Abstract
Purpose:
To evaluate the effect of the spatially constrained incoherent motion (SCIM) method on improving the precision and robustness of fast and slow diffusion parameter estimates from diffusion-weighted MRI in liver and spleen in comparison to the independent voxel-wise intravoxel incoherent motion (IVIM) model.
Methods:
We collected diffusion-weighted MRI (DW-MRI) data of 29 subjects (5 healthy subjects and 24 patients with Crohn’s disease in the ileum). We evaluated parameters estimates’ robustness against different combinations of b-values (i.e., 4 b-values and 7 b-values) by comparing the variance of the estimates obtained with the SCIM and the independent voxel-wise IVIM model. We also evaluated the improvement in the precision of parameter estimates by comparing the coefficient of variation (CV) of the SCIM parameter estimates to that of the IVIM.
Results:
The SCIM method was more robust compared to IVIM (up to 70% in liver and spleen) for different combinations of b-values. Also, the CV values of the parameter estimations using the SCIM method were significantly lower compared to repeated acquisition and signal averaging estimated using IVIM, especially for the fast diffusion parameter in liver (CVIV IM = 46.61 ± 11.22, CVSCIM = 16.85 ± 2.160, p < 0.001) and spleen (CVIV IM = 95.15 ± 19.82, CVSCIM = 52.55 ± 1.91, p < 0.001).
Conclusions:
The SCIM method characterizes fast and slow diffusion more precisely compared to the independent voxel-wise IVIM model fitting in the liver and spleen.
Keywords: diffusion-weighted imaging, intravoxel incoherent motion, spatially constrained estimation
1. INTRODUCTION
Diffusion-weighted MRI (DW-MRI) is a noninvasive imaging technique sensitive to the diffusivity of water molecules at the cellular and subcellular levels. Quantitatively, the decay of water signal with b-factor in DW-MRI images is commonly modeled with a monoexponential function associated with the so-called apparent diffusion coefficient (ADC) as the decay rate parameter.1 The bright signal in high b-value images and the low ADC parameter estimates have previously been viewed as potential biomarkers for identifying regions with restricted diffusion in some diseases, e.g., liver cirrhosis and Crohn’s disease.2–4
Depending on acquisition parameters, the ADC value can reflect a combination of slow diffusion associated with the free diffusion of water molecules observed in the b-values range of 200–800 mm2/s and fast diffusion observed in the b-values range of 0–200 mm2/s.5 In contrast, the intravoxel incoherent motion (IVIM) biexponential model aims to separate these fast and slow diffusion components by introducing a biexponential model of the signal decay as a function of the b-values.6,7 For example, in the context of DW-MRI analysis of cirrhotic liver, some studies show the reduction of the fast diffusion parameter, which is related to the reduction of the mean capillary segment length and average blood velocity in cirrhotic liver.8 Further, the slow and fast diffusion parameters were significantly decreased in patients with type 2 diabetes with steatosis compared with nonsteatosis.9 In the context of Crohn’s disease, Freiman et al. show that the decreased ADC values are related to decreases in the fast diffusion component rather than to decreases in the slow diffusion component.10
While quantitative analysis of fast and slow diffusion from DW-MRI data have recently shown promise as quantitative imaging biomarkers for various clinical applications in the body, the clinical utility of IVIM parametric imaging with DW-MRI is diminished by a lack of verified methods for producing reliable estimates of both fast and slow diffusion parameters from the DW-MRI signal.11 First, the fast and slow diffusion parameter estimates can be sensitive to the choice of b-values used in the DW-MRI acquisition.12 Cohen et al. reported that the fast diffusion in the liver would be underestimated using IVIM if only a few low b-values are included in the parameter estimation.13 Therefore, it can be very challenging to compare data obtained with different sets of b-values to conduct multicenter trials and to establish standardized guidelines. Further, reliable estimation of both fast and slow diffusivity from the DW-MRI signal is challenging with the commonly used IVIM model11 due to (a) the nonlinearity of the IVIM model, and (b) the low signal-to-noise ratio (SNR) observed in body DW-MRI.
Several studies utilized the average of multiple DW-MRI images acquired with each b-value11 to increase SNR in DW-MRI. This approach, however, increases the acquisition time, which is especially problematic in evaluating children and adolescent patients who may have difficulty remaining still in the scanner for a long MR examination. An alternative is to average the image in a region of interest (ROI) or use the regional estimation as a prior for voxel-wise parameter estimates.14 However, these parameter estimates may not reflect critical heterogeneous environments, such as regions of chronic inflammation in the bowel or focal lesions in the liver.15
The spatially constrained incoherent motion (SCIM) method and the fusion bootstrap moves (FBM) solver were recently introduced to robustly obtain precise fast and slow diffusion parameter estimates from DW-MRI data.16 The SCIM method extends the IVIM model by simultaneously estimating fast and slow diffusion parameters for all voxels with a spatial homogeneity constraint to improve the precision of the fast and slow diffusion parameter estimates compared to the IVIM model fitting. However, a detailed analysis of improvements in parameter estimates compared to those achieved with high SNR images acquired with repeated imaging or with applying spatial smoothing to the IVIM parametric maps is still required to determine its potential utilization for clinical applications.
It is the goal of this study to examine the improvement achieved by utilizing the SCIM method for quantitative analysis of DW-MRI data and to evaluate any improvements that result from the SCIM method compared to repeated imaging with the commonly used IVIM model fitting. We will especially look at the robustness and precision of parameters in different tissue types, i.e., liver and spleen. This study extends our preliminary evaluation17 which was focused on the analysis of group differences in Crohn’s disease and quantifies (1) the improvement in parameter estimates’ robustness to different choices of b-values and (2) the improvement in parameter estimates precision in different tissue types, i.e., liver and spleen.
