Abstract
Myoepithelial (ME) tumors of soft tissue and bone display a heterogeneous histologic spectrum and in about half of the cases harbor EWSR1 gene rearrangements. Despite rare case reports, the prevalence of FUS gene abnormalities and its related fusion partners remains undetermined among ME tumors. Therefore, we screened 66 EWSR1-negative ME tumors for FUS abnormalities by fluorescence in situ hybridization (FISH). In an index FUS-rearranged case, 3'-Rapid Amplification of cDNA Ends (RACE) was applied to identify the fusion partner. Results were further confirmed by RT-PCR, followed by FISH screening the entire cohort of FUS-rearranged and EWSR1-positive ME lesions lacking a known fusion partner. The correlation between genotype and clinicopathological features was also investigated. As a result, six (9%) FUS-rearranged cases were identified, spanning divergent age groups, tumor locations, and morphologic features. A novel FUS-KLF17 fusion was identified by 3’RACE in an 11 year-old girl with a foot lesion associated with locoregional metastases. Three additional cases with FUS-KLF17 fusions were identified and one KLF17 rearrangement (6.3%) was found among the 16 EWSR1-positive cases tested. The KLF17-related ME tumors affected younger patients and often exhibited trabecular growth in a myxohyaline stroma, but this genotype did not correlate with a malignant phenotype. In conclusion, a small subset of ME tumors harbor FUS rearrangements, two thirds of them being associated with KLF17 fusion. FUS FISH analysis is recommended in EWSR1-negative lesions in which a ME diagnosis is suspected. KLF17 is also a rare gene fusion partner to EWSR1-rearranged ME tumors.
Keywords: myoepithelial tumor, FUS, KLF17, EWSR1
INTRODUCTION
Myoepithelial (ME) tumors of soft tissue and bone are rare tumors resembling their counterparts in salivary gland (Kilpatrick et al., 1997; Hornick and Fletcher, 2003; Rekhi et al., 2012). They show a wide morphologic spectrum with occasional duct formation or chondroid differentiation, but consistently co-express epithelial markers and S100 protein. While pleomorphic adenomas of salivary gland frequently exhibit PLAG1 and HMGA2 abnormalities (Kas et al., 1997; Martins et al., 2005; Persson et al., 2009), PLAG1 rearrangements were mainly identified in a subset of cutaneous and superficial soft tissue ME tumors, often displaying ductal structures, consistent with benign mixed tumors (Bahrami et al., 2012; Antonescu et al., 2013). Instead, most deep-seated soft tissue and bone ME tumors harbor different molecular alterations. A handful of early case reports described EWSR1 gene rearrangements, including t(1;22)(q23;q12) resulting in an EWSR1-PBX1 fusion and t(19;22)(q13;q12) involving an EWSR1-ZNF444 fusion (Gleason and Fletcher, 2007; Brandal et al., 2008; Brandal et al., 2009). A subsequent detailed molecular study of 66 ME tumors from soft tissue, bone, and lung revealed EWSR1 rearrangements occurred in 45% of cases, with different fusion partners including POU5F1, PBX1, and ZNF444 (Antonescu et al., 2010). The EWSR1-positive ME tumors were often seen in children and young adults, involving soft tissue of the extremities. Five ME tumors with EWSR1-POU5F1 fusion shared a predominantly nested growth pattern with distinctive clear cell morphology. EWSR1-PBX1 translocations were found in 5 ME tumors associated with a bland sclerotic appearance and EWSR1-ZNF444 was identified in a pulmonary ME tumor. More recently our group described novel EWSR1-PBX3 gene fusions in 3 cases of ME tumors, occurring with predilection in skeletal locations (Agaram et al., 2014). Of the 30 EWSR1-negative ME tumors, only one pulmonary case showed FUS gene rearrangement (3%). Two more recent case reports described EWSR1-ATF1 and FUS-POU5F1 fusion transcripts in a pelvic ME tumor of 57 year-old woman and in a sacral lesion of a 54 year-old man, respectively (Flucke et al., 2012; Puls et al., 2014).
As a significant proportion of ME tumors lack EWSR1 and PLAG1 alterations, we sought to investigate the prevalence of FUS rearrangement by FISH and to analyze the clinicopathologic features of such lesions in a larger cohort of ME tumors. Additionally, we performed 3'-RACE in a FUS-rearranged ME tumor in order to identify possible novel gene partners and to determine their recurrent potential among FUS and EWSR1-positive ME tumors.
MATERIALS AND METHODS
Tumor Samples and Patients Cohort
We retrieved 66 cases of ME tumors lacking EWSR1 and PLAG1 gene rearrangements, 30 of which were included in our earlier study (Antonescu et al., 2010). The cases were selected from the MSKCC Surgical Pathology and consultation files of the senior authors (CRA, CDF). The slides and corresponding immunohistochemical stains were re-reviewed to confirm the diagnosis according to the standard criteria: tumors were composed of epithelioid, plasmacytoid, or spindled cells forming solid, reticular, or trabecular architectures in a variably myxoid or hyalinized stroma. All tumors showed immunohistochemical evidence of cytokeratin or epithelial membrane antigen (EMA) and S100 protein reactivity, as previously described (Hornick and Fletcher, 2003). The study was approved by the Institutional Review Board 02-060.
