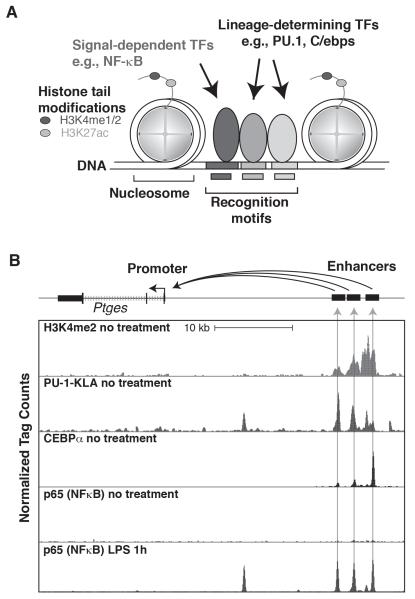
Figure 1. Enhancers regulate cell-specific and signal-dependent gene expression.
A. Typical features of enhancers. DNA is packaged in the nucleus in nucleosomes, in which DNA makes approximately two turns around a histone octomer, usually consisting of two tetramers of histone H2A, H2B, H3 and H4. Enhancers consist of collections of recognition motifs for sequence-specific transcription factors (indicated by boxes in the DNA). Specific enhancers become selected for activity when the correct combinations of sequence-specific transcription factors are co-expressed. Lineage-determining transcription factors play key roles in enhancer selection in specific cell types by collaborating with each other (e.g., PU.1 and C/EBP factors in macrophages) and with other sequence-specific transcription factors (not shown). Binding of LDTFs results in nucleosome remodeling and accessibility to signal-dependent transcription factors (e.g., NFκB). Active enhancers exhibit mono and/or dimethylation of histone H3 at lysine 4 (H3K4me1/2) and acetylation of histone H3 at lysine 27 (H3K27ac). B. Actions of a signal dependent transcription factor at an enhancer primed by PU.1 and C/EBPα in macrophages. The panel illustrates a region of the genome containing the Ptges gene, which is transcribed from right to left. Tracks of ChIP-Seq data for H3K4me2, PU.1, C/EBPα and p65 are shown. Peaks for H3K4me2, PU.1 and C/EBPα are present under no treatment conditions, with regions containing all three marks labeled as enhancers, to the right. The Ptges gene is expressed at very low levels under no treatment conditions. Treatment with LPS leads to nuclear entry of p65, which co-localizes with pre-existing PU.1 and C/EBPα, resulting in enhancer activation and high levels of Ptges expression.
A major effort of our laboratory has been to investigate mechanisms by which macrophages select their specific complements of enhancers from the genome. We gained important insights into this question by studying the genomic binding sites of PU.1, a lineage determining transcription factor that is required for the development of both macrophages and B cells 19. Using ChIP-Sequencing, we found that PU.1 bound to different regions of the genome in macrophages and B cells that were associated with macrophage or B cell-specific programs of gene expression, respectively. The basis for cell specific binding was shown to result from collaborative interactions between PU.1 and alternative transcription factors that were differentially expressed in each cell type. C/EBP factors were found to be important collaborators with PU.1 in macrophages. Regions of the genome containing closely spaced recognition motifs for PU.1 and C/EBPs became occupied by combinations of these factors in macrophages, while PU.1 did not bind to these regions in B cells. Conversely, EBF and Oct factors were important collaborators with PU.1 in B cells (Figure 2). Regions of the genome containing closely spaced recognition motifs for PU.1 and Oct or EBF became occupied by these factors in B cells, but were not occupied by PU.1 in macrophages. Thus, different combinations of factors functioned to prime distinct genomic regions for enhancer activity. Importantly, these primed regions of the genome were the major binding sites for signal-dependent transcription factors, such as NFκB and nuclear receptors 19, 20 (Figure 1B, 2). These SDTFs in turn recruited additional factors, including histone acetyltransferases (HATs), that transformed poised enhancers to active enhancers 20. Thus even though SDTFs can be broadly expressed and can respond to similar signals, they can direct very different transcriptional responses because they are directed to cell-specific enhancers. A similar hierarchical relationship for LDTFs and SDTFs was found in regulatory T cells, embryonic stem cells, and dendritic cells 21-23.