1.A. Theory
The IVIM model assumes a biexponential function for the DW-MRI signal decay,
| (1) |
where sv,i is the expected signal of voxel v at b-value bi, sv,0 is the baseline signal (without any diffusion effect), fv is the fast diffusion fraction, and and Dv are the fast and slow diffusion components, respectively. However, fitting the IVIM model to each voxel independently resulted in imprecise and unreliable parameter estimates due to the combination of the model nonlinearity and the low SNR in the DW-MRI images.
The SCIM method fits the IVIM model to all voxels simultaneously by introducing a spatial dependency constraint that describes the expected homogeneity of signal decay parameters in spatially related voxels. Parameter estimates are obtained by maximizing the posterior probability associated with the maps given the observed signal s and the spatial homogeneity constraint,
| (2) |
By representing the spatial homogeneity constraint in the form of a Markov random field (MRF),18 the posterior probability can be decomposed into the product of clique potentials,
| (3) |
where represents the probability of voxel v to have the DW-MRI signal sv given the model parameters θv, vp ∼ vq is the collection of the voxels in a clique containing voxel vp, and p(θvp, θvq) is the spatial homogeneity prior in the model. The parametric map is then estimated by minimizing the following energy functional:
| (4) |
where is the negative logarithm of the signal likelihood distribution, , and ψ(θvp, θvq) is a spatial homogeneity term defined as
| (5) |
where α ≥ 0 weights the amount of spatial homogeneity enforced by the model and W is a diagonal weighting matrix that accounts for the different scales of the parameters in θv. We set the rescaling matrix W diagonal to (1.0,0.001,0.0001,0.01) and the weighting parameter α to 0.01 as used by Freiman et al.16 The fusion bootstrap moves solver16 is then utilized to minimize the energy functional in Eq. (4).
2. MATERIALS AND METHODS
We carried out the study according to a protocol approved by our Institutional Review Board. First, we acquired DW-MRI images of 5 healthy volunteers (2 males and 3 females; mean age 25.4 yr; range: 21–37 yr) who underwent research abdominal MRI studies between January 2013 and May 2013. We carried out the MRI imaging studies of the abdomen and pelvis using a 1.5-T MRI scanner (Magnetom Avanto, Siemens Medical Solutions, Erlangen, Germany). We performed free-breathing single-shot SE echo-planar imaging using the following parameters: repetition/echo time (TR/TE) = 6800/59 ms; acquisition (and reconstruction) ; field of view = 300 × 260 mm; slice thickness/gap = 5/0.5 mm; 40 axial slices; and 7 b-values = 5, 50, 100, 200, 400, 600, 800 s/mm2. We used a tetrahedral gradient scheme19 to acquire four images in multiple directions at each b-value with an overall scan acquisition time of 4 min. Diffusion trace-weighted images at each b-value were then generated using geometric averages of the images acquired in each diffusion sensitization direction.20 This method allows us to acquire high b-value images with a shorter TE than required by the three-scan trace-weighted imaging method, has an inherently high SNR, and demonstrates reduced motion-related artifacts for measurements of averaged diffusivity.19 We repeated the imaging acquisition six times to acquire six DW-MRI datasets, each with low SNR (DW-MRIlow). As all six images for each subject were acquired during one imaging session, the acquired images had acceptable initial alignments which were visually checked. We then averaged the six DW-MRI datasets using geometric averaging to achieve high SNR DW-MRI images (DW-MRIhigh). Finally, using the itksnap software tool,21 we annotated three circular regions of interest of radius 5 mm in the liver, including ROI 1 in the lower right lobe, ROI 2 in the upper right lobe, ROI 3 in the left lobe, and two circular regions of interest of radius 5 mm in the spleen, including ROI 4 in the lower spleen and ROI 5 in the upper spleen (Fig. 1). We selected the ROIs while avoiding large vessels.
FIG. 1.

Five ROIs selected in the liver and spleen were overlaid on the DW-MRI with b-values (a) and (d) 5 s/mm2, (b) and (e) 100 s/mm2, and (c) and (f) 600 s/mm2. (a)–(c) ROI 1 (lower right lobe) in purple and ROI 4 (lower spleen) in cyan and (d)–(f) ROI 2 (upper right lobe) in blue, ROI 3 (left lobe) in pink, and ROI 5 (upper spleen) in orange.
In addition, we acquired DW-MRI data from 24 patients with confirmed Crohn’s disease in the ileum (15 males and 9 females; mean age 14.7 yr; range: 5–24 yr) who underwent a clinically indicated MRI study between January 1, 2011 and October 31, 2011 in our outpatient MRI department. The liver and spleen of all the subjects were normal. We carried out MR imaging studies of the abdomen and pelvis using an identical protocol as described above. The clinical acquisition protocol included polyethylene glycol administration for bowel distention. We used the same scanner as that used for the healthy volunteers to avoid interscanner variability and acquired all images from each subject in one session. We estimated the model parameters using the IVIM model and the SCIM method with an in-house software package written in matlab ® (R2010b; The MathWorks, Natick, MA) and c++. To achieve more robust parameter estimates with the independent voxel-wise fitting of the IVIM model, we used a two-phase approach as follows. We used the segmented least-squares method22,23 to obtain initial estimates of the IVIM model parameters by applying the following 3 steps: 1. the value of the D parameter was estimated using the monoexponential function fitted to SI of b-values 200–800 s/mm2, 2. then, the value of the parameter f was estimated by extrapolating SI to b = 0 s/mm2 and calculating the relative difference between the extrapolated and measured signal at b-value = 0 s/mm2, 3. the value of the D* parameter was calculated by fitting the biexponential model to the observed data with D and f values fixed. Next, we used the initial estimates to initialize the BOBYQA nonlinear solver24 that fit the biexponential model to the data for D, D*, and f simultaneously.
