Abstract
This study evaluated the relative mRNA expression levels of nerve growth factor (NGF) and the p75 neurothrophin receptor (p75NTR) in different histological stages of human liver disease. Fifty-one liver biopsy specimens obtained from patients with hepatitis B virus (n = 6), hepatitis C virus (n = 28), and non-viral hepatitis – (n = 9) and standard histological liver (n = 8) as controls (CT) were subjected to qPCR and histopathological exams. Our data revealed a significant difference in the NGF expression levels between the three patient groups and the Control group. p75NTR expression levels in the HCV and NVH groups were higher than those observed in the HBV and Control groups. In cases of liver cirrhosis, higher p75NTR mRNA expression was observed, whereas NGF was expressed at higher levels in patients with hepatic fibrosis. NGF expression was lower in the F1 liver fibrosis stage, and p75NTR receptor expression continuously and proportionately increased compared to the increase in the degree of fibrosis and was significantly higher in livers in fibrosis stages 3 and 4. The hepatic levels of NGF and p75NTR were decreased and increased, respectively, relative to the stage of inflammatory activity. A positive correlation between p75NTR and NGF gene expression was observed in livers with mild to moderate fibrosis, though not in cases of severe fibrosis and cirrhosis.
Conclusion
Our results demonstrate that the course of chronic liver disease can be regulated by NGF and p75NTR, which function by decreasing or inhibiting hepatocyte regeneration and proliferation.
Introduction
Viral hepatitis is one of the great pandemics of our time. Hepatitis B virus (HBV) and hepatitis C virus (HCV) are responsible for the vast majority of chronic liver diseases worldwide; therefore, they are important public health problems [1]. Approximately 170 million people, 3% of the world population, are carriers of chronic HCV infection, with different patterns of geographic distribution, while approximately 7% of the world population is chronically infected with HBV [1,2]. Due to persistent liver inflammation, the entire liver tissue undergoes a high rate of cellular destruction and regeneration, which results in an increased risk of developing conditions such as cirrhosis and hepatocellular carcinoma [3]. Liver injury triggered by HBV and HCV infection is primarily mediated by the host immune response to viral proteins expressed in infected hepatocytes and, to a lesser degree, the cytopathic effect directly caused by the virus [4].
As a consequence of chronic liver injury, quiescent hepatic stellate cells (HSCs), which store fat and vitamin A, undergo differentiation to an activated myofibroblastic phenotype under the action of pro-inflammatory cytokines, increasing synthesis levels of extracellular matrix components (i.e., collagens, elastin, proteoglycans and constituent proteins) [5,6]. This HSC activation is accompanied by a reorganization and expression of cytoskeletal proteins, such as α-smooth muscle actin (α-SMA), that acquire pro-fibrogenic properties [7]. The excessive deposition of extracellular matrix is the result of an imbalance between fibrogenesis and fibrolysis in the liver, and the proportion of deposited extracellular matrix becomes greater than the amount removed, especially collagen types I and II, proteoglycans, and glycoproteins [7].
Fibrosis is perceived as an initially beneficial physiological mechanism to limit the extent of the inflammatory process; however, if the fibrosis persists and becomes aggressive, it becomes pathological, leading to the distortion of the hepatic architecture [8]. Knowing the stage of liver fibrosis is essential for prognosis and to determine the appropriate antiviral therapy [9]. Patients without fibrosis or who manifest it at a minimum degree seem to progress slowly, and treatment could possibly be delayed or may be unnecessary. However, patients with significant degrees of fibrosis (septal or bridging) almost invariably progress to cirrhosis; in such cases, antiviral treatments should be strongly considered [10,11].
Nerve growth factor (NGF), discovered by Levi-Montalcini et al. in the 1950s, was originally characterized according to its capacity to stimulate the growth, differentiation, survival and maintenance of neurons during development and after injury [12,13]. NGF is a member of the neurotrophin (NT) family, which also includes brain-derived neurotrophic factor (BDNF), NT-3 and NT-4/5; NGF is the prototype and best-characterized member, both structurally and functionally [14]. The cellular response to NGF is determined by the combination of receptor expression: TrkA intermediates survival and differentiation, while p75 neurotrophin receptor (p75NTR), in the absence of TrkA, generally indicates the start of apoptosis [14]. Studies have shown that the components of the neurotrophin axis, NGF and p75NTR, are respectively expressed in hepatocytes and activated HSCs in normal and fibrotic liver in both humans and rats [15–19]. Studies have demonstrated that HSCs express p75NTR and have undergone apoptosis in tissue culture in response to stimulation with recombinant NGF, suggesting that p75NTR is a new marker of activated HSCs and that signaling by binding of this receptor may provide a selective apoptosis mechanism for HSCs [15–17].
Concerning the information already described about the involvement of NGF and P75NTR as immunological components which target apoptosis and inflammatory processes, we wondered if NGF and P75NTR expression could have some influence on liver tissue of HCV and HBV infected subjects. This study aimed to quantify NGF and P75 NTR gene expression levels in liver biopsy specimens of patients with chronic hepatitis by HBV, HCV, and non-viral causes (NVH—non-viral hepatitis) to relate their possible roles with the clinical development of these infections and the various stages of the fibrosis process and hepatic inflammatory activity according to the METAVIR classification.