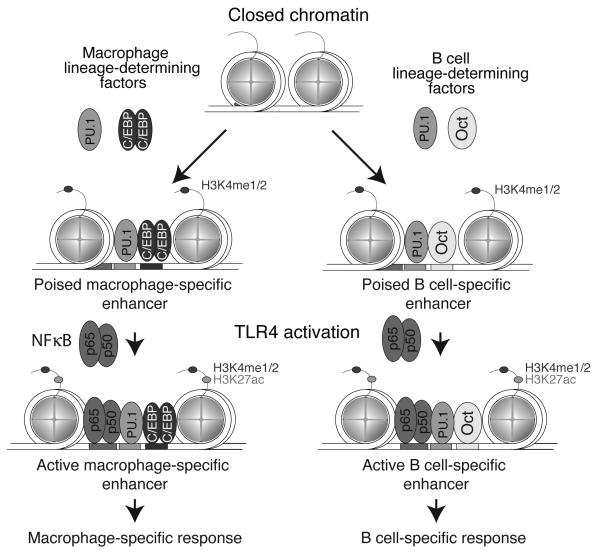
Figure 2. PU.1-dependent selection of macrophage and B cell-specific enhancers.
Regions of the genome containing closely spaced recognition motifs for PU.1 and other macrophage LDTFs become occupied by these factors in macrophages, but not B cells. Conversely, regions of the genome containing closely spaced recognition motifs for PU.1 and other B cell LDTFs become occupied by these factors in B cells, but not macrophages. Genomic regions that become primed by the binding of LDTFs in each cell type provide open regions of chromatin that are accessible by signal-dependent transcription factors, such as NFκB (exemplified by p50/p65 heterodimers). Because PU.1 primes different enhancer landscapes in each cell type dependent on distinct collaborative interactions with alternate transcription factors, the binding sites for the signal-dependent TFs are also cell type specific and result in corresponding cell-specific actions on nearby genes.
Observations made through the comparison of macrophages and B cells led us to wonder whether similar mechanisms would underlie the distinct transcriptional programs of resident peritoneal macrophages and microglia (Figure 3A). To address this question, we initially performed ChIP-Seq experiments for H3K4me2, histone H3 acetylated at lysine 20 (H3K27ac) and PU.1 in LPMs, SPMs and MG. H3K4me2 marks both enhancers and promoters and is present at both primed and active regulatory elements 20, 24, while H3K27ac is a mark associated with active promoters and enhancers25. We found that similar to patterns of gene expression, MG and LPMs exhibited a much more different pattern of H3K4me2-marked regions than observed for the comparison of LPMs and SPMs 9. The differences observed for MG and LPMs were predominantly at enhancer-like regions. Of 8439 promoters, only 329 showed > 4-fold differences between cell types. In contrast, of ~50,000 distal regions marked by H3K4me2, ~12,500 showed > 4-fold differences. Overlaying H3K27ac data on regions marked by H3K4me2 allowed estimation of the extent to which both common and cell-specific enhancers were active. We found evidence that in addition to activation of MG-specific or LPM-specific enhancers, many enhancers that were primed in all three cell types (based on H3K4me2) were only activate in one or two cell types (based on H3K27ac). For example, an enhancer adjacent to the Rarb gene, encoding a retinoic acid receptor and also a retinoic acid (RA) responsive gene, exhibited H3K4me2 in MG, LPMs and SPMs, but was almost exclusively marked by H3K27ac in LPMs and SPMs. This is significant from the standpoint of understanding the effect of environment on gene expression, because omentum-derived RA has recently been found to be an important inducer of the LPM phenotype by stimulating retinoic acid receptor (RAR)-dependent transcription 5. Our findings are consistent with RA being an important environmental factor in the peritoneum, but not in the brain, and acting on a common poised enhancer to selectively induce Rarb expression in the peritoneal population of macrophages.
Figure 3.
