Abstract
Background/Aims
In the present study, we examined whether DNA methylation of the brain-derived neurotrophic factor (BDNF) promoter is associated with the manifestation and clinical presentation of Alzheimer's disease (AD).
Methods
Of 20 patients with AD and 20 age-matched normal controls (NCs), the DNA methylation of the BDNF promoter (measured using peripheral blood samples) was completely analyzed in 12 patients with AD and 6 NCs. The resulting methylation levels were compared statistically. Next, we investigated the correlation between the DNA methylation levels and the clinical presentation of AD.
Results
The total methylation ratio (in %) of the 20 CpG sites was significantly higher in the AD patients (5.08 ± 5.52%) than in the NCs (2.09 ± 0.81%; p < 0.05). Of the 20 CpG sites, the methylation level at the CpG4 site was significantly higher in the AD subjects than in the NCs (p < 0.05). Moreover, the methylation level was significantly and negatively correlated with some neuropsychological test subscores (registration, recall, and prehension behavior scores; p < 0.05).
Conclusion
These results suggest that the DNA methylation of the BDNF promoter may significantly influence the manifestation of AD and might be associated with its neurocognitive presentation.
Key Words: Brain-derived neurotrophic factor, BDNF, DNA methylation, Alzheimer's disease, Epigenetics, Neurotrophic factors
Introduction
Alzheimer's disease (AD) is a progressive neurodegenerative disorder that is mainly characterized by memory disorder, visuospatial impairment, attentional impairment, and executive dysfunction as its core symptoms, leading to neuropsychiatric problems [1,2,3]. The deposition of causative proteins, such as amyloid-beta (Aβ) and tau (τ), in the pathogenesis of AD has been reported previously and has been focused on as a possible therapeutic target [4,5]. Since the 1990s, apolipoprotein E (APOE) has been reported as a strong candidate gene for sporadic AD, and this speculation has been verified in a large sample genome-wide association study [6,7]. The pathophysiological mechanism responsible for the effect of APOE polymorphisms on degenerative alterations in AD patients has been shown to involve APOE4-mediated neurovascular vulnerability, leading to neural loss or dysfunction [8]. Moreover, the CpG methylation alterations of the amyloid precursor protein (APP), microtubule-associated protein tau (MAPT), and glycogen synthase kinase-3β (GSK3B) genes in neuronal cells have been reported as pathogenic mechanisms of sporadic AD [9]. Thus, among some epigenetic markers, such DNA methylations have been implicated as crucial biomarkers of AD [10].
Brain-derived neurotrophic factor (BDNF) promotes the development and survival or the maintenance function of neural cells [11]. From a neuroprotective viewpoint, BDNF secretion within the synapse cleft is modulated by an activity-dependent change and long-term potentiation, which influences cognitive function and protects against neural cell death caused by the aggregation of Aβ and τ proteins in the AD pathology through the regulation of neural plasticity [12]. As a representative BDNF functional SNP, Val66Met influences intracellular BDNF trafficking and secretory regulation and is linked to cognitive functions or neuropsychiatric symptoms in AD patients [13,14,15]. This SNP (Val66Met) might also be a biomarker regulating or reflecting the clinical presentations and prognosis of AD patients.
In some previous studies, DNA methylation alterations of the BDNF promoter region (evaluated using postmortem or peripheral blood samples) have been reported in patients with neuropsychiatric disorders [16,17,18]. Hypermethylation of the BDNF promoter region in postmortem AD brain tissues has been described, supporting a reduction in the mRNA or protein levels in certain brain areas (hippocampus or frontal cortex) [16,19,20]. Such studies imply that BDNF methylation might be a useful surrogate marker of the neurodegenerative process that occurs in AD. However, the association between the BDNF DNA methylation levels and the clinical presentation of AD (e.g., cognitive function, neuropsychiatric symptoms, and plasma BDNF levels) has not been previously investigated.
In the present cross-sectional study, we hypothesized that DNA methylation in the BDNF promoter region might be a biomarker for the manifestation of AD or its relevant clinical presentations during the neurodegenerative course. Accordingly, we compared the DNA methylation levels of the exon I promoter region in BDNF between AD patients and age-matched normal controls (NCs). Then we investigated the correlation between the DNA methylation levels of BDNF and some clinical presentations using cognitive and neuropsychiatric test batteries.
