SUMMARY
Most of our knowledge on the biological role of the testis-specific Na,K-ATPase alpha 4 isoform derives from studies performed in non-human species. Here, we studied the function of human Na,K-ATPase alpha 4 after its expression in transgenic mice. Using a bacterial artificial chromosome (BAC) construct, containing the human ATP1A4 gene locus, we obtained expression of the human α4 transgene specifically in mouse sperm, enriched in the sperm flagellum. The expressed, human alpha 4 was active, and compared to wild-type sperm, those from transgenic mice displayed higher Na,K-ATPase alpha 4 activity and greater binding of fluorescently labeled ouabain, which is typical of the alpha 4 isoform. The expression and activity of endogenous alpha 4 and the other Na,K-ATPase alpha isoform present in sperm, alpha 1, remained unchanged. Male mice expressing the human ATP1A4 transgene exhibited similar testis size and morphology, normal sperm number and shape, and no changes in overall fertility compared to wild-type mice. Sperm carrying the human transgene exhibited enhanced total motility and an increase in multiple parameters of sperm movement, including higher sperm hyperactive motility. In contrast, no statistically significant changes in sperm membrane potential, protein tyrosine phosphorylation, or spontaneous acrosome reaction were found between wild-type and transgenic mice. Altogether, these results provide new genetic evidence for an important role of human Na,K-ATPase alpha 4 in sperm motility and hyperactivation, and establishes a new animal model for future studies of this isoform.
Keywords: ion transport, ouabain, sperm motility, sperm capacitation
INTRODUCTION
Na,K-ATPase, is a plasma-membrane enzyme that catalyzes the active transport of Na+ and K+ between the cell and its environment, using the energy from the hydrolysis of ATP (Kaplan 2002). Na,K-ATPase activity is required for many different functions in the cell, including maintenance of cell ionic homeostasis, cell resting membrane potential, and the secondary transport of other ions, solutes, and water across the cell surface (Blanco 2005). Na,K-ATPase is an oligomer formed from the association of two main proteins, a catalytic α subunit and an accompanying β polypeptide (Morth et al. 2011). Na,K-ATPase has multiple molecular variants derived from four different α isoforms (α1, α2, α3, and α4) and three distinct β isoforms (β1, β2, and β3), which are used differentially among tissues of many species, including humans (Blanco and Mercer 1998; Geering 2001; Mobasheri et al. 2000). Each α and β subunit assemble in different combinations to generate a series of Na,K-ATPase isoenzymes (Blanco and Mercer 1998), each of which exhibit a particular pattern of expression that is cell- and tissue-specific (Jewell et al. 1992). While Na,K-ATPase α1 and β1 are found in nearly every cell, the expression of other α and β polypeptides is more restricted. For example, α2 is expressed in skeletal muscle, heart, brain, and adipocytes; α3 is present in nervous tissue; and α4 – the most restricted in terms of tissue distribution, – is found only in the testis (Blanco and Mercer 1998; McDermott et al. 2012; Shamraj and Lingrel 1994). The β2 isoform is abundant in the pineal gland, skeletal muscle, and nervous tissues, while β3 is expressed in retina, liver, lung, and testis (Blanco 2005; Mobasheri et al. 2000). The different Na,K-ATPase isoforms have peculiar functional properties that mainly depend on which α- subunit incorporated into the enzyme (Crambert et al. 2000; Geering 2008; Martin 2005). In this manner, the presence of different variants of the Na,K-ATPase is a mechanism that cells use to adapt Na+ and K+ transport for their specific physiologic needs (Blanco 2005).
Na,K-ATPase α4 is expressed in male germ cells of the rat and mouse testis after meiosis, and remains enriched in spermatozoa, specifically at the sperm flagellum (Blanco et al. 2000; Woo et al. 2000). Sperm also express the ubiquitous α1 isoform and both the β1 and β3 subunits (Wagoner et al. 2005). Na,K-ATPase α4 exhibits affinities for Na+, K+, ATP, and the inhibitor ouabain, which is distinct from the other Na,K-ATPases; this latter attribute has been used as a tool to distinguish the function of α4 from the other Na,K-ATPase α isoforms (Blanco et al. 1999; Woo et al. 1999). This distinct pharmacology has linked Na,K-ATPase α4 activity to sperm motility and sperm flagellar beat (Jimenez et al. 2012; Jimenez et al. 2010; Wagoner et al. 2005; Woo et al. 2000; Woo et al. 2002). Recent studies in which the α4 isoform was either over-expressed or knocked-out in genetically modified mice, further established the role of α4 in sperm motility, and showed that this isoform is important for male fertility (Jimenez et al. 2011a; Jimenez et al. 2011b).
Most functional studies of Na,K-ATPase α4 have been conducted on protein from rat, mouse, and bull (Blanco et al. 2000; Jimenez et al. 2011a; Newton et al. 2010; Wagoner et al. 2005; Woo et al. 2000); little experimental evidence exists for human Na,K-ATPase α4 activity. As in other mammals, Human α4 is present in mature human testis and its expression is developmentally coincident with sexual maturation (Hlivko et al. 2006; Shamraj and Lingrel 1994). Human α4 localizeds to the sperm flagellum, and it has enzymatic properties that are similar to those of its mouse and rat orthologs, (Sanchez et al. 2006). In addition, in vitro treatment of human sperm with ouabain, at concentrations that selectively inhibit Na,K-ATPase α4, decreases human sperm motility (Sanchez et al. 2006).
While some progress has been made in characterizing the expression and enzymatic properties of the endogenous Na,K-ATPase α4 in human sperm, a more comprehensive understanding of the biological relevance of this testis-specific protein in different aspects of sperm physiology is necessary before results of Na,K-ATPase α4 research can be applied to the clinic. In the present work, we investigated the in vivo role of human Na,K-ATPase α4 using transgenic mice engineered to express the human α4 protein, via a bacterial- artificial-chromosome (BAC) construct. Our results show that human Na,K-ATPase α4 is efficiently and specifically expressed as a functional protein in the mouse testis localizes to the sperm flagellum, as does the endogenous mouse α4 isoform; and enhances the total and hyperactive motility of mouse spermatozoa. In contrast, human ATP1A4 expression had no significant effect on plasma membrane potential, capacitation dependent protein phosphorylation or spontaneous acrosome reaction of mouse sperm. These results demonstrate the functional relevance of human Na,K-ATPase α4. In addition, our transgenic strategy establishes a novel mouse model that will be useful for future studies related to human Na,K-ATPase α4.
RESULTS
Human Na,K-ATPase α4 is expressed in transgenic mice
To understand the biological relevance of human Na,K-ATPase α4 in vivo, we engineered transgenic mice over-expressing this protein via a BAC construct. Our decision to utilize a BAC was prompted by several advantages that this approach offers compared to classical transgenic techniques, including reduced positional effects on gene expression associated with integration in the genome. As BACs carry extended promoter regions and regulatory elements that are important for proper spatial and temporal expression of the desired gene, expression of genes on the BAC more effectively mimic the expression pattern of the endogenous genes. Finally, BACs reduce artifacts associated with multiple-copy integration of smaller transgenes by alternative methods. (Yang and Gong 2005). As the genomic elements required for human α4 expression are unknown, we include the whole chromosomal locus to ensure that, all the regulatory elements required for the expression of the gene are present. The BAC contained a portion of human chromosome 1q23, with the full sequence of the ATP1A4 gene and the predicted promoter, which is believed to drive specific expression of the transgene in male germ cells (Keryanov and Gardner 2002; Rodova et al. 2006). The BAC construct used also contains flanking genes that are expressed in somatic cells, non-coding regions, and the pBACe3.6 vector (Supplemental Fig. 1). One of the flanking genes encodes Na,K-ATPase α2 (ATP1A2), which is closely linked to the ATP1A4 locus. ATP1A2 is muscle- and glial cell-specific, and was not exogenously expressed in the testis of transgenic mice (data not shown).