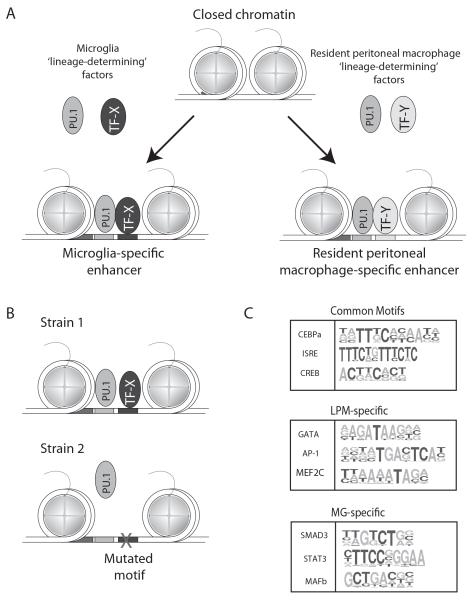
PU.1-depedent selection of microglia and large peritoneal macrophage-specific enhancers. Panel A. PU.1 is hypothesized to localize to microglia-specific enhancers by collaborating with transcription factors selectively expressed in microglia (e.g., TF-X), and conversely to localize to resident peritoneal-specific enhancers by collaborating with transcription factors selectively expressed in these cells (e.g., TF-Y). Panel B. Discovery of motifs for collaborative partners of PU.1 using natural genetic variation. Mutations in the recognition motif for a collaborative factor (e.g., TF-X) that abolish its binding also abolishes the collaborative binding of PU.1. Panel C. Common, LPM-specific and MG-specific motifs identified as recognition sites for collaborative binding partners of PU.1 by analysis of PU.1 binding in LPMs and MG derived from C57BL/6J, Spret and NOD mice.
The finding that microglia and LPMs exhibit activation of both common and distinct enhancers led us to consider the possibility that, like differences between macrophages and B cells, PU.1 would collaborate with alternative sets of transcription factors to prime subset-specific enhancers (Figure 3A). We therefore performed ChIP-Seq for PU.1 in microglia and large peritoneal macrophages. In addition to binding to a common set of genomic locations, we found that PU.1 localized to thousands of different regions of the genome in each macrophage subset 9. We performed de novo motif analysis of the MG-specific and LPM-specific binding sites to identify transcription factor recognition motifs co-localizing with PU.1 at these locations. As expected, an identical consensus PU.1 motif was found at both MG-specific and LPM-specific binding sites, analogous to our earlier findings at macrophage and B cell–specific binding sites. In contrast, the other sequence motifs co-localizing with PU.1 binding were completely different in MG and LPMs. Recognition elements for C/EBPs, AP-1 factors, GATA factors and retinoic acid receptors were highly enriched near PU.1 binding sites in LPMs, while recognition motifs for HIC3, Mef2, and unknown factor and Smad proteins were co-enriched near MG-specific PU.1 binding sites. These findings are of significance because of the recent discovery of the importance of retinoic acid and GATA6 in the development of LPMs 5, 26, 27 and the recently established requirement of brain-derived TGFβ, which regulates gene expression through SMAD transcription factors, in MG development 28, 29. Collectively, the factors recognizing motifs co-enriched with PU.1 are putative LDTFs and/or SDTFs that serve to drive LPM-specific or MG-specific enhancer selection and activity.
Natural genetic variation enables discovery of collaborating transcription factors
To further investigate the mechanisms establishing MG and LPM-specific enhancers, we exploited the natural genetic variation provided by inbred laboratory and wild-derived strains of mice on the binding of PU.1 in each cell type. Because binding of PU.1 to chromatin requires collaborative interactions with other transcription factors, single nucleotide polymorphisms can disrupt PU.1 binding not only by occurring in the PU.1 recognition motif, but also by disrupting the recognition motif for a nearby collaborative factor 30 (Figure 3B). Therefore, if PU.1 binding is lost at a genomic region at which there is no alteration in the PU.1 binding site, the loss of binding is likely due to a mutation in the recognition motif for a collaborative factor. Regions of the genome exhibiting strain-specific binding of PU.1 can be systematically interrogated for such mutations. The ability of this approach to identify motifs for collaborative transcription factors is dependent on the number of informative events of strain specific binding of PU.1 at locations at which the PU.1 recognition motif is intact.