Materials and Methods
Study population
The study group consisted of 51 individuals, consecutive cases of chronic carriers of HBV (n = 6), HCV (n = 28) and NVH (n = 9) (including non-alcoholic liver disease, autoimmune hepatitis, and primary biliary cirrhosis, among others), seen at the Hepatology Outpatient Service of the Hospital of Holy House of Mercy Foundation of Pará (Fundação Santa Casa de Misericórdia do Pará—FSCMPA) and a Control group (n = 8) consisting of patients who underwent conventional biliary cholecystectomy without necro-inflammatory liver changes at the Surgery Service of João de Barros Barreto University Hospital (Hospital Universitário João de Barros Barreto) at the Federal University of Pará (Universidade Federal do Pará—UFPA).
All selected patients were clinically evaluated and were subjected to further investigation consisting of blood, biochemical, serological (HBsAg, HBeAg, anti-HBeAg, anti-HBctotal and anti-HCV), and virological (quantitative HCV-RNA and genotyping) tests, ultrasound and endoscopy, in addition to liver biopsy. These data were transcribed from clinical records to the study database. The criteria for inclusion of subjects in the study were 18 years of age or older, both genders, HBsAg carriers for more than 6 months, positive HCV-RNA carriers, with or without high values of alanine aminotransferase (ALT) and gamma-glutamyl transferase (GGT). Subjects who did not meet the aforementioned requirements and those patients co-infected with hepatitis D virus (HDV) and/or human immunodeficiency virus-1 (HIV-1) and patients who used or were using specific antiviral therapies against HBV or HCV were excluded from the research. This study was submitted and approved by the Research Ethics Committee of Holy House of Mercy Foundation of Pará, protocol Nos. 117/2009 and 684.432/2014, following the Guidelines and Rules for Research Involving Humans (Resolution 196 of the National Council of Health [Conselho Nacional de Saúde]). All subjects who agreed to participate in the study signed an Informed Consent Form (ICF).
Obtained samples
Liver biopsy specimens were obtained from patients with medical indications for investigation of changes in the liver parenchyma. The biopsies were performed with a Tru-Cut needle and were guided by ultrasound. Each sample was divided into two parts, one of which was subjected to histopathological examination after hematoxylin-eosin (HE), chromotrope aniline blue (CAB), Gomori’s reticulin and Shikata’s orcein staining at the Department of Anatomic Pathology, UFPA. The diagnosis followed the classification of the Brazilian Society of Hepatology [20] and the French classification METAVIR [21], scoring the activity of the portal and periportal inflammatory infiltrates from 0 to 3 and any structural changes from 0 to 4. The other part of each biopsy specimen was sent for genetic study at the Laboratory of Virology/ICB/UFPA and was stored at −70°C until the time of use. Blood samples were also collected in vacuum tubes containing EDTA as an anticoagulant, and the plasma was separated by centrifugation and stored at −20°C until the time of use to assess ALT, aspartate aminotransferase (AST), GGT, alpha-fetoprotein and viral markers.
RNA extraction
The fragment of liver tissue was kept in 500 μL of RNA Later Tissue Collection solution for RNA preservation and was subsequently extracted using a Norgen Biotek Corporation kit according to the manufacturer's recommended protocol. The sample purity was determined by the ratio between the absorbance measurements at 260 and 280 nm. A good extraction was considered in which the absorbance ratio ranged from 1.6 to 1.8. To observe the RNA purity and integrity, samples were loaded on 1% agarose gel in 1X TAE buffer stained with ethidium bromide (0.5 μg/mL) and subjected to electrophoresis at 80 V for 90 minutes. The observed bands correspond to the 28S, 18S and 5S rRNAs. The 28S and 18S major bands should appear on the gel at a 2:1 ratio. The 5S band should appear as faint as possible, indicating a low level of RNA degradation. To calculate the concentration, RNA was quantified by spectrophotometric reading on a Qubit 2.0 Fluorometer using Qubit RNA Assay Kits according to the manufacturer's protocol. For each sample, the concentration was adjusted to 50 ng/μL, and the sample was kept at −70°C until transcription.
Reverse transcription (cDNA)
RNA samples were transcribed into complementary DNA (cDNA) using the High-Capacity cDNA Reverse Transcription kit (without inhibitor) (Life Technologies, Carlsbad, CA, USA). The cDNA reaction was prepared with a final volume of 20.0 μL containing 4.2 μL H2O, 2.0 μL buffer, 2.0 μL random primers, 0.8 μL dNTP Mix (100 mM), 1.0 μL reverse transcriptase (RT) enzyme (provided in the kit), and 10.0 μL extracted RNA. Subsequently, the mixture was placed in a Perkin-Elmer thermocycler machine (Cetus Corp., USA) and was submitted to cycling at 25°C for 10 minutes, 37°C for 120 minutes and 85°C for 5 minutes.
mRNA quantification by Real-Time PCR (qPCR)
Real-time PCR (qPCR) was performed in 96-well plates using TaqMan reagents (Applied Biosystems, USA) in a Step One Plus machine (Life Technologies, Carlsbad, CA, USA). Expression assays for the NGF and p75 NTR genes, with (glyceraldehyde-3-phosphate dehydrogenase) GAPDH as a reference gene, were performed in separate wells (singleplex) for both the patient and Control groups with primers obtained commercially from Life Technologies (Carlsbad, CA, USA). Each reaction consisted of 15 μL 2X TaqMan Universal PCR Master Mix, 1.5 μL 20X TaqMan Gene Expression Assay (NGF Hs00171458_m1 and p75NTR Hs00609977_m1), 3 μL cDNA and 10.5 μL RNase-free water. The GAPDH gene (P/N 4326317E, Life Technologies, CA, USA) was used as a reference gene (endogenous control) to normalize the qPCR reactions.