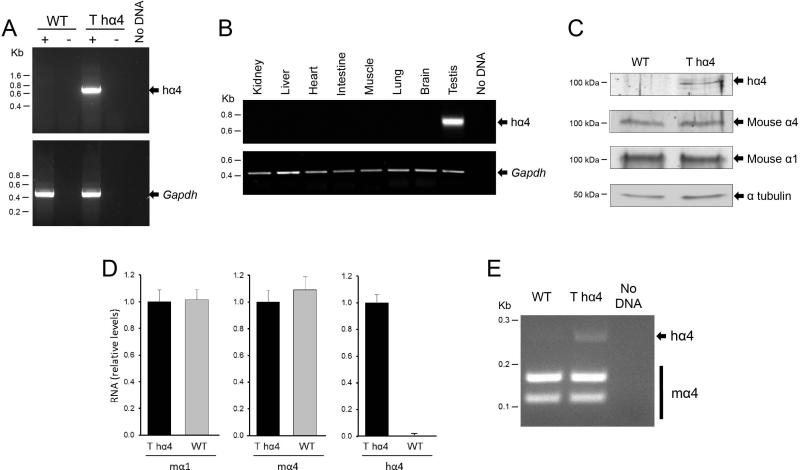
Expression of human ATP1A4 in the mice was confirmed by reverse-transcriptase -PCR (RT-PCR) on RNA isolated from testis samples, using human ATP1A4- specific primers. A 748-bp DNA fragment of human ATP1A4 was specifically expressed in the transgenic mice; no band was detected in the samples from wild- type mice or in which reverse transcriptase was omitted (Fig. 1A). Tissue expression of the transgene was assessed by, RT-PCR on RNA from different organs of the transgenic mice. This revealed the specific expression of human ATP1A4 RNA in the testis, with no transcript detectable in any of the other major tissues tested (Fig 1B). This spatial expression agreed with the normal distribution of ATP1A4 RNA (McDermott et al. 2012; Shamraj and Lingrel 1994).
Figure 1.
Human ATP1A4 expression in transgenic mice. A: Analysis of wild type (WT) and transgenic human ATP1A4 mice (T hα4). RNA from mouse testis was isolated and subjected to RT-PCR to amplify the human ATP1A4 mRNA (hα4) and mouse Gapdh, at 748 bp and 430 bp respectively. Samples Amplification reactions were carried out in the presence (+) or absence (−) of reverse transcriptase, to ensure the absence of genomic DNA contamination. A sample without cDNA (No DNA) was included as a negative control. B: Human ATP1A4 expression in different tissues from the transgenic mice. RT-PCR was performed on RNA samples of the indicated tissues, following the steps mentioned in A. Gapdh was used as a loading control. C: Immunoblot analysis of transgenic human α4 expression. Sperm from the cauda epididymis of wild-type and transgenic mice were lysed and subjected to SDS-PAGE and immunoblot. Anti-human and anti-mouse α4 generated in chicken, and anti-mouse α1 generated in mouse were used to identify the corresponding polypeptides; α-tubulin detection was used as a loading control. D: Relative levels of mouse and human ATP1A4 RNA (mα4 and hα4, respectively) in WT and T hα4 mice. Transcript abundance was performed using qPCR, as described in the Methods section. E: Comparison of RNA levels for the mouse and human 4ATP1A4 transcripts in WT and T hα4 mice. RNA was isolated and subjected to RT-PCR, and the products were digested with EcoRV to differentiate human from mouse orthologs. The bottom two bands (156 and 109 bp) correspond to the EcoRV-cleaved mouse isoform, while the top band (265 bp) corresponds to the human sequence.
We used immunoblot analysis of testis proteins to determine if the human ATP1A4 transgene was appropriately expressed at the protein level, using chicken antibodies against the human α4 polypeptide (Sanchez et al. 2006). The anti-human α4 antibody, identified a band with a molecular weight corresponding to the human α4 isoform in samples from the transgenic animals, but not in those from wild-type mice (Fig 1C). The absence of α4 in wild- type mice supports the lack of anti-human α4 antibody cross-reactivity to endogenous mouse α4 polypeptide. Conversely, expression of the endogenous mouse α4 and α1 polypeptides, the only two Na,K-ATPase α isoforms expressed in mouse testis (Blanco 2005), was detected using mouse anti-α4 and anti-α1 antisera; both isoforms were , detected at similar levels in samples from wild-type and transgenic mice compared to the housekeeping protein α-tubulin (Fig 1C). Thus, the human α4 polypeptide is expressed in transgenic mice, and this process does not interfere with the expression of endogenous Na,K-ATPase α4 and α1 isoforms.
We used two different methods to estimate the efficiency of human ATP1A4 expression in the transgenic males. On the one hand, we used quantitative reverse-transcriptase PCR (qPCR) to assess the relative levels of the endogenous mouse Atp1a1 and Atp1a4 transcripts versus the human ATP1A4 RNA, in both wild-type and transgenic mice. As expected, the relative transcript levels for the mouse Na,K-ATPase α1 and α4 were similar in both mouse lines, while human ATP1A4 RNA was only found in the transgenic mice (Fig 1D). Alternatively, we used sequence differences between the mouse and human orthologs to discriminate which animals expressed the human versus endogenous isoforms. Standard RT-PCR was performed from testis RNA of wild-type and transgenic animals, using forward and reverse primers shared by the mouse and human ATP1A4 genes. The amplified products were then digested to completion with EcoRV, which cleaves the amplified mouse, but not the human fragment. While both wild-type and transgenic mice showed the expected digested bands (156 and 109 bp) of the mouse Atp1a4 product, an uncleaved band (265 bp), corresponding to human ATP1A4 was detected only in the transgenic mice (Fig. 1E). Densitometry analysis of the endogenous versus exogenous gene transcripts indicated that message of endogenous Atp1a4 was 11.5±1.1-fold higher than the human transgene. Altogether, our results show that this BAC construct can be used to express human α4 in mice, and that the expression levels are modest compared to the endogenous gene.
Mice expressing human Na,K-ATPase α4 appeared normal and did not show differences in testis size, gross testis morphology (data not shown), or histology compared to wild-type mice (Fig 2A). Mice expressing human ATP1A4 produced sperm that was normal in number and shape. When mated, the transgenic animals presented normal fertility, with a number of mice per litter of 7.0±1.8 and 7.5±1.8 for transgenic and wild-type mice, respectively.
Figure 2.
Testis morphology and human Na,K-ATPase α4 expression in testis and sperm from wild-type and transgenic mice. A: Testes were dissected, fixed, and embedded in paraffin. Sections of 10-μm thickness were stained with hematoxylin and-eosin, and examined under 20X and 40X magnifications. Scale bars, 50 μm (top panels) and 20 μm (bottom panels). B: Testis sections (10 μm) from wild-type and transgenic mice were fixed, permeabilized, and subjected to immunohistochemistry, using the anti-human α4 antibody (hα4 antibody). Samples were incubated with a secondary goat anti-chicken antibody conjugated to Alexa Fluor 488. DAPI was used to label cell nuclei. Scale bars, 50 μm. L, Leydig cells; S, Sertoli cells; G, spermatogonia; and Sp, spermatozoa. C: Isolated caudal epididymal sperm were collected, plated onto glass cover slips, and subjected to immunocytochemistry. Cells were labeled with the anti-human α4 antibody and goat anti-chicken secondary antibody conjugated to Alexa Fluor 488. Cells were mounted, and DAPI was included for nuclei staining. Cells are shown at a magnification of 20X (left C panels) or 60X (right C panels). Scale bar, 10 μm.
Immunofluorescence of tissue sections from the gonad were performed to determine the presence of human α4 in the testis. Samples were probed with the anti-human α4 antibody, which did not label testis from the wild-type mice (Fig. 2B, upper left panel). Anti-human α4 staining was detected in the testis of transgenic mice, being particularly prominent in the seminiferous epithelium and spermatozoa. No label was observed outside of the tubules, where Leydig cells are located (Fig 2B, bottom left panel). Further immunocytochemical analysis of sperm taken from the cauda of the epididymis demonstrated the lack of human α4 in spermatozoa from wild-type mice (Fig 2C, upper panels), versus its presence in sperm from transgenic mice. The human α4 ortholog was detected in the sperm flagellum, localizing to the midpiece of the sperm tail (Fig 2C, bottom panels). Altogether, these results indicate that transgenic mice express the human Na,K-ATPase α4 polypeptide in spermatozoa.
Human α4 is functional in transgenic mice
We measured Na,K-ATPase activity in spermatozoa collected from the cauda epididymis to determine if the human α4 isoform is functional in transgenic mice. The inhibition constant for ouabain is much lower for both mouse and human Na,K-ATPase α4 (in the nanomolar range) than for the mouse α1 isoform (in the micromolar range) (Blanco et al. 2000; Sanchez et al. 2006), which for the discrimination of human and mouse α4 activity from that of mouse α1 (Wagoner et al. 2005). For example, 10−6 M ouabain can completely inhibit α4 activity, without a significant effect on α1, whereas 10−3 M ouabain inactivates both the α1 and α4 isoforms (Wagoner et al. 2005). Therefore, the hydrolysis of ATP corresponding to the α4 isoform can be calculated by subtracting the Na,K-ATPase activity measured at 10−6 M from that in the absence of ouabain. The difference in activity between samples containing 10−6 M and 10−3 M ouabain reflects α1-dependent hydrolysis of ATP. The total Na,K-ATPase activity in sperm from mice expressing human α4 was significantly higher than that of wild-type sperm (Fig 3A, bars labeled “Total”). Individual analysis of α4 and α1 activities indicated that this boost in ATP hydrolysis was due to the selective increase in α4 activity (Fig 3A, bars labeled “α4”). In contrast, the activity of the α1 isoform remained unchanged in wild-type and transgenic mouse sperm (Fig 3A, bars labeled “α1”). Thus, human α4 is catalytically competent in mouse sperm.