To explore the feasibility of this approach, we performed ChIP-Seq for PU.1 in MG and LPMs derived from NOD mice and SPRET mice. Compared to the C57BL/6J reference strain, NOD mice exhibit ~5 million SNPs, while Spret mice exhibit ~40 million SNPs 31. We found thousands of strain-specific PU.1 binding sites in each cell type at which the PU.1 site was not mutated, providing a large number of informative events for analysis. We systematically evaluated these genomic regions for mutations in the recognition motifs for the 100 most highly expressed transcription factors in MG or LPMs. Of these, mutations in more than 20 of these motifs were found to be significantly associated with strain-specific binding of PU.1 9. Importantly, this analysis recovered recognition motifs for C/EBPs and AP-1 factors, which we previously established as collaborative factors with PU.1 through gain and loss of expression studies (Figure 3C). In addition, we identified a dozen motifs that were associated with LPM-specific binding of PU.1, and four motifs associated with MG-specific binding. Motifs associated with LPM-specific binding included recognition elements for GATA factors, KLF4 and MEF2c, while motifs associated with MG-specific binding of PU.1 included recognition motifs for MAFb and SMAD3 (Figure 3C). These findings therefore confirmed the results of motif enrichment analysis as well as identified motifs for factors not previously known to contribute to macrophage development.
With these findings in hand, we then revisited the question of how environment controls LPM and MG-specific gene expression. Recent studies of LPMs indicate their maturation and function is dependent on omentum derived retinoic acid that is an activating ligand for RARβ. RARβ in turn induces the expression of GATA6 5. Conversely, TGFβ signaling in the brain is essential for maintenance of MG 28, 29. Although an important role for RARβ has been established, we found that mRNAs encoding all three nuclear receptors for retinoic acid (Rara, Rarb and Rarg) are highly expressed in LPMs compared to MG, while the main receptors for TGFβ signaling (Tbfbr1, Tgfbr2) are preferentially expressed in MG compared to LPMs. Remarkably, all three Rar genes were found to be significantly down-regulated when LPMs were transferred to culture conditions for 7 days. In addition, nearly two thirds of the transcription factors recognizing sequence motifs identified by analyzing effects of genetic variation were also down regulated under these conditions, including GATA6 9.
We then treated LPMs in culture with retinoic acid for seven days to determine the extent to which the in vivo LPM phenotype could be maintained. Notably, expression of approximately half of the genes that exhibit LPM-specific expression compared to MG was maintained by retinoic acid treatment. In contrast, less than 5% of the genes exhibiting common expression between LPMs and MG were RA-dependent. Although Rarb expression was maintained by RA treatment, expression of Rara and Rarb was not, indicating that environmental factors other than RA are required. Similarly, of the additional collaborative factors identified by analysis of effects of genetic variation, Gata6, Bhlhe40 and Tfeb were maintained by RA treatment, while the remaining environment-dependent factors were not. Conversely, we also treated these LPMs in vitro with TGFβ to determine the extent to which their gene expression program could be converted to that of MG. TGFβ induced expression of about half of the genes that are normally highly specific for MG, but only 4% of the genes that are commonly expressed between these cell types. Therefore RA and TGFβ primarily regulate genes that specify LPM and MG phenotypes, respectively 9.
Effects of removal from the in vivo environment and treatment with RA or TGFβ on gene expression were also mirrored at the level of enhancers. Removal of LPMs from the peritoneal cavity resulted in the loss of H3K4me2/H3K27ac at approximately half of the enhancer like regions in the cell. Of the lost enhancers, approximately one third could be maintained by RA treatment. Conversely, treatment of LPMs in vitro with TGFβ for 7 days resulted in partial reprogramming of the enhancer landscape towards that of MG.