2.A. Study design
We evaluated the improvement achieved by using the SCIM method compared to the commonly used IVIM model fitting in three experiments. We first evaluated the robustness of the fast and slow diffusion parameter estimates obtained by the SCIM and IVIM methods using different b-value combinations of DW-MRI data in 5 healthy subjects (group 1) and 24 Crohn’s disease patients (group 2).
We then examined the precision of the fast and slow diffusion parameter estimates calculated using SCIM by comparing the coefficient of variation of diffusion parameter estimates for low SNR DW-MRI data with estimates using voxel-wise fitting for high SNR DW-MRI data obtained by averaging multiple images.
Finally, we examined the improvement in the precision of the fast and slow diffusion parameter estimates obtained with the SCIM method vs those obtained by (1) independent voxel-wise fitting of the IVIM model and (2) independent voxel-wise fitting of the IVIM model with additional spatial smoothing of the resulted parametric maps on low SNR DW-MRI data. We describe each experiment in detail next.
2.B. Robustness of parameter estimates using 4 and 7 b-values
The goal of this experiment was to determine whether the SCIM method improves the robustness of fast and slow diffusion parameter estimates using different combinations of b-values compared to those typically used with the IVIM model fitting for quantitative DW-MRI data analysis. To this end, we estimated the fast and slow diffusion parameters at each voxel using the IVIM and SCIM methods with different combinations of 4 b-values and with all the b-values (7 b-values) as follows:
4 b-values: 5 s/mm2, one out of 50, 100 s/mm2, one out of 200, 400 s/mm2, and one out of 600, 800 s/mm2, in total 8 combinations.
7 b-values: 5, 50, 100, 200, 400, 600, and 800 s/mm2, in total one combination.
Next, we calculated the variation (standard deviation), , at voxel v in each ROI in the liver and spleen,
| (6) |
where is the parameter estimates at voxel v using the b-value combination b and B is the set of all b-values combinations, i.e., 4 and 7 b-value combinations. Last, we determined whether the SCIM method yielded fast and slow diffusion parameter estimates that were more robust to different combinations of b-values by comparing the variation in values of the SCIM and independent voxel-wise IVIM model fitting parameter estimates, respectively, using a two-tailed, paired Student’s t-test with p ≤ 0.05 indicating a significant difference.
2.C. Precision of parameter estimates using SCIM vs multiple DW-MR images averaging
In the next experiment, we compared the precision of fast and slow diffusion parameter estimates obtained with the SCIM method from low SNR 1 NEX images to the precision obtained with IVIM model fitting from high SNR 6 NEX images in the healthy subjects. We averaged the multiple 1 NEX DW-MRI images (DW-MRIlow) of the subjects, each with low SNR to achieve high SNR DW-MRI images (DW-MRIhigh). We measured the coefficient of variation (CV) values of the diffusion parameters from (1) DW-MRIhigh using the IVIM model (IVIMhigh), (2) DW-MRIlow using the IVIM model (IVIMlow), and (3) DW-MRIlow using the SCIM method (SCIMlow), that is,
| (7) |
where is the ith parameter estimates at voxel v using the wild bootstrap analysis.16,25 The wild bootstrap resampling is defined as
| (8) |
where is the resampled measures at the b-value bi, is the rescaled version of the raw residual between the observed signal (mv,i) and the expected signal (sv,i), and tv,i is a two-point Rademacher distributed random variable with and defined for each b-value separately. Resampling and model fitting are repeated for a large number of fixed repetitions to obtain a large set of diffusion models. We calculated the CV values using the wild bootstrap analysis at each voxel separately and then averaged the CV values at each ROI. The lower CV values illustrate the more precise parameter estimates. Following this, we examined the amount of reduction and the statistical difference in the CV values of the IVIMhigh, IVIMlow, and SCIMlow estimates obtained from the subjects using a two-tailed paired Student’s t-test (p ≤ 0.05). We separated the analysis for each tissue type, as their average parameter values are different as reported previously.16
2.D. Precision of parameter estimates using SCIM vs smoothing of parameter maps
We examined the effect of the spatial prior of the SCIM method on the parameter estimation by calculating the parameter maps using: (1) independent voxel-wise fitting of the IVIM model, (2) independent voxel-wise fitting of the IVIM model with spatial smoothing of the parametric maps (IVIMσ), and (3) simultaneous voxel-wise fitting of the parameters with spatial homogeneity constraint using the SCIM method from the low SNR DW-MRI data. We calculated the CV values at each voxel separately using the wild bootstrap method.16,25 To avoid significantly blurring the parameter map and suppressing the details, we used a Gaussian filter with variance of half a voxel, to smooth the IVIM parameter maps (σ2 = 1 mm). We then examined the amount of reduction and the statistical difference in the CV values of the IVIM, IVIMσ and SCIM estimates obtained from the subjects using a two-tailed paired Student’s t-test (p ≤ 0.05).