Figure 3.
Human Na,K-ATPase α4 isoform expressed in transgenic mice sperm is catalytically competent. A: Na,K-ATPase assays. Caudal epididymal sperm was collected, homogenized, and total Na,K-ATPase activity was determined as the Na+-and K+-dependent hydrolysis of ATP. Activity of the α4 isoform was associated with a sensitivity to 10−6 M ouabain, whereas activity of α1 was estimated as the difference in specific ATP hydrolysis between 10−6 M and 10−3 M ouabain. B: Ouabain-binding assays. Epididymal sperm from wild-type and transgenic mice was labeled with the fluorescent ouabain derivative, bodipy-ouabain. Fluorescence was determined at an excitation of 488 nm by flow cytometry. In both A and B, bars represent the mean ± standard errors of three experiments. Values significantly different from the wild type are indicated with an asterisks * P< 0.05.
Additional information was obtained through binding studies using the Na,K-ATPase-specific ligand ouabain. We determined the ability of sperm from wild-type and transgenic mice to bind bodipy-ouabain, a fluorescent derivative of ouabain. We used relatively low concentrations of bodipy-ouabain (10−8 M), which thespecifically associates with the high-affinity α4, but not the low-affinity α1 isoform (Blanco et al. 1999), to measure Na,K-ATPase α4 ouabain-binding kinetics. As expected, sperm from wild-type mice bound bodipy ouabain, via endogenous Na,K-ATPase α4 (Fig 3B). Importantly, sperm from transgenic mice showed an ~40% increase in bodipyouabain binding compared to sperm from wild-type mice – a difference that represents binding to the exogenously expressed human α4 isoform. Altogether, these results demonstrate that human α4 produced in transgenic mice is functional and able to hydrolyze ATP, and retain the characteristic high-ouabain-affinity of the α4 isoform.
Human α4 enhances mouse sperm motility and hyperactivity during capacitation
Previous studies have shown that pharmacological inhibition of Na,K-ATPase α4 with ouabain, inhibits the motility of sperm from different species (Jimenez et al. 2010; Sanchez et al. 2006; Woo et al. 2000). To determine the role of the human α4 isoform in sperm function, we first assessed motility in caudal epididymal sperm from wild-type and human α4 transgenic mice for 2 h under non-capacitating conditions. By computer assisted sperm analysis (CASA), total sperm motility remained relatively constant throughout the measurement period for gametes from of sperm from wild-type and transgenic mice (Fig 4). Sperm expressing human α4, however, always exhibited significantly higher total motility at all the time points tested (Fig 4, open circles).
Figure 4.
Human Na,K-ATPase α4 enhances flagellar beat of mouse sperm. Sperm from the cauda of epididymides from wild- type and transgenic mice were collected in modified Tyrode's medium with the addition of calcium, and sperm motility was determined using CASA. Different parameters of sperm movement were analyzed during a total period of 2 h, including: (A) total motility, (B) progressive motility, (C) straight line velocity, (D) curvilinear velocity, (E) average path velocity, (F) amplitude of lateral head displacement, (G) linearity and (H) beat cross frequency. Symbols represent wild-type (filled circles), or transgenic mouse sperm (open circles) samples. Values are the mean ± standard errors of three experiments. Values significantly different from wild-type are indicated with an asterisks (P values ranging between < 0.05 and < 0.001).
Sperm from human α4 transgenic mice also showed a consistent increase progressive motility; straight-line, curvilinear, and average path velocities; and amplitude of lateral head displacement, linearity, and beat cross frequency (Fig 4B-H). These experiments together demonstrate that increased expression of human α4 enhances flagellar movement of mouse sperm, augmenting not only total motility, but also different aspects of sperm movement.
Before they can fertilize an egg, sperm need to undergo capacitation, a series of changes in the cells that, includes a specific pattern of motility or hyperactivation (Suarez 2008; Visconti et al. 2011). To determine if human α4 plays a role in mouse sperm hyperactive motility, we incubated wild-type and transgenic sperm for different times in medium supporting capacitation, and then measured hypermotility using CASA. Sperm from wild-type mice showed a modest increase in hyperactive motility after incubation while sperm from the transgenic mice showed a time-dependent increase in hyperactive motility that was significantly higher at all the time points studied (Fig 5A). Thus, human Na,K-ATPase α4 participates in sperm hypermotility.
Figure 5.
A: Expression of human Na,K-ATPase α4 enhances sperm hyperactive motility in transgenic mice. Epididymal sperm from wild-type (WT) and transgenic mice (T hα4) was incubated at 37°C for the indicated times in modified Tyrode's medium supplemented with calcium, bicarbonate and bovine serum albumin to support capacitation. Hyperactive motility was determined using CASA. Sperm from transgenic (open circles) and, wild-type (folled circles) mice are shown. Each value is the mean ± standard errors of six experiments. Values significantly different from wild-type are indicated with an asterisks, (* P< 0.001). B : Plasma membrane potential of sperm from WT and T hα4 mice. Membrane potential was determined in epididymal sperm in modified Tyrode's medium using the fluorescent indicator DiSC3(5). Bars represent the mean ± standard errors of 12 and 8 experiments for WT and T hα4, respectively. P= 0.125.
Sperm hyperactivation is also accompanied by hyperpolarization of the sperm plasma membrane (Chavez et al. 2013; Hernandez-Gonzalez et al. 2006; Lopez-Gonzalez et al. 2014). In previous reports, we observed that α4 activity helps maintain the membrane potential (Jimenez et al. 2012). Using the fluorescent indicator DiSC3(5), as previously described (Hernandez-Gonzalez et al. 2006), we were able to investigate the effect of human α4 expression on membrane potential in sperm from wild-type and transgenic mice. Compared to wild-type samples, sperm expressing human α4 maintained a lower membrane potential –although the differences were not statistically significant (P= 0.125) (Fig 5B). Therefore, the expression and activity of human α4 cannot further hyperpolarize the sperm plasma membrane.
Altogether, these physiology data suggest that the activity of human Na,K-ATPase α4 is important for supporting sperm flagellar beating under non-capacitated conditions, and the hyperactive pattern of motility that is characteristic of capacitated sperm.
Human α4 does not affect intracellular signaling events associated with sperm capacitation
Another event associated with sperm capacitation is the phosphorylation of a series of proteins in tyrosine residues (Ickowicz et al. 2012; Salicioni et al. 2007; Visconti et al. 2011). We therefore explored protein tyrosine phosphorylation during capacitation to determine the role of human α4 isoform in sperm. Epididymal caudal sperm from wild-type and transgenic mice were incubated in non-capacitating and capacitating media for different times, and the pattern of protein tyrosine phosphorylation was determined after lysis and immunoblotting with the anti-phosphotyrosine antibody 4G10. The levels of protein phosphorylation were low in sperm from both wild-type and transgenic mice after 0, 1, and 2 h of incubation under non-capacitating conditions. In contrast, phosphotyrosine levels significantly increased after incubating the cells for 1 and 2 h in medium supporting capacitation. The enhancement and pattern of protein phosphorylation following sperm capacitation was similar in wild-type and transgenic mice (Fig 6). Thus, expression of human Na,K-ATPase α4 did not change protein tyrosine phosphorylation signaling in the host mouse sperm.
Figure 6.
Human Na,K-ATPase α4 does not affect capacitation-dependent protein tyrosine phosphorylation in sperm. Epididymal sperm from wild- type and transgenic mice were incubated in modified Tyrode's medium under non-capacitating and capacitating conditions for the indicated times. After cell lysis and SDS-PAGE separation, proteins were immunobloted for phosphotyrosine, using the 4G10 antibody. The upper panel shows the quantified densitometry analysis of six different experiments; the bottom panel shows a representative immunoblot. Data were expressed relative to the non-capacitated, 0 time point after correcting for loading with tubulin. Bars are the means ± standard errors of six experiments. Values significantly different from the non-capacitated, wild-type sperm at time 0 are indicated with an asterisk, s P values ranging between < 0.05 and < 0.001. NC, non-capacitated, C, capacitated.