Conclusions and future directions
Our findings suggest a hierarchical model by which LDTFs and SDTFs regulate macrophage identity and tissue-specific phenotypes 9, 19, 30 (Figure 4). We propose that a core set of LDTFs, exemplified by PU.1, play essential roles in priming an enhancer landscape that is common to many types of tissue macrophages. Tissue-specific signals activate different sets of primed enhancers to direct different programs of gene expression. For example, while the enhancers in the vicinity of Rarb are primed in all macrophage subsets, they are only activated in the peritoneal cavity in response to local retinoic acid. Similarly, enhancers in the vicinity of Cx3cr1 are primed in all macrophage subsets, but these enhancers are selectively activated in MG in response to local TGFβ. Direct actions of these signaling molecules on common poised enhancers account for part of the different patterns of gene expression observed in LPMs and MG. Importantly, however, our findings also indicate that actions of environmental factors on common poised enhancers leads to the differential expression of alternative transcription factors that collaborate with PU.1 to select subset-specific enhancers. Thus, environment drives tissue-specific programs of gene expression through both direct and indirect activation of subset-specific enhancers.
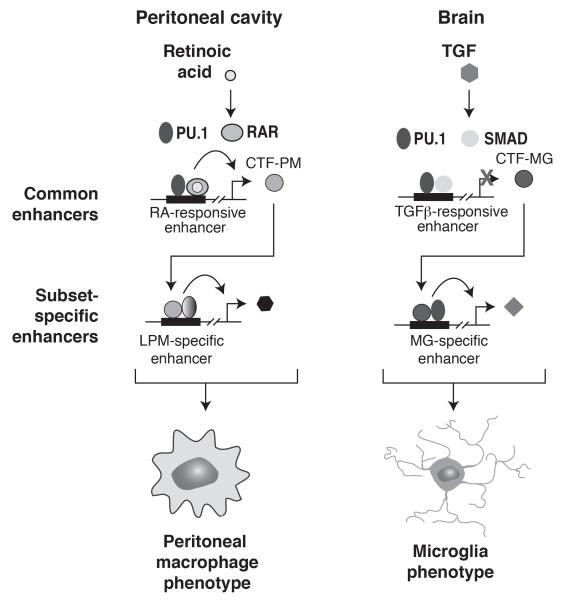
Figure 4. Model for selection and activation of enhancers that specify microglia and large peritoneal macrophage gene expression.
Environment-specific signals, such as retinoic acid and TGFβ act on a common set of enhancers to drive gene expression. Because retinoic acid is present in the peritoneal cavity, but not the brain, retinoic acid-responsive enhancers are only activated in peritoneal macrophages. Because TGFβ signaling is most active in the brain, TGFβ-responsive enhancers are preferentially activated in microglia. Retinoic acid responsive genes include genes encoding transcription factors that collaborate with PU.1 to establish LPM-specific enhancers. Conversely, TGFβ-responsive genes include genes encoding transcription factors that collaborate with PU.1 to establish MG-specific enhancers. The combination of direct target genes and indirect target genes resulting from environment-specific selection and activation of enhancers contributes to peritoneal macrophage-specific and microglia-specific phenotypes.
We speculate that this hierarchical mechanism of enhancer selection and activation operates in other macrophage subsets, and probably other tissue-specific cell types. The discoveries of TGFβ and retinoic acid as key regulators of in vivo macrophage phenotypes resulted from loss of function studies, but these findings also independently emerged from the analysis of tissue macrophage enhancers. We speculate that this approach could be used to identify other classes of signaling molecules, including lipids, in other tissue environments. The strong influence of environment on enhancer landscapes and gene expression suggest that the diverse phenotypes of macrophages observed within atherosclerotic lesions reflect, at least in part, substantial differences in microenvironment 6. There are many factors that are present in lesions that affect macrophage phenotypes when tested in vitro, including oxidized lipoproteins, cytokines, cholesterol crystals, apoptotic cells, etc. While the present studies were performed using hundreds of thousands of cells to obtain robust ChIP-Seq data sets, recent innovations in genomic technologies now enable these types of experiments to be performed with tens of thousands of cells or less 32, 33. It is thus now possible to begin to interrogate many populations of interest within in vivo environments, including the artery wall, and to ask not only how tissue specific enhancer landscapes are established, but also effects of pathogenic stimuli and therapeutic interventions. It is worth noting here that emerging classes of small molecules that act on proteins that write, read and erase histone modifications, such as HDAC inhibitors and acetylated histone tail mimetics, exert many of their effects at enhancers 20, 34. Because each cell type selects a different complement of enhancers from the genome to establish its identity and regulatory potential, each cell type will also respond in a unique way to drugs that alter enhancer function. Recent genetic and pharmacologic studies suggest that these interventions may have promise in the prevention and treatment of cardiovascular disease 35, 36.