The thermocycling conditions were 1 cycle of 2 min at 50°C followed by 10 min at 95°C, 40 cycles at 95°C for 15 sec and 60°C for 1 min. The relative expression level of each gene is presented as a multiple of the respective gene normalized to the Control sample.
In the standardization of the qPCR reactions, cDNAs and probes (endogenous and target genes) were titrated, aiming to calculate the amplification efficiencies of the reactions. For standardization, different cDNA concentrations were tested (neat and 4 dilutions of factor 2–1:2, 1:4, 1:8 and 1:16). All reactions were performed in triplicate. The same cDNA was simultaneously analyzed on plates (at different dilutions) with different probes to produce an efficiency curve to validate the 2−ΔΔCT analysis method.
Relative quantification (RQ) of target genes and the calculation of the confidence interval were performed using the Comparative CT (ΔΔCT) method or the 2−ΔΔ method, a method for RQ comparison of the exponential phase threshold without using the standard curve, where ΔΔCT = ΔCTsample- ΔCTreference (Life Technologies, Foster City, CA, USA).
Statistical procedures
All expression results are depicted in median values, and serum dosages are shown as averages. For statistical analysis, GraphPad Prism 5.0 [22] and BioEstat 5.0 [23] software programs were used. The differences between the groups were analyzed with the Kruskal-Wallis test and the Mann-Whitney U-test, as appropriate. Relationships between two variables were determined by Spearman correlation analysis. The significance level was established at 5% (p-value ≤ 0.05).
Results
In the three patient groups and the Control group, the ALT and GGT levels, the stage of liver fibrosis, and inflammatory activity were assessed (Table 1). Mean ALT levels were higher in the groups of patients with HCV (89 IU/L) and NVH (97 IU/L); the same was observed for GGT (78 IU/L and 179 IU/L, respectively). Comparisons of ALT values revealed significant differences between HBV vs. HCV (p = 0.005), HBV vs. NVH (p = 0.012), HCV vs. NVH (p = 0.033), HCV vs. Control (p = 0.004) and NVH vs. Control (p = 0.0007) groups.
Table 1. Clinical and laboratory information study participants with and without HBV and HCV, with different degrees of disease.
| Variables | HBV chronic hepatitis | HCV chronic hepatitis | Non-viral chronic hepatitis | Normal control |
|---|---|---|---|---|
| Study subjects (n) | 6 | 28 | 9 | 8 |
| Gender (F/M) | 4/2 | 16/12 | 5/4 | 5/3 |
| ALT (UI/L) Mean ± SD | 35.8 ± 60.8 | 89 ± 97.1 | 97 ± 76.21 | 27 ± 8.11 |
| Median | 30.5 | 56 | 96 | 27 |
| GGT (UI/L) Mean ± SD | 30 ± 14.7 | 78 ± 98.8 | 170 ± 141.77 | 29 ± 10.74 |
| Median | 20.5 | 55 | 155 | 29 |
| Fibrosis Stage a | ||||
| F0 (%) | 1 (16.7) | 2 (7.1) | 1 (16.7) | 8 (100.0) |
| F1 (%) | 4 (66.6) | 13 (46.5) | 2 (33.3) | - |
| F2 (%) | - | 7 (25.0) | - | - |
| F3 (%) | - | 3 (10.7) | 1 (16.7) | - |
| F4 (%) | 1 (16.7) | 3(10.7) | 2 (33.3) | - |
| Inflammation Stage b | ||||
| A0 (%) | 1 (16.7) | 4 (14.3) | 1 (16.7) | 8 (100.0) |
| A1 (%) | 4 (66.6) | 13 (46.4) | 4 (66.6) | - |
| A2 (%) | 1 (16.7) | 11 (39.3) | 1 (16.7) | - |
ALT: alanine aminotransferase; GGT: gamma-glutamyl transferase;
aFibrosis Stages—F0: no fibrosis; F1: portal without septa; F2: portal with some septa; F3: many septa without cirrhosis; F4: cirrhosis;
bInflammation Stages—A0: absent; A1: mild; A2: moderate.
Comparisons of GGT serum levels revealed significant differences among all groups (HBV vs. HCV [p = 0.016], HBV vs. NVH [p = 0.0003], HCV vs. NVH [p = 0.003], HCV vs. Control [p = 0.021] and NVH vs. Control [p = 0.0003]). Comparisons of ALT and GGT levels between HBV and Control groups were not significant (p > 0.05).
Among HCV patients, 53.6% were classified as F0 and F1 stages according to fibrosis level. Among patients with NVH, 50% were classified as F0 and F1 fibrosis stages and 50% were classified as stages F3 and F4, whereas among HBV patients, only 16.7% were classified in the F4 stage of fibrosis.
Evaluation of the degree of inflammatory response revealed that 83.3% of HBV and NVH patients were in the A1 and A2 stages, similar to what was observed among HCV patients (85.7%). In the Control group, 100% were classified as A0 stage (Table 1).