Patients and Methods
Participants
Forty peripheral blood samples were provided by 3 institutions: 20 AD patients (8 males and 12 females; mean age: 66.5 ± 5.07 years) were recruited out of consecutive memory clinic outpatients who visited the Jikei University Hospital (Tokyo) or the Jikei University Kashiwa Hospital (Kashiwa city), and 20 NCs (8 males and 12 females; mean age: 66.5 ± 5.12 years) were recruited from Juntendo University Hospital (Tokyo). Genomic DNA was obtained from mononuclear cells in the peripheral blood samples using the Qiagen DNA extraction kit [14]. All AD patients were diagnosed as having probable AD based on the National Institute of Neurology and Communicative Disorder and Stroke/Alzheimer Disease and Related Disorder Association (NINCDS/ADRDA) criteria [21]. The neuropsychiatric symptoms in the AD patients were assessed based on information obtained from a structured interview with each patient's caregiver by the same geriatric psychiatrists using the behavioral pathology in AD (Behave-AD) scale [22]. Two neuropsychological tests, the Frontal Assessment Battery (FAB) reflecting executive function and the Mini-Mental State Examination (MMSE) reflecting global cognitive function, were also administered to the AD patients by a clinical psychologist [23,24]. NCs with a past history of treatment for any psychiatric disorder were excluded.
This study was approved by the Ethics Committee of the Jikei University School of Medicine (Tokyo and Kashiwa city) and the Juntendo University School of Medicine (Tokyo). Written informed consent was obtained from both the patients and their caregivers.
Analysis and Measurement of DNA Methylation Levels in the BDNF Promoter Region
According to the enrollment protocol, each DNA sample (1 µg) was subjected to bisulfite treatment with the MethylEasy™ Xceed Rapid DNA Bisulfite Modification Kit (Takara Bio Inc.). Using the bisulfite-treated DNA as a template, the BDNF promoter region was amplified by nested polymerase chain reaction (PCR) with TaKaRa EpiTaq HS (Takara Bio Inc.). The PCR products were purified using the QIAquick PCR purification kit (Qiagen), and the restriction sites PstI and BglII were added to the second PCR product. The detailed conditions are described below: a total volume of 50 µl containing 0.25 µl of TaKaRa EpiTaq HS (5 U/µl), 5 µl of 10× EpiTaq PCR buffer (Mg+ free), 5 µl of 25 mM MgCl2, 6 µl of dNTP mixture, 4 µl of template DNA, 0.4 µl of PCR forward primer (50 µM), 0.4 µl of PCR reverse primer (50 µM), and 28.95 µl of PCR-grade water. The primers used for the first PCR were as follows: 5′-GGGTTGAGGTTGGGGTTGGA-3′ (BDNF_P_F1 forward primer) and 5′-CTCCTCCAAACCTTCCCTCCTCTACTTC-3′ (BDNF_P_R1 reverse primer). The thermal profile consisted of 30 cycles at 98°C for 10 s, at 55°C for 30 s, and at 72°C for 60 s. The primers used for the second PCR were as follows: 5′-ATCCTGCAGGTTTTAGGGGAGGAGGTGAGGATAGG-3′ (Pst_BDNF_P_F2 forward primer) and 5′-TCCAGATCTCCCCAAAAATAACTCCAAATC-3′ (BDNF_P_Bgl_R2 reverse primer). The thermal profile was followed by 40 cycles at 98°C for 10 s, at 53°C for 30 s, and at 72°C for 60 s.
According to the standard technique, purified PCR products were digested with PstI and BglII and inserted into the Litmus 28i vector (New England Biolabs). The plasmids were transformed into Stbl3 chemically competent Escherichia coli cells (Invitrogen), and colonies were obtained after overnight cultivation. Colony PCR was performed using KOD FX Neo (Toyobo) in a total volume of 50 µl containing 1 µl of KOD FX Neo (1 U/µl), 25 µl of 2× PCR buffer, 10 µl of 2 mM dNTP mixture, 0.2 µl of PCR forward primer (50 µM), 0.2 µl of PCR reverse primer (50 µM), and 13.6 µl of PCR-grade water. About 40 positive colonies were selected. The primers used for the colony PCR were as follows: 5′-CGCCAGGGTTTTCCCAGTCACGAC-3′ (M13 primer P7) and 5′-AGCGGATAACAATTTCACACAGGAAAC-3′ (M13 primer P8). The thermal profile was 94°C for 2 min, followed by 30 cycles at 98°C for 10 s and at 68°C for 30 s.