Acknowledgements
These studies were primarily supported by NIH grants DK091183, CA17390, HL088083 and DK063491 and GM085764.
References
- 1.Geissmann F, Manz MG, Jung S, Sieweke MH, Merad M, Ley K. Development of monocytes, macrophages, and dendritic cells. Science. 2010;327:656–661. doi: 10.1126/science.1178331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gordon S, Pluddemann A, Martinez Estrada F. Macrophage heterogeneity in tissues: Phenotypic diversity and functions. Immunological reviews. 2014;262:36–55. doi: 10.1111/imr.12223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wynn TA, Chawla A, Pollard JW. Macrophage biology in development, homeostasis and disease. Nature. 2013;496:445–455. doi: 10.1038/nature12034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ghosn EE, Cassado AA, Govoni GR, Fukuhara T, Yang Y, Monack DM, Bortoluci KR, Almeida SR, Herzenberg LA, Herzenberg LA. Two physically, functionally, and developmentally distinct peritoneal macrophage subsets. Proc Natl Acad Sci U S A. 2010;107:2568–2573. doi: 10.1073/pnas.0915000107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Okabe Y, Medzhitov R. Tissue-specific signals control reversible program of localization and functional polarization of macrophages. Cell. 2014;157:832–844. doi: 10.1016/j.cell.2014.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Colin S, Chinetti-Gbaguidi G, Staels B. Macrophage phenotypes in atherosclerosis. Immunological reviews. 2014;262:153–166. doi: 10.1111/imr.12218. [DOI] [PubMed] [Google Scholar]
- 7.Wang Z, Gerstein M, Snyder M. Rna-seq: A revolutionary tool for transcriptomics. Nature reviews. Genetics. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morozova O, Marra MA. Applications of next-generation sequencing technologies in functional genomics. Genomics. 2008;92:255–264. doi: 10.1016/j.ygeno.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 9.Gosselin D, Link V, Romanoski CE, Fonseca GJ, Eichenfield DZ, Spann NJ, Stender JD, Chun HB, Garner H, Geissmann F, Glass CK. Environmnent drives selection and function of enhancers controlling tissue-specific macrophage identities. Cell. 2014;159:1–14. doi: 10.1016/j.cell.2014.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Steimle V, Otten LA, Zufferey M, Mach B. Complementation cloning of an mhc class ii transactivator mutated in hereditary mhc class ii deficiency (or bare lymphocyte syndrome) Cell. 1993;75:135–146. [PubMed] [Google Scholar]
- 11.Witmer-Pack MD, Hughes DA, Schuler G, Lawson L, McWilliam A, Inaba K, Steinman RM, Gordon S. Identification of macrophages and dendritic cells in the osteopetrotic (op/op) mouse. J Cell Sci. 1993;104(Pt 4):1021–1029. doi: 10.1242/jcs.104.4.1021. [DOI] [PubMed] [Google Scholar]
- 12.Schulz C, Gomez Perdiguero E, Chorro L, Szabo-Rogers H, Cagnard N, Kierdorf K, Prinz M, Wu B, Jacobsen SE, Pollard JW, Frampton J, Liu KJ, Geissmann F. A lineage of myeloid cells independent of myb and hematopoietic stem cells. Science. 2012;336:86–90. doi: 10.1126/science.1219179. [DOI] [PubMed] [Google Scholar]
- 13.Greter M, Lelios I, Pelczar P, Hoeffel G, Price J, Leboeuf M, Kundig TM, Frei K, Ginhoux F, Merad M, Becher B. Stroma-derived interleukin-34 controls the development and maintenance of langerhans cells and the maintenance of microglia. Immunity. 2012;37:1050–1060. doi: 10.1016/j.immuni.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Y, Szretter KJ, Vermi W, Gilfillan S, Rossini C, Cella M, Barrow AD, Diamond MS, Colonna M. Il-34 is a tissue-restricted ligand of csf1r required for the development of langerhans cells and microglia. Nat Immunol. 