The NGF and p75 NTR mRNA expression levels were measured, and the values are expressed as fold changes relative to the reference calibrator.
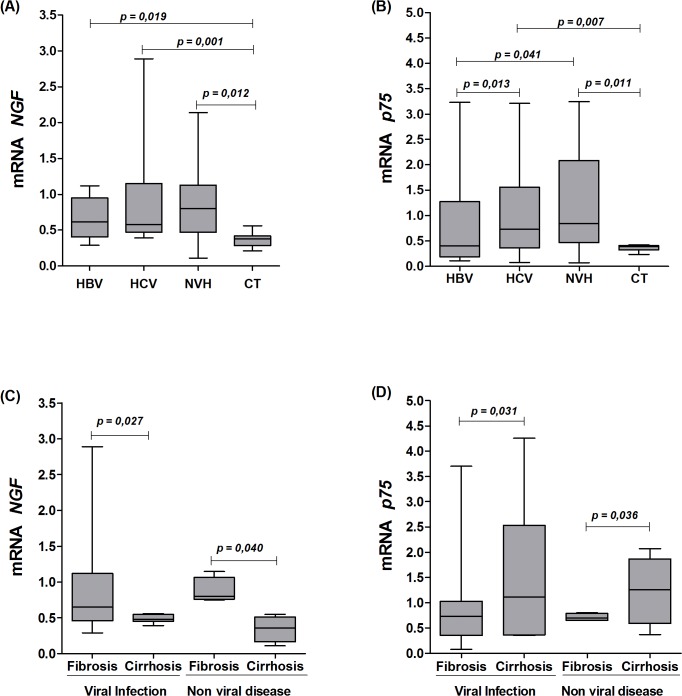
Fig. 1A shows that NGF mRNA levels were significantly higher in patients with HBV, HCV, and NVH when compared to the Control group. The same pattern was observed for p75 NTR mRNA expression (Fig. 1B). In the latter case, an increasing trend in mRNA expression was observed (in the ascending order of HBV < HCV < NVH); the differences between the HCV and NVH groups were statistically significant.
Fig 1. mRNA levels of NGF in the liver tissues.
A: mRNA levels of NGF in the liver tissues of subgroups with HBV, HCV and NVH and the Control group (CT). B: mRNA levels of p75NTR in the liver tissues of subgroups with HBV, HCV and NVH and the Control group (CT). C, D: mRNA levels of NGF and p75 NTR between the groups with hepatic fibrosis and cirrhosis due to viral and non-viral causes.
When mRNA levels were assessed according to the clinical conditions of liver fibrosis (without cirrhosis) or cirrhosis (Fig. 1C and 1D), the NGF mRNA levels were higher in the group with fibrosis than in the group with cirrhosis (p = 0.027 and p = 0040), whereas p75 NTR expression levels were the opposite, with higher expression in patients with cirrhosis than in the group with liver fibrosis; these differences were significant (p = 0.031 and p = 0.036), regardless of viral or non-viral disease.
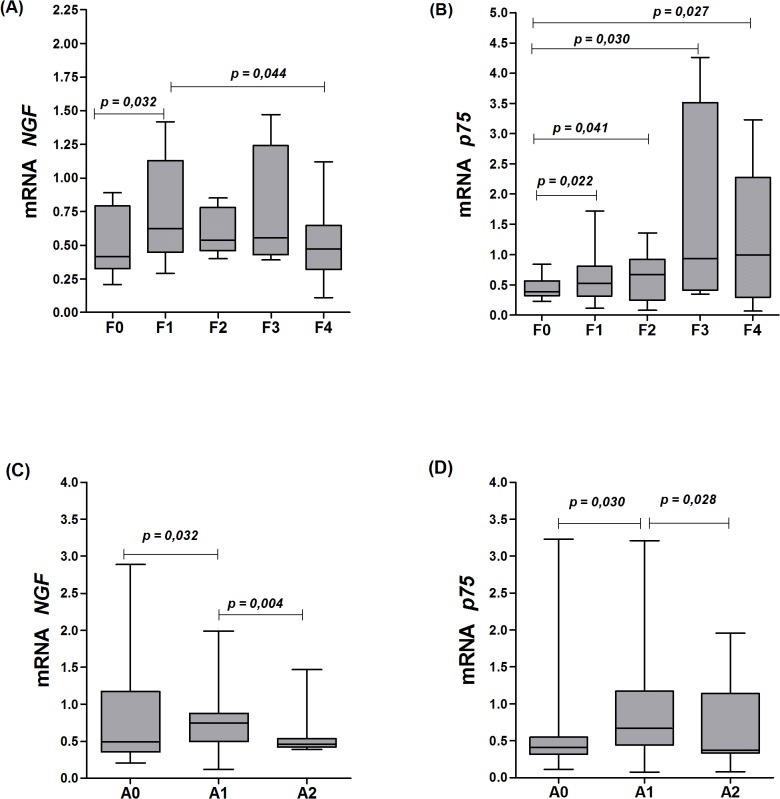
The grouping of all subjects (HBV, HCV and NVH) for the purpose of comparative analyses of fibrosis stage and hepatic inflammation and NGF and p75 NTR expression levels is depicted in Fig. 2. Regarding the degree of fibrosis, NGF was most expressed in F1 to F3 fibrosis, with lower expression in F0 and F4 groups (Fig. 1A). p75 NTR had lower mRNA expression levels in tissues without fibrosis (F0), with a sequential and a significant increase from F1 to F4 fibrosis stages (Fig. 2B).
Fig 2. mRNA levels of NGF according to the clinical conditions of liver.
A-D: mRNA levels of NGF and p75 NTR according to the stage of fibrosis (F0 to F4) and inflammatory activity (A0 to A2) in the livers of all individuals with liver disease in the study population.
According to the inflammatory process, NGF mRNA expression levels were higher in tissues at the A0 stage, and values decreased significantly to the A2 stage (Fig. 2C). Furthermore, p75 NTR mRNA expression levels were higher in liver samples with A1 and A2 stage inflammatory activities and lower in liver samples without inflammation (stage A0) (Fig. 2D).
Significant associations were observed between high ALT levels (p = 0.041) and normal GGT levels (p = 0.028) with increased hepatic expression of NGF, while p75NTR was expressed at higher levels (p = 0.012) in the livers of patients with high GGT levels (Table 2).
Table 2. Comparison of median values of relative NGF and p75 NTR gene expression with serum levels of liver enzymes in individuals with and without HBV and HCV, with different degrees of the disease.
| Gene | Normal ALT | High ALT | p | Normal GGT | High GGT | p |
|---|---|---|---|---|---|---|
| (n = 26) | (n = 17) | (n = 19) | (n = 24) | |||
| NGF | 0.58 | 0.77 | 0.041 a | 0.80 | 0.55 | 0.028 b |
| p75 NTR | 0.82 | 0.56 | 0.55 c | 0.62 | 0.81 | 0.012 d |
ALT: alanine aminotransferase (normal: 14 to 55 UI/L); GGT: gamma-glutamyl transferase (normal: < 50 UI/L);
aNGF (normal vs. high ALT);
bNGF (normal vs. high GGT);
c p75NTR (normal vs. high ALT);
d p75NTR (normal vs. high GGT).
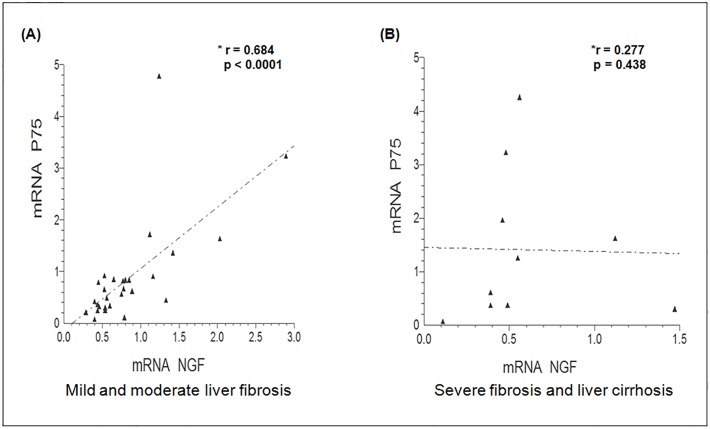
A positive correlation was observed between p75 NTR and NGF mRNA expression levels in liver tissue (Fig. 3A) with mild to moderate fibrosis (p <0.0001); however, this correlation was not observed in tissues with severe liver fibrosis and cirrhosis (Fig. 3B).
Fig 3. Correlation of p75 NTR and NGF mRNA levels with fibrosis and cirrhosis stages.
A, B: Spearman correlation analysis with mild to moderate fibrosis stages and severe fibrosis and cirrhosis stages.
Discussion
In this study, we found that the NGF and p75 NTR mRNA expression levels in liver tissues in chronic HBV, HCV and NVH carriers were significantly higher when compared with normal liver biopsies. Our results partially disagree with in vitro studies and studies of experimentally induced liver fibrosis [16,17,20], which showed the expression of these genes only in fibrous liver tissue or after partial hepatectomy. Our results corroborate the findings of Trim et al. [15] and Kendall et al. [19], who used immunohistochemistry in normal liver to identify perisinusoidal cells with morphologies consistent with HSCs and p75NTR positivity.
We observed that NGF mRNA expression was higher in subjects with hepatic fibrosis without cirrhosis compared to the group with cirrhosis, suggesting that NGF is expressed by hepatocytes in a state of regeneration and proliferation, while it is clear that NGF mRNA levels are lower in cirrhotic liver, where this regeneration process is reduced or absent [24,25]. In contrast to NGF, p75 NTR mRNA expression was higher in subjects with cirrhosis compared to patients with fibrosis only, with a correlation between p75 NTR expression and the reactivity of activated HSCs [26], similar to other in vitro experimental fibrosis studies [15–19].
As expected, there was an association between higher NGF mRNA expression levels and high ALT and normal GGT serum concentrations, while p75 NTR was associated with high GGT concentrations. ALT is commonly high in chronic hepatocellular damage; however, with progression to fibrosis, ALT activity is typically reduced [27,28]. Although some patients with normal liver enzymes had severe fibrosis or even cirrhosis, most of them had a moderate liver disease caused by HCV.
A common feature of the study population was chronic liver injury with the absence of any prior drug therapy. Thus, we have evaluated the NGF and p75 NTR mRNA expression levels in relation to the degree of fibrosis and hepatic inflammatory activity, showing associations between the expression levels of these two genes and the progression of liver injury. In the first stage of liver fibrosis and inflammation (F1/A1), NGF mRNA levels were significantly higher than in other liver injury stages, and p75NTR mRNA levels were lower. This result maybe is due to the fact that in the early stages of liver fibrosis, activation of human HSCs which is mediated by p75NTR stimulates the release of growth factors such as hepatocyte growth factor (HGF), and stimulates hepatocyte regeneration and proliferation [25,29].
We observed a direct, progressive increase in p75 NTR mRNA levels with the progression from fibrosis to cirrhosis, demonstrating that activated HSCs, mediated by p75NTR expression, can mediate both the start and end of liver regeneration. In the early stages of liver regeneration, activated HSCs contain high HGF levels; this strongly mitogenic growth factor can substitute for the effects of transforming growth factor (TGF)-β1, the major hepatocyte antiproliferative factor produced by these cells [30]. In contrast, in the terminal stages of liver scarring, activated HSCs produce high TGF-β1 levels, triggering the start of an autocrine signaling cycle that perpetuates the activation of HSCs and becomes a barrier to liver regeneration [30].
Differing from our study, some results with experimental injuries induced by carbon tetrachloride (CCl4) and in vitro studies strongly suggest the pro-apoptotic role of NGF, directly regulating apoptosis of activated HSCs and mediated by p75NTR, which would represent a potential determining factor in liver fibrosis resolution. However, these results are best characterized only in self-limited injuries [16]. A positive correlation between p75NTR and NGF expression was identified in the early stages of liver fibrosis, characterizing a likely paracrine action of activated HSCs mediated by p75NTR in the compensatory regeneration of hepatocytes via NGF; however, this association was not sustained under severe liver fibrosis and cirrhosis conditions. In the livers of patients with cirrhosis caused by HCV, Novo et al. found that activated HSCs are resistant to most pro-apoptotic stimuli, including NGF neurotrophin due to overexpression of the Bcl-2 anti-apoptotic marker present in liver tissue; this characteristic may play a key role in the progression of fibrosis in chronic liver diseases [31].
Therefore, we suggest that, in accordance with the profile of the study population, our gene expression results could be also the product of other stimuli in livers damaged by viral persistence of HCV, HBV and other chronic infections because HSCs express receptors for HCV, such as CD80 and LDL (low-density lipoprotein) receptor, allowing an increase in the viral infection rate. The expression of non-structural proteins and core HCV proteins induces stellate cell proliferation and the release of inflammatory signals [32,33]. In nonalcoholic fatty liver disease, adipokines may mediate hepatic fibrogenesis [34,35]. As an example of these adipokines, leptin, a circulating adipogenic hormone, promotes fibrogenesis in HSCs [36–38]. Equally important for the development of liver fibrosis is insulin resistance, which is related to steatosis [39,40] and oxidative stress, is associated with ethanol metabolism, and is an important stimulus for hepatic fibrogenesis [41].
Taken as a whole, our results demonstrate that in the course of chronic liver disease in the study population, activated HSCs are negatively regulated by p75NTR-mediated inhibition of hepatocyte proliferation, which can be modulated by viral and non-viral components and supported by the persistence of aggression to the liver. The degree of histological fibrosis is an important marker of the disease stage [10,11], as the natural history of hepatitis B and C involves the gradual progression of hepatic fibrosis, which can eventually lead to cirrhosis, where most related complications occur [3]. In this context, the individual knowledge of p75NTR expression levels would be a potent marker for the progression of hepatic fibrosis. However, we understand that it would be necessary to understand the gene expression mechanisms of this neurotrophin and its receptor subsequent to already established antiviral therapies. Finally, regarding the small number of subjects enrolled in the present study, specifically in the HBV group, which could not represent the molecular pathogenesis of the fibrotic transition to severe stages, we suggest that a follow up study should be conducted with a large number of patients, aiming to confirm our results.
Acknowledgments
We thank all of the individuals who participated in this research.
Data Availability
The raw data belongs to the Virus Laboratory of the Federal University of Para, which is part of the National Network of CD4 + and CD8 + T Cell Count and Viral Load Plasma managed by the Brazilian Ministry of Health. The data cannot be accessed without the approval of the Ethical Committee of Federal University of Para and the Brazilian Ministry of Health, as there are ethical restrictions on the data, given that the research deals with information starting from biological samples taken from patients. Interested researchers should send requests to access the data to the Corresponding Author at vallinoto@ufpa.br or vallinoto@me.com, and the Corresponding Author will reach out to the Brazilian Ministry of Health and to the Ethical Committee of Federal University of Para for permission.