The final PCR product was treated using EXOSAP-IT (Affymetrix, Inc.), and direct nucleotide sequencing with an ABI 3730 DNA analyzer [sequencing reactions were performed using BigDye Terminator v3.1 Cycle Sequencing Kits (Applied Biosystems)] was performed to identify the DNA sequences. The primers used for the sequences were as follows: 5′-CGCCAGGGTTTTCCCAGTCACGAC-3′ (M13 primer P7), 5′-GTAAAACGACGGCCAGT-3′ (M13 primer M3), 5′-AGCGGATAACAATTTCACACAGGAAAC-3′ (M13 primer P8), and 5′-CAGCTATGACCATGATTACG-3′ (M13 primer RV2). All sequence analyses were performed using DNA Sequencing Analysis software v5.3.1 (Applied Biosystems). The DNA methylation data were analyzed using the quantification tool for methylation analysis (QUMA; Riken, Japan; http://quma.cdb.riken.jp/).
Statistical Analysis
SPSS 19.0J for Windows (SPSS Japan Inc.) was used for all statistical analyses. First, we evaluated the data normality of each variable and the hypothesis that the distributions of all variables would be rejected by the Shapiro-Wilk test (p < 0.05), except for age, when the AD and NC groups were compared. Thus, nonparametric statistics were used for most of the following data evaluations, except for the age comparison. We compared the DNA methylation levels (in %) of the BDNF promoter region between the AD and NC groups using the Mann-Whitney test. The sex ratio (female to male) between the AD and NC groups was assessed using the χ2 test. Only the age comparison between the AD and NC groups was performed using the t test. Moreover, we examined the correlations between the methylation levels (in %) of significant sites, as detected based on comparisons between the groups, and the clinical presentations [age, duration of illness in months, MMSE total and subtest scores, FAB total and subtest scores, Behave-AD total score and each of the 7 subscale scores, Clinical Dementia Rating Scale sum of boxes (CDR SB), and the plasma BDNF concentration (in pg/ml)] using the Spearman rank correlation coefficient. A p value <0.05 was considered statistically significant.
Results
Patient Characteristics
Of the 40 DNA samples available, the DNA methylation levels were completely analyzed in 18 subjects (12 AD patients and 6 NCs). Among the two groups, the sex and age ratios were not significantly different (table 1). All 12 AD patients had taken acetylcholine esterase inhibitors (AchEI) (11 patients had taken donepezil and 1 had taken galantamine) for over 6 months.
Table 1.
Subject characteristics of the AD and NC groups
| NC (n = 6) | AD (n = 12) | t/×2 | p value | |
|---|---|---|---|---|
| Males/females | 4/2 | 4/8 | 1.8a | 0.18 |
| Age, years | 64.33 ±6.53 | 65.67± 5.18 | 0.473b | 0.642 |
| Age at onset, years | - | 61.90 ± 6.11 | ||
| Duration of illness, months | - | 44.25 ± 26.06 | ||
| MMSE score | - | 18.83 ± 7.94 | ||
| FAB score | - | 10.33 ± 3.87 | ||
| Behave-AD score | - | 5.25 ± 3.98 | ||
| CDR SB | - | 6.04 ± 3.88 |
All values are given as means ± SD except for the sex ratio.
The sex ratio was analyzed using the ×2 test.
Age was compared using the t test.
Comparison of DNA Methylation Levels in the BDNF Promoter Region
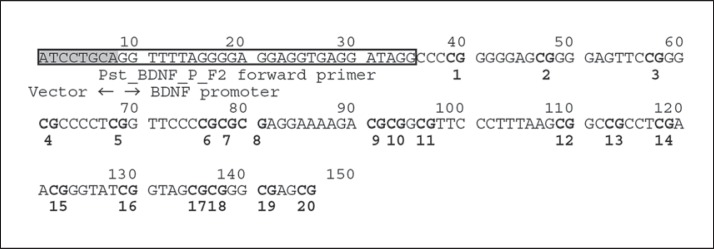
Within the promoter region of exon I, 20 CpG sites with sequences that had been verified by an analysis of the complementary strand were targeted for statistical comparison. The targeted regions, consisting of about 120 bp, were positioned from the terminal point of the forward primer (GenBank: AF411339; at position 53144) for direct nucleotide sequencing (fig. 1). The total methylation ratio of the 20 CpG sites was calculated and compared between the AD and NC groups, and a significantly higher value was seen in the AD group (5.08 ± 5.52%) than in the NCs (2.09 ± 0.81%; p = 0.041; table 2). Moreover, among the 20 CpG sites, the DNA methylation level at the CpG4 site (fig. 1) was significantly higher in the AD patients than in the NCs (p = 0.014; table 2). However, no significant differences in the methylation levels at the other CpG sites were observed (table 2).