Human α4 does not affect spontaneous acrosome reaction
An event that follows sperm capacitation is the release of acrosome contents (Ickowicz et al. 2012; Okabe 2013; Tulsiani and Abou-Haila 2011). To examine if the human α4 isoform is involved in the acrosome exocytosis, caudal epididymal sperm were placed in noncapacitating medium, or incubated in medium supporting capacitation for different times, followed by Coomassie-blue staining of the cells to evaluate the status of the acrosome. Exocytosis of the acrosome increased with sperm capacitation in a time-dependent manner, but there was no difference observed in acrosomal content between sperm from wild-type and transgenic mice (Fig 7). These results suggest that expression of human α4 does not affect the spontaneous release of acrosome content by mouse sperm.
Figure 7.
Human Na,K-ATPase α4 expression does not affect spontaneous acrosome reactions in transgenic mice. Epididymal sperm from wild type (WT) and transgenic mice (T hα4) were incubated in modified Tyrode's medium that does not support capacitation (non-capacitated, NC), or in modified Tyrode's medium supplemented with calcium, bicarbonate, and bovine serum albumin to support capacitation (C). After incubation at 37°C for the indicated times, the acrosome status of the cells was measured using Coomassie blue G-250 stain. A minimum of 350 cells was scored for each time point per experiment. Acrosome-reacted spermatozoa with no label were expressed as a percentage of the total amount of cells counted. Bars are the means ± standard errors of three experiments.
DISCUSSION
Previous in vitro studies in our laboratory have used ouabain as a tool to decipher the function of Na,K-ATPase α4 in human sperm (Sanchez et al. 2006); here, we took an alternative approach to understand the role of human Na,K-ATPase α4. As BAC constructs were successfully used in transgenic experiment, mostly in somatic cells but also germ cells (Giraldo and Montoliu 2001; Perry et al. 2001; Zhang et al. 2006), we created a transgenic mouse carrying a BAC construct that contained the entire genomic locus and predicted promoter of the human ATP1A4 gene. This BAC construct provided the endogenous genomic regulatory elements necessary to direct tissue-specific expression of Na,K-ATPase human α4 in mouse testis and spermatozoa, yielding functionally active protein that resulted in phenotypic changes in sperm. These genetically modified animals provided a unique opportunity to assess, in a more comprehensive manner, the in vivo role of human α4, without the complications associated with undesired effects of pharmacological inhibition with ouabain (Sanchez et al. 2006).
Compared to classical transgenesis constructs, such as driving expression of rat Na,K-ATPase α4 under the protamine 1 promoter (Jimenez et al. 2011b), our present approach with the BAC allowed us to test how the non-coding regions of the human α4 locus contributed to α4 synthesis in mice. Our RT-PCR and immunoblot analysis showed that the human α4 promoter could drive expression in the murine host under the same testis-specific pattern as the native mouse α4 (McDermott et al. 2012). Compared to the endogenous murine ortholog, expression of human ATP1A4 was low –, at least at the RNA level, although this could depend on species differences in the ATP1A4 promoter. At present, information on the structure and function of the ATP1A4 promoter is very limited: The size and structure of the ATP1A4 promoter is unknown, and only a small region of ~1 kb upstream of the translation-initiation site has been studied (Keryanov and Gardner 2002; Rodova et al. 2006). An alignment of the known mouse and human ATP1A4 proximal promoter region shows an overall identity of ~48.3% (Supplemental Figure 2), an amount that could result in species-dependent of transcriptional activation versus repression of the gene. In addition, differences in the stability/degradation of ATP1A4 mRNA could account for the modest abundance of this transcript in murine tissue. Yet, the human ATP1A4 promoter is still transcribed in mice, possibly . because of their shared transcriptional binding sites: Both human and mouse proximal promoters contain consensus cyclic AMP response element (CRE)-like sites, albeit not at the same positions (Supplemental Figure 2). We previously showed that a CRE-like motif in the proximal human ATP1A4 promoter can activate the its transcription in vitro (Rodova et al. 2006). In this manner, CRE-like sites could be a common regulator of ATP1A4 expression among species that utilize it. The mouse model that we have generated could therefore be a valuable tool for future studies directed to understand the in vivo transcriptional regulation of ATP1A4.
The amount of human Na,K-ATPase α4 protein produced in transgenic mice, and its relation to the endogenous murine ortholog, could not be adequately estimated, since human and mouse α4 are recognized by species specific antibodies that could, have different affinities for their respective epitopes. Immunoblot analysis did, however, show that the level abundance of human α4 protein is moderate, suggesting that feedback mechanisms may autoregulate the quantity of protein produced. The larger genomic sequences contained in our BAC construct, as opposed to smaller promoters classically used in transgenic constructs, may account for this more refined regulation since its controlled output can minimize the impact of transgene expression on that of the endogenous genes. This model is in agreement with our findings that human α4 expression did not alter the levels of the endogenous α1 or α4 isoforms, at the RNA or protein levels.
Our immunocytochemical analysis confirmed expression of human α4 in mouse spermatozoa, and revealed its localization in the sperm tail. This exogenous α4 isoform is enriched in the midpiece of the mouse sperm flagellum, which agrees with the location reported for the native α4 in rat, mouse, and human sperm (Sanchez et al. 2006; Wagoner et al. 2005; Woo et al. 1999). Thus, the membrane-targeting mechanisms present in mouse sperm can efficiently deliver human α4 to the appropriate membrane domain in sperm.
The total Na,K-ATPase activity in spermatozoa from the transgenic mice was higher than that of wild-type mice, in agreement with the supplemental expression of human α4. As shown by selective inhibition of Na,K-ATPase activity with ouabain, transgenic mice exhibited greater ouabain sensitivity regarding the hydrolysis of ATP, which is characteristic of the α4 isoform, but displayed no changes in ouabain resistance for the, α1 isoform (Blanco and Mercer 1998). As the α4 orthologs of Na,K-ATPase among different species has a similar, nanomolar affinity to ouabain (Blanco et al. 1999; Blanco et al. 2000; Woo et al. 1999; (Sanchez et al. 2006), Sanchez et al. 2006), it is not possible to discriminate within the individual contribution of human α4 from the endogenous α4 contribution to the measured enzymatic activity. Comparison of the activity associated with the wild-type versus the transgenic mouse sperm, however, demonstrates that, human α4 nearly doubled the activity associated with the ouabain-sensitive population of Na,K-ATPase; similar increase in Na,KATPase activity was found when the rat α4 isoform was expressed in mouse sperm (Jimenez et al. 2011b). Moreover, we found that sperm from the transgenic mice displayed higher specific binding to bodipy-ouabain, further revealing that the α4 transgene is expressed and functionally competent. Bodipy-ouabain is a polar compound that cannot cross the cell plasma membrane, therefore it is used as a specific marker for the Na,K-ATPase present at the cell plasma membrane (Kaplan 2002). The enhanced binding of bodipy-ouabain that we observed in sperm from transgenic mice thus, indicates that the α4 transgene is properly localized at the cell surface, where its activity can contribute to sperm physiology.
Expression of the human ATP1A4 transgene did not change the activity of the Na,K-ATPase α1 isoform, suggesting that the net increase in α4 does not modify the abundance of the endogenous α1 isoform. Indeed, previous work has shown that the enzymatic and ion transport properties, cell localization, and function of α1 and α4 are distinct (Blanco 2005; Blanco et al. 1999; Wagoner et al. 2005), wherein only α4, Is involved in sperm-specific processes (Jimenez et al. 2011a; Jimenez et al. 2010). Our current results agree with these findings that the abundance and activity of α1 and α4 are independent, and support the notion that Na,K-ATPase α1 and α4 expression and function are regulated via separate mechanisms and that these isoforms have distinct roles in sperm.
Transgenic adult mice expressing exogenous human α4 protein are indistinguishable from wild-type mice, exhibiting an overall normal reproductive phenotype and reproductive characteristics. The presence of human α4, however, increased the motility of mouse sperm. The use of pharmacological inhibition with ouabain or of genetic deletion, or over-expression of α4 in mice has shown a role for α4 in sperm motility (Jimenez et al. 2011a; Woo et al. 2000). Our current data provide more direct evidence that the expression and activity of human α4 in transgenic mice is associated with sperm motility, enhancing total flagellar beat. In addition, human α4 increases progressive motility; straight line, curvilinear, and average path velocities; and amplitude of lateral head displacement, linearity, and beat cross frequency. These contributions of human α4 to sperm motility complement previous reports of decreased sperm flagellar motion in human sperm after inhibition of α4 with ouabain. In the context of these transgenic sperm, ouabain did not produce significant changes in other parameters of sperm motility, as previously observed (Sanchez et al. 2006). This discrepancy could be related to differences in the regulation of the molecular motors in human and mouse sperm or in the sensitivity of the different approaches used to determine the various parameters of sperm motion. In any case, our current results show that human α4 has a net-positive effect on sperm flagellar beat.