Funding Statement
This study was funded by the National Council for Scientific and Technological Development (Conselho Nacional de Desenvolvimento Científico e Tecnológico—CNPq) and by the Dean of Research and Graduate Studies Office (Pró-Reitoria de Pesquisa e Pós-Graduação—PROPESP/UFPA). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Aguilera Guirao A, Romero Yuste S, Regueiro BJ. Epidemiology and clinical manifestations of viral hepatitis. Enferm Infecc Microbiol Clin. 2006;24: 264–276. [DOI] [PubMed] [Google Scholar]
- 2. Alter MJ. Epidemiology of hepatitis C virus infections. World J Gastroenterol. 2006;13: 2436–2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mita E, Hayashi N, Iio S, Takehara T, Hijioka T, Kasahara A, et al. Role of Fas ligand in apoptosis induced by hepatitis C virus infection. Biochem Biophys Res Commun. 1994;204: 468–474. [DOI] [PubMed] [Google Scholar]
- 4. Guicciardi ME, Gores GJ. Apoptosis: a mechanism of acute and chronic liver injury. Gut 2005;54: 1024–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Malhi H, Guicciardi ME, Gores GJ. Hepatocyte death: a clear and present danger. Physiol Rev. 2010; 90: 1165–1194. doi: 10.1152/physrev.00061.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Benyon RC, Iredale JP. Is liver fibrosis reversible? Gut 2000;46: 443–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gäbele E, Brenner DA, Rippe RA. Liver fibrosis: signals leading to the amplification of the fibrogenic hepatic stellate cell. Front Biosci. 2003;8: d69–d77. [DOI] [PubMed] [Google Scholar]
- 8. Zaman A, Rosen HR, Ingram K, Corless CL, Oh E, Smith K. Assessment of FIBROSpect II to detect hepatic fibrosis in chronic hepatitis C patients. Am J Med. 2007;120: 280.e9–14. [DOI] [PubMed] [Google Scholar]
- 9. Bedossa P. Intraobserver and interobserver variations in liver biopsy interpretation in patients with chronic hepatitis C. The French METAVIR Cooperative Study Group. Hepatol. 1994;20: 15–20. [PubMed] [Google Scholar]
- 10. Poynard T, Yuen MF, Ratziu V, Lai CL. Viral hepatitis C. Lancet 2003;362: 2095–2100. [DOI] [PubMed] [Google Scholar]
- 11. Ryder SD, Irving WL, Jones DA, Neal KR, Underwood JC, Trent Hepatitis C Study Group. Progression of hepatic fibrosis in patients with hepatitis C: a prospective repeat liver biopsy study. Gut 2004;53: 451–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cohen S, Levi-Montalcini R, Hamburger V. A nerve growth-stimulating factor isolated from sarcomas 37 and 180. Proc Natl Acad Sci U S A. 1954;40: 1014–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Levi-Montalcini R, Hamburger V. Selective growth stimulating effects of mouse sarcoma on the sensory and sympathetic nervous system of the chick embryo. J Exp Zool. 1951;116: 321–361. [DOI] [PubMed] [Google Scholar]
- 14. Sofroniew MV, Howe CL, Mobley WC. Nerve growth factor signaling, neuroprotection, and neural repair. Annu Rev Neurosci. 2001;24: 1217–1281. [DOI] [PubMed] [Google Scholar]
- 15. Trim N, Morgan S, Evans M, Issa R, Fine D, Afford S, et al. Hepatic stellate cells express the low affinity nerve growth factor receptor p75 and undergo apoptosis in response to nerve growth factor stimulation. Am J Pathol. 2000;156: 1235–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Oakley F, Trim N, Constandinou CM, Ye W, Gray AM, Frantz G, et al. Hepatocytes express nerve growth factor during liver injury: evidence for paracrine regulation of hepatic stellate cell apoptosis. Am J Pathol. 2003;16: 1849–1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Passino MA, Adams RA, Sikorski SL, Akassoglou K. Regulation of hepatic stellate cell differentiation by the neurotrophin receptor p75NTR. Science 2007;315: 1853–1856. [DOI] [PubMed] [Google Scholar]
- 18. Asai K, Tamakawa S, Yamamoto M, Yoshie M, Tokusashi Y, Yaginuma Y, et al. Activated hepatic stellate cells overexpress p75NTR after partial hepatectomy and undergo apoptosis on nerve growth factor stimulation. Liver Int. 2006;26: 595–603. [DOI] [PubMed] [Google Scholar]
- 19. Kendall TJ, Hennedige SA, Aucott RL, Hartland SNN, Vernon MA, Benyon RC, et al. p75 Neurotrophin receptor signaling regulates hepatic myofibroblast proliferation and apoptosis in recovery from rodent liver fibrosis. Hepatol. 2009;49: 901–910. [DOI] [PubMed] [Google Scholar]
- 20. Gayotto LC. Comitê SBP-SBH. Visão histórica e consenso nacional sobre a classificação das hepatites crônicas [Historical vision and national consensus on the classification of chronic hepatitis]. Project of the Liver Pathology Club of the Brazilian society of pathology approved by the Brazilian Society of Hepatology. GED Gastroenterol Endosc Dig. 2000;19: 137–140. [Google Scholar]
- 21. Bedossa P, Poynard T. An algorithm for the grading of activity in chronic hepatitis C. The METAVIR Cooperative Study Group. Hepatol. 1996;24: 289–293. [DOI] [PubMed] [Google Scholar]
- 22. Ayres M, Ayres M, Ayres DL, Santos AS. BioEstat 5.0: Aplicações estatísticas nas áreas de ciências biológicas e médicas [BioEstat 5.0: statistical applications in biological and medical sciences] Belém: Sociedade Civil Mamirauá, 2008. [Google Scholar]
- 23.GraphPad Software Inc. Prism (data analysis software system), version 5. Software e Guia do Usuário, 2007.