Fig. 1.
BDNF promoter region examined in the DNA methylation analysis. The target region used for the DNA methylation analysis contains 20 CpG sites, shown in bold type. The vector region has been shaded. The Pst_BDNF_P_F2 forward primer is shown in the box. The targeted regions, consisting of about 120 bp, were positioned from the terminal point of the forward primer (GenBank: AF411339; at position 53144) for direct nucleotide sequencing.
Table 2.
Comparison of CpG méthylation (in %) among the AD and NC groups
| NC (n = 6) |
AD (n = 12) |
Mann-Whitney Z score | p value | |||
|---|---|---|---|---|---|---|
| mean ± SD | median | mean ± SD | median | |||
| Total methylation ratio | 2.09 ± 0.81 | 2.03 | 5.08 ± 5.52 | 3.28 | 14 | 0.041* |
| CpG1 | 1.53 ± 1.75 | 1.09 | 3.49 ± 6.81 | 0.00 | 3.15 | 0.616 |
| CpG2 | 1.11 ± 2.72 | 0.00 | 2.59 ± 6.02 | 0.00 | 32.5 | 0.653 |
| CpG3 | 0.00 ± 0.00 | 0.00 | 2.64 ± 5.97 | 0.00 | 27 | 0.194 |
| CpG4 | 0.00 ± 0.00 | 0.00 | 4.85 ± 6.91 | 3.35 | 12 | 0.014* |
| CpG5 | 0.00 ± 0.00 | 0.00 | 2.10 ± 5.73 | 0.00 | 27 | 0.194 |
| CpG6 | 1.54 ± 2.46 | 0.00 | 1.83 ± 6.34 | 0.00 | 28 | 0.249 |
| CpG7 | 1.75 ± 1.97 | 1.39 | 1.16 ± 1.76 | 0.00 | 30 | 0.523 |
| CpG8 | 3.55 ± 6.86 | 0.00 | 7.31 ± 18.36 | 0.00 | 33 | 0.749 |
| CpG9 | 4.84 ± 3.19 | 5.26 | 4.23 ± 7.28 | 0.00 | 24 | 0.239 |
| CpG10 | 1.84 ± 3.16 | 0.00 | 1.29 ± 2.04 | 0.00 | 34 | 0.823 |
| CpG11 | 1.80 ± 2.02 | 1.39 | 13.02 ± 29.16 | 1.52 | 33 | 0.764 |
| CpG12 | 9.04 ± 9.57 | 7.98 | 20.39 ± 33.12 | 3.81 | 35.5 | 0.962 |
| CpG13 | 1.79 ± 3.14 | 0.00 | 8.29 ± 24.30 | 0.00 | 36 | 1 |
| CpG14 | 1.18 ± 1.84 | 0.00 | 5.42 ± 11.37 | 0.00 | 30 | 0.523 |
| CpG15 | 2.00 ± 3.11 | 0.00 | 0.68 ± 1.67 | 0.00 | 28 | 0.304 |
| CpG16 | 3.69 ± 3.49 | 3.33 | 4.73 ± 5.44 | 3.42 | 35 | 0.924 |
| CpG17 | 2.27 ± 5.57 | 0.00 | 3.83 ± 3.23 | 4.10 | 22 | 0.161 |
| CpG18 | 2.60 ± 4.07 | 0.00 | 2.89 ± 5.13 | 0.00 | 36 | 1 |
| CpG19 | 0.58 ± 1.41 | 0.00 | 3.30 ± 4.35 | 0.00 | 24.5 | 0.199 |
| CpG20 | 1.52 ± 2.44 | 0.00 | 6.85 ± 10.23 | 0.00 | 28 | 0.394 |
The CpG methylation levels in the BDNF promoter region were analyzed using the Mann-Whitney test.
p < 0.05.