Fluorescence in situ Hybridization (FISH)
FISH for FUS and EWSR1 break-apart assay was applied on formalin-fixed and paraffin-embedded 4-micron sections in all cases. FISH was performed by applying custom probes using bacterial artificial chromosomes (BACs), covering and flanking the FUS and EWSR1 gene. Additional BAC clones were designed flanking the novel gene identified in one case by 3’RACE assay. BAC clones were chosen according to UCSC genome browser (http://genome.ucsc.edu), see Supplementary Table 1. The BAC clones were obtained from BACPAC sources of Children's Hospital of Oakland Research Institute (CHORI) (Oakland, CA)(http://bacpac.chori.org). DNA from individual BACs was isolated according to the manufacturer’s instructions, labeled with different fluorochromes in a nick translation reaction, denatured, and hybridized to pretreated slides. Slides were then incubated, washed, and mounted with DAPI in an antifade solution, as previously described (Antonescu et al., 2010). The genomic location of each BAC set was verified by hybridizing them to normal metaphase chromosomes. Two hundred successive nuclei were examined using a Zeiss fluorescence microscope (Zeiss Axioplan, Oberkochen, Germany), controlled by Isis 5 software (Metasystems, Newton, MA). A positive score was interpreted when at least 20% of the nuclei showed a break-apart signal. Nuclei with incomplete set of signals were omitted from the score.
3'- RACE and Reverse Transcription-PCR (RT-PCR)
Total RNA was extracted from the frozen tissue available in the index case (case #1), using Trizol reagent according to the manufacturer's instructions (Invitrogen, Carlsbad, CA). RNA quality was determined by Eukaryote Total RNA Nano Assay and cDNA quality was tested for PGK housekeeping gene (247 bp amplified product). One microgram of total RNA was prepared for 3'-RACE cDNA synthesis by using the SMARTer™ RACE cDNA Amplification Kit (Clontech, Mountain View, CA). Reverse transcription was initiated at the poly(A) tail of mRNA with 3'-RACE CDS Primer A for making 3'-RACE-Ready cDNA according to the manufacturer's manual. The first round PCR was done by Clontech Advantage 2 PCR Enzyme System kit with the Universal Primer A and FUS_Ex1/2 Fwd primer (5'-CCTCAAACGATTATACCCAACAAGC-3'), followed by nested PCR using the Nested Universal Primer A and FUS_Ex4 Fwd primer (5'-CCAATCGTCTTACGGGCAG CAG-3'). The PCR products were analyzed by electrophoresis, TA cloning, and direct sequencing. Subsequent RT-PCR was performed to validate the RACE results using Clontech Advantage 2 PCR Enzyme System kit for 33 cycles at a 64.5°C annealing temperature, using the above FUS_Ex1/2 Fwd primer and KLF17_Ex2 Rev (5'-GGCTGCTCTGGTAGAAATG GG-3') primer. The amplicon was confirmed by Sanger sequencing. Frozen tissue was available from an additional tumor (case #2) and the FUS-KLF17 fusion transcript was confirmed by RT-PCR using similar primers as above. The cDNA synthesis was performed with SuperScript® III First-Strand Synthesis Kit (Invitrogen), followed by a PCR step. The PCR amplicons were confirmed by direct sequencing.
RESULTS
Clinical and Pathologic Characteristics of FUS-Positive ME Tumors
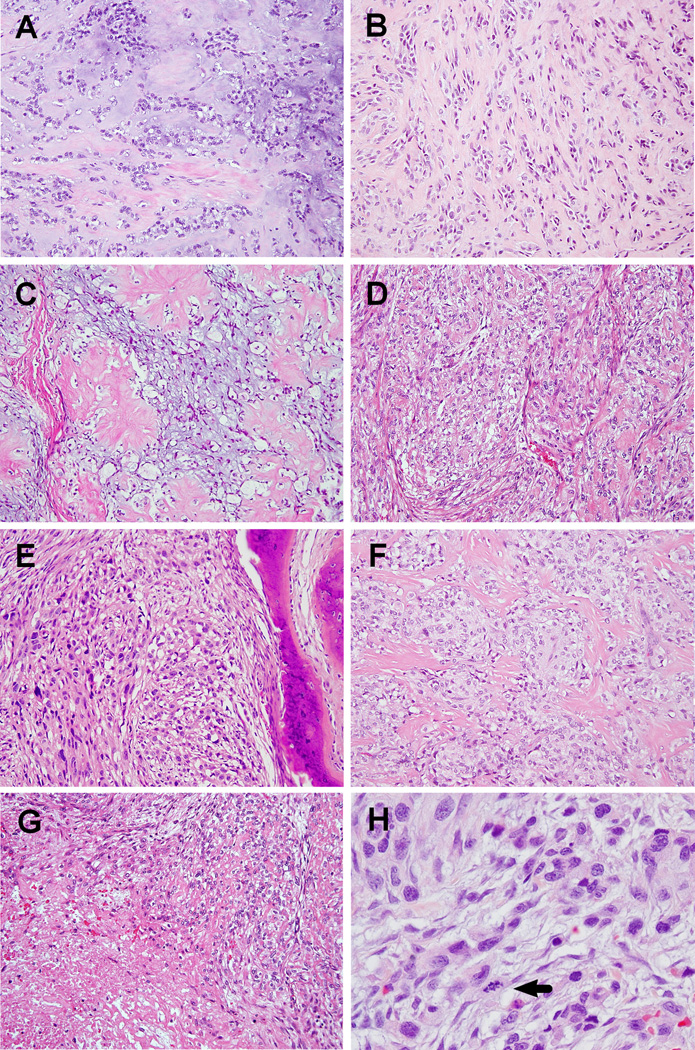
A total of 6 FUS-rearranged cases (9%) were identified from the 66 ME tumors lacking abnormalities in EWSR1 or PLAG1 genes. There were 4 males and 2 females with a mean age at diagnosis of 28 years (range 8–50; median 32). Most tumors occurred in soft tissue, including extremities, trunk and neck. One lesion involved the tibial periosteum and one was cutaneous. Only one case arose in a visceral location, involving the lung. The tumors demonstrated a broad histologic spectrum, as summarized in Table 1. The tumors were composed of a mixture of spindled, ovoid, or epithelioid cells, with amphophilic to eosinophilic cytoplasm. Their growth patterns varied from trabecular or reticular within a myxohyaline or hyaline background in 3 cases (Cases #1–3, Figs. 1A–C), to a solid sheet-like arrangement with scant stromal component in 2 cases (Cases #4 and 5, Figs. 1D, 1E). The remaining case displayed a nested architecture separated by sclerotic septa (Case #6, Fig. 1F). Focal microcystic change was noted in myxoid areas of case #1. Case #4 showed prominent cytoplasmic vacuolation, imparting a chordoma-like appearance, while case #3 had focal clear cell changes. No ductal differentiation or osteochondroid matrix formation was present in any of the cases.