- 24. Taub R. Liver regeneration: from myth to mechanism. Nat Rev Mol Cell Biol. 2004;5: 836–847. [DOI] [PubMed] [Google Scholar]
- 25. Bataller R, Brenner DA. Hepatic stellate cells as a target for the treatment of liver fibrosis. Semin Liver Dis. 2001;21: 437–451. [DOI] [PubMed] [Google Scholar]
- 26. Pinzani M. Novel insights into the biology and physiology of the Ito cell. Pharmacol Ther. 1995;66: 387–412. [DOI] [PubMed] [Google Scholar]
- 27. Williams AL, Hoofnagle JH. Ratio of serum aspartate to alanine aminotransferase in chronic hepatitis. Relationship to cirrhosis. Gastroenterol. 1998;95: 734–739. [DOI] [PubMed] [Google Scholar]
- 28. Sheth SG, Flamm SL, Gordon FD, Chopra S. AST/ALT ratio predicts cirrhosis in patients with chronic hepatitis C virus infection. Am J Gastroenterol. 1998;93: 44–48. [DOI] [PubMed] [Google Scholar]
- 29. Uyama N, Shimahara Y, Kawada N, Seki S, Okuyama H, Limuro Y, et al. Regulation of cultured rat hepatocyte proliferation by stellate cells. J Hepatol. 2002;36: 590–599. [DOI] [PubMed] [Google Scholar]
- 30. Oakley F, Teoh V, Ching-A-Sue G, Bataller R, Colmenero J, Josson JR, et al. Angiotensin II activates I kappaB kinase phosphorylation of RelA at Ser 536 to promote myofibroblast survival and liver fibrosis. Gastroenterol. 2009;136: 2334–2344.e.1. doi: 10.1053/j.gastro.2009.02.081 [DOI] [PubMed] [Google Scholar]
- 31. Novo E, Marra F, Zamara E, Valfrè di Bonzo L, Monitillo L, Cannito S, et al. Overexpression of Bcl-2 by activated human hepatic stellate cells: resistance to apoptosis as a mechanism of progressive hepatic fibrogenesis in humans. Gut 2006;55: 1174–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bataller R, Paik YH, Lindquist JN, Lemasters JJ, Brenner DA. Hepatitis C virus core and nonstructural proteins induce fibrogenic effects in hepatic stellate cells. Gastroenterol. 2004;126: 529–540. [DOI] [PubMed] [Google Scholar]
- 33. Shin JY, Hur W, Wang JS, Jang JW, Kim CW, Bae SH, et al. HCV core protein promotes liver fibrogenesis via up-regulation of CTGF with TGF-beta1. Exp Mol Med. 2005;37: 138–145. [DOI] [PubMed] [Google Scholar]
- 34. London RM, George J. Pathogenesis of NASH: animal models. Clin Liver Dis. 2007;11: 55–74, viii. [DOI] [PubMed] [Google Scholar]
- 35. Ikejima K, Okumura K, Kon K, Takei Y, Sato N. Role of adipocytokines in hepatic fibrogenesis. J Gastroenterol Hepatol. 2007;22(Suppl 1): S87–S92. [DOI] [PubMed] [Google Scholar]
- 36. Ikejima K, Takei Y, Honda H, Hirose M, Yoshikawa M, Zhang YJ, et al. Leptin receptor-mediated signaling regulates hepatic fibrogenesis and remodeling of extracellular matrix in the rat. Gastroenterol. 2002;122: 1399–1410. [DOI] [PubMed] [Google Scholar]
- 37. Lin S, Saxena NK, Ding X, Stein LL, Anania FA. Leptin increases tissue inhibitor of metalloproteinase I (TIMP-1) gene expression by a specificity protein 1/signal transducer and activator of transcription 3 mechanism. Mol Endocrinol. 2006;20: 3376–3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Leclercq IA, Farrell GC, Schriemer R, Robertson GR. Leptin is essential for the hepatic fibrogenic response to chronic liver injury. J Hepatol. 2002;37: 206–213. [DOI] [PubMed] [Google Scholar]
- 39. Kamada Y, Tamura S, Kiso S, Matsumoto H, Saji Y, Yoshida Y, et al. Enhanced carbon tetrachloride-induced liver fibrosis in mice lacking adiponectin. Gastroenterol. 2003;125: 1796–1807. [DOI] [PubMed] [Google Scholar]
- 40. Lonardo A, Adinolfi LE, Loria P, Carulli N, Ruggiero G, Day CP. Steatosis and hepatitis C virus: mechanisms and significance for hepatic and extrahepatic disease. Gastroenterol. 2004;126: 586–597. [DOI] [PubMed] [Google Scholar]
- 41. Parola M, Robino G. Oxidative stress-related molecules and liver fibrosis. J Hepatol. 2001;35: 297–306. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The raw data belongs to the Virus Laboratory of the Federal University of Para, which is part of the National Network of CD4 + and CD8 + T Cell Count and Viral Load Plasma managed by the Brazilian Ministry of Health. The data cannot be accessed without the approval of the Ethical Committee of Federal University of Para and the Brazilian Ministry of Health, as there are ethical restrictions on the data, given that the research deals with information starting from biological samples taken from patients. Interested researchers should send requests to access the data to the Corresponding Author at vallinoto@ufpa.br or vallinoto@me.com, and the Corresponding Author will reach out to the Brazilian Ministry of Health and to the Ethical Committee of Federal University of Para for permission.