Correlations between the CpG4 Methylation Level and the Characteristics of the AD Patients
We examined the associations between the CpG4 methylation level and the patient characteristics (age, duration of illness, MMSE, FAB, Behave-AD, CDR SB, and the plasma BDNF level) using the Spearman rank correlation coefficient. A significant positive correlation was found between the CpG4 methylation level and the duration of illness (ρ = 0.588; p = 0.044), but no other correlations with the other characteristics were seen (table 3).
Table 3.
Correlations between CpG4 methylation levels and patient characteristics in the 12 AD patients
| ρ | p value | |
|---|---|---|
| Age | -0.384 | 0.218 |
| Duration of illness | 0.588 | 0.044* |
| MMSE score | -0.391 | 0.209 |
| FAB score | -0.308 | 0.329 |
| Behave-AD score | 0.156 | 0.629 |
| CDR SB | 0.541 | 0.07 |
| Plasma BDNF levels | 0.055 | 0.880 |
ρ = Spearman's correlation coefficient.
p < 0.05.
Correlations between the CpG4 Methylation Level and the FAB or MMSE Subscale Scores in the AD Patients
A significant negative correlation was found between the CpG4 methylation level and the prehension behavior score (ρ = −0.642; p = 0.024) among the 6 FAB subtests as well as between the registration score (ρ = −0.651; p = 0.022) and the recall score (ρ = −0.663; p = 0.019) among the 11 MMSE subtest scores (tables 4, 5).
Table 4.
Correlations between CpG4 methylation levels and FAB subtest scores in the 12 AD patients
| ρ | p value | |
|---|---|---|
| Similarities | 0.182 | 0.571 |
| Lexical fluency | -0.248 | 0.437 |
| Motor series | -0.352 | 0.262 |
| Conflicting instructions | -0.539 | 0.07 |
| Go/no-go | -0.122 | 0.706 |
| Prehension behavior | -0.642 | 0.024* |
ρ = Spearman's correlation coefficient.
p < 0.05.
Table 5.
Correlations between CpG4 methylation levels and MMSE subtest scores in the 12 AD patients
| ρ | p value | |
|---|---|---|
| Orientation to time | -0.123 | 0.703 |
| Orientation to place | -0.424 | 0.169 |
| Registration | -0.651 | 0.022* |
| Attention and calculation | -0.319 | 0.313 |
| Recall | -0.663 | 0.019* |
| Naming | -0.400 | 0.197 |
| Repetition | -0.443 | 0.149 |
| 3-stage command | 0.117 | 0.717 |
| Reading | -0.400 | 0.197 |
| Writing | -0.539 | 0.070 |
| Copying | -0.235 | 0.463 |
ρ = Spearman's correlation coefficient.
p < 0.05.
Correlations between the CpG4 Methylation Level and the Behave-AD Subscale Scores in the AD Patients
No significant correlations between the CpG4 methylation level and any of the Behave-AD subscale scores were found (table 6).
Table 6.
Correlations between CpG4 methylation levels and Behave-AD subtest scores in the 12 AD patients
| ρ | p value | |
|---|---|---|
| Delusions | -0.398 | 0.200 |
| Hallucinations | - | |
| Activity disturbances | 0.146 | 0.651 |
| Aggressiveness | 0.130 | 0.687 |
| Diurnal rhythm disturbances | - | |
| Affective disturbances | -0.308 | 0.330 |
| Anxiety/phobias | -0.313 | 0.322 |
ρ = Spearman's correlation coefficient.
Discussion
In the present study, we found that the DNA methylation levels of the BDNF promoter region were higher in AD patients than in NCs. Moreover, the methylation level at the CpG4 site was significantly correlated with some clinical presentations examined using cognitive and neuropsychiatric test batteries, such as illness and neuropsychological subtest scores. However, no other significant correlations between the CpG4 methylation levels and other factors (e.g., neuropsychiatric symptoms, plasma BDNF levels, and CDR SB) were observed.
Our results support those of previous studies showing that elevated levels of methylation in AD patients lead to reductions in the mRNA or protein expression levels of BDNF [16,19]. Some other studies investigating methylation alterations in patients with major depressive disorder or bipolar disorder have shown higher methylation levels of the exon I promoter region in BDNF compared with the levels in NCs [16,17,18,25,26]. The present study also found a higher methylated average score (in %) for most of the CpG sites (16 of the 20 CpG sites) in the AD patients than in the NCs (table 2). Future studies involving a larger sample are needed to reach a definite conclusion.