Table 1.
Clinical and pathological features of myoepithelial tumors with FUS and/or KLF17 translocations.
| Case | Age (years)/ Sex |
Site | Malignant Potential |
Morphological Features | Nuclear pleomorphism |
Necrosis (%) |
Mitosis (/10 HPF) |
Genetic alteration |
Metastasis |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 11/F | Foot/soft tissue | Malignant | Trabecular, myxohyaline stroma | none | 0 | 5 | FUS-KLF17 | Thigh |
| 2 | 8/M | Tibia/periosteum | Benign | Trabecular, myxohyaline stroma | none | 0 | 0 | FUS-KLF17 | None |
| 3 | 37/M | Back/skin | Benign | Trabecular, myxohyaline stroma | none | 0 | 0 | FUS-KLF17 | None |
| 4 | 30/M | Lung | Benign | Cellular nesting and trabecular | Mild | 10 | 0 | FUS-KLF17 | None |
| 5 | 34/M | Chest/soft tissue | Malignant | Cellular nesting and trabecular | Severe | 40 | 4 | FUS-rearranged | None |
| 6 | 50/F | Neck/soft tissue | Benign | Nesting, sclerosing stroma | none | 10 | 0 | FUS-rearranged | None |
| 7 | 20/M | Foot/soft tissue | Benign | Trabecular, myxohyaline stroma | none | 0 | 0 | EWSR1-KLF17 | None |
Figure 1. Pathologic findings of FUS-rearranged ME tumors.
Case #1 (A, 200×), #2 (B, 200×), and #3 (C, 200×) showing cords or clusters of tumor cells set in variably myxohyaline background. Prominent cytoplasmic vacuolation was noted in case #3. Both case #4 (D, 200×) and case #5 (E, 200×) showed higher cellularity with less stromal component; nuclear atypia was evident in both cases. Nests of tumor cells separated by sclerotic bands in case #6 (F, 200×). Tumors exhibiting focal necrosis (G, case #4, 200×). Malignant ME tumor with significant nuclear pleomorphism and increased mitotic activity (H, case #5, 400×).
Two of the cases were designated as malignant (cases #1 and 5). Case #1 had brisk mitotic activity (5/10 HPFs) without significant nuclear pleomorphism, and designated as malignant due to presentation with loco-regional metastatic disease. Case #5 showed significant nuclear atypia, increased mitotic activity (4/10 HPFs), and obvious necrosis (Fig. 1H). This lesion occurred in chest wall soft tissue and was associated with rib invasion (Fig. 1E). The remaining 4 cases with no nuclear atypia or increased mitotic activity were diagnosed as benign in spite of two cases showing focal necrosis.
The fusion negative ME group encompassed 38 benign and 22 malignant ME tumors with a male to female ratio of 1.6 and an average age of 43.2 years (median 45, range 2 months to 85 years) after excluding 3 cases without available data. The most common location was lower extremity (20), followed by trunk (13), viscera (10), upper extremity (9), head & neck (4), and body cavity (4).
Novel FUS-KLF17 fusions identified in more than half of FUS-rearranged ME tumors
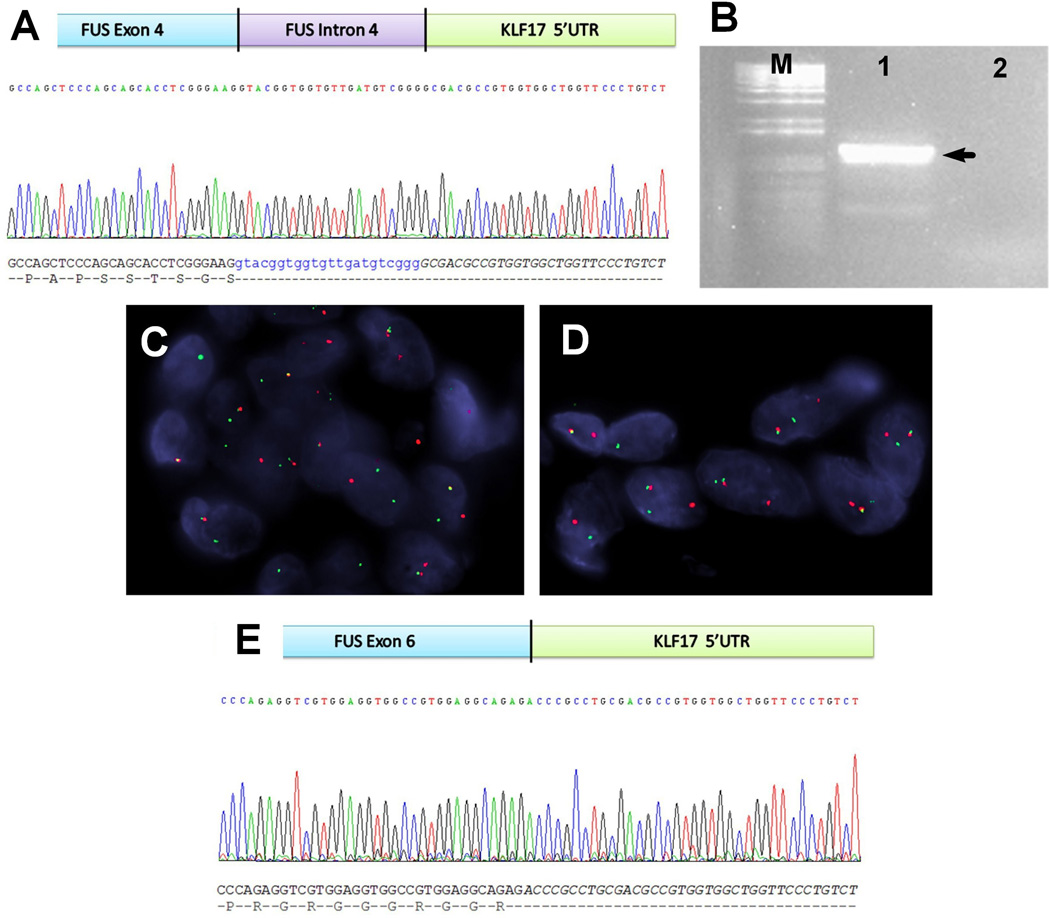
From the index case, a 11 year-old girl (case #1) who presented with a 6 cm foot mass and multiple small nodules in her lower leg and thigh, frozen tissue was available to perform 3'-RACE PCR. The amplified sequence revealed a chimeric transcript composed of FUS exon 4, followed by a small fragment of FUS intron 4, fused with the 5'-untranslated region (5'-UTR) of KLF17 (Fig. 2A). Subsequent RT-PCR (Fig. 2B) using primers flanking this breakpoint confirmed the fusion transcript by Sanger sequencing. This result was further validated by FISH analysis, using KLF17 specific break-apart probes (Figs. 2C, 2D). FISH screening for KLF17 abnormalities was performed in the remaining 5 FUS-rearranged ME tumors. Three additional cases (cases # 2–4) were found to have FUS-KLF17 fusions. In case # 2 frozen tissue was also available to perform RT-PCR analysis, which showed FUS exon 6 fused to the 5'-UTR of KLF17 gene (Figs. 2E).
Figure 2. Novel FUS-KLF17 and EWSR1-KLF17 gene fusions in ME soft tissue tumors.
The 3'-RACE disclosed a transcript of exon 4 and partial intron 4 of FUS fused with 5'-UTR of KLF17 (A), which was further confirmed by RT-PCR (M: size maker, lane 1: product, lane 2: negative control; the approximate size, 1100 bp) (B). FISH assays revealed split signals of FUS (C) and KLF17 (D) probes in the index case (case #1). The RT-PCR and Sanger sequencing (E) of case #2 identified a variant of fusion pattern involving exon 6 of FUS and 5'-UTR of KLF17.
ME tumors harboring FUS-KL17 occurred in 3 males and one female, with a mean age of 21.5 years (range 8–37; median 20.5). The 2 pediatric tumors arose in the lower extremity, while the lesions occurring in adult patients were located in the lung and the skin of the back. Both pediatric FUS-KLF17 ME tumors were composed of cords or nests of epithelioid to spindled tumor cells embedded in a myxoid or hyaline stroma (Fig. 1A, 1B). One pediatric tumor (case #1) demonstrated brisk mitotic activity (Fig. 1A) and developed multiple soft tissue metastases in the ipsilateral thigh. The cutaneous lesion (case #3) showed epithelioid cells with prominent cytoplasmic vacuoles, reminiscent of "physaliferous cells", arranged in trabeculae or clusters in an alternating myxoid and sclerotic stroma (Fig. 1C). The pulmonary ME tumor (case #4) showed increased cellularity (Fig. 1D), focal nuclear atypia and necrosis (Fig. 1G); however, no increased mitotic activity was noted.
Rare EWSR1-KLF17 Gene Fusions Identified in ME Tumors
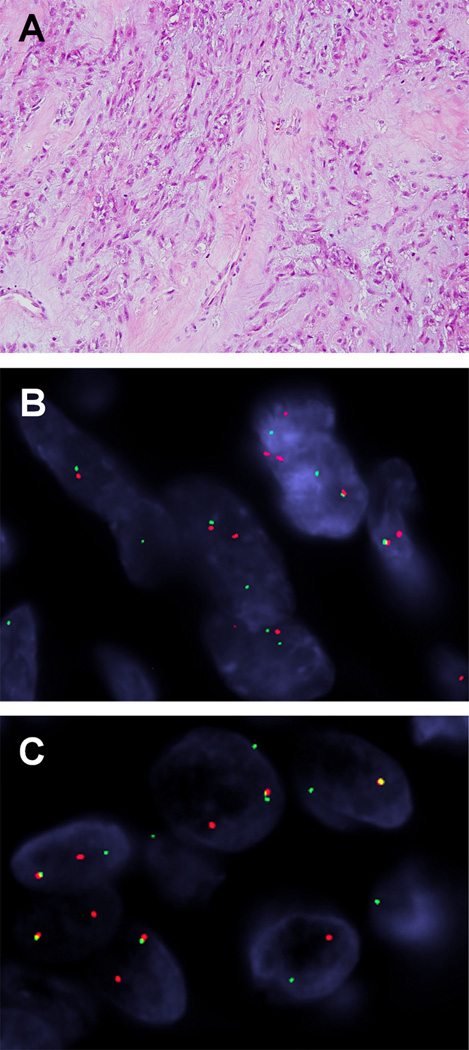
We additionally tested 19 EWSR1-rearranged ME cases lacking a known gene partner, including 9 soft tissue ME cases from our previous study (Antonescu et al., 2010) and 4 cases of cutaneous syncytial myoepithelioma described in another study (Jo et al., 2013). Excluding three failed cases, only one tumor (case #7) showed an EWSR1-KLF17 fusion among the 16 cases tested (Fig. 3B, 3C). This case occurred in the left foot soft tissue of a 20 year-old male. Microscopically, the tumor had a lobular architecture and consisted of interconnecting cords or clusters of epithelioid to spindled cells in a myxohyaline stroma (Fig. 3A). No mitotic activity, nuclear atypia, or necrosis was observed in this case.
Figure 3. Morphologic appearance and FISH findings of EWSR1-KLF17 positive ME tumor.
Histologically, the spindled or ovoid tumor cells formed cords or clusters in myxohyaline stroma (A). FISH demonstrated break-apart signals involving EWSR1 (B) and KLF17 genes (C).
Discussion
Since first being described by Dr. Stout in 1959 as cutaneous mixed tumors of the salivary gland type (Stout and Gorman, 1959), the subsequent studies of ME tumors of soft tissue and bone suggested a heterogeneous histologic spectrum (Kilpatrick et al., 1997; Hornick and Fletcher, 2003). The critical denominator among these different lesions is the consistent immunoexpression of cytokeratin/EMA and S100 protein, in conjunction with variable degrees of calponin, GFAP, smooth muscle actin, and p63 immunoreactivity, which indicates myoepithelial differentiation (Kilpatrick et al., 1997; Hornick and Fletcher, 2003; Rekhi et al., 2012). Genetically, the most common abnormality is EWSR1 gene rearrangement, present in approximately half of ME tumors (Antonescu et al., 2010; Flucke et al., 2011), which is seen most often among deep soft tissue, bone or lung lesions. The most common gene fusions are EWSR1-POU5F1 and EWSR1-PBX1, each accounting for 17% of EWSR1-rearranged tumors (Antonescu et al., 2010). Alternative fusions have been documented mainly as case reports or small case series, including EWSR1-ZNF444, EWSR1-ATF1, and EWSR1-PBX3 (Brandal et al., 2009; Antonescu et al., 2010; Flucke et al., 2012; Agaram et al., 2014). Recently, a distinctive variant of cutaneous ME tumor, designated as syncytial myoepithelioma, was found to have a higher percentage (82%) of EWSR1 gene rearrangement (Jo et al., 2013). However, despite an extensive FISH work-up for known gene candidates, no fusion partner was detected.
The second common molecular finding in ME lesions is the presence of PLAG1 gene rearrangements, detected in 37% of cases lacking EWSR1 and FUS abnormalities, with a rare case demonstrating LIFR-PLAG1 fusion (Antonescu et al., 2013). All 13 PLAG1-rearranged ME cases were benign and occur in dermis or subcutis and 12 of them showed tubulo-ductal differentiation. Among 11 ME tumors tested, Bahrami et al. found 5 cutaneous and 3 soft tissue cases exhibiting PLAG1 rearrangement, which were associated with prominent ductal structures and PLAG1 immunoreactivity (Bahrami et al., 2012). Conversely, a different study showed that cutaneous mixed tumors could also harbor EWSR1 abnormalities (Flucke et al., 2011). No HMGA2 gene abnormalities were discovered so far in soft tissue or bone ME tumors (Hallor et al., 2008; Antonescu et al., 2013).
FUS rearrangement in ME tumors has only rarely been documented in literature. One such example was described in a pulmonary lesion of a 30 year-old man (Antonescu et al., 2010), also included in our current study (case #4). The second case is that of an intraosseous sacral ME tumor from a 54 year-old man (Puls et al., 2014). Morphologically, this latter case was composed of epithelioid cells arranged in a reticular and nested pattern in a densely hyalinized matrix. The tumor showed mild nuclear pleomorphism and a mitotic count of 3/10 HPFs. Cytogenetic analysis revealed a 46,XY,del(6)(p21),der(16)t(6;16)(p21;p11) karyotype. The SNP array confirmed a 6p22.3-6p21.2 deletion, affecting the POU5F1 gene, and a 16p deletion, with one of the breakpoints in the FUS gene. Subsequent FISH and RT-PCR confirmed a FUS-POU5F1 transcript involving FUS exon 5 and POU5F1 intron 1 and exon 2.
The current work investigates the prevalence of FUS gene abnormalities and its potential fusion partners in a large cohort of 66 EWSR1 and/or PLAG1-negative ME tumors. Our results from combined FISH and 3’RACE/RT-PCR assays found 6 FUS-rearranged ME tumors (9%). As seen with most EWSR1-positive lesions, the FUS-rearranged ME tumors had wide clinical presentations, morphologic patterns, and consistently lacked ductal differentiation or osteochondroid matrix formation. Remarkably, 4 of the 6 FUS-rearranged ME tumors showed novel FUS-KLF17 fusion. KLF17 (Krüppel-like factor 17), a.k.a. ZNF393 (Zinc Finger Protein 393), located at chromosome 1p34.1 region, is the human homologue to murine Zfp393 and encodes a protein containing 389 amino acids (van Vliet et al., 2006). The Sp/KLF family consists of 25 known members and is characterized by a highly conserved C-terminal DNA-binding domains having three C2H2 Krüppel-like zinc motifs, which bind G/C-rich sites in DNA such as GC-box and CACCC-box sequences (Swamynathan, 2010). Thereby, KLF17 protein acts as a transcription factor involved in regulation of different genes. In breast cancer, KLF17 protein acts as a negative regulator of epithelial-mesenchymal transition and metastasis (Gumireddy et al., 2009), by binding its 5'-CACCC-3' sequence to the ID1 promoter, a key metastasis regulator in breast cancer, and repressing its expression. Similarly reduced KLF17 levels correlates with tumor growth, distant metastasis and poor prognosis in lung carcinoma, hepatocellular carcinoma, gastric cancer, and papillary thyroid carcinoma (Cai et al., 2012; Liu et al., 2013; Peng et al., 2014; Ye et al., 2014). In endometrioid-type uterine cancer, KLF17 was implicated in epithelial to mesenchymal transition via direct activation of TWIST1 and its overexpression was associated with high tumor grade and loss of hormone receptors (Dong et al., 2014). KLF17 may also function as a germ cell-specific transcription factor involving in spermatid differentiation and oocyte development (Yan et al., 2002). As the KLF17 breakpoint in ME tumor was located in the 5'-UTR, which is a critical region controlling mRNA translation, degradation, and localization (Mignone et al., 2002), the subsequent FUS-KLF17 or EWSR1-KLF17 chimeric transcript may dysregulate the translation or function of the KLF17 protein.
Half of the FUS-KLF17-positive ME tumors occurred in children, involving the soft tissue of lower extremities and exhibited uniform epithelioid to spindled cell morphology, arranged in trabecular or reticular patterns in a myxohyaline stroma. In contrast, EWSR1-KLF17 gene fusion was found only in one of the 16 EWSR1-rearranged cases tested. This case occurred in the lower extremity soft tissue of a young patient and demonstrated classic morphologic features similar to those of FUS-KLF17 fusion positive ME tumors.
Based on their similar function, it is not surprising that EWSR1 and FUS can be involved interchangeably as oncogenic fusion partners in ME neoplasms. The FUS (fused in sarcoma) gene, also known as TLS (translocated in liposarcoma), is a member of the TET family together with EWSR1 (EWS RNA-binding protein 1 or Ewing sarcoma breakpoint region 1) and TATA-binding protein-associated factor 15 (TAF15) (Aman et al., 1996; Morohoshi et al., 1998; Paronetto, 2013). Similar to EWSR1 gene, the FUS gene participates in multiple chimeric transcripts not only in mesenchymal tumors but also in hematologic malignancies (Ichikawa et al., 1994). There are many notable examples where EWSR1 and FUS can substitute for one another as fusion partners in sarcomas, such as myxoid liposarcoma (FUS/EWSR1-DDIT3), low-grade fibromyxoid sarcoma (EWSR1/FUS related fusions either with CREB3L2 or CREB3L1), and angiomatoid fibrous histiocytoma (EWSR1/FUS-ATF1) (Rabbitts et al., 1993; Waters et al., 2000; Panagopoulos et al., 2004; Mertens et al., 2005). However, there is rarely equal involvement, with either EWSR1 or FUS prevalence predominating in a particular type of sarcoma, while, in some sarcoma types only one of the genes is implicated in their pathogenesis (i.e. clear cell sarcoma, desmoplastic small round cell tumor) (Fisher, 2014). In keeping with these results, the EWSR1-related fusions are more frequent events compared to FUS alterations in ME neoplasms. One possible explanation why FUS-KLF17 fusions outnumber EWSR1-KLF17 fusions in ME tumors is that both FUS and KLF17 have the same orientation (centromeric) and their fusion can be achieved by a simple balanced translocation.
In summary, we identified a small subset (9%) of FUS-rearranged cases from studying a large cohort of EWSR1-negative ME tumors. The FUS-rearranged ME tumors spanned a heterogeneous clinical and pathologic spectrum, including both benign and malignant/metastatic examples. The most common fusion partner was KLF17 seen in 4 cases (67%), resulting in novel FUS-KLF17 fusions, which occurred mainly in the deep soft tissue of extremities in children. Their morphology showed epithelioid to ovoid cells arranged in a trabecular growth in a myxohyaline stroma. Thus, FUS FISH analysis is recommended in EWSR1-negative lesions in which a ME diagnosis is suspected. Additionally, KLF17 was found as a rare fusion partner with EWSR1, with only one of the 16 (6%) EWSR1-positive ME cases lacking a known gene partner being positive for this gene fusion.
Supplementary Material
Acknowledgments
Supported in part by: P01CA47179 (CRA), P50CA140146-01 (CRA), Cycle for Survival (CRA)
Footnotes
Conflict of interest: none
REFERENCES
- Agaram NP, Chen HW, Zhang L, Sung YS, Panicek D, Healey JH, Nielsen GP, Fletcher CD, Antonescu CR. EWSR1-PBX3: A novel gene fusion in myoepithelial tumors. Genes Chromosomes Cancer. 2014 doi: 10.1002/gcc.22216. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aman P, Panagopoulos I, Lassen C, Fioretos T, Mencinger M, Toresson H, Hoglund M, Forster A, Rabbitts TH, Ron D, Mandahl N, Mitelman F. Expression patterns of the human sarcoma-associated genes FUS and EWS and the genomic structure of FUS. Genomics. 1996;37:1–8. doi: 10.1006/geno.1996.0513. [DOI] [PubMed] [Google Scholar]
- Antonescu CR, Zhang L, Chang NE, Pawel BR, Travis W, Katabi N, Edelman M, Rosenberg AE, Nielsen GP, Dal Cin P, Fletcher CD. EWSR1-POU5F1 fusion in soft tissue myoepithelial tumors. A molecular analysis of sixty-six cases, including soft tissue, bone, and visceral lesions, showing common involvement of the EWSR1 gene. Genes Chromosomes Cancer. 2010;49:1114–1124. doi: 10.1002/gcc.20819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonescu CR, Zhang L, Shao SY, Mosquera JM, Weinreb I, Katabi N, Fletcher CD. Frequent PLAG1 gene rearrangements in skin and soft tissue myoepithelioma with ductal differentiation. Genes Chromosomes Cancer. 2013;52:675–682. doi: 10.1002/gcc.22063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahrami A, Dalton JD, Krane JF, Fletcher CD. A subset of cutaneous and soft tissue mixed tumors are genetically linked to their salivary gland counterpart. Genes Chromosomes Cancer. 2012;51:140–148. doi: 10.1002/gcc.20938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandal P, Panagopoulos I, Bjerkehagen B, Gorunova L, Skjeldal S, Micci F, Heim S. Detection of a t(1;22)(q23;q12) translocation leading to an EWSR1-PBX1 fusion gene in a myoepithelioma. Genes Chromosomes Cancer. 2008;47:558–564. doi: 10.1002/gcc.20559. [DOI] [PubMed] [Google Scholar]
- Brandal P, Panagopoulos I, Bjerkehagen B, Heim S. t(19;22)(q13;q12) Translocation leading to the novel fusion gene EWSR1-ZNF444 in soft tissue myoepithelial carcinoma. Genes Chromosomes Cancer. 2009;48:1051–1056. doi: 10.1002/gcc.20706. [DOI] [PubMed] [Google Scholar]
- Cai XD, Zhou YB, Huang LX, Zeng QL, Zhang LJ, Wang QQ, Li SL, Feng JQ, Han AJ. Reduced expression of Kruppel-like factor 17 is related to tumor growth and poor prognosis in lung adenocarcinoma. Biochem Biophys Res Commun. 2012;418:67–73. doi: 10.1016/j.bbrc.2011.12.129. [DOI] [PubMed] [Google Scholar]
- Dong P, Kaneuchi M, Xiong Y, Cao L, Cai M, Liu X, Guo SW, Ju J, Jia N, Konno Y, Watari H, Hosaka M, Sudo S, Sakuragi N. Identification of KLF17 as a novel epithelial to mesenchymal transition inducer via direct activation of TWIST1 in endometrioid endometrial cancer. Carcinogenesis. 2014;35:760–768. doi: 10.1093/carcin/bgt369. [DOI] [PubMed] [Google Scholar]
- Fisher C. The diversity of soft tissue tumours with EWSR1 gene rearrangements: a review. Histopathology. 2014;64:134–150. doi: 10.1111/his.12269. [DOI] [PubMed] [Google Scholar]
- Flucke U, Mentzel T, Verdijk MA, Slootweg PJ, Creytens DH, Suurmeijer AJ, Tops BB. EWSR1-ATF1 chimeric transcript in a myoepithelial tumor of soft tissue: a case report. Hum Pathol. 2012;43:764–768. doi: 10.1016/j.humpath.2011.08.004. [DOI] [PubMed] [Google Scholar]
- Flucke U, Palmedo G, Blankenhorn N, Slootweg PJ, Kutzner H, Mentzel T. EWSR1 gene rearrangement occurs in a subset of cutaneous myoepithelial tumors: a study of 18 cases. Mod Pathol. 2011;24:1444–1450. doi: 10.1038/modpathol.2011.108. [DOI] [PubMed] [Google Scholar]
- Gleason BC, Fletcher CD. Myoepithelial carcinoma of soft tissue in children: an aggressive neoplasm analyzed in a series of 29 cases. Am J Surg Pathol. 2007;31:1813–1824. doi: 10.1097/PAS.0b013e31805f6775. [DOI] [PubMed] [Google Scholar]
- Gumireddy K, Li A, Gimotty PA, Klein-Szanto AJ, Showe LC, Katsaros D, Coukos G, Zhang L, Huang Q. KLF17 is a negative regulator of epithelial-mesenchymal transition and metastasis in breast cancer. Nat Cell Biol. 2009;11:1297–1304. doi: 10.1038/ncb1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallor KH, Teixeira MR, Fletcher CD, Bizarro S, Staaf J, Domanski HA, von Steyern FV, Panagopoulos I, Mandahl N, Mertens F. Heterogeneous genetic profiles in soft tissue myoepitheliomas. Mod Pathol. 2008;21:1311–1319. doi: 10.1038/modpathol.2008.124. [DOI] [PubMed] [Google Scholar]
- Hornick JL, Fletcher CD. Myoepithelial tumors of soft tissue: a clinicopathologic and immunohistochemical study of 101 cases with evaluation of prognostic parameters. Am J Surg Pathol. 2003;27:1183–1196. doi: 10.1097/00000478-200309000-00001. [DOI] [PubMed] [Google Scholar]
- Ichikawa H, Shimizu K, Hayashi Y, Ohki M. An RNA-binding protein gene, TLS/FUS, is fused to ERG in human myeloid leukemia with t(16;21) chromosomal translocation. Cancer Res. 1994;54:2865–2868. [PubMed] [Google Scholar]
- Jo VY, Antonescu CR, Zhang L, Dal Cin P, Hornick JL, Fletcher CD. Cutaneous syncytial myoepithelioma: clinicopathologic characterization in a series of 38 cases. Am J Surg Pathol. 2013;37:710–718. doi: 10.1097/PAS.0b013e3182772bba. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kas K, Voz ML, Roijer E, Astrom AK, Meyen E, Stenman G, Van de Ven WJ. Promoter swapping between the genes for a novel zinc finger protein and beta-catenin in pleiomorphic adenomas with t(3;8)(p21;q12) translocations. Nat Genet. 1997;15:170–174. doi: 10.1038/ng0297-170. [DOI] [PubMed] [Google Scholar]
- Kilpatrick SE, Hitchcock MG, Kraus MD, Calonje E, Fletcher CD. Mixed tumors and myoepitheliomas of soft tissue: a clinicopathologic study of 19 cases with a unifying concept. Am J Surg Pathol. 1997;21:13–22. doi: 10.1097/00000478-199701000-00002. [DOI] [PubMed] [Google Scholar]
- Liu FY, Deng YL, Li Y, Zeng D, Zhou ZZ, Tian DA, Liu M. Down-regulated KLF17 expression is associated with tumor invasion and poor prognosis in hepatocellular carcinoma. Med Oncol. 2013;30:425. doi: 10.1007/s12032-012-0425-3. [DOI] [PubMed] [Google Scholar]
- Martins C, Fonseca I, Roque L, Pereira T, Ribeiro C, Bullerdiek J, Soares J. PLAG1 gene alterations in salivary gland pleomorphic adenoma and carcinoma ex-pleomorphic adenoma: a combined study using chromosome banding, in situ hybridization and immunocytochemistry. Mod Pathol. 2005;18:1048–1055. doi: 10.1038/modpathol.3800386. [DOI] [PubMed] [Google Scholar]
- Mertens F, Fletcher CD, Antonescu CR, Coindre JM, Colecchia M, Domanski HA, Downs-Kelly E, Fisher C, Goldblum JR, Guillou L, Reid R, Rosai J, Sciot R, Mandahl N, Panagopoulos I. Clinicopathologic and molecular genetic characterization of low-grade fibromyxoid sarcoma, and cloning of a novel FUS/CREB3L1 fusion gene. Lab Invest. 2005;85:408–415. doi: 10.1038/labinvest.3700230. [DOI] [PubMed] [Google Scholar]
- Mignone F, Gissi C, Liuni S, Pesole G. Untranslated regions of mRNAs. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-3-reviews0004. REVIEWS0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morohoshi F, Ootsuka Y, Arai K, Ichikawa H, Mitani S, Munakata N, Ohki M. Genomic structure of the human RBP56/hTAFII68 and FUS/TLS genes. Gene. 1998;221:191–198. doi: 10.1016/s0378-1119(98)00463-6. [DOI] [PubMed] [Google Scholar]
- Panagopoulos I, Storlazzi CT, Fletcher CD, Fletcher JA, Nascimento A, Domanski HA, Wejde J, Brosjo O, Rydholm A, Isaksson M, Mandahl N, Mertens F. The chimeric FUS/CREB3l2 gene is specific for low-grade fibromyxoid sarcoma. Genes Chromosomes Cancer. 2004;40:218–228. doi: 10.1002/gcc.20037. [DOI] [PubMed] [Google Scholar]
- Paronetto MP. Ewing sarcoma protein: a key player in human cancer. Int J Cell Biol. 2013;2013:642853. doi: 10.1155/2013/642853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng JJ, Wu B, Xiao XB, Shao YS, Feng Y, Yin MX. Reduced Kruppel-like factor 17 (KLF17) expression correlates with poor survival in patients with gastric cancer. Arch Med Res. 2014;45:394–399. doi: 10.1016/j.arcmed.2014.06.005. [DOI] [PubMed] [Google Scholar]
- Persson F, Andren Y, Winnes M, Wedell B, Nordkvist A, Gudnadottir G, Dahlenfors R, Sjogren H, Mark J, Stenman G. High-resolution genomic profiling of adenomas and carcinomas of the salivary glands reveals amplification, rearrangement, and fusion of HMGA2. Genes Chromosomes Cancer. 2009;48:69–82. doi: 10.1002/gcc.20619. [DOI] [PubMed] [Google Scholar]
- Puls F, Arbajian E, Magnusson L, Douis H, Kindblom LG, Mertens F. Myoepithelioma of Bone with a Novel FUS-POU5F1 Fusion Gene. Histopathology. 2014 doi: 10.1111/his.12517. [DOI] [PubMed] [Google Scholar]
- Rabbitts TH, Forster A, Larson R, Nathan P. Fusion of the dominant negative transcription regulator CHOP with a novel gene FUS by translocation t(12;16) in malignant liposarcoma. Nat Genet. 1993;4:175–180. doi: 10.1038/ng0693-175. [DOI] [PubMed] [Google Scholar]
- Rekhi B, Sable M, Jambhekar NA. Histopathological, immunohistochemical and molecular spectrum of myoepithelial tumours of soft tissues. Virchows Arch. 2012;461:687–697. doi: 10.1007/s00428-012-1335-7. [DOI] [PubMed] [Google Scholar]
- Stout AP, Gorman JG. Mixed tumors of the skin of the salivary gland type. Cancer. 1959;12:537–543. doi: 10.1002/1097-0142(195905/06)12:3<537::aid-cncr2820120313>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- Swamynathan SK. Kruppel-like factors: three fingers in control. Hum Genomics. 2010;4:263–270. doi: 10.1186/1479-7364-4-4-263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Vliet J, Crofts LA, Quinlan KG, Czolij R, Perkins AC, Crossley M. Human KLF17 is a new member of the Sp/KLF family of transcription factors. Genomics. 2006;87:474–482. doi: 10.1016/j.ygeno.2005.12.011. [DOI] [PubMed] [Google Scholar]
- Waters BL, Panagopoulos I, Allen EF. Genetic characterization of angiomatoid fibrous histiocytoma identifies fusion of the FUS and ATF-1 genes induced by a chromosomal translocation involving bands 12q13 and 16p11. Cancer Genet Cytogenet. 2000;121:109–116. doi: 10.1016/s0165-4608(00)00237-5. [DOI] [PubMed] [Google Scholar]
- Yan W, Burns KH, Ma L, Matzuk MM. Identification of Zfp393, a germ cell-specific gene encoding a novel zinc finger protein. Mech Dev. 2002;118:233–239. doi: 10.1016/s0925-4773(02)00258-7. [DOI] [PubMed] [Google Scholar]
- Ye WC, Gao L, Huang J, Fang XM, Xie G. Suppressed Kruppellike factor 17 expression induces tumor proliferation, metastasis and a poor prognosis in papillary thyroid carcinoma. Mol Med Rep. 2014;10:2087–2092. doi: 10.3892/mmr.2014.2429. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.