Sperm from transgenic mice exhibit enhanced hyperactive motility compared to wild- type mice, suggesting that activity of the human α4 isoform is involved in this capacitation-dependent pattern of sperm motion (Suarez 2008). Previous studies in our laboratory have shown that over-expression of rat α4 positively affects different patterns of sperm motion, including hyperactivated motility (Jimenez et al. 2010), which is further associated with membrane hyperpolarization (Chavez et al. 2013; Hernandez-Gonzalez et al. 2006). Yet, we found no statistically significant differences in membrane potential between sperm from human ATP1A4 transgenic and wild-type mice, which is distinct from our previous measurement of a slight, albeit significant, hyperpolarized membrane potential in sperm that over-express rat α4 (Jimenez et al. 2010). Our present data suggests that human α4 may be producing changes in membrane potential that are too small to be we detected with the fluorescence method that we used. Alternatively, as ion transporters in addition to Na,KATPase are required to maintain the distribution of transmembrane electrical charges that contribute to the membrane potential, it is plausible that human α4 may not functionally interact with endogenous murine ion transporters as efficiently as the native or the rat α4 does, resulting in reduced hyperpolarization. Indeed, human α4 may provide mouse sperm with the ability to sustain membrane potential, which enhances sperm movement under both non-capacitating and capacitating conditions. The similarities in sperm phenotype between the human and rat ATP1A4 transgenic mouse models reveal the conservation of α4 function across species and highlight the biological relevance of this ion transporter in sperm.
At present, it is not known why expression of additional Na,K-ATPase α4 enhances sperm motility. Besides contributing to sperm membrane potential, the Na+ and K+ transport catalyzed by Na,K-ATPase α4 is indirectly involved in maintaining sperm pH and intracellular calcium levels (Jimenez et al. 2012; Jimenez et al. 2010), which both help regulate. sperm motility (Florman et al. 2010; Lishko et al. 2012; Navarro et al. 2008; Nishigaki et al. 2014). It is therefore plausible that the exogenous human α4 assists the endogenous Na,K-ATPase by preventing cell acidification and by regulating intracellular calcium, thus providing the circumstances necessary to maintain cell conditions that are optimal for enhanced sperm flagellar beat.
Despite the stimulation of sperm motility and hyperactivity by human α4, the transgenic mice exhibit normal fertility. This unaffected phenotype would be expected since the female limits fertility and litter size after in vivo mating. Additional experiments are required to determine if increased abundance of α4 results in enhanced fertilizing capabilities in sperm.
Our results also show that expression of human Na,K-ATPase α4 does not modify the capacitation-dependent level and pattern of protein phosphorylation in sperm. Therefore, α4 does not appear to significantly influence protein tyrosine phosphorylation levels or kinetics during sperm capacitation. Alternatively, protein phosphorylation may already reach maximal levels during capacitation in wild-type mice, preventing the detection of any further increase that may be associated with human Na,KATPase α4 activity. In addition, expression of human α4 did not alter the probability of spontaneous acrosome reactions, which is consistent with reports that rat and mouse Na,K-ATPase α4 are not directly involved in the sperm acrosome reaction (Jimenez et al. 2011b). Thus, the impact of human α4 activity appears to be limited to early stages of the complex process of sperm capacitation.
In conclusion, our work provides new evidence for the role for the human Na,K-ATPase α4 isoform in sperm motility and capacitation. The transgenic mouse that we engineered establishes a new animal model to further study the mechanisms of action of human Na,K-ATPase α4 in sperm physiology. Future research should be directed to explore the potential for using human α4 as a biomarker for male fertility, in cases of idiopathic male infertility, or as a target for reversible male contraception.
MATERIALS AND METHODS
All experimental protocols involving animals in this work were approved by the University of Kansas Medical Center Institutional Animal Care and Use Committee.
Preparation of Transgenic Animals
The transgenic mouse over-expressing human ATP1A4 was generated using BAC RP11-536C5 (CHORI), which contains the entire genomic locus and predicted promoter region of the gene (Keryanov and Gardner 2002), as well as other genomic flanking regions, in the pBACe3.6 vector (Supplemental Fig. 1). This BAC was linearized with AscI, microinjected into the pronucleus of one-cell hybrid FVB x C57Bl/6 embryos, and surgically transferred into pseudo-pregnant recipient female mice following standard procedures (Kumar et al. 2009). Resulting mice were screened for the transgene, and a founder line was established.
Genotype analysis was performed on tail biopsies by PCR on isolated genomic DNA using the REDExtract-N-Amp Tissue PCR Kit (Sigma, St. Louis, MO). The presence of human ATP1A4 was detected using primers specific to this isoform (5’-ACACAGCTGACAAGTGCAGGAGTT and 5’-AGGAAGGTGGGAAATGCAGTGAGA generates a 930-bp product).
Sperm and Testis Preparation
Spermatozoa from wild-type and transgenic mice were isolated from the cauda of 12-week-old mouse epididymides after swim up, as previously described (Jimenez et al. 2010). Sperm was resuspended in modified Tyrode's medium, containing 100 mM NaCl, 4.7 mM KCl, 1.2 mM KH2PO4, 1.2 mM MgSO4, 5.5 mM glucose, 0.8 mM pyruvic acid, 4.8 mM lactic acid, and 20 mM Hepes (pH 7.4). Cells were then counted and used for the different assays. In some experiments, sperm was capacitated in modified Tyrode's medium supplemented with 1.7 mM CaCl2, 25 mM sodium bicarbonate, and 0.5% bovine serum albumin (BSA).
Testes from wild-type and transgenic mice were used for histological and morphological analysis. Testes were dissected and fixed with buffered formalin phosphate (Fisher Scientific, Pittsburgh, PA). After embedding in paraffin, the tissue was cut into 10-μm sections, treated with xylene and ethanol to remove the paraffin, and stained with hematoxylin and eosin.
Reverse-transcriptase-PCR analysis
Total testis RNA was prepared using Tri Reagent Solution, according to the supplier specifications (Life Technologies, Grand Island, NY). A total of 10.0 μg RNA was incubated in 100 μL of 1× DNase buffer with 1 U RNase-free DNase I (Roche, Indianapolis, IN) at 37 °C for 20 minutes. Complementary DNA was prepared by reverse transcription of 1.0 μg of total RNA using SuperScript II Reverse Transcriptase (Invitrogen), as described previously (Wagoner et al. 2005). The resulting first-strand cDNA was amplified using primers specific for human ATP1A4 (sense, 5’-CCATAGCCACCAAAGGGCAA and antisense, 5’-CAGCCAACCATAGCCCAAGA) and for mouse glyceraldehyde phosphate dehydrogenase (Gapdh) (sense, 5’-GGTATCGTGGAAGGACTCATGAC and antisense, 5-ATGCCAGTGAGCTTCCCGTTCAGC). Amplified DNA fragments were identified by electrophoresis in 1% agarose gels stained with ethidium bromide. Expected sizes for the amplification are 748 and 450 bp, respectively.
Primer sets were specifically designed for qPCR using the RealTime PCR Tool (IDT). These target genes included: NM_144699.3 human ATP1A4 (sense, 5′-TGTAAGCAACTGTTCGGAGG and antisense, 5′-GACAGTACGATGCTCAGGTAG); NM_013734.1 mouse Atp1a1 (sense, 5′-CTTTCTTATCCTACTGCCCCG and antisense, 5′-ATAATGAGCTTCCGCACCTC),; NM_144900.2 mouse Atp1a4 (sense, 5′-GTGGACTTGACCAAGGGTCT and antisense, 5′-ACAGTAGGAGGGAGAAGCCA); and NM_008907.1 mouse peptidylprolyl isomerase A (Ppia) (sense. 5′-TGTGCCAGGGTGGTGACTTT and antisense, 5′-CGTTTGTGTTTGGTCCAGCAT) as a reference gene for the mouse testis (Gong et al. 2014). All primer sets used had squared correlation coefficients (r2) at >0.99, and a single peak in their dissociation curve.
The amplification program used for qPCR analysis was 95° for 10 min, and 40 cycles of 15 s at 95° and 60 s at 60°. Each amplification reaction was followed by a dissociation curve to ensure a single peak. qPCR was performed on an ABI Prism 7900HT Sequence Detection System (ABI). Reactions contained 5 μL Power Sybr Green PCR Master Mix (Life Technologies), 1 μL cDNA template, 90 nM of each primer, and H2O to bring the final volume to 10 μL. Each reaction was performed in triplicate and repeated 3 times. Normalized comparisons between our transgenic and wild- type testis cDNA were determined by using the ΔΔCT method (Livak and Schmittgen 2001). The ratio of the gene of interest between the transgenic relative to our wild- type samples was determined by taking 2ΔΔCT.