Among the neuropsychological subscores, memory function (registration and recall) or sensitivity to environmental cues (prehension behavior) were significantly and negatively correlated with the CpG4 methylation level (tables 4, 5). Furthermore, the duration of illness was significantly and positively correlated with the CpG4 methylation level. The acceleration of the methylation of CpG islands in BDNF is caused by aging, inflammatory-associated, and psychogenic stress, which may attenuate its neuroprotective effect against the accumulation of Aβ and τ linking to synaptic or neuronal loss during the course of degeneration [12,17,27,28]. Neurotoxic exposure following the accumulation of Aβ can cause secondary epigenetic alterations via oxidative stress [28,29,30,31]. Therefore, a longer duration of illness might have influenced the elevation in the methylation levels of BDNF in the AD patients. Some previous studies using AD animal models have demonstrated that BDNF mRNA expression and its methylation levels within the hippocampus influence memory function, and the present study also supports a significant association between a BDNF hypermethylated state and memory impairment in AD patients [12,20,29,32]. The hypermethylation of BDNF and the reduction in the protein levels in the AD prefrontal cortex have been shown in previous reports [16,19,23], while the comparatively simple task of prehension behavior among the 6 FAB subtests was significantly and negatively correlated with the CpG4 methylation levels in the present study. Moreover, the duration of illness was significantly and negatively correlated with prehension behavior (ρ = −0.654; p = 0.021) but not registration (ρ = −0.538; p = 0.071) or recall (ρ = −0.346; p = 0.270). Memory impairment is the most distinct core symptom among several cognitive dysfunctions observed in AD patients and presents at the initial stage of the illness. Thus, a reduction in the registration and recall scores might appear irrespective of the duration of illness [1,2]. On the other hand, CpG4 hypermethylation in the frontal cortex may reflect a longer duration of illness and might cause a reduction in prehension behavior. However, the correlations between other FAB subtest scores reflecting relatively complex executive functions, such as conceptualization, sensitivity to interference, and inhibitory control, and the BDNF methylation levels should be investigated in future studies.
The present study has some limitations. Firstly, no significant correlations between the BDNF methylation and plasma BDNF levels or the Behave-AD score, reflecting neuropsychiatric problems, were found. The peripheral BDNF levels are influenced by the secretion from platelets and other comparatively transient sources of neuropsychiatric stress [33,34]. The evidence obtained in the present study cannot explain the direct neurobiological or chemical mechanisms by which BDNF DNA methylation influences neuropsychological presentations. Other intermediate phenotypic information, such as neuroimaging or neurophysiological data, is needed in future studies. Moreover, the methylation levels are influenced by psychotropic agents (antidepressants or mood stabilizers), and the serum BDNF levels are also associated with AchEI usage [35,36,37]. In the present study, AchEIs had been taken by all AD patients, and 3 AD patients had been treated with antidepressants (2 patients had taken paroxetine and 1 patient had taken fluvoxamine); these medications might have modified the elevation in DNA methylation. Secondly, the sample size in this study is comparatively small; therefore, some type II errors may exist. However, we found a significant association between the methylation levels and cognitive function or AD manifestations, and the effect sizes or p values were about the same as those presented in previous reports [16,27]. Of the 40 DNA samples in the present study, only 18 samples were completely analyzed. Therefore, more stable experimental results are needed in the future. Thirdly, the present study used peripheral blood samples, not brain tissue, similar to previous reports that investigated peripheral blood samples and found significant methylation alterations [17,18].
In conclusion, despite some limitations of the present cross-sectional study, we observed elevated methylation levels in the BDNF promoter region in AD patients, and these alterations were associated with some neurocognitive functions. In particular, an increase in BDNF methylation may influence memory impairment, which is a distinct core symptom of AD that leads to dementia. Such associations between details of the clinical presentation and BDNF methylation in AD patients have not been previously reported. Our results may contribute to the elucidation of neurodegenerative mechanisms and a better understanding of the actual roles of the methylation of the BDNF gene in the progression of AD and its possible role as a functional surrogate marker. Therefore, a larger sample should be added to the present study, and the data should be reevaluated in the future.
Acknowledgement
The present work was supported by a Grant-in-Aid for Scientific Research (grant No. 25893251) from the Ministry of Education, Culture, Sports, Science, and Technology, Japan, The Jikei University Research Fund, and The Ministry of Education, Culