Abstract
Circular RNAs (circRNAs) have re-emerged as an interesting RNA species. Here, by deep RNA profiling in different mouse tissues, we observed that circRNAs are significantly enriched in brain.and a disproportionate fraction of them is derived from host genes that code for synaptic proteins. Moreover, based on separate profiling of the RNAs localized in neuronal cell bodies and neuropil, on average, circRNAs are more enriched in the neuropil than their host gene mRNA isoforms. Using high resolution in situ hybridization we, for the first time, visualized circRNA punctae in the dendrites of neurons. Consistent with the idea that circRNAs might regulate synaptic function, during development, many circRNAs change their abundance abruptly at a time corresponding to synaptogenesis. In addition, following a homeostatic downscaling of neuronal activity many circRNAs exhibit significant up or down-regulation. Together, our data indicate that brain circRNAs are positioned to respond to and regulate synaptic function.
Introduction
Cellular RNAs can exist in both linear and circular forms. Whereas a linear RNA possesses distinct 5′ and 3′ termini reflecting the start and end sites of transcription, the latter is formed by the covalent joining the 5′ end of one exon with the 3′ end of another, so-called “head-to-tail splicing” resulting in a circular transcript. Recently, the deep sequencing of ribosome RNA (rRNA)-depleted RNA, combined with computational tools, has led to the identification of thousands of new circRNAs in organisms ranging from Archaea to human 1-3. For many years, no clear function was attributed to any of the circRNAs, but it was recently demonstrated that two previously identified circRNAs can serve as miRNA “sponges” 4,5, sequestering miRNAs and preventing their interactions with target mRNAs. Although this observation offers one model for circRNA function, circRNAs represent a heterogeneous group of transcripts that likely also affect cellular function via as yet undiscovered and likely diverse mechanisms. In addition to miRNA regulation, it has been proposed, for example, that circRNAs could sequester RNA-binding proteins (RBPs) and thereby could regulate the intracellular transport of associated RBPs or RNAs 6,7.
The regulation of cellular function and protein translation by RNA-mediated mechanisms is exploited in many polarized cells to functionalize cellular compartments. This is particularly evident in neurons, where the complex morphology and distal location of synapses mandate a high degree of local regulation 8. Indeed, localized protein-synthesis has been observed in both dendrites and growing axons, owing to the localization of translational machinery and over 2000 mRNAs in each compartment 9,10. In recent years, other types of RNA species and RNA-based regulation have been identified in neurons including miRNAs and lncRNAs 11,12. Here we examined the expression pattern of circRNAs and find that circRNAs are enriched in the brain, relative to other tissues. We discovered that a disproportionate fraction of circRNAs is derived from host genes that code for synaptic proteins. Using PacBio sequencing of RT-PCR products for a subset of circRNA candidates, we identified rolling circle sequences indicating, for the first time, a true circular structure. At the same time, we determined the full-length sequences of these circRNAs and demonstrate the alternative usage of internal exons from the circRNAs with the same head-to-tail junctions. Furthermore, based on the separate profiling of the RNAs localized in neuronal cell bodies and synaptic processes (axons and dendrites), we found that, on average, circRNAs are more enriched in synaptic processes than their linear isoforms. Using high resolution in situ hybridization we visualized a subset of circRNAs directly in the dendrites of neurons. Finally, we show that the abundance of several circRNAs changes at developmental stages that correspond to synapse formation and also following homeostatic plasticity.
Results
CircRNAs are enriched in brain
To systematically determine the tissue-specific expression pattern of mammalian circRNAs, we deep-sequenced rRNA-depleted total RNA samples from different mouse tissues, including brain, liver, lung, heart and testis (Figure 1A). From two biological replicate experiments, in each tissue, we obtained a minimum of 16 × 106 reads and a maximum of 21 × 106 reads with mappable reads ranging from 88.7 to 96.1%, depending on the tissue (Table 1). Reads that map directly to reference genome sequences or canonical exon-exon junctions can be derived from either linear mRNAs or circRNAs and therefore were used to estimate the expression of the total transcriptional output (hereafter referred to as TTO) of the corresponding gene loci. To specifically identify circRNAs we used the remaining reads that spanned the 5′ and 3′ splicing sites of exon(s) of individual genes, but in reverse order (head-to-tail junction reads, see Figure 1A and Methods). From the five tissues, we detected a total of 13011 unique circRNAs.
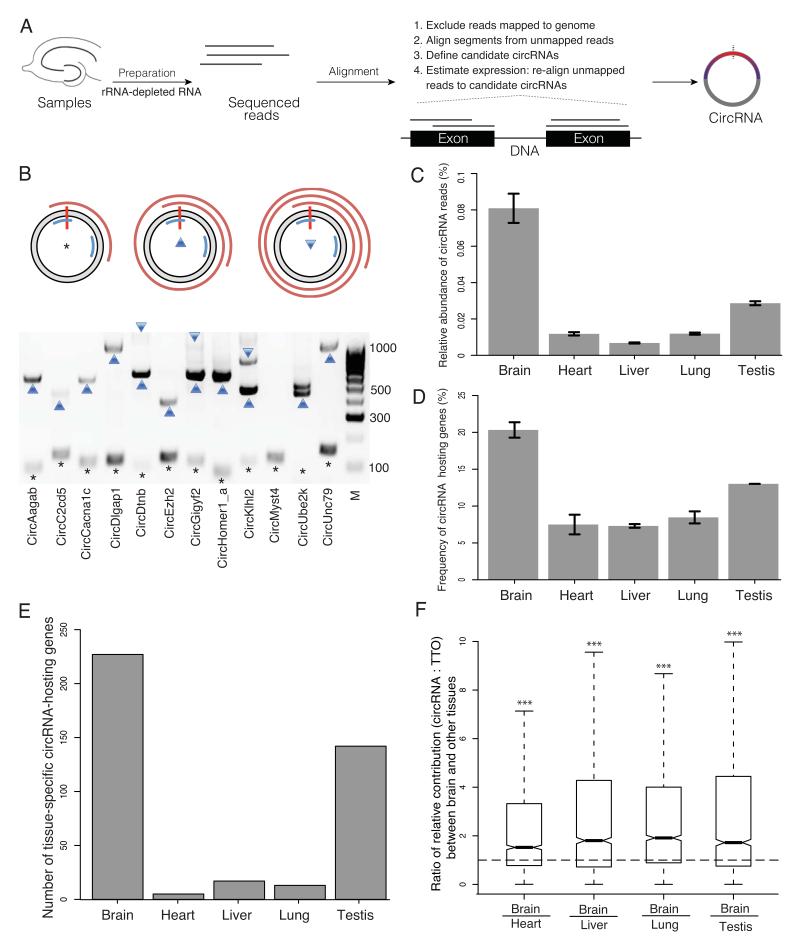
Figure 1. Profiling of circRNAs across tissues reveals enrichment in brain.
A. Experiment and analysis pipeline. B. The rolling cycle cDNA products from circRNAs. The grey ring represents a circRNA with the red vertical bar marks the head-to-tail junction. Two blue arcs mark the PCR primers. The red spirals on the outside represent PCR products that were deep sequenced by PacBio technology. The asterisk, upward triangle and downward triangle symbols on the gel image denote the 0-cycle, 1-cycle and 2-cycle RT products identified by PacBio sequencing, respectively. Eleven out of 12 circRNAs tested generated rolling cycle products (the exception was circMyst4). C. The percentage of circular junction reads from all the reads mapped on the genome is shown for different tissues, with the highest value (0.75-0.87%) in brain, followed by testis (0.28-0.29%). D. The percentage of genes that produced circRNAs from all the expressed genes is shown across different tissues, with the highest value (20-21%) in brain, followed by testis (13%). E. The number of circRNA host genes that are exclusively expressed in one tissue is shown across different tissues, with the highest value in brain, F. The relative contribution of circRNA to the total transcription output of the same gene locus i.e. the ratio between the abundance of each circRNA and the total transcriptional output (TTO) of the hosting gene loci (measured in TPM, transcripts per million) is significantly higher in brain compared to all other tissues, The ratios of the relative contribution between brain and other four tissues are significantly larger than one (two-sided one-sample t-test, *** p < 2.2E-16). C.-F. Tissues were obtained from two animals.
Table 1.
Summary of RNA-seq results for five different mouse tissues.
| Brain | Heart | Liver | Lung | Testis | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Replicate1 | Replicate2 | Replicate1 | Replicate2 | Replicate1 | Replicate2 | Replicate1 | Replicate2 | Replicate1 | Replicate2 | |
| Total No. reads | 19794174 | 19164999 | 16507635 | 19876852 | 21001677 | 19056514 | 20390058 | 18406517 | 19940919 | 20222389 |
| No. reads mapped on genome | 18765595 | 18283420 | 14636897 | 19049052 | 20195791 | 18322339 | 19595737 | 17282775 | 18802286 | 19198630 |
| No. reads mapped on intronic regions | 6884089 | 6971714 | 3028058 | 4629968 | 4184807 | 3926699 | 9071821 | 7834607 | 4611925 | 5042735 |
| No. reads mapped on intergenic regions | 2125517 | 1862307 | 2353147 | 1381043 | 1196394 | 1141757 | 1854519 | 2078503 | 3909966 | 3954450 |
| No. reads mapped on protein coding genes | 8290663 | 8085776 | 7775596 | 11156730 | 13316937 | 12017101 | 7516840 | 6480473 | 9450865 | 9429187 |
| No. protein coding genes expressed (TPM > 5) | 11781 | 11763 | 10044 | 9824 | 8261 | 8248 | 11987 | 11882 | 10665 | 10684 |
| No. circular junction reads | 16573 | 14068 | 1646 | 2393 | 1442 | 1231 | 2453 | 2024 | 5606 | 6053 |
| No. circRNA species | 6186 | 5664 | 989 | 1315 | 912 | 816 | 1556 | 1320 | 2943 | 3093 |
| No. circRNA-hosting protein coding genes | 2569 | 2386 | 707 | 869 | 676 | 637 | 1116 | 955 | 1480 | 1482 |
We validated the authenticity of identified circRNAs by three independent methods. First, as circRNAs usually do not possess a poly(A) tail, their representation should be depleted in a poly(A)-enriched sequencing library. As shown in Figure S1A, compared with rRNA-depleted total RNA sequencing, poly(A) RNA sequencing produced a much lower number of sequencing reads derived from our circRNA population. Second, compared with linear RNAs, circRNAs are endowed with a strong resistance to exonuclease RNAse R. We therefore quantified the RNAse R resistance of 20 candidate circRNAs and all of them showed > 5 times higher stability than the linear transcripts following RNase R treatment (Figure S1B). Third, we deep sequenced the cDNA products derived from 12 candidate circRNAs. For 11 of them, we observed the cDNA reads corresponding to the rolling cycle RT products (Figure 1B), which could not be detected in the cDNAs from any linear forms. This serves as direct evidence for the circular nature of the circRNA structure, and to our knowledge, is the first time that the full sequences of the circRNAs have been identified. Interestingly, for two circRNAs (circDtnb and circEzh2), in addition to the ‘canonical′ forms that encompass all of the annotated exons between the two involved in the back-splicing, we also observed additional circular isoforms that consisted of the same junction sequences, but with one internal exon skipped (circDtnb) or one unannotated exon inserted (circEzh2) (Table S1, S2).
Although circRNAs were observed in all the tissues we examined, their abundance was clearly highest in brain (Figure 1C, see also Figure S1C), where twenty percent of the protein-coding genes produce circRNAs (Figure 1D; Table 1). Two factors contributed to the higher abundance of circRNAs in brain: i) many host genes that produce circRNAs are expressed exclusively in brain (Figure 1E, see also Figure S1C). ii) on average, when a host gene is expressed in brain as well as other tissue(s), the relative contribution of circRNAs (defined as the ratio of TPM between a circRNA and the TTO of its host gene locus, where TPM is a measure of relative transcript abundance defined as Transcripts Per Million) is significantly higher in brain compared to all other tissues (Figure 1F, see also Figure S1C). We also reanalyzed published RNA-seq datasets from different rat tissues of rat13 and observed a similar enrichment of circRNAs in brain (Figure S1D).Together, our data and analyses demonstrate that circRNAs, as a group, are enriched in brain.
circRNAs: synaptic gene origin and dendritic localization
Is the likelihood that a given linear transcript will be spliced to produce a circRNA related to the host gene function? To address this, we conducted a Gene Ontology analysis of the genes that give rise to brain-expressed circRNAs (see methods). Interestingly, several functional groups related to synaptic function such as synapse, synapse part, presynaptic active zone, presynaptic membrane and postsynaptic density were significantly enriched (Figure 2A). This enrichment was present regardless of the expression level of the host genes (Figure S2A).
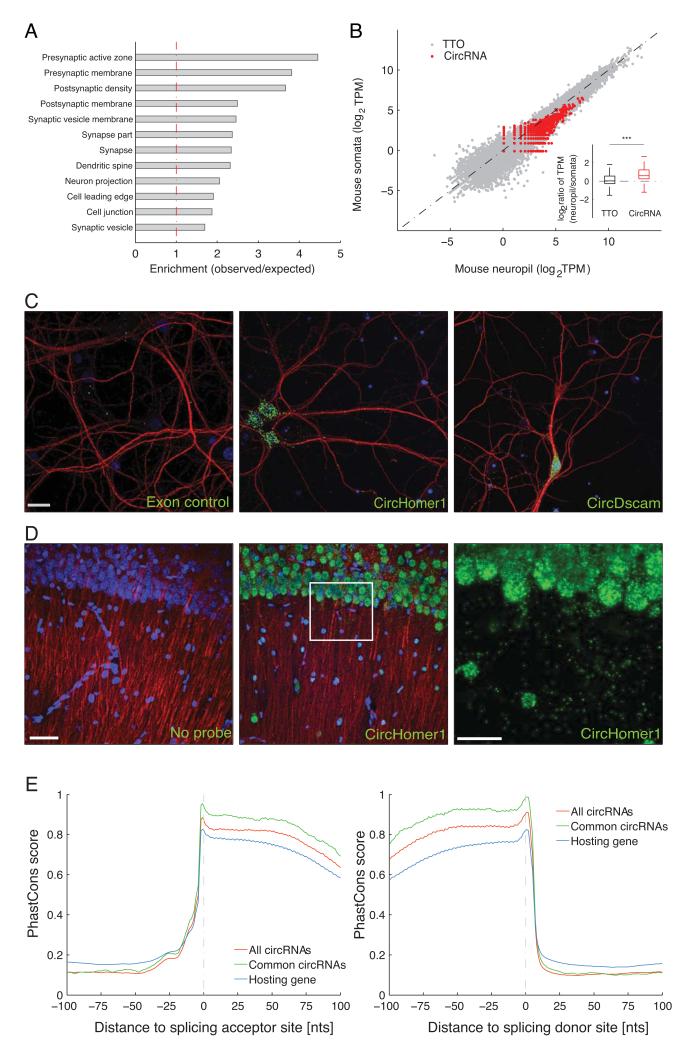
Figure 2. Brain expressed circRNAs derived from genes coding for synaptic proteins and are enriched in synaptic tissues.
A. Gene Ontology enrichment analysis of the genes producing brain expressed circRNAs. Functional groups related to synaptic function were overrepresented in the genes producing brain circRNAs. B. The abundance of circRNA and total transcriptional output of protein-coding gene loci (measured in TPM) were compared between neuropil (X-axis) and the somatic layer of the hippocampus (Y axis) in mouse. Each red dot represents one circRNA, and each dark dot represents one protein-coding gene locus. Inset shows that the abundance of circRNAs, but not total transcriptional output, is significantly higher in the synaptosome and neuropil fractions (two-sided, unpaired Student’s T-test, *** p < 2.2E-16). C-D. High resolution in situ hybridization experiments in cultured hippocampal neurons (C) or hippocampal slices (D) using probe sets designed to detect the indicated circRNA (green). In each case, many circRNA-positive particles are apparent in the cell bodies (nuclei stained with DAPI, blue), but also in the dendritic processes, detected using an anti-MAP2 antibody (red). A control (exon) probe designed to detect non-contiguous regions of two exons that could not form head-to-tail junction (see Methods) yielded just a few background particles. Scale bars = 20, 50 and 75 microns, in C and D and D-enlarged, respectively. For cells, one representative image from 3 - 43 images is shown, for slices one image from 4 images is shown. E. The exonic sequences around the splicing sites (Left: splicing acceptor; right splicing donor) involved in the formation of mouse circRNA head-to-tail junctions (red) are more conserved than those from the same gene locus, but not involved (blue). Y-axis denotes the average PhastCons score. X-axis marks the distance to the splicing site (negative and positive values means upstream and downstream, respectively). Importantly, the exonic sequences around the circRNA junctions common in mouse and rat (purple) are even more conserved, almost reaching the maximum PhastCons score.
Given the enrichment of host genes with synapse related functions, we next examined whether the circRNAs are enriched in synaptic tissue. To address this, we prepared synaptosomes, a biochemically purified preparation that is enriched in synapses 14,15 or microdissected the synaptic neuropil from the hippocampus, a brain structure that exhibits robust synaptic plasticity and is important for learning and memory 16. We then compared the abundance of circRNAs in these compartments (synaptosomes or neuropil) to that observed in a whole hippocampal homogenate or a microdissected layer comprising primarily hippocampal neuronal somata. We found that most circRNAs are indeed enriched in either or both of the two synaptic fractions examined (Figure 2B and Figure S2B) and the overlap between the two synaptic fractions was statistically significant (Fig S2D; p-value < 2.2E-16, Fisher Exact Test). The same pattern of results was observed when the tissue was obtained from rat (Figure S2C) and there was substantial overlap between the circRNAs identified in mouse and rat (Figure S2E).
In order to validate the enrichment of circRNAs in the biochemically or manually dissected synaptic compartments, we directly visualized the circRNAs in cultured hippocampal neurons using high-resolution in situ hybridization 9 optimized for the detection of circRNAs (see Methods, Figure S3A-D). Using an in situ probe set designed for the detection of circHomer1_a, a circRNA derived from the synaptic scaffolding molecule Homer1 transcript 17,we detected circHomer1_a particles in the cell body of neurons and also in the dendrites, visualized using an antibody (anti-MAP2) (Figure 2C). In contrast, use of an exon control probe (using a sequence from two exons that do not form a circRNA) set resulted in a dramatically reduced signal with only one or two particles evident in the vicinity of the cell bodies and nuclei (Figure 2C, Figure S3B). Use of additional probes developed for the detection of circRNAs derived from synapse-related genes including circDscam, circKlhl2, circElavl3, circNlgn1, circGigyf2, circNbea and circRmst resulted in a similar pattern with abundant particles present in the cell body and distributed particles observed throughout the dendritic arbor (Figure 2C, Figure S3E-J). In hippocampal slices, in situ hybridization using a circHomer1_a probe revealed substantial expression of this circRNA in both somata and neuropil layers of CA1 hippocampal region (Figure 2D). In addition to the exon control, scrambled probe and no probe control experiments (Figure 2C, Figure S3C,D), we validated the specificity of our circRNA in situ hybridization by comparing the signal intensity of circRmst, circKlhl2 and circGigyf2 in brain, liver and lung (Figure S3, S4). Consistent with the RNA-seq data, the in situ hybridization data revealed only background levels of expression of circRmst and circKlhl2 in liver and lung while these circRNAs were clearly evident in hippocampal neurons. In contrast, circGigyf2 was expressed in all examined tissues as expected from RNA-seq data (Figure S4). To test if the circRNA localization can mimic that of its host transcript, we performed in situ hybridization of circRims1 and its host mRNA Rims1 in cultured hippocampal neurons. While signals for the circRNA and mRNA were apparent in both cell body and dendrites, they clearly did not co-localize (Figure S5). Given the anticipated diversity of circRNA populations, however, one must be open to counter-examples of co-localization of circRNA and mRNA when more cases are examined.
Absence of miRNA/RBP binding or translation into protein
Recent studies of two individual circRNAs suggested that they function as miRNA “sponges”, sequestering miRNAs 4,5. Using a bioinformatics approach, we estimated the potential of the brain circRNA population identified here to serve as miRNA sponges, and concluded that as a general class, the brain circRNAs do not exhibit a greater capacity to serve as miRNA sponges than linear mRNAs (Figure S6A), consistent with recent analyses from other groups 18.
We also examined the possibility that brain circRNAs might function to bind or sequester RNA-binding-proteins (RBPs). For this purpose, we predicted the binding sites of 38 RBPs based on their binding sequence motifs deposited in the database RBPDB (Methods). As shown in Figure S6B, circRNAs are of lower RBP binding density, when compared to either the coding sequence or the 3′UTR of protein-coding genes. This trend is consistently observed for circRNAs of different abundances (Figure S6B). Thus, these data indicate, based on nucleotide sequence alone, that circRNAs as a group are no more likely to bind to RBPs than linear mRNAs.
As we observed that neuronal circRNAs are mostly composed of protein-coding exons, we investigated their potential to be translated into peptides. Using a large mass spectrometry (MS) dataset obtained from hippocampal neurons we searched for peptides predicted by circular junctions but were unsuccessful. The inability to detect a circRNA-derived peptide, however, could be due to the well-known low detection sensitivity of MS-based shotgun proteomics approach. Therefore, we further studied the association of circRNAs with ribosomes. First, we performed ribosome profiling 19 on rat brain. Similar to what was recently reported for circRNAs from a human cell line 18, in our rat brain samples, we did not detect a single ribosome-protected fragment (RPF) that mapped to a circRNA head-to-tail junction and thus could serve as evidence for circRNA translation. This negative finding could be due to the short read length of RPFs and more importantly, the ribosome might only associate with the sequences outside the junction. To circumvent this limitation, we performed polysome profiling on mouse brain. As shown in Figure S6C and D, in contrast to mRNAs, circRNAs are enriched in the nonribosomal RNA fraction and strongly depleted in the ribosome/polysome bound fractions. Together, these results demonstrate that circRNAs, as a group, are unlikely to be translated into peptides.
Conservation of circRNA para-junctional sequences
As functionally important elements are often evolutionary conserved, we examined the sequence conservation around the mouse circRNA junctions. As shown in Figure 2E, compared to splicing sites from the same hosting genes that are not involved in forming head-to-tail junctions, the exonic sequences around head-to-tail junctions are more conserved. Moreover, those sequences around common head-to-tail junctions detected in both mouse and rat are even more conserved, almost reaching the maximum PhastCons score. The observation that exonic sequences around circRNA junctions are of ultra evolutionary conservation strongly suggests their potential functional relevance.
Expression of circRNAs in brain during development
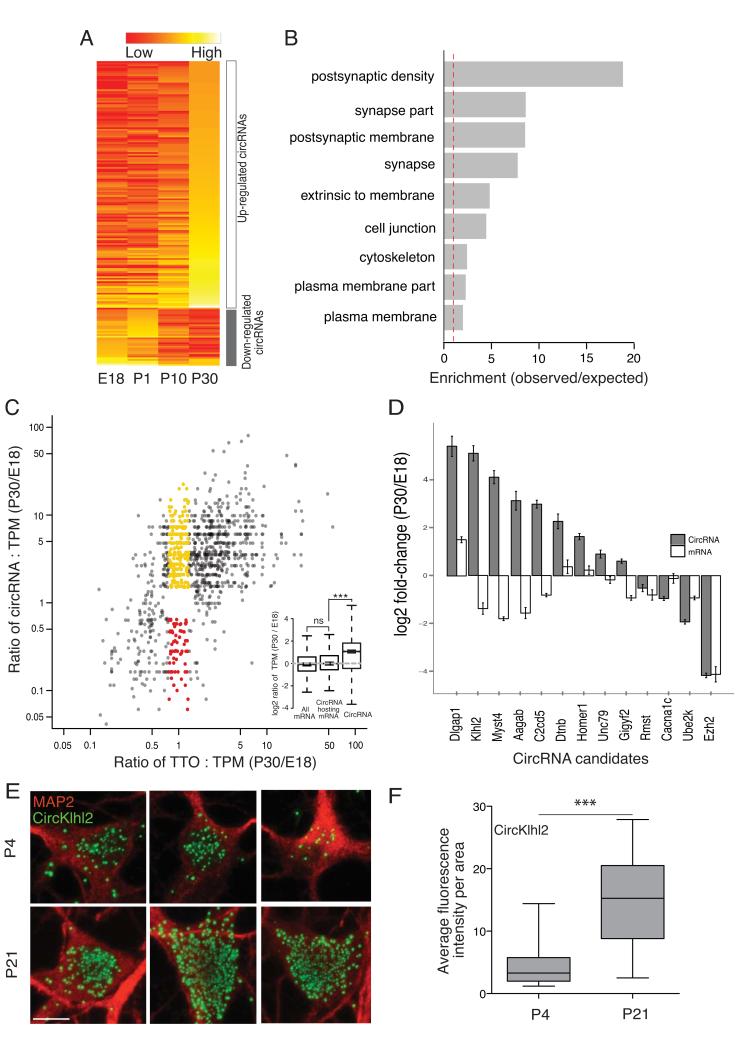
To determine whether the expression of circRNAs is developmentally regulated in brain we profiled the circRNA population in the hippocampus over several stages: embryonic (E18), early postnatal (P1), postnatal at the beginning of synapse formation (P10) and late postnatal hippocampus following the establishment of mature neural circuits (P30) (Figure S7). As shown in Figure 3A, there was a clear shift in the circRNA expression pattern associated with the onset of synaptogenesis at P10. Interestingly, the circRNAs that were consistently up-regulated during hippocampal development were produced from the gene loci that also code for proteins enriched with synapse-related functions (Figure 3B). In contrast, no enrichment of any functional categories could be observed for the gene loci showing the opposite (down-regulated) circRNA dynamic expression pattern. We next examined the relationship between the expression of a circRNA and its linear host comparing the earliest (E18) and latest (P30) developmental stages. We found that many circRNAs change their expression independent of their host transcripts during synaptogenesis (Figure 3C). Thirteen circRNA/mRNA pairs with different expression patterns were validated using quantitative PCR (Figure 3D). Dlgap1, whose protein product is a core component of postsynaptic density (PSD), showed an over 20-fold increase in circRNA expression at P30 when compared with E18, whilst the mRNA expression increased only less than 4-fold. Genes such as Myst4, Klhl2, and Aagab dramatically increased their circRNA expression over development while their mRNA expression significantly decreased. In contrast, Cacna1c showed substantial decreases in circRNA expression along developmental stages, while the mRNA remained unchanged. Using high-resolution in situ hybridization in cultured hippocampal neurons, we further validated the developmental regulation of a circRNA that exhibited strong up-regulation during development circKlhl2 (Figure 3E). Analysis of the average fluorescence intensity at an early and late developmental stage (neurons cultured beginning at P1, days in vitro = 4 or 21) revealed a significant enhancement of the circKlhl2 expression levels (Figure 3F). Thus, taken together the data from high-throughput sequencing, quantitative PCR and in situ hybridization, indicate that the expression of circRNAs is developmentally regulated in neurons and that many circRNAs change their expression independent of their host linear transcripts.
Figure 3. Regulated expression of brain circRNAs during development.
A. Heatmap of circRNA expression across four different developmental stages showing the regulation of several circRNA clusters between P0 and P10- the time at which synapses typically form. The abundance of circRNAs across four developmental stages is depicted on a scale from red (low) to yellow (high). A developmentally down-regulated cluster consisting of 43 circRNAs exhibited an early peak expression at E18 or P0 and then declined over subsequent developmental time points. A developmentally up-regulated cluster, consisting of 181 circRNAs, exhibited increasing expression that peaked at P10 or P30. B. The significantly enriched GO terms (p-value < 0.05 in either cluster). The host genes with circRNAs that exhibited peak expression associated with the time of synapse formation were enriched for synaptic function whereas the other group (down-regulated) did not exhibit significant enrichment of any GO terms. C. Fold change of both circRNA abundance (Y-axis) and the total transcriptional output (TTO) of their gene loci (X-axis) between stage E18 and P30. Each dot represents one circRNA. Dots in red and yellow highlight circRNAs that changed significantly whilst the total transcriptional output of their host loci was not substantially altered. Inset shows that while most circRNA-hosting genes do not change much in abundance compared to all genes (two-sided unpaired Student-t test, ns for p = 0.09709), circRNAs are significantly up-regulated (two-sided unpaired Student’s-t test, *** p < 2.2E-16). Six or seven mice were pooled in each of two replicates of E18. Three or four mice were pooled in each of two replicates of P30. D. The expression change for both circRNA and mRNA was validated using quantitative PCR for 13 circRNAs including Homer1, Dlgap1, Rmst, Myst4 and Ezh2. Error bars represent standard deviation. E–F. Validation of circRNA expression changes over developmental stages using high resolution in situ hybridization for circKlhl2 (green) at two time points -days in culture 4 (n = 26) or 21 (n = 24). circKlhl2 expression was significantly up-regulated between these developmental stages (two-sided unpaired Student’s t-test with Welch’s correction, *** p < 0.0001). The outline of the neuronal somata was identified using an anti-MAP2 antibody (red). Scale bar = 10 microns.
Neuronal plasticity changes circRNA expression
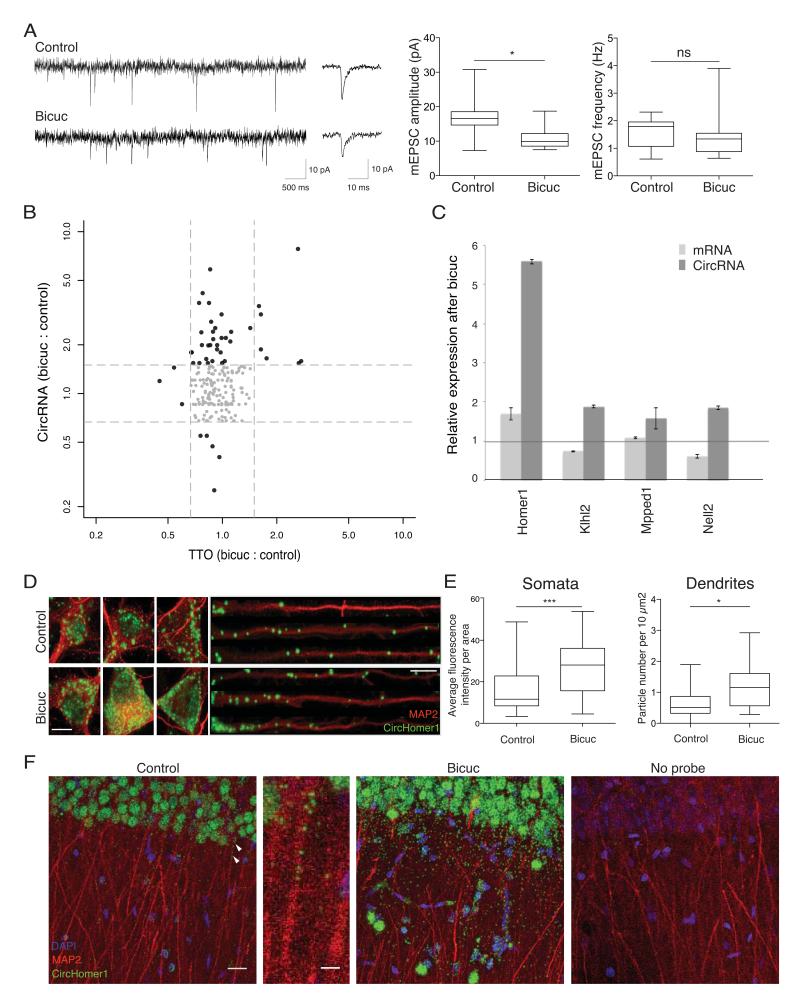
If circRNAs regulate synaptic function, then their expression levels might be modulated by alterations in neuronal activity and plasticity. We induced homeostatic synaptic plasticity in cultured hippocampal neurons by manipulating neuronal activity using bicuculline, an antagonist to the GABAA receptor. Treatment with bicuculline enhances excitatory neuronal network activity leading to a homeostatic decrease in the mini excitatory postsynaptic current (mEPSC) amplitude, without a change in mEPSC frequency (Figure 4A) 20. Following induction of homeostatic plasticity, the circRNA population exhibited dynamic behavior - the expression of 37 circRNAs was enhanced (Figure 4B) while five were reduced. In contrast, most of their linear host transcripts showed no significant change in expression level (Figure 4B). We validated the plasticity-induced changes in four circRNA candidates using quantitative PCR (Figure 4C). We also visualized directly the circRNA expression changes after homeostatic plasticity for additional candidates using in situ hybridization. CircHomer1_a was significantly up-regulated in primary hippocampal neurons (Figure 4D, E) and hippocampal slices (Figure 4F). Taken together, these data show that circRNA expression levels are regulated by neural plasticity, suggesting their importance in regulating synaptic transmission and/or local translation.
Figure 4. Regulation of circRNAs by homeostatic plasticity.
A. Electrophysiology traces of miniature excitatory postsynaptic current (mEPSC) from control cultures and cultures treated with bicuculline for 12 hrs. Representative recordings (left), the average mEPSC waveform and quantification of mEPSC amplitudes and frequency (right). B. Expression changes of circRNA (Y-axis) and TTO (X-axis) after homeostatic plasticity. Each dot represented one circRNA. Grey dots represent circRNAs in which expression remained largely the same (less than 30% change) for both circRNA and TTO. C. Quantitative PCR validation of expression change for both circRNAs and their cognate host mRNAs following plasticity (error bars represent standard deviation). D. Validation of circRNA expression changes following homeostatic plasticity using high resolution in situ hybridization for circHomer1_a in control or bicuculine-treated neurons. Dendrites were identified using an anti-MAP2 antibody. Scale bar = 10 microns. E. circHomer1_a expression was significantly up-regulated in both the neuronal somata (n for control = 34; n for bicuculline = 43) and dendrites (n for control =12; n for bicuculline=13) following homeostatic plasticity. Primary hippocampal cell cultures were prepared from 10-20 animals. F. circHomer1_a expression was significantly upregulated in hippocampal slices following homeostatic plasticity. From left to right, control slice, zoom of a region of the control slice (indicated by arrows) showing presence of several circHomer1_a in a continuous stretch of dendrites, bicuculline-treated slices, and no probe control. Scale bars = 20 microns and 5 microns, from left to right. Bicuculline-treatment resulted in a significant upregulation of circHomer_1 in both stratum pyramidale (somatic layer) and stratum radiatum (neuropil layer), (n = 3 slices each for control and bicuculline treatment, p <0.0005 for somata and p<0.0011 for neuropil).
Discussion
Eukaryotic circRNAs are a class of low-abundance but biochemically stable cellular RNAs that possess neither a 5′ nor a 3′ end. The property of circularity has contributed to their relative anonymity (until recently), as most of the transcriptome-wide studies begin with the purification of a poly(A)+ RNA fraction. Like other recent studies 2,5,18,21 we sequenced and analyzed rRNA-depleted samples that allow one to analyze circRNAs and their linear host transcripts in a quantitative manner. While in previous studies circRNA identification relied on available genome annotation2,5, we set up a computational pipeline that does not rely on exon annotations or assume canonical splice sites, and can thus identify circRNAs derived from previously unannotated exons and transcripts. This allowed us to identify the circRNAs in rat, which, to date, has a relatively incomplete transcriptome annotation.
We validated our findings with several independent approaches. Most importantly, using PacBio deep sequencing of the cDNA products derived from candidate circRNAs, we observed reads that correspond to the rolling cycle RT products, which i) serves as direct evidence for the circular nature of the RNA and ii) provides the full length sequences of the circRNAs. Interestingly, based on the sequences, we, for the first time, identified circRNA isoforms with the same head-to-tail junctions, but different internal exon composition. As observed in this study and other previous reports 2,5,18,21, multiple circRNAs with different junctions could form from the same gene loci. The identification of circRNA isoforms with the same junction, but different internal exons, adds another layer to circRNA diversity. The fact that the internal exon composition cannot be simply predicted using junction exons necessitates the experimental determination of full-length sequence of a circRNA before any further functional investigation.
We found that circRNAs are most abundant in brain, consistent with a recent report analyzing circRNAs in fly heads 22. Furthermore, the brain-expressed circRNAs are derived from gene loci that also code for proteins enriched for synapse-related functions. To examine whether circRNAs themselves might be associated with the function of synapses, we studied both synaptosomes and the microdissected neuropil from hippocampal slices and found in these samples a relative enrichment of circRNAs, compared to their host linear transcripts. In addition, for the first time, we visualized individual circRNA species directly both in vitro and in vivo (in hippocampal slices). We observed the localization of circRNAs both in the cell body and the dendrites of neurons, similar to what has been observed for both mRNAs 9 and other regulatory RNAs like miRNAs 23.
These findings are interesting in the light of recent debate in the field concerning the question of circRNA function. Thus far, clear functions have been established for two circRNAs 4,5. Recently, Guo et al suggested that the majority of circRNAs are mere side-products of pre-mRNA splicing 18. Other studies suggested that circRNAs may have biological functions based on the observation that even lowly expressed circRNAs are regulated 24. As conservation in evolution often implies functionality, we analyzed the conservation of mouse circRNA sequences across vertebrates. Compared to splicing sites not involved in circRNA biogenesis, the exonic sequences around the circRNA head-to-tail junctions showed higher conservation (Figure 2G). Moreover, we analyzed the overlap of circRNAs detected in rat and mouse, and found 23.6% of the circRNAs identified in mouse neuropil were also expressed in rat neuropil (Figure S2E). This observation is in line with a recent study in which 20% of mouse circRNAs were detected in human cell lines 18 but higher than in another study in which only 4% of the mouse circRNAs were identified in human samples 5. The difference might be explained by different sampling depths, as most identified circRNAs are expressed at low levels and thus might ‘stochastically′ escape detection. Indeed as shown in Figure S2F, the circRNAs detected in both mouse and rat samples are clearly of much higher abundance than those detected in only one sample. This further suggests that circRNAs are conserved, and our observation of a 23% overlap between mouse and rat circRNAs may prove to be an underestimate. More importantly, the exonic sequences around head-to-tail junctions detected in both mouse and rat are extremely conserved, strongly suggesting their potential functional relevance (Figure 2E).
Here we present evidence for the developmental regulation of circRNAs in neurons. Importantly, there are many circRNAs whose abundance changes independent of the host linear transcript, suggesting a circRNA-specific regulation of biogenesis and/or turnover (Fig. 3C,D) The development of the CNS/brain involves neuronal maturation, neurite outgrowth and synaptogenesis. Non-coding RNAs such as miRNAs and lncRNAs have emerged as key players in regulating these developmental processes 25,26 Recently, for example, the lncRNA RMST was identified as a factor important for neuronal differentiation as well as a co-regulator of SOX2, a mediator of neural stem cell fate 27. We identified a set of circRNAs that are differentially expressed in the mouse hippocampus at different developmental stages (E18 to P30). Intriguingly, a circRNA that was significantly down-regulated at later stages arises from the host gene coding for Rmst, thus supporting a potential function of circRNAs in brain development. In contrast, the expression of circKlhl2 was increased at P30 (P21) compared to E18 (P4) indicating a putative role of this circRNA during synaptogenesis or when mature synapses have formed. In summary, we showed a shift in the expression pattern for a large set of circRNAs associated with the onset of synaptogenesis, indicating role of circRNAs in hippocampal development.
The brain is the most plastic organ and its circuits undergo tight regulation and modification throughout the entire lifespan of animals. Both the stability and flexibility of neuronal networks is central to all behavior, including learning and memory. Experience-dependent alterations in the connectivity of neural networks can result in plasticity of intrinsic excitability and synaptic strength. We induced homeostatic plasticity by treating cultured hippocampal neurons with the GABAA receptor antagonist bicuculline and observed a dynamic change in circRNA expression. Interestingly, a circRNA (circHomer1_a) derived from the Homer1 linear transcript was the most significantly up-regulated circRNA after plasticity. The Homer1 protein plays a major role in the organization of the postsynaptic density. It is known that neuronal activity causes an increase in expression of an immediate early gene variant of Homer1, Homer1a, whereas the expression of Homer1b/c is relatively unchanged 28. Such imbalanced regulation of different Homer1 isoforms contributes to homeostatic downscaling such that the truncated protein encoded by Homer1a interferes with the native interaction between mGluR and Homer1b/c-encoded functional scaffold protein 29. Interestingly, the head-to-tail junction formed in circHomer1_a uses the splicing donor of intron 5, which is only required to splice the Homer1b/c transcripts, but not Homer1a. Therefore, the biogenesis of circHomer1_a could compete with that of Homer1b/c mRNA. Upregulation of circHomer1_a could then prevent the potential overexpression of Homer1b/c, which would otherwise be detrimental to homeostatic synaptic downscaling. It is therefore conceivable in this case that transcriptional upregulation, the predominant usage of an upstream polyadenylation signal and circRNA biogenesis work together to achieve the same goal, i.e., to reduce the interaction of surface mGluR and scaffold Homer protein. However, it should be noted that only a few circRNAs showed co-regulation with their host genes; more common was the observation that circRNAs exhibited changes independent of the cognate mRNA following plasticity.
Finally, as a heterogeneous group of transcripts, it is very likely that circRNAs affect cellular and neuronal function via a diverse set of mechanisms. The different datasets accumulated in this study should serve as a rich resource for future functional research, where genetic perturbation of specific circRNAs followed by careful phenotypic examination in different in vitro and/or in vivo neuronal systems will be needed to shed more light on circRNA function in the nervous system and specifically to address their role in learning and memory.
Materials and Methods
Tissue collection and hippocampal microdissections
Wild type C57B6 or C57BL/6J-Etv1-ARRAy TS88 (male and female) mice and Sprague Dawley (male) rats were housed in standard cages and fed standard lab chow and water ad libitum. All animal work was performed following regulations of German animal welfare law. For the developmental studies, hippocampi were dissected from mice at the age of E18, P1, P10 and P30. For tissue profiling two wild type C57B6 male mice at the age of 20 weeks were used to dissect the brain, liver, lung, heart and testes. To profile distinct sub-neuronal compartments (somata and neuropil), C57BL/6J-Etv1-ARRAy TS88 male and female mouse and rat male hippocampal slices (500 μm) were prepared from four to five week old animals and microdissected as described previously for rat 9. All tissues were collected in RNA later (Ambion) and subsequently lysed in Trizol (Invitrogen) to extract RNA following the manufacturer′s instructions.
Preparation of synaptosomes
Synaptosomes were purified from 20 hippocampi of adult mice (4 weeks old) as previously described 15. The hippocampi were homogenized in ice-cold sucrose buffer (320 mM sucrose, 5 mM HEPES, pH 7.4) with a 15-ml Teflon-glass tissue grinder and homogenized with eight even strokes. The homogenate was subjected to three differential centrifugations (1,000 g for 10 min, supernatant was further subjected to centrifugation at 12,000 g for 10 min followed by 13,000 g for 10 min) before applying one gradient centrifugation (3%, 10%, 15% and 23% PercollPlus (GE He al thecare) in sucrose buffer at 31,000 g for 5 min). The latter resulted in separation into five different fractions. The fraction at the interface of the 15% and 23% Percoll contained the most pure synaptosomes, as verified by Western blot (enrichment of AMPA receptor subunit, GluR4, substantial depletion of glial fibrillary marker protein GFAP), and was therefore used for all experiments performed in this study.
Polysome profiling
Mouse brains were collected, snap frozen in liquid nitrogen and stored in −80 °C. Frozen mouse whole brain was pulverized under liquid nitrogen and the powder lysed in 1 ml of lysis buffer (10 mM HEPES pH 7.4,150 mM KCl, 10 mM MgCl2, 1 % NP40, 0.5 mM DTT,100 μg /ml cycloheximide). After lysing the cells by passing them several times through a 26-gauge needle, the nuclei and membrane debris were removed by centrifugation (13000 rpm, 10 min, at 4°C). The supernatant was layered onto a 10 mL linear sucrose gradient (10%-50% [w/v], supplemented with 10 mM HEPES pH 7.4, 150 mM KCl,10 mM MgCl2,0.5 mM DTT,100 μg/ml cycloheximide), and centrifuged in a SW41Ti rotor (Beckman) for 120 min at 36,000 rpm at 4 °C. Fractions were collected from Gradient Fractionator (Biocomp) and digested with 200 μg proteinase K in 1% SDS and for 30 min at 42°C. RNAs were recovered by extraction with an equal volume of acid phenol-chloroform (pH 4.5), followed by ethanol precipitation, then converted to double stranded library using TruSeq Stranded Total RNA Sample Preparation kit (Illumina) and sequenced using Illumina HiSeq2000 platform.
Pharmacological treatments
Rat primary hippocampal neurons were prepared by pooling hippocampi from 10 to 20 male and female pups and grown in 6 cm petri dishes at a density of 400K. At DIV28, neurons were treated with bicuculline (Tocris, 40 μM) or a water control for 12 hrs. Subsequently, cells were scraped using Trizol (Invitrogen) followed by RNA extraction. Rat (4 week-old, male, Sprague Dawley) hippocampal slices (500 μm) were treated with bicuculline (Tocris, 40 μM) or water control for 4 hrs on filter paper in a recovery chamber. Subsequently, slices were fixed and re-sectioned (30 μm) using a vibratome.
RNA-seq
Ribosomal RNA was depleted from total RNA from different rat and mouse samples using the RiboZero Gold kit (Epicentre Bio-technologies). Poly(A) RNA was enriched using oligo-dT beads (Invitrogen). RNA-seq library was then generated from either rRNA-depleted or poly(A) enriched RNA using Illumina stranded RNA Sample Prep kit according to the manufacturer′s instruction, and was subsequently sequenced for 150 nt from single end on an Illumina HiSeq 2000.
Total Transcriptional Output (TTO) estimation
After removing the Illumina sequencing adaptor at 3′ end, the reads were aligned to the mouse (mm9) or rat (rn5.0) genome reference sequences using Tophat2, allowing up to six mismatches. Cufflinks (v2.21) 30 was then used to estimate the total transcriptional output based on Ensembl gene annotation for mouse (mm9, version67) or rat (rno5.0, version72). Since most circRNAs are derived from protein coding genes and lincRNAs (Table S5), genes annotated as “protein-coding” or “lincRNA” were retained for further analysis. To compare gene expression between two samples, we converted the FPKM (Fragments per kilobase per million) to TPM (Transcripts per million) using the following formula: TPM = FPKM * 1,000,000 / (sum_of_FPKM) 31.
CircRNA identification and quantification
For each sample, the unmapped reads were aligned to the respective genome reference sequences by BWA 32 in local mode (with parameters: -mem -k16). Partial alignments of segments within a single read that mapped to i) regions on the same chromosome and no more than 1Mb away from each other ii) on the same strand iii) but in reverse order, were retained as candidates supporting head-to-tail junction. The strength of potential splicing sites supported by these candidate head-to-tail junction reads was then estimated using MaxEntScan 33. The exact junction site was determined by selecting the donor and accepter sites with the highest splicing strength score. Candidate circRNAs were reported if the head-to-tail junction was supported by at least two reads and the splicing score was greater than or equal to 10. To estimate the expression of circRNA, we re-aligned all the unmapped reads to the circRNA candidates. As for most of the circRNAs there is no direct evidence for their exact sequence, we filled in the sequence using existing exon annotation. Sequence at the 5′ end was concatenated to the 3′ end to form circular junctions. Reads that mapped to the junction (with an overhang of at least 5nt) were counted for each candidate. TPM (Transcripts Per Million) was calculated for each circRNA candidate, where the effective length was calculated as: (sequencing length – 2 * 6). The analysis pipeline with a detailed description is publicly available at (https://code.google.com/p/acfs/)
RNase R treatment & Quantitative PCR
Mouse or rat brain DNase1-treated total RNA (1ug) was incubated 15 min at 37 °C with or without 3 U/μg of RNaseR (Epicentre Bio-technologies). RNA was subsequently purified by phenol-chloroform extraction. Reverse transcription (RT) was performed using random hexamers and reverse transcriptase (SSIII, Invitrogen). Quantitative PCR (qPCR) was done using SYBR green master mix (Roche). For circRNA transcripts, one primer was designed to anneal at the circular junction whereas the other was within the circRNA transcript. For linear transcripts, both primers were designed to amplify the sequence that is not part of any circRNA derived from the same gene locus. All qPCR primers are listed in Supplementary Table S3.
PacBio sequencing of RT-PCR products
The RT-PCR products obtained from mouse brain and rat brain samples, as described in the previous section, were directly sequenced using PacBio RS system as previously described 34.
Processing of PacBio sequencing data
Circular consensus reads (CCS-reads) obtained from PacBio sequencing were aligned to custom database (consisting of sequences from both linear mRNAs and circRNAs) using Blast (parameters: -evalue 1E-10 -word_size 9). We reported the alignments with identity higher than 95% for both linear and rolling-cycle products using an in-house perl script.
Conservation analyses
The positions of rat circRNAs were converted to mouse (mm9) genome coordinates using the UCSC liftOver tool, then were intersected with mouse circRNA using BEDTools. To examine the evolutionary conservation of the sequences around mouse circRNA junctions, we downloaded PhastCons scores for alignment of 29 vertebrate genomes with mouse (mm9) from (http://hgdownload.soe.ucsc.edu/goldenPath/mm9/phastCons30way/vertebrate/). To rule out possible biases, we compared the sequences around the splicing sites involved against those not involved in the formation of head-to-tail junction from the same gene locus.
Gene Ontology enrichment analysis
We performed Gene Ontology enrichment analysis using DAVID (http://david.abcc.ncifcrf.gov) 35,36. The background gene set consists of all expressed genes (TTO > 0.01) and the test gene set consists of all expressed circRNA-hosting genes. In Figure S2A, we chose a background gene set consisting of the 1000, 2000, 3000, 5000, 10000 most highly expressed genes (top1k, top2k, top3k, top5k, top10k), and then tested for the enrichment of GO terms for the circRNA-hosting genes within the same gene set, respectively.
High-resolution in situ hybridization in primary cells and slices
Dissociated rat hippocampal neurons were prepared and maintained as described previously 37. Four week-old rats were perfused with 1x PBS and 4% paraformaldehyde solution, the lung and liver were dissected, sliced to 1 mm, and fixed for 3 h. Lung and liver cells were dissociated and plated to poly-L-lysine coated dishes as described 38. We performed in situ hybridization using the QuantiGene ViewRNA™ miRNA ISH Cell Assay for Fluorescence miRNA and RNA In Situ Hybridization (RNA FISH) with custom-made probes targeting the circRNA exon junctions or the cognate mRNA (see Table S4 for circRNA probe sequences). Cultured neurons (DIV 26-28 or DIV 4/21 for developmental studies) were fixed for 1 h at room temperature using a 4% paraformaldehyde solution (4% paraformaldehyde, 5.4% Glucose, 0.01 M sodium metaperiodate in lysine-phosphate buffer). The in situ hybridization was performed following the manufacturer′s protocol omitting the dehydration/rehydration step as well as the protease treatment. Dendrites were stained using an anti-Map2 antibody (Millipore AB5622, 1:1000 dilution). Liver cells were stained with anti-Albumin antibody (Abcam ab106582, 1:50 dilution) and lung cells with anti-Heme Oxygenase 1 antibody (Abcam ab13243, 1:200 dilution). For in situ hybridization in hippocampal slices, slices were dissected and fixed overnight in 4% PFA in PBS solution at 4 °C, gently shaking. Hippocampal slices were embedded in 4% low melt agarose and 30 μm sections were prepared using a vibratome (Leica VT 1200S). The in situ hybridization was performed as described above with a few modifications from the manufacturer′s protocol: the slices were post-fixed for 10-15 min at RT, washed with PBS and incubated for 15 min with the detergent solution. After completion of the in situ hybridization the slices were blocked for 1 hour at RT in 1% BSA/1%TritonX in PBS. Slices were incubated with the primary antibody anti-Map2 (Millipore AB5622, 1:1000 dilution) over night at 4°C to stain dendrites, washed three times with 1x PBS and incubated with the secondary antibody (Invtrogen, A11008, Alexa 488-goat anti-rabbit; 1:1000 dilution) and DAPI (1:1000 dilution) to visualize nuclei for 2 hours at RT.
Image acquisition and processing
Confocal microscopy was performed using a Zeiss LSM780 confocal laser fluorescence microscope system. Image analysis was done from Z-stacks of confocal image series of 10-30 confocal planes taken at 35-50 μm intervals using a 40x oil immersion objective. For analysis, conducted in a non-blind manner, cell bodies were circumscribed and dendrites straightened using the software ImageJ. To quantify the in situ signal in cell bodies, the average fluorescence intensity was measured per Z-stack and normalized to the area of the circumscribed cell body. In dendrites the particle abundance was determined using a custom MATLAB script, then the number of particles was normalized to the area of the straightened dendrite. The statistical significance of the in situ data was tested using either an unpaired t-test or Mann-Whitney U test according the normality of the distribution pre-tested using Lilliefors test.
Electrophysiology
Whole-cell patch-clamp recordings were made with an Axopatch 200B amplifier from cultured hippocampal neurons (DIV 28-29) bathed in HBS containing 119 mM NaCl, 5 mM KCl, 2 mM CaCl2, 2 mM MgCl2, 30 mM glucose, 10 mM HEPES [pH 7.4; ~310 mOsm] plus 1 μM TTX and 20 μM bicuculline. Whole-cell pipette internal solution contained 120 mM potassium gluconate, 20 mM KCl, 0.1 mM EGTA, 2 mM MgCl2, 2 mM adenosine triphosphate, 0.4 mM guanosine triphosphate, 10 mM HEPES (pH 7.2; ~300 mOsm) and the pipette resistances ranged from 4 to 6 MΩ. Bicuculline was added in conditioned media for 12 hrs and the media was replaced with the HBS 15 min prior to recording. Neurons were voltage clamped at -70 mV while the series resistance was left uncompensated during the recordings. mEPSCs were analyzed offline using Stimfit 39 software by employing a template-matching algorithm. Recordings were started 5 min after patching and the recording duration usually ranged from 5 to 10 min. Statistical differences between experimental conditions were determined by Mann-Whitney U test.
Potential miRNA binding sites
To quantify miRNA binding sites, exonic sequences within each circRNA were concatenated using Ensembl annotation, and the number of predicted miRNA binding sites (7mer-m8) 40 for all miRNA (deposited in miRBase version19) 41 was counted. As a control, the same procedure was performed on CDS and 3′UTR of the protein-coding genes.
Potential RNA-binding protein (RBP) binding sites
We predicted the RBP binding sites based on their sequencing motifs deposited in RBPDB 42. Predicted RBP binding sites on circRNAs were compared to those on CDS and 3′UTR of protein-coding genes.
Potential translatability of circRNAs
To estimate the translational capacity of circRNAs, we studied their association with ribosome complexes. We performed polysome profiling on mouse brain and ribosome footprinting on rat brain. Sequencing reads from four fractions of mouse brain (free, 60S, 80S and polysome) and ribosome-protected fragments (RPFs) of rat brain were aligned to circRNAs using BWA, and reads spanning the circular junctions were counted and converted to TPM as described above.
To investigate potential peptides arising from circRNA candidates, a liquid chromatography mass spectrometry sequencing was conducted on total lysate from 21 days old primary neurons without any pharmacological or electrophysiological treatment. The genomic representation of circRNA candidates was translated in 6 potential frames (3 frames per strand) and the position of the circRNA junction was recorded. This custom database was merged together with the rat protein RefSeq database and was used as a template for peptide matching with Mascot. A custom Perl script was used to identify peptides crossing the circular junction position – such peptides could only arise from circRNA translation.
Statistical analysis
For statistical analysis, two-sided one-sample t-test (Figure 1F), two-sided unpaired t-test with Welch’s correction (Figure 2B, 3C, 3F) and two-sided Mann-Whitney U test (Figure 4A, 4E) were performed. In Figure 1F, 2B, 3C, whiskers show extreme data points no more than 1.5 times the interquartile range; and in Figure 3F, 4A, 4E whiskers show minimum to maximum. The statistical significance of the in situ data was tested using either an unpaired t-test or Mann-Whitney U test since the normality of the distribution was pretested using Lilliefors test. No statistical methods were used to predetermine sample sizes, but our sample sizes are similar to those generally employed in the field. Data collection and analysis were not performed blind to the conditions of the experiments and no randomization of data was performed.
Supplementary Material
Acknowledgements
We thank Ina Bartnik, Nicole Fuerst and Anja Staab for the preparation of cultured hippocampal neurons and Mirjam Feldkamp and Claudia Langnick for their excellent technical assistance on sequencing. We thank Christoph Schanzenbaecher and Julian Langer for generating mass spectrometry data and for the bioinformatic evaluation of circRNA translation. Erin Schuman is funded by the Max Planck Society, an Advanced Investigator award from the European Research Council, DFG CRC 902: Molecular Principles of RNA-based Regulation; DFG CRC 1080: Molecular and Cellular Mechanisms of Neural Homeostasis; and the DFG Cluster of Excellence for Macromolecular Complexes, Goethe University, Frankfurt. As part of the Berlin Institute for Medical Systems Biology at the MDC, the research group of Wei Chen is funded by the Federal Ministry for Education and Research (BMBF) and the Senate of Berlin, Berlin, Germany (BIMSB 0315362A, 0315362C). Hongyu Liu, Tao Chen and Wei Sun are supported by the Chinese Scholarship Council (CSC).
Footnotes
Accession code. The sequencing data have been deposited at Gene Expression Omnibus (GEO) under accession number GSE61991.
The authors have no competing financial interest.
A supplementary methods checklist is available.
References
- 1.Danan M, Schwartz S, Edelheit S, Sorek R. Transcriptome-wide discovery of circular RNAs in Archaea. Nucleic Acids Res. 2012;40:3131–3142. doi: 10.1093/nar/gkr1009. doi:gkr1009 [pii] 10.1093/nar/gkr1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One. 2012;7:e30733. doi: 10.1371/journal.pone.0030733. doi:10.1371/journal.pone.0030733 PONE-D-11-22239 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang PL, et al. Circular RNA is expressed across the eukaryotic tree of life. PLoS One. 2014;9:e90859. doi: 10.1371/journal.pone.0090859. doi:10.1371/journal.pone.0090859 PONE-D-13-49149 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hansen TB, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. doi:nature11993 [pii] 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 5.Memczak S, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. doi:nature11928 [pii] 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 6.Hentze MW, Preiss T. Circular RNAs: splicing’s enigma variations. EMBO J. 2013;32:923–925. doi: 10.1038/emboj.2013.53. doi:emboj201353 [pii] 10.1038/emboj.2013.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32:453–461. doi: 10.1038/nbt.2890. doi:nbt.2890 [pii] 10.1038/nbt.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hanus C, Schuman EM. Proteostasis in complex dendrites. Nat Rev Neurosci. 2013;14:638–648. doi: 10.1038/nrn3546. doi:nrn3546 [pii] 10.1038/nrn3546. [DOI] [PubMed] [Google Scholar]
- 9.Cajigas IJ, et al. The local transcriptome in the synaptic neuropil revealed by deep sequencing and high-resolution imaging. Neuron. 2012;74:453–466. doi: 10.1016/j.neuron.2012.02.036. doi:S0896-6273(12)00286-3 [pii] 10.1016/j.neuron.2012.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zivraj KH, et al. Subcellular profiling reveals distinct and developmentally regulated repertoire of growth cone mRNAs. J Neurosci. 2010;30:15464–15478. doi: 10.1523/JNEUROSCI.1800-10.2010. doi:30/46/15464 [pii] 10.1523/JNEUROSCI.1800-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huntzinger E, Izaurralde E. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat Rev Genet. 2011;12:99–110. doi: 10.1038/nrg2936. doi:nrg2936 [pii] 10.1038/nrg2936. [DOI] [PubMed] [Google Scholar]
- 12.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. doi:10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu Y, et al. A rat RNA-Seq transcriptomic BodyMap across 11 organs and 4 developmental stages. Nature communications. 2014;5:3230. doi: 10.1038/ncomms4230. doi:10.1038/ncomms4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carlin RK, Grab DJ, Cohen RS, Siekevitz P. Isolation and characterization of postsynaptic densities from various brain regions: enrichment of different types of postsynaptic densities. J Cell Biol. 1980;86:831–845. doi: 10.1083/jcb.86.3.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dunkley PR, Jarvie PE, Robinson PJ. A rapid Percoll gradient procedure for preparation of synaptosomes. Nat Protoc. 2008;3:1718–1728. doi: 10.1038/nprot.2008.171. doi:nprot.2008.171 [pii] 10.1038/nprot.2008.171. [DOI] [PubMed] [Google Scholar]
- 16.Squire LR. Memory and the hippocampus: a synthesis from findings with rats, monkeys, and humans. Psychol Rev. 1992;99:195–231. doi: 10.1037/0033-295x.99.2.195. [DOI] [PubMed] [Google Scholar]
- 17.Brakeman PR, et al. Homer: a protein that selectively binds metabotropic glutamate receptors. Nature. 1997;386:284–288. doi: 10.1038/386284a0. doi:10.1038/386284a0. [DOI] [PubMed] [Google Scholar]
- 18.Guo JU, Agarwal V, Guo H, Bartel DP. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014;15:409. doi: 10.1186/s13059-014-0409-z. doi:10.1186/s13059-014-0409-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ingolia NT, Brar GA, Rouskin S, McGeachy AM, Weissman JS. The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nat Protoc. 2012;7:1534–1550. doi: 10.1038/nprot.2012.086. doi:nprot.2012.086 [pii] 10.1038/nprot.2012.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Turrigiano GG, Leslie KR, Desai NS, Rutherford LC, Nelson SB. Activity-dependent scaling of quantal amplitude in neocortical neurons. Nature. 1998;391:892–896. doi: 10.1038/36103. doi:10.1038/36103. [DOI] [PubMed] [Google Scholar]
- 21.Jeck WR, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–157. doi: 10.1261/rna.035667.112. doi:rna.035667.112 [pii] 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Westholm JO, et al. Genome-wide Analysis of Drosophila Circular RNAs Reveals Their Structural and Sequence Properties and Age-Dependent Neural Accumulation. Cell reports. 2014;9:1966–1980. doi: 10.1016/j.celrep.2014.10.062. doi:10.1016/j.celrep.2014.10.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tai HC, Schuman EM. MicroRNA: microRNAs reach out into dendrites. Curr Biol. 2006;16:R121–123. doi: 10.1016/j.cub.2006.02.006. doi:S0960-9822(06)01116-X [pii] 10.1016/j.cub.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 24.Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO. Cell-type specific features of circular RNA expression. PLoS Genet. 2013;9:e1003777. doi: 10.1371/journal.pgen.1003777. doi:10.1371/journal.pgen.1003777 PGENETICS-D-13-00715 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guttman M, et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295–300. doi: 10.1038/nature10398. doi:nature10398 [pii] 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mercer TR, et al. Long noncoding RNAs in neuronal-glial fate specification and oligodendrocyte lineage maturation. BMC Neurosci. 2010;11:14. doi: 10.1186/1471-2202-11-14. doi:1471-2202-11-14 [pii] 10.1186/1471-2202-11-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ng SY, Bogu GK, Soh BS, Stanton LW. The long noncoding RNA RMST interacts with SOX2 to regulate neurogenesis. Mol Cell. 2013;51:349–359. doi: 10.1016/j.molcel.2013.07.017. doi:S1097-2765(13)00543-1 [pii] 10.1016/j.molcel.2013.07.017. [DOI] [PubMed] [Google Scholar]
- 28.Bottai D, et al. Synaptic activity-induced conversion of intronic to exonic sequence in Homer 1 immediate early gene expression. J Neurosci. 2002;22:167–175. doi: 10.1523/JNEUROSCI.22-01-00167.2002. doi:22/1/167 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hu JH, et al. Homeostatic scaling requires group I mGluR activation mediated by Homer1a. Neuron. 2010;68:1128–1142. doi: 10.1016/j.neuron.2010.11.008. doi:S0896-6273(10)00915-3 [pii] 10.1016/j.neuron.2010.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roberts A, Pimentel H, Trapnell C, Pachter L. Identification of novel transcripts in annotated genomes using RNA-Seq. Bioinformatics. 2011;27:2325–2329. doi: 10.1093/bioinformatics/btr355. doi:10.1093/bioinformatics/btr355. [DOI] [PubMed] [Google Scholar]
- 31.Li B, Dewey CN. RSEM: accurate transcript quantification from RNASeq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. doi:1471-2105-12-323 [pii] 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. doi:btp324 [pii] 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yeo G, Burge CB. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J Comput Biol. 2004;11:377–394. doi: 10.1089/1066527041410418. doi:10.1089/1066527041410418. [DOI] [PubMed] [Google Scholar]
- 34.Sun W, et al. Ultra-deep profiling of alternatively spliced Drosophila Dscam isoforms by circularization-assisted multi-segment sequencing. EMBO J. 2013;32:2029–2038. doi: 10.1038/emboj.2013.144. doi:10.1038/emboj.2013.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. doi:10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 36.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. doi:10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aakalu G, Smith WB, Nguyen N, Jiang C, Schuman EM. Dynamic visualization of local protein synthesis in hippocampal neurons. Neuron. 2001;30:489–502. doi: 10.1016/s0896-6273(01)00295-1. doi:S0896-6273(01)00295-1 [pii] [DOI] [PubMed] [Google Scholar]
- 38.Yamashita K, Kusakabe M, Sano M. A simple and rapid method of dissociating hepatocytes from fixed liver of the mouse. Stain technology. 1981;56:29–33. doi: 10.3109/10520298109067271. [DOI] [PubMed] [Google Scholar]
- 39.Schlogl A, Jonas P, Schmidt-Hieber C, Guzman SJ. Stimfit: A Fast Visualization and Analysis Environment for Cellular Neurophysiology. Biomed Tech (Berl) 2013 doi: 10.1515/bmt-2013-4181. doi:10.1515/bmt-2013-4181/j/bmte.2013.58.issue-s1-G/bmt-2013-4181/bmt-2013-4181.xml [pii] [DOI] [PubMed] [Google Scholar]
- 40.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. doi:S0092-8674(09)00008-7 [pii] 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Griffiths-Jones S. The microRNA Registry. Nucleic Acids Res. 2004;32:D109–111. doi: 10.1093/nar/gkh023. doi:10.1093/nar/gkh023 32/suppl_1/D109 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cook KB, Kazan H, Zuberi K, Morris Q, Hughes TR. RBPDB: a database of RNA-binding specificities. Nucleic Acids Res. 2011;39:D301–308. doi: 10.1093/nar/gkq1069. doi:gkq1069 [pii] 10.1093/nar/gkq1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.