An additional approach was used to quantify the ratio of endogenous to exogenous α4. For this, testis RNA was isolated and subjected to RT-PCR, as described above, using primers common to mouse and human ATP1A4 genes (sense, 5’-GTCTCCAGAGGGTTCTCATG and antisense, 5’-GAGGCCAAGAGCTCCAAGAT). Samples were then subjected to total digestion with EcoRV, which cleaves the amplified mouse, but not the human DNA fragment, releasing products of 156 and 109 bp. Densitometry of the resultant bands was then determined and expressed as a ratio of the endogenous α4 bands to the human product using a BioRad Gel Doc XR+ Imaging System and Image Lab 3.0 software (BioRad).
Immunoblot analysis
Tissue samples were homogenized in RIPA buffer (1% NP-40, 0.25% NaDOC, 1 mM Na3VO4, 1 mM NaF, 1 mM EDTA, 150 mM NaCl, 10 μM leupeptin, 10 μg/ml apotinin, 1mM PMSF, and 50mM Tris (pH 7.4)) and centrifuged; resulting supernatant was used for immunoblot analysis. Samples were subjected to SDS-polyacrylamide gel electrophoresis (SDS-PAGE) (7.5% gel) and immunoblotting, as previously described (Wagoner et al. 2005). The mouse Na,K-ATPase α1 and α4 isoforms were identified using antisera generated against specific regions of the polypeptides in rabbit and chicken, respectively. Human Na,K-ATPase α4 was identified using antisera generated against specific regions of the α4 polypeptide in chicken. Mouse anti-α-tubulin (1:1000 dilution) (Sigma-Aldrich, St. Louis, MO) was used as a loading control. Horseradish peroxidase-conjugated secondary antibodies (Jackson ImmunoResearch Laboratories, Inc.) and chemiluminescence was used for detection.
Immunofluorescence
Testes were dissected and fixed with buffered formalin phosphate (Fisher Scientific, Pittsburgh, PA). After embedding in paraffin, the tissue was cut into 10-μm sections, treated with xylene and ethanol to remove the paraffin, and then rehydrated. Samples were permeabilized with 0.3% Triton X-100, and auto- fluorescence was quenched with 100 mM ammonium chloride. Sections were block for 2 hour at room temperature with 0.2% BSA and 2% normal goat serum in phosphate-buffered saline (PBS). Then, the primary antibody against the human Na,K-ATPase α4 isoform was applied and incubated overnight 4°C, followed by three 15-min washes in PBS. Samples were then treated with secondary antibody conjugated to Alexa Fluor 488 (Molecular Probes, Eugene, OR). After washing, as above, samples were mounted on slides using SlowFade mounting solution (Molecular Probes, Eugene, OR), containing 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI) to stain the cell nuclei. Digital images were obtained using a Nikon Eclipse 80i microscope equipped with an Olympus DP72 camera.
For sperm immunocytochemistry, cells were isolated from cauda epididymides in modified Tyrode's medium and plated on glass cover-slips. Cells were fixed with 4% paraformaldehyde, and permeabilized with 0.3% Triton X-100 in PBS. Immunofluorescence of the samples was performed and analyzed as described above.
Na,K-ATPase and ouabain-binding Assays
Na,K-ATPase activity was assayed on sperm homogenates by, determining the initial rate of release of 32Pi from γ[32P]-ATP, as previously described (Jimenez et al. 2010). The ATPase activity of 10 μg of total protein per sample was measured in a final volume of 0.25 mL in medium containing 120 mM NaCl, 30 mM KCl, 3 mM MgCl2, 0.2 mM EGTA, 30 mM Tris-HCl (pH 7.4), and 3 mM ATP with 0.2 μCi γ[32P]-ATP, in the presence or absence ouabain at the indicated concentrations. Curve fitting of the experimental data were performed using a Marquardt least-squares non-linear regression program (Sigma Plot; Jandel Scientific, San Rafael, CA).
Ouabain binding capacity of spermatozoa from wild-type and transgenic mice was measured using bodipy-ouabain (Invitrogen). Sperm was placed in modified Tyrode's medium at a concentration of 2 × 106 cells/mL, and 10−8 M bodipy-ouabain was added. Samples containing 1 mM unlabeled ouabain were used as a control for the specificity of bodipy-ouabain binding. After incubation for 20 min at 37°C, cells were washed twice for 5 min in modified Tyrode's medium. Finally, fluorescence was measured at 488 nm, using an LSRII flow cytometer (BD Biosciences).
Sperm-motility Assays
Approximately 3 × 106 cells from wild-type or transgenic mice were used to determine sperm motility, as previously described (Jimenez et al. 2010). Samples were analyzed by CASA, using the Minitube Sperm-Vision Digital Semen Evaluation system (version 3.5, Penetrating Innovations, Verona, WI). Total sperm motility and different parameters of sperm movement were analyzed using analytical setup parameters as defined before (Jimenez et al. 2010). Briefly, an average of 200 cells/field were captured, at a rate of 30 frames per field, and a total of 10 fields in each sample were analyzed. The setup parameters included a cell identification area between 15 and 900 µm2, minimum motility speed corresponding to a straight line velocity of 5 µm/s and progressive motility at a straight line velocity of more than 20 µm/s. Hyperactivity was measured using curvilinear velocity (VCL) and linearity (LIN) as parameters for vigour and progression, respectively. These conditions were based on a previously reported bivariate analysis, which defined the percent of hyperactivated spermatozoa as the number of cells that exceeded the 90th percentile for VCL and fell below the 10th percentile for LIN, divided by the number of tracked cells, multiplied by 100 (Cancel et al. 2000). The cut-off levels for VCL and LIN were set as higher than 180 μm/s and lower than 32% respectively.
Sperm membrane potential determination
Membrane potential was measured using the fluorescent indicator DiSC3(5), as previously described (Espinosa and Darszon 1995; Plasek and Hrouda 1991; Zeng et al. 1995). Briefly, a total of 10 × 106 cells/ml were resuspended in modified Tyrode's medium and treated with 1 µM DiSC3(5) for 3 min at 37°C. Sperm was further incubated for another 2 min with m-chlorophenyldrazone (CCCP), at a final concentration of 1 µM, to block mitochondrial membrane potential. Then, 2.5 ml of the suspension was transferred into a 37°C cuvette with gentle stirring. Fluorescence was recorded at an excitation/emission wavelength of 620/670 nm. Calibration of the fluorescence changes into mV was performed in the same sample, by adjusting the membrane potential of the cells with the K+ ionophore valinomycin, as previously described (Espinosa and Darszon 1995). Membrane potential was calculated from the distribution of K+ in the cells, using the Nernst equilibrium (Espinosa and Darszon 1995; Hernandez-Gonzalez et al. 2006; Plasek and Hrouda 1991; Zeng et al. 1995).
Phosphorylation Analysis
Aliquots of 5 × 105 spermatozoa from wild-type and transgenic mice were incubated for various time points at 37°C in a humidified incubator with 5% CO2 in noncapacitating and capacitating media. After incubation, sperm was centrifuged at 5 min at 10,000 rpm, and resuspended in lysis buffer (1 mM orthovanadate, 1 mM EDTA, 1 mM NaF, 1% NP-40, 1% deoxicolate, 0.1% SDS, and 1 × Phosphostop (Roche, Indianapolis, IN)). Samples were subjected to SDS-PAGE (7.5% gel) and immunoblotted, as previously described (Wagoner et al. 2005). Briefly, the membrane was blocked using 5% BSA in Tris-buffered saline with 0.5% Tween-20 (TBS-T) for 1 hour. Phosphorylation was detected by probing overnight at 4°C with anti-phosphotyrosine antibody (4G10 antibody; 1:1000 dilution in TBS-T Millipore, Billerica, MA. Blots were then wash 3 times with TBS-T, and then incubated for 1 hr with donkey anti-mouse IgG conjugated to horseradish peroxidase 1:1000 dilution in TBS-T. Following extensive washing, positive bands were detected using chemiluminescence. To ensure equal protein loading, membranes were stripped and re-probed with a polyclonal anti-α-tubulin antibody (1:1000 dilution) (Sigma-Aldrich). Phosphorylation intensities of the samples were analyzed with Pro-Gel Analyzer 4.5, and reported relative to the non-capacitated samples.
Acrosome reaction assays
Approximately 2 × 106 spermatozoa from wild-type and transgenic mice were incubated at different time points in non-capacitating and capacitating media at 37°C in a humidified incubator with 5% CO2. After incubation, samples were fixed with 10% paraformaldehyde for 10 minutes. Cells were pelleted at 3,000 rpm, and then resuspend in PBS. Samples were smeared on glass slides and allowed to air dry. Slides were stained with Commassie Blue G-250 (0.22% Commassie Blue G-250 in 50% methanol and 10% glacial acetic acid) for 1 minute. Slides were washed extensively with water, and then mounted with Permount (Fisher Scientific, Pittsburgh, PA ). A minimum of 350 cells was scored for each time point per experiment. Acrosome-reacted spermatozoa with no blue label were expressed as a percentage of the total number of cells counted.