2012;13:753–760. doi: 10.1038/ni.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Andersson R, Gebhard C, Miguel-Escalada I, Hoof I, Bornholdt J, Boyd M, Chen Y, Zhao X, Schmidl C, Suzuki T, Ntini E, Arner E, Valen E, Li K, Schwarzfischer L, Glatz D, Raithel J, Lilje B, Rapin N, Bagger FO, Jorgensen M, Andersen PR, Bertin N, Rackham O, Burroughs AM, Baillie JK, Ishizu Y, Shimizu Y, Furuhata E, Maeda S, Negishi Y, Mungall CJ, Meehan TF, Lassmann T, Itoh M, Kawaji H, Kondo N, Kawai J, Lennartsson A, Daub CO, Heutink P, Hume DA, Jensen TH, Suzuki H, Hayashizaki Y, Muller F, Consortium F, Forrest AR, Carninci P, Rehli M, Sandelin A. An atlas of active enhancers across human cell types and tissues. Nature. 2014;507:455–461. doi: 10.1038/nature12787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Levine M. Transcriptional enhancers in animal development and evolution. Current biology : CB. 2010;20:R754–763. doi: 10.1016/j.cub.2010.06.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shlyueva D, Stampfel G, Stark A. Transcriptional enhancers: From properties to genome-wide predictions. Nature reviews. Genetics. 2014;15:272–286. doi: 10.1038/nrg3682. [DOI] [PubMed] [Google Scholar]
- 18.Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, Snyder M. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and b cell identities. Molecular cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaikkonen MU, Spann NJ, Heinz S, Romanoski CE, Allison KA, Stender JD, Chun HB, Tough DF, Prinjha RK, Benner C, Glass CK. Remodeling of the enhancer landscape during macrophage activation is coupled to enhancer transcription. Molecular cell. 2013;51:310–325. doi: 10.1016/j.molcel.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garber M, Yosef N, Goren A, Raychowdhury R, Thielke A, Guttman M, Robinson J, Minie B, Chevrier N, Itzhaki Z, Blecher-Gonen R, Bornstein C, Amann-Zalcenstein D, Weiner A, Friedrich D, Meldrim J, Ram O, Cheng C, Gnirke A, Fisher S, Friedman N, Wong B, Bernstein BE, Nusbaum C, Hacohen N, Regev A, Amit I. A high-throughput chromatin immunoprecipitation approach reveals principles of dynamic gene regulation in mammals. Molecular cell. 2012;47:810–822. doi: 10.1016/j.molcel.2012.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mullen AC, Orlando DA, Newman JJ, Loven J, Kumar RM, Bilodeau S, Reddy J, Guenther MG, DeKoter RP, Young RA. Master transcription factors determine cell-type-specific responses to tgf-beta signaling. Cell. 2011;147:565–576. doi: 10.1016/j.cell.2011.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Samstein RM, Arvey A, Josefowicz SZ, Peng X, Reynolds A, Sandstrom R, Neph S, Sabo P, Kim JM, Liao W, Li MO, Leslie C, Stamatoyannopoulos JA, Rudensky AY. Foxp3 exploits a pre-existent enhancer landscape for regulatory t cell lineage specification. Cell. 2012;151:153–166. doi: 10.1016/j.cell.2012.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.He HH, Meyer CA, Shin H, Bailey ST, Wei G, Wang Q, Zhang Y, Xu K, Ni M, Lupien M, Mieczkowski P, Lieb JD, Zhao K, Brown M, Liu XS. Nucleosome dynamics define transcriptional enhancers. Nature genetics. 2010;42:343–347. doi: 10.1038/ng.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, Boyer LA, Young RA, Jaenisch R. Histone h3k27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gautier EL, Shay T, Miller J, Greter M, Jakubzick C, Ivanov S, Helft J, Chow A, Elpek KG, Gordonov S, Mazloom AR, Ma’ayan A, Chua WJ, Hansen TH, Turley SJ, Merad M, Randolph GJ, Immunological Genome C Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nat Immunol. 