3. RESULTS
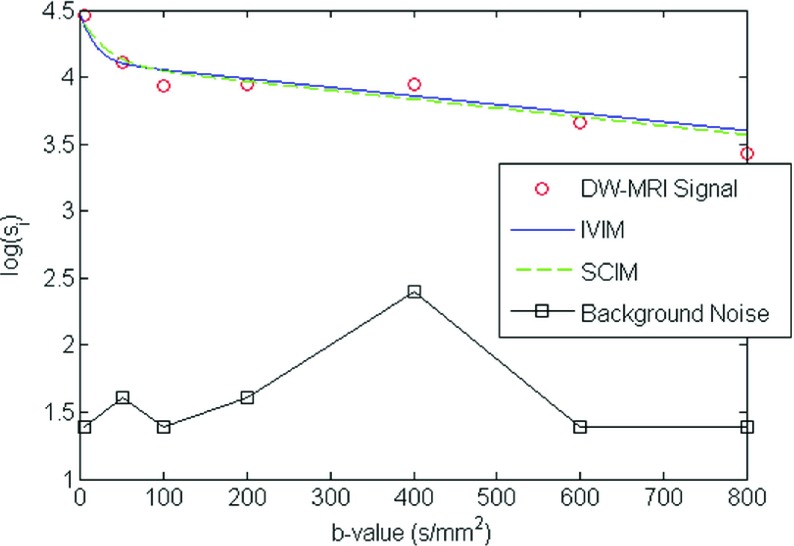
The running time required to reconstruct the parametric maps on a single processor machine Intel® Xeon® at 2.40 GHz and cache size = 12 MB were 0.137 and 14.21 ms/voxel using the IVIM and the SCIM methods, respectively. Figure 2 illustrates the DW-MRI signal decay estimated using SCIM and IVIM, along with the signal decay at 7 b-values for a representative subject in group 1. The SCIM method shows a better fit (CVD-IVIM = 0.4611, CVD-SCIM = 0.4736; CVD*-IVIM = 0.7404 and CVD*-SCIM = 0.4233; CVf-IVIM = 0.4636, CVf-SCIM = 0.3986, RMSEIV IM = 0.0952, RMSESCIM = 0.0865, F-statistic = 1.280, f-test p < 0.771) compared with IVIM for this subject. However, the improvement was not significant.
FIG. 2.
Representative example of the observed DW-MRI signal and the estimated models calculated by the IVIM and SCIM estimators from all 7 b-values (5, 50, 100, 200, 400, 600, and 800).
Table I presents the average and standard deviation of the parameter estimates using both the IVIM and SCIM methods in five regions of interest in the liver and spleen over the healthy and CD subject groups. The difference between parameters D and f estimated by IVIM and SCIM in all five ROIs was not found to be significant.
TABLE I.
The average and standard deviation (mean ± SD) of the incoherent motion parameter estimates over the healthy and CD subject groups in three ROIs in liver and two ROIs in spleen. D and D∗ are in units of μm2/ms. Significant values in bold.
| ROI 1: Liver lower right lobe | ROI 2: Liver lower right lobe | ROI 3: Liver left lobe | |||||||
|---|---|---|---|---|---|---|---|---|---|
| IVIM | SCIM | P | IVIM | SCIM | P | IVIM | SCIM | P | |
| D | 1.01 ± 0.29 | 1.03 ± 0.26 | 0.511 | 1.05 ± 0.31 | 1.05 ± 0.26 | 0.809 | 1.11 ± 0.52 | 1.12 ± 0.48 | 0.701 |
| D∗ | 54.6 ± 55.1 | 28.3 ± 17.7 | <0.001 | 44.2 ± 51.5 | 21.8 ± 16.3 | <0.001 | 34.3 ± 46.0 | 18.8 ± 18.9 | <0.001 |
| f | 0.25 ± 0.18 | 0.25 ± 0.17 | 0.923 | 0.23 ± 0.17 | 0.24 ± 0.15 | 0.507 | 0.32 ± 0.21 | 0.32 ± 0.19 | 0.841 |
| ROI 4: Lower spleen | ROI 5: Upper spleen | |||||
|---|---|---|---|---|---|---|
| IVIM | SCIM | P | IVIM | SCIM | P | |
| D | 1.65 ± 0.52 | 1.66 ± 0.50 | 0.785 | 0.75 ± 0.28 | 0.74 ± 0.27 | 0.437 |
| D∗ | 35.5 ± 49.4 | 21.8 ± 27.2 | <0.001 | 32.5 ± 50.6 | 15.0 ± 19.0 | <0.001 |
| F | 0.27 ± 0.21 | 0.27 ± 0.20 | 0.722 | 0.13 ± 0.15 | 0.14 ± 0.14 | 0.445 |
3.A. Robustness of parameter estimates using 4 and 7 b-values
Table II summarizes the average variation (standard deviation) of the incoherent motion parameter estimates against all combinations of b-values for both groups in the liver and spleen. This shows how robustly each model estimates the incoherent motion parameters with respect to different combination of b-values. The SCIM method reduced the variation of all signal decay model parameters compared to IVIM. Most notable is a reduction of around 70% in the average variation of the fast diffusion decay rate parameter (D*) in all regions of interest in the spleen and liver.
TABLE II.
The average variation of the incoherent motion parameter estimates for all combinations of b-values among both group 1 and 2 in three ROI’s in liver and two ROI’s in spleen. D and D∗ are in units of μm2/ms. Significant values in bold.
| ROI 1: Liver lower right lobe | ROI 2: Liver lower right lobe | ROI 3: Liver left lobe | |||||||
|---|---|---|---|---|---|---|---|---|---|
| IVIM | SCIM | P | IVIM | SCIM | P | IVIM | SCIM | P | |
| D | 0.33 | 0.30 | 0.071 | 0.35 | 0.32 | 0.134 | 0.68 | 0.61 | 0.096 |
| D∗ | 40.6 | 13.0 | <0.001 | 39.4 | 8.98 | <0.001 | 35.3 | 9.14 | <0.001 |
| F | 0.11 | 0.08 | <0.001 | 0.11 | 0.09 | <0.001 | 0.16 | 0.14 | <0.001 |
| ROI 4: Lower spleen | ROI 5: Upper spleen | |||||
|---|---|---|---|---|---|---|
| IVIM | SCIM | P | IVIM | SCIM | P | |
| D | 2.14 | 0.37 | 0.017 | 0.32 | 0.31 | 0.609 |
| D∗ | 33.3 | 12.2 | <0.001 | 35.9 | 9.79 | <0.001 |
| F | 0.12 | 0.09 | <0.001 | 0.11 | 0.09 | 0.020 |
3.B. Precision of parameter estimates using SCIM vs multiple DW-MR image averaging
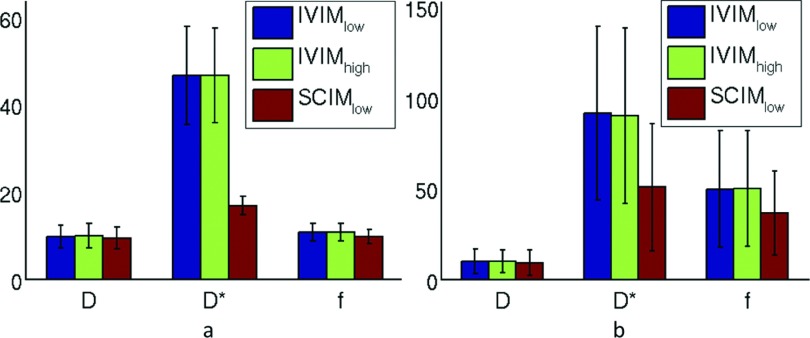
Figure 3 presents the CV values of the parameter estimates for group 1 in the liver and spleen. The reduction in the CV achieved using the IVIM model with high SNR images (IVIMhigh) compared to that achieved with the IVIM model with the low SNR images (IVIMlow) was not significant, however. In contrast, estimating the signal decay model parameters using low SNR images with the SCIM method (SCIMlow) reduced the CV of the parameters estimates by up to 65% and 45% in the liver and spleen, respectively. The reduction in the CV of parameter D* of the SCIMlow compared to those of the IVIMlow and IVIMhigh was significant in the liver (p < 0.001).
FIG. 3.
Bar plot of the CV of the incoherent motion parameters as estimated from group1 in (a) liver and (b) spleen.
3.C. Precision of parameter estimates using SCIM vs smoothing of parameter maps
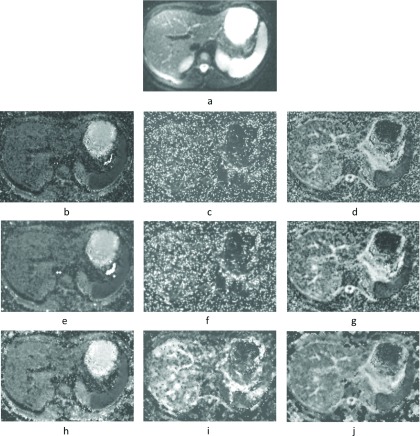
Figure 4 depicts (a) a representative DW-MR image of the liver and spleen along with representative parametric maps produced using (b)–(d) the IVIM, (e)–(g) the IVIM maps smoothed with a Gaussian filter (σ2 = 1 mm), and (h)–(j) the SCIM maps. The SCIM method produces smoother D* and f parameter maps compared with the IVIM model.
FIG. 4.
(a) The DW-MR image illustrates the liver and spleen of a subject in group 2, (b)–(d) D, D*, and f parameter maps estimated using the IVIM model, (e)–(g) the corresponding IVIM parameter maps smoothed with a Gaussian filter (σ2 = 1 mm), and (h)–(j) D, D*, and f parameter maps estimated using the SCIM method.
Figure 5 depicts the improvement in the precision of the parameter estimates achieved by using SCIM compared to these of IVIM and IVIMσ for group 2 in the liver and spleen. The SCIM method reduced the CV of the parameter estimates up to 52% and 47% in liver and spleen, respectively, compared to IVIM, and the reduction in the CV of parameters f and D* was significant in the liver and spleen (p < 0.001). The CV values achieved using the SCIM method were still up to 38% and 37% lower than those achieved by the IVIMσ in liver and spleen, respectively. However, the reduction was significant only for the parameter D* (p < 0.001).
FIG. 5.
Bar plot of the CV of the incoherent motion parameters as estimated from group 2 in (a) liver and (b) spleen using the IVIM method, the IVIM method while smoothing the parameter maps (IVIMσ), and the SCIM method.
4. DISCUSSION
Quantitative analysis of DW-MRI images has the potential to provide quantitative and objective insight into tissue physiology and to serve as a biomarker in many clinical applications.26–28 Specifically, the intravoxel incoherent motion model of the DW-MRI signal decay aims to distinguish between a fast diffusion component observed in the b-values range of about 0–200 s/mm2 and a slow diffusion component observed in the b-values range of about 200–800 s/mm2 of the signal decay.6,7
The high sensitivity of the model parameter estimates to the b-values used in the acquisition and the low precision of the incoherent motion parameters estimates demonstrated by the independent voxel-wise intravoxel incoherent motion model has hampered its utilization in clinical studies and patient management.11 There are currently several approaches that can increase the reliability of quantitative DW-MRI analysis to varying degrees. Commonly, researchers use an approximation of the nonlinear DW-MRI signal decay by a log-linear model with the ADC as the decay rate parameter.29 Unfortunately, this simplified model precludes the independent characterization of slow diffusion and fast diffusion components—a process essential to accurately quantifying biological phenomena taking place inside the body.
Koh et al. recommended increasing the DW-MRI SNR by acquiring multiple DW-MRI images from the patient, averaging the images at each b-value, and using the averaged DW-MRI signal to estimate IVIM model parameters.11 However, this approach requires substantially increased acquisition times—an undesirable solution, especially in children who generally have difficulty remaining still for long periods of time. Zhang et al.30 improved the SNR by averaging the DW-MRI signals over a ROI in the kidney, in order to achieve more reliable parameter estimation in their DW-MRI acquisition optimization study. However, by averaging the signal over a ROI, the estimated parameters do not reflect critical heterogeneous environments, such as regions with abnormality.
Several researchers proposed to improve the reliability of the IVIM parameter fitting by optimizing the set of b-values used in the acquisition. Zhang et al.30 optimized a set of b-values for estimating the IVIM model parameters in the kidneys. Merisaari et al.31 optimized b-value distribution for the ADC and IVIM models using simulations and repeated DW-MRI examinations with b-values up to 2000 s/mm2. Pang et al.12 evaluated the effect of different b-values on IVIM parameter estimation and demonstrated that the parameter estimates highly depend on the b-values, and that those estimated without higher b-values correlate best with DCE-MRI results. Furthermore, Jambor et al.31 reported on an optimal b-value distribution in terms of measurement reliability and repeatability, which forms three clusters at low, middle, and high b-values, that improves parameter estimation by minimizing the convergence of the total error function.
Bayesian model fitting is another approach to increase the reliability of the parameter estimation in DW-MR images. Neil et al.32 suggested a Bayesian model fitting approach to increase the reliability of IVIM parameter estimates by calculating the probability distribution function of each parameter rather than by calculating point estimates as is done using maximum-likelihood estimators. However, this method considers the information at each voxel independently, effectively ignoring its spatial context. Moreover, it requires numerical integration of the marginal posterior probabilities over the possible ranges of parameter values, which are sensitive both to discretization/sampling effects and to the chosen integration limits.33 Recently, Orton et al.14 introduced a Bayesian shrinkage prior (BSP) approach for regional IVIM to reduce heterogeneity and uncertainty of the IVIM parameter estimates. However, this model does not utilize the local spatial information but assumes the true parameter distribution is Gaussian.
In contrast, the SCIM method integrates the spatial information of DW-MR images in a neighborhood to reliably estimate the diffusion parameters. The SCIM method utilizes a Markov random field to adaptively select the neighboring voxels [Eq. (2)] and optimize the energy functional [Eq. (4)], which is different than the spatial smoothing of the DWI or parameter maps and done without considering the optimization of the energy functional. Introducing a spatial homogeneity prior, as we have done may cause blurring. To minimize this blurring effect we used the L1 norm as our regularization prior, which better preserves discontinuities compared to the commonly used L2 norm. In addition, the smoothing prior considers only a very limited neighborhood around the voxel while the radiologic evaluation is carried out on larger regions. Therefore, we do not expect significant adverse effects. However, this needs to be verified for specific clinical applications and warrants future study.
In this work, we examined whether the use of a spatial homogeneity prior in the form of the SCIM method substantially improves the robustness and reliability of fast and slow diffusion parameter estimates from DW-MRI data. As opposed to previously proposed methods, the SCIM method does not require additional images, averaging the signal over some ROI or a specific set of b-values to improve the robustness of the fast and slow diffusion parameter estimates. In fact, the SCIM method adaptively selects the neighboring voxels in a MRF and chooses between different parameter combinations proposed by the bootstrap process using a graph min-cut algorithm to avoid local minima and to lower the total energy, which is not considered using spatial smoothing of the DWI or parameter maps.
Our first experiment shows that the fast and slow diffusion parameter estimates with the SCIM method are more robust to different sets of b-values. First, we have shown that by using the SCIM method, we reduced the variation in the parameter estimates using different combinations of b-values by up to 70% for the D* parameter in the data in the liver and spleen. These results indicate that by using the SCIM method for fast and slow diffusion parameter estimates, we may increase its robustness to different choices of b-values and thus make fast and slow diffusion parameters ready for use in multiinstitutional studies and standard clinical care. These results also illustrate that the need for multiple DW-MRI images to improve SNR can be avoided using the SCIM method, which could in turn accelerate the image acquisition process.
Our second experiment shows that by using the SCIM method, the coefficient of variation of the fast and slow diffusion parameter estimates can be reduced by about 50% compared to parameter estimates obtained from the same images using the voxel-wise independent IVIM model fitting. Moreover, the SCIM method provides more precise estimates from low SNR images compared to those obtained with the IVIM model fitting from high SNR images. Therefore, it can be useful in shortening the overall time required to obtain DW-MRI data that are suitable for quantitative analysis.
The third experiment reveals that the SCIM method yields more precise parametric maps compared with the IVIM model fitting even when applying a spatial smoothing to the IVIM maps, especially for f and D* parameters (up to 38%). In general, we expect close water molecule diffusivity in homogeneous areas, keeping in mind, there is no abnormal tissue in these subjects. This should result in close parameter estimates in each neighborhood and smoother parameter maps, which cannot be seen in the IVIM results, especially for parameters f and D* as reported in the previous studies.34 Please note that further smoothing of the maps, i.e., σ2 > 1 mm, would blur the edges, which obscures the boundaries between different anatomic areas.
Our study had limitations. First, this study population was limited by the number of patients and by their age range. Consequently, our assessment of DW-MRI data was restricted to 29 subjects. Second, although we showed the robustness of the SCIM method to variations in the b-value, the DW-MRI acquisition protocols for this study were limited to those protocols routinely used in our institution. As a result, we employed a fixed set of b-values for the subjects in group 2 who underwent abdominal scans. An additional study is warranted to evaluate the impact of the b-values selected (i.e., by means of values and number of b-values) on estimated parameters. Third, the literature is inconsistent about the definition of the fast-diffusion exponential in IVIM, whether it is D*22,23 or (D* + D).5,35 We used in our experiments the definition of Luciani et al.;35 however, as our contribution is focused on the spatially constrained estimation method and its numerical solution, it can be applied seamlessly with the definition of Pekar et al.23
In conclusion, we show that the SCIM model can increase the robustness of fast and slow diffusion parameter estimates without the need for additional acquisition time.
ACKNOWLEDGMENTS
Research reported in this publication is supported in part by the National Institute of Diabetes and Digestive and Kidney Diseases of the National Institutes of Health under Award No. R01DK100404. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. This investigation is also supported in part by research grants from the Translational Research Program at Boston Children’s Hospital. The senior author, Moti Freiman, is also supported in part by a Research Fellow Award from the Crohn’s and Colitis Foundation of America.