Statistical Analysis
Statistical significance of differences between controls and ouabain- treated samples was determined by the Student's t-test, using Sigma Plot software (Jandel Scientific, San Rafael, CA). Statistical significance was defined as P < 0.05.
Supplementary Material
Supplemental Figure 1. Schematic representation of the BAC used to generate the human ATP1A4 mouse. The BAC contained 170.8 kb of human chromosome 1q23 and the vector pBACe3.6 (11.6 kb). The chromosome region contained the ATP1A4 and its proximal promoter region, as well as genes for the muscle specific Na,K-ATPase α2 (ATP1A2), phosphoprotein enriched in astrocytes PEA15, calsequestrin 1 (CASQ1), immunoglubolin member 8 (IGSF8) and potassium inwardly rectifying channel 9 (KCNJ9) of somatic cells. The AscI restriction site, located in the pBACe3.6 vector and used to linearize the construct, is shown.
Supplemental Figure 2. Sequence alignment of the proximal promoter regions of the human and mouse ATP1A4 orthologs. Alignment was performed using Clustal O(1.2.1.). Base-pair positions are shown in the right margin. Asterisks indicate residues that are identical. The ATG start site is indicated, and putative CRE-like sites are highlighted.
ACKNOWLEDGMENTS
We would like to thank Melissa Larson, from the Transgenic and Gene-targeting Institutional Facility at University of Kansas, for helping us with the generation of the transgenic mice used in this work.
Grant sponsor: NIH
Grant number: 5U01HD080423
Abbreviations
- ATP1A1/4
Na,K-ATPase alpha 4/1
- BAC
bacterial artificial chromosome
- CASA
computer assisted sperm analysis
- qPCR
quantitative reverse-transcriptase polymerase chain reaction
- RT-PCR
reverse transcriptase polymerase chain reaction
REFERENCES
- Blanco G. Na,K-ATPase subunit heterogeneity as a mechanism for tissue-specific ion regulation. Seminars in nephrology. 2005;25(5):292–303. doi: 10.1016/j.semnephrol.2005.03.004. [DOI] [PubMed] [Google Scholar]
- Blanco G, Melton RJ, Sanchez G, Mercer RW. Functional characterization of a testes-specific alpha-subunit isoform of the sodium/potassium adenosinetriphosphatase. Biochemistry. 1999;38(41):13661–13669. doi: 10.1021/bi991207b. [DOI] [PubMed] [Google Scholar]
- Blanco G, Mercer RW. Isozymes of the Na-K-ATPase: heterogeneity in structure, diversity in function. The American journal of physiology. 1998;275(5 Pt 2):F633–650. doi: 10.1152/ajprenal.1998.275.5.F633. [DOI] [PubMed] [Google Scholar]
- Blanco G, Sanchez G, Melton RJ, Tourtellotte WG, Mercer RW. The alpha4 isoform of the Na,K-ATPase is expressed in the germ cells of the testes. The journal of histochemistry and cytochemistry : official journal of the Histochemistry Society. 2000;48(8):1023–1032. doi: 10.1177/002215540004800801. [DOI] [PubMed] [Google Scholar]
- Cancel AM, Lobdell D, Mendola P, Perreault SD. Objective evaluation of hyperactivated motility in rat spermatozoa using computer-assisted sperm analysis. Hum Reprod. 2000;15(6):1322–1328. doi: 10.1093/humrep/15.6.1322. [DOI] [PubMed] [Google Scholar]
- Chavez JC, de la Vega-Beltran JL, Escoffier J, Visconti PE, Trevino CL, Darszon A, Salkoff L, Santi CM. Ion permeabilities in mouse sperm reveal an external trigger for SLO3-dependent hyperpolarization. PloS one. 2013;8(4):e60578. doi: 10.1371/journal.pone.0060578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crambert G, Hasler U, Beggah AT, Yu C, Modyanov NN, Horisberger JD, Lelievre L, Geering K. Transport and pharmacological properties of nine different human Na, K-ATPase isozymes. The Journal of biological chemistry. 2000;275(3):1976–1986. doi: 10.1074/jbc.275.3.1976. [DOI] [PubMed] [Google Scholar]
- Espinosa F, Darszon A. Mouse sperm membrane potential: changes induced by Ca2+. FEBS letters. 1995;372(1):119–125. doi: 10.1016/0014-5793(95)00962-9. [DOI] [PubMed] [Google Scholar]
- Florman HM, Jungnickel MK, Sutton KA. Shedding light on sperm pHertility. Cell. 2010;140(3):310–312. doi: 10.1016/j.cell.2010.01.035. [DOI] [PubMed] [Google Scholar]
- Geering K. The functional role of beta subunits in oligomeric P-type ATPases. Journal of bioenergetics and biomembranes. 2001;33(5):425–438. doi: 10.1023/a:1010623724749. [DOI] [PubMed] [Google Scholar]
- Geering K. Functional roles of Na,K-ATPase subunits. Current opinion in nephrology and hypertension. 2008;17(5):526–532. doi: 10.1097/MNH.0b013e3283036cbf. [DOI] [PubMed] [Google Scholar]
- Giraldo P, Montoliu L. Size matters: use of YACs, BACs and PACs in transgenic animals. Transgenic research. 2001;10(2):83–103. doi: 10.1023/a:1008918913249. [DOI] [PubMed] [Google Scholar]
- Gong ZK, Wang SJ, Huang YQ, Zhao RQ, Zhu QF, Lin WZ. Identification and validation of suitable reference genes for RT-qPCR analysis in mouse testis development. Molecular genetics and genomics : MGG. 2014 doi: 10.1007/s00438-014-0877-6. [DOI] [PubMed] [Google Scholar]
- Hernandez-Gonzalez EO, Sosnik J, Edwards J, Acevedo JJ, Mendoza-Lujambio I, Lopez-Gonzalez I, Demarco I, Wertheimer E, Darszon A, Visconti PE. Sodium and epithelial sodium channels participate in the regulation of the capacitation-associated hyperpolarization in mouse sperm. The Journal of biological chemistry. 2006;281(9):5623–5633. doi: 10.1074/jbc.M508172200. [DOI] [PubMed] [Google Scholar]
- Hlivko JT, Chakraborty S, Hlivko TJ, Sengupta A, James PF. The human Na,KATPase alpha 4 isoform is a ouabain-sensitive alpha isoform that is expressed in sperm. Molecular reproduction and development. 2006;73(1):101–115. doi: 10.1002/mrd.20383. [DOI] [PubMed] [Google Scholar]
- Ickowicz D, Finkelstein M, Breitbart H. Mechanism of sperm capacitation and the acrosome reaction: role of protein kinases. Asian journal of andrology. 2012;14(6):816–821. doi: 10.1038/aja.2012.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jewell EA, Shamraj OI, Lingrel JB. Isoforms of the alpha subunit of Na,K-ATPase and their significance. Acta physiologica Scandinavica Supplementum. 1992;607:161–169. [PubMed] [Google Scholar]
- Jimenez T, McDermott JP, Sanchez G, Blanco G. Na,K-ATPase alpha4 isoform is essential for sperm fertility. Proceedings of the National Academy of Sciences of the United States of America. 2011a;108(2):644–649. doi: 10.1073/pnas.1016902108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez T, Sanchez G, Blanco G. Activity of the Na,K-ATPase alpha4 isoform is regulated during sperm capacitation to support sperm motility. Journal of andrology. 2012;33(5):1047–1057. doi: 10.2164/jandrol.111.015545. [DOI] [PubMed] [Google Scholar]
- Jimenez T, Sanchez G, McDermott JP, Nguyen AN, Kumar TR, Blanco G. Increased expression of the Na,K-ATPase alpha4 isoform enhances sperm motility in transgenic mice. Biology of reproduction. 2011b;84(1):153–161. doi: 10.1095/biolreprod.110.087064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez T, Sanchez G, Wertheimer E, Blanco G. Activity of the Na,K-ATPase alpha4 isoform is important for membrane potential, intracellular Ca2+, and pH to maintain motility in rat spermatozoa. Reproduction. 2010;139(5):835–845. doi: 10.1530/REP-09-0495. [DOI] [PubMed] [Google Scholar]
- Kaplan JH. Biochemistry of Na,K-ATPase. Annual review of biochemistry. 2002;71:511–535. doi: 10.1146/annurev.biochem.71.102201.141218. [DOI] [PubMed] [Google Scholar]
- Keryanov S, Gardner KL. Physical mapping and characterization of the human Na,K-ATPase isoform, ATP1A4. Gene. 2002;292(1-2):151–166. doi: 10.1016/s0378-1119(02)00647-9. [DOI] [PubMed] [Google Scholar]
- Kumar TR, Larson M, Wang H, McDermott J, Bronshteyn I. Transgenic mouse technology: principles and methods. Methods in molecular biology. 2009;590:335–362. doi: 10.1007/978-1-60327-378-7_22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lishko PV, Kirichok Y, Ren D, Navarro B, Chung JJ, Clapham DE. The control of male fertility by spermatozoan ion channels. Annual review of physiology. 2012;74:453–475. doi: 10.1146/annurev-physiol-020911-153258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lopez-Gonzalez I, Torres-Rodriguez P, Sanchez-Carranza O, Solis-Lopez A, Santi CM, Darszon A, Trevino CL. Membrane hyperpolarization during human sperm capacitation. Molecular human reproduction. 2014;20(7):619–629. doi: 10.1093/molehr/gau029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin DW. Structure-function relationships in the NA+,K+-pump. Seminars in nephrology. 2005;25(5):282–291. doi: 10.1016/j.semnephrol.2005.03.003. [DOI] [PubMed] [Google Scholar]
- McDermott JP, Sanchez G, Chennathukuzhi V, Blanco G. Green fluorescence protein driven by the Na,K-ATPase alpha4 isoform promoter is expressed only in male germ cells of mouse testis. Journal of assisted reproduction and genetics. 2012;29(12):1313–1325. doi: 10.1007/s10815-012-9876-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mobasheri A, Avila J, Cozar-Castellano I, Brownleader MD, Trevan M, Francis MJ, Lamb JF, Martin-Vasallo P. Na+, K+-ATPase isozyme diversity; comparative biochemistry and physiological implications of novel functional interactions. Bioscience reports. 2000;20(2):51–91. doi: 10.1023/a:1005580332144. [DOI] [PubMed] [Google Scholar]
- Morth JP, Pedersen BP, Buch-Pedersen MJ, Andersen JP, Vilsen B, Palmgren MG, Nissen P. A structural overview of the plasma membrane Na+,K+-ATPase and H+-ATPase ion pumps. Nature reviews Molecular cell biology. 2011;12(1):60–70. doi: 10.1038/nrm3031. [DOI] [PubMed] [Google Scholar]
- Navarro B, Kirichok Y, Chung JJ, Clapham DE. Ion channels that control fertility in mammalian spermatozoa. The International journal of developmental biology. 2008;52(5-6):607–613. doi: 10.1387/ijdb.072554bn. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton LD, Krishnakumar S, Menon AG, Kastelic JP, van der Hoorn FA, Thundathil JC. Na+/K+ATPase regulates sperm capacitation through a mechanism involving kinases and redistribution of its testis-specific isoform. Molecular reproduction and development. 2010;77(2):136–148. doi: 10.1002/mrd.21114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishigaki T, Jose O, Gonzalez-Cota AL, Romero F, Trevino CL, Darszon A. Intracellular pH in sperm physiology. Biochemical and biophysical research communications. 2014 doi: 10.1016/j.bbrc.2014.05.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okabe M. The cell biology of mammalian fertilization. Development. 2013;140(22):4471–4479. doi: 10.1242/dev.090613. [DOI] [PubMed] [Google Scholar]
- Perry AC, Rothman A, de las Heras JI, Feinstein P, Mombaerts P, Cooke HJ, Wakayama T. Efficient metaphase II transgenesis with different transgene archetypes. Nature biotechnology. 2001;19(11):1071–1073. doi: 10.1038/nbt1101-1071. [DOI] [PubMed] [Google Scholar]
- Plasek J, Hrouda V. Assessment of membrane potential changes using the carbocyanine dye, diS-C3-(5): synchronous excitation spectroscopy studies. Eur Biophys J. 1991;19(4):183–188. doi: 10.1007/BF00196344. [DOI] [PubMed] [Google Scholar]
- Rodova M, Nguyen AN, Blanco G. The transcription factor CREMtau and cAMP regulate promoter activity of the Na,K-ATPase alpha4 isoform. Molecular reproduction and development. 2006;73(11):1435–1447. doi: 10.1002/mrd.20518. [DOI] [PubMed] [Google Scholar]
- Salicioni AM, Platt MD, Wertheimer EV, Arcelay E, Allaire A, Sosnik J, Visconti PE. Signalling pathways involved in sperm capacitation. Society of Reproduction and Fertility supplement. 2007;65:245–259. [PubMed] [Google Scholar]
- Sanchez G, Nguyen AN, Timmerberg B, Tash JS, Blanco G. The Na,K-ATPase alpha4 isoform from humans has distinct enzymatic properties and is important for sperm motility. Molecular human reproduction. 2006;12(9):565–576. doi: 10.1093/molehr/gal062. [DOI] [PubMed] [Google Scholar]
- Shamraj OI, Lingrel JB. A putative fourth Na+,K(+)-ATPase alpha-subunit gene is expressed in testis. Proceedings of the National Academy of Sciences of the United States of America. 1994;91(26):12952–12956. doi: 10.1073/pnas.91.26.12952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez SS. Control of hyperactivation in sperm. Hum Reprod Update. 2008;14(6):647–657. doi: 10.1093/humupd/dmn029. [DOI] [PubMed] [Google Scholar]
- Tulsiani DR, Abou-Haila A. Molecular events that regulate mammalian fertilization. Minerva ginecologica. 2011;63(2):103–118. [PubMed] [Google Scholar]
- Visconti PE, Krapf D, de la Vega-Beltran JL, Acevedo JJ, Darszon A. Ion channels, phosphorylation and mammalian sperm capacitation. Asian journal of andrology. 2011;13(3):395–405. doi: 10.1038/aja.2010.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagoner K, Sanchez G, Nguyen AN, Enders GC, Blanco G. Different expression and activity of the alpha1 and alpha4 isoforms of the Na,K-ATPase during rat male germ cell ontogeny. Reproduction. 2005;130(5):627–641. doi: 10.1530/rep.1.00806. [DOI] [PubMed] [Google Scholar]
- Woo AL, James PF, Lingrel JB. Characterization of the fourth alpha isoform of the Na,K-ATPase. The Journal of membrane biology. 1999;169(1):39–44. doi: 10.1007/pl00005899. [DOI] [PubMed] [Google Scholar]
- Woo AL, James PF, Lingrel JB. Sperm motility is dependent on a unique isoform of the Na,K-ATPase. The Journal of biological chemistry. 2000;275(27):20693–20699. doi: 10.1074/jbc.M002323200. [DOI] [PubMed] [Google Scholar]
- Woo AL, James PF, Lingrel JB. Roles of the Na,K-ATPase alpha4 isoform and the Na+/H+ exchanger in sperm motility. Molecular reproduction and development. 2002;62(3):348–356. doi: 10.1002/mrd.90002. [DOI] [PubMed] [Google Scholar]
- Yang XW, Gong S. An overview on the generation of BAC transgenic mice for neuroscience research. Current protocols in neuroscience / editorial board, Jacqueline N Crawley [et al] Chapter 5:Unit 5 20. 2005 doi: 10.1002/0471142301.ns0520s31. [DOI] [PubMed] [Google Scholar]
- Zeng Y, Clark EN, Florman HM. Sperm membrane potential: hyperpolarization during capacitation regulates zona pellucida-dependent acrosomal secretion. Dev Biol. 1995;171(2):554–563. doi: 10.1006/dbio.1995.1304. [DOI] [PubMed] [Google Scholar]
- Zhang H, Barnoski BL, Sol-Church K, Stabley DL, Martin-Deleon PA. Murine Spam1 mRNA: involvement of AU-rich elements in the 3′UTR and antisense RNA in its tight post-transcriptional regulation in spermatids. Molecular reproduction and development. 2006;73(2):247–255. doi: 10.1002/mrd.20400. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Schematic representation of the BAC used to generate the human ATP1A4 mouse. The BAC contained 170.8 kb of human chromosome 1q23 and the vector pBACe3.6 (11.6 kb). The chromosome region contained the ATP1A4 and its proximal promoter region, as well as genes for the muscle specific Na,K-ATPase α2 (ATP1A2), phosphoprotein enriched in astrocytes PEA15, calsequestrin 1 (CASQ1), immunoglubolin member 8 (IGSF8) and potassium inwardly rectifying channel 9 (KCNJ9) of somatic cells. The AscI restriction site, located in the pBACe3.6 vector and used to linearize the construct, is shown.
Supplemental Figure 2. Sequence alignment of the proximal promoter regions of the human and mouse ATP1A4 orthologs. Alignment was performed using Clustal O(1.2.1.). Base-pair positions are shown in the right margin. Asterisks indicate residues that are identical. The ATG start site is indicated, and putative CRE-like sites are highlighted.