2012;13:1118–1128. doi: 10.1038/ni.2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rosas M, Davies LC, Giles PJ, Liao CT, Kharfan B, Stone TC, O’Donnell VB, Fraser DJ, Jones SA, Taylor PR. The transcription factor gata6 links tissue macrophage phenotype and proliferative renewal. Science. 2014;344:645–648. doi: 10.1126/science.1251414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Butovsky O, Jedrychowski MP, Moore CS, Cialic R, Lanser AJ, Gabriely G, Koeglsperger T, Dake B, Wu PM, Doykan CE, Fanek Z, Liu L, Chen Z, Rothstein JD, Ransohoff RM, Gygi SP, Antel JP, Weiner HL. Identification of a unique tgf-beta-dependent molecular and functional signature in microglia. Nat Neurosci. 2014;17:131–143. doi: 10.1038/nn.3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Makwana M, Jones LL, Cuthill D, Heuer H, Bohatschek M, Hristova M, Friedrichsen S, Ormsby I, Bueringer D, Koppius A, Bauer K, Doetschman T, Raivich G. Endogenous transforming growth factor beta 1 suppresses inflammation and promotes survival in adult cns. J Neurosci. 2007;27:11201–11213. doi: 10.1523/JNEUROSCI.2255-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heinz S, Romanoski CE, Benner C, Allison KA, Kaikkonen MU, Orozco LD, Glass CK. Effect of natural genetic variation on enhancer selection and function. Nature. 2013;503:487–492. doi: 10.1038/nature12615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Keane TM, Goodstadt L, Danecek P, White MA, Wong K, Yalcin B, Heger A, Agam A, Slater G, Goodson M, Furlotte NA, Eskin E, Nellaker C, Whitley H, Cleak J, Janowitz D, Hernandez-Pliego P, Edwards A, Belgard TG, Oliver PL, McIntyre RE, Bhomra A, Nicod J, Gan X, Yuan W, van der Weyden L, Steward CA, Bala S, Stalker J, Mott R, Durbin R, Jackson IJ, Czechanski A, Guerra-Assuncao JA, Donahue LR, Reinholdt LG, Payseur BA, Ponting CP, Birney E, Flint J, Adams DJ. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature. 2011;477:289–294. doi: 10.1038/nature10413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nature methods. 2013;10:1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lara-Astiaso D, Weiner A, Lorenzo-Vivas E, Zaretsky I, Jaitin DA, David E, Keren-Shaul H, Mildner A, Winter D, Jung S, Friedman N, Amit I. Immunogenetics. Chromatin state dynamics during blood formation. Science. 2014;345:943–949. doi: 10.1126/science.1256271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li P, Spann NJ, Kaikkonen MU, Lu M, Oh da Y, Fox JN, Bandyopadhyay G, Talukdar S, Xu J, Lagakos WS, Patsouris D, Armando A, Quehenberger O, Dennis EA, Watkins SM, Auwerx J, Glass CK, Olefsky JM. Ncor repression of lxrs restricts macrophage biosynthesis of insulin-sensitizing omega 3 fatty acids. Cell. 2013;155:200–214. doi: 10.1016/j.cell.2013.08.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brown JD, Lin CY, Duan Q, Griffin G, Federation AJ, Paranal RM, Bair S, Newton G, Lichtman AH, Kung AL, Yang T, Wang H, Luscinskas FW, Croce KJ, Bradner JE, Plutzky J. Nf-kappab directs dynamic super enhancer formation in inflammation and atherogenesis. Molecular cell. 2014 doi: 10.1016/j.molcel.2014.08.024. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hoeksema MA, Gijbels MJ, Van den Bossche J, van der Velden S, Sijm A, Neele AE, Seijkens T, Stoger JL, Meiler S, Boshuizen MC, Dallinga-Thie GM, Levels JH, Boon L, Mullican SE, Spann NJ, Cleutjens JP, Glass CK, Lazar MA, de Vries CJ, Biessen EA, Daemen MJ, Lutgens E, de Winther MP. Targeting macrophage histone deacetylase 3 stabilizes atherosclerotic lesions. EMBO molecular medicine. 2014;6:1124–1132. doi: 10.15252/emmm.201404170. [DOI] [PMC free article] [PubMed] [Google Scholar]