REFERENCES
- 1.Le Bihan D., Breton E., Lallemand D., Grenier P., Cabanis E., and Laval-Jeantet M., “MR imaging of intravoxel incoherent motions: Application to diffusion and perfusion in neurologic disorders,” Radiology 161, 401–407 (1986). 10.1148/radiology.161.2.3763909 [DOI] [PubMed] [Google Scholar]
- 2.Kiryu S., Dodanuki K., Takao H., Watanabe M., Inoue Y., Takazoe M., Sahara R., and Unuma K., “Free-breathing diffusion-weighted imaging for the assessment of inflammatory activity in Crohn’s disease,” J. Magn. Reson. Imaging 29, 880–886 (2009). 10.1002/jmri.21725 [DOI] [PubMed] [Google Scholar]
- 3.Oto A., Kayhan A., Williams J. T., Fan X., Yun L., Arkani S., and Rubin D. T., “Active Crohn’s disease in the small bowel: Evaluation by diffusion weighted imaging and quantitative dynamic contrast enhanced MR imaging,” J. Magn. Reson. Imaging 33, 615–624 (2011). 10.1002/jmri.22435 [DOI] [PubMed] [Google Scholar]
- 4.Oussalah A., Laurent V., Bruot O., Bressenot A., Bigard M. A., Regent D., and Peyrin-Biroulet L., “Diffusion-weighted magnetic resonance without bowel preparation for detecting colonic inflammation in inflammatory bowel disease,” Gut 59, 1056–1065 (2010). 10.1136/gut.2009.197665 [DOI] [PubMed] [Google Scholar]
- 5.Lemke A., Laun F. B., Simon D., Stieltjes B., and Schad L. R., “An in vivo verification of the intravoxel incoherent motion effect in diffusion-weighted imaging of the abdomen,” Magn. Reson. Med. 64, 1580–1585 (2010). 10.1002/mrm.22565 [DOI] [PubMed] [Google Scholar]
- 6.Le Bihan D., “Intravoxel incoherent motion perfusion MR imaging: A wake-up call,” Radiology 249, 748–752 (2008). 10.1148/radiol.2493081301 [DOI] [PubMed] [Google Scholar]
- 7.Le Bihan D., Breton E., Lallemand D., Aubin M. L., Vignaud J., and Laval-Jeantet M., “Separation of diffusion and perfusion in intravoxel incoherent motion MR imaging,” Radiology 168, 497–505 (1988). 10.1148/radiology.168.2.3393671 [DOI] [PubMed] [Google Scholar]
- 8.Guiu B. and Cercueil J.-P., “Liver diffusion-weighted MR imaging: The tower of Babel?,” Eur. Radiol. 21, 463–467 (2011). 10.1007/s00330-010-2017-y [DOI] [PubMed] [Google Scholar]
- 9.Guiu B., Petit J.-M., Capitan V., Aho S., Masson D., Lefevre P.-H., Favelier S., Loffroy R., Vergès B., and Hillon P., “Intravoxel incoherent motion diffusion-weighted imaging in nonalcoholic fatty liver disease: A 3.0-T MR study,” Radiology 265, 96–103 (2012). 10.1148/radiol.12112478 [DOI] [PubMed] [Google Scholar]
- 10.Freiman M., Perez-rossello J. M., Callahan M. J., Bittman M., Mulkern R. V., Bousvaros A., and Warfield S. K., “Characterization of fast and slow diffusion from diffusion-weighted MRI of pediatric Crohn’s disease,” J. Magn. Reson. Imaging 37, 156–163 (2013). 10.1002/jmri.23781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koh D.-M., Collins D. J., and Orton M. R., “Intravoxel incoherent motion in body diffusion-weighted MRI: Reality and challenges,” Am. J. Roentgenol. 196, 1351–1361 (2011). 10.2214/ajr.10.5515 [DOI] [PubMed] [Google Scholar]
- 12.Pang Y., Turkbey B., Bernardo M., Kruecker J., Kadoury S., Merino M. J., Wood B. J., Pinto P. A., and Choyke P. L., “Intravoxel incoherent motion MR imaging for prostate cancer: An evaluation of perfusion fraction and diffusion coefficient derived from different b-value combinations,” Magn. Reson. Med. 69, 553–562 (2013). 10.1002/mrm.24277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cohen A. D., Schieke M. C., Hohenwalter M. D., and Schmainda K. M., “The effect of low b-values on the intravoxel incoherent motion derived pseudodiffusion parameter in liver,” Magn. Reson. Med. 73, 306–311 (2015). 10.1002/mrm.25109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Orton M. R., Collins D. J., Koh D. M., and Leach M. O., “Improved intravoxel incoherent motion analysis of diffusion weighted imaging by data driven Bayesian modeling,” Magn. Reson. Med. 71, 411–420 (2014). 10.1002/mrm.24649 [DOI] [PubMed] [Google Scholar]
- 15.Lee Y., Lee S. S., Kim N., Kim E., Kim Y. J., Yun S.-C., Kühn B., Kim I. S., Park S. H., and Kim S. Y., “Intravoxel incoherent motion diffusion-weighted MR imaging of the Liver: Effect of triggering methods on regional variability and measurement repeatability of quantitative parameters,” Radiology 274, 405–415 (2015). 10.1148/radiol.14140759 [DOI] [PubMed] [Google Scholar]
- 16.Freiman M., Perez-Rossello J. M., Callahan M. J., Voss S. D., Ecklund K., Mulkern R. V., and Warfield S. K., “Reliable estimation of incoherent motion parametric maps from diffusion-weighted MRI using fusion bootstrap moves,” Med. Image Anal. 17, 325–336 (2013). 10.1016/j.media.2012.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Freiman M., Voss S. D., Mulkern R. V., Perez-Rossello J. M., Callahan M. J., and Warfield S. K., “Reliable assessment of perfusivity and diffusivity from diffusion imaging of the body,” inMedical Image Computing and Computer-Assisted Intervention–MICCAI 2012 (Springer-Verlag, Berlin, Heidelberg, 2012), pp. 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Winkler G., Image Analysis Random Fields and Markov Chain Monte Carlo Methods: A Mathematical Introduction (Springer-Verlag, Berlin, Heidelberg, 2003). [Google Scholar]
- 19.Conturo T. E., McKinstry R. C., Akbudak E., and Robinson B. H., “Encoding of anisotropic diffusion with tetrahedral gradients: A general mathematical diffusion formalism and experimental results,” Magn. Reson. Med. 35, 399–412 (1996). 10.1002/mrm.1910350319 [DOI] [PubMed] [Google Scholar]
- 20.Mulkern R. V., Vajapeyam S., Robertson R. L., Caruso P. A., Rivkin M. J., and Maier S. E., “Biexponential apparent diffusion coefficient parametrization in adult vs newborn brain,” Magn. Reson. Imaging 19, 659–668 (2001). 10.1016/s0730-725x(01)00383-6 [DOI] [PubMed] [Google Scholar]
- 21.Yushkevich P. A., Piven J., Hazlett H. C., Smith R. G., Ho S., Gee J. C., and Gerig G., “User-guided 3D active contour segmentation of anatomical structures: Significantly improved efficiency and reliability,” Neuroimage 31, 1116–1128 (2006). 10.1016/j.neuroimage.2006.01.015 [DOI] [PubMed] [Google Scholar]
- 22.Cho G. Y., Moy L., Zhang J. L., Baete S., Lattanzi R., Moccaldi M., Babb J. S., Kim S., Sodickson D. K., and Sigmund E. E., “Comparison of fitting methods and b-value sampling strategies for intravoxel incoherent motion in breast cancer,” Magn. Reson. Med.(2014) (E-pub ahead of print), see http://onlinelibrary.wiley.com/doi/10.1002/mrm.25484/abstract 10.1002/mrm.25484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pekar J., Moonen C. T., and van Zijl P., “On the precision of diffusion/perfusion imaging by gradient sensitization,” Magn. Reson. Med. 23, 122–129 (1992). 10.1002/mrm.1910230113 [DOI] [PubMed] [Google Scholar]
- 24.Powell M. J., “Developments of NEWUOA for minimization without derivatives,” IMA J. Numer. Anal. 28, 649–664 (2008). 10.1093/imanum/drm047 [DOI] [Google Scholar]
- 25.Davidson R. and Flachaire E., “The wild bootstrap, tamed at last,” J. Econometrics 146, 162–169 (2008). 10.1016/j.jeconom.2008.08.003 [DOI] [Google Scholar]
- 26.Lewis J. D., “The utility of biomarkers in the diagnosis and therapy of inflammatory bowel disease,” Gastroenterology 140, 1817–1826 (2011). 10.1053/j.gastro.2010.11.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qayyum A., “Diffusion-weighted imaging in the abdomen and pelvis: Concepts and applications,” Radiographics 29, 1797–1810 (2009). 10.1148/rg.296095521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rimola J., Rodríguez S., García-Bosch O., Ordás I., Ayala E., Aceituno M., Pellisé M., Ayuso C., Ricart E., and Donoso L., “Magnetic resonance for assessment of disease activity and severity in ileocolonic Crohn’s disease,” Gut 58, 1113–1120 (2009). 10.1136/gut.2008.167957 [DOI] [PubMed] [Google Scholar]
- 29.Stejskal E. O. and Tanner J. E., “Spin diffusion measurements: Spin-echo in the presence of a time dependent field gradient,” J. Chem. Phys. 42, 288–292 (1965). 10.1063/1.1695690 [DOI] [Google Scholar]
- 30.Zhang J. L., Sigmund E. E., Rusinek H., Chandarana H., Storey P., Chen Q., and Lee V. S., “Optimization of b-value sampling for diffusion-weighted imaging of the kidney,” Magn. Reson. Med. 67, 89–97 (2012). 10.1002/mrm.22982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jambor I., Merisaari H., Aronen H. J., Järvinen J., Saunavaara J., Kauko T., Borra R., and Pesola M., “Optimization of b-value distribution for biexponential diffusion-weighted MR imaging of normal prostate,” J. Magn. Reson. Imaging 39, 1213–1222 (2014). 10.1002/jmri.24271 [DOI] [PubMed] [Google Scholar]
- 32.Neil J. J. and Bretthorst G. L., “On the use of Bayesian probability theory for analysis of exponential decay date: An example taken from intravoxel incoherent motion experiments,” Magn. Reson. Med. 29, 642–647 (1993). 10.1002/mrm.1910290510 [DOI] [PubMed] [Google Scholar]
- 33.Behrens T., Woolrich M., Jenkinson M., Johansen-Berg H., Nunes R., Clare S., Matthews P., Brady J., and Smith S., “Characterization and propagation of uncertainty in diffusion-weighted MR imaging,” Magn. Reson. Med. 50, 1077–1088 (2003). 10.1002/mrm.10609 [DOI] [PubMed] [Google Scholar]
- 34.Andreou A., Koh D., Collins D., Blackledge M., Wallace T., Leach M., and Orton M., “Measurement reproducibility of perfusion fraction and pseudodiffusion coefficient derived by intravoxel incoherent motion diffusion-weighted MR imaging in normal liver and metastases,” Eur. Radiol. 23, 428–434 (2013). 10.1007/s00330-012-2604-1 [DOI] [PubMed] [Google Scholar]
- 35.Luciani A., Vignaud A., Cavet M., Tran Van Nhieu J., Mallat A., Ruel L., Laurent A., Deux J.-F., Brugieres P., and Rahmouni A., “Liver cirrhosis: Intravoxel incoherent motion MR imaging—Pilot study 1,” Radiology 249, 891–899 (2008). 10.1148/radiol.2493080080 [DOI] [PubMed] [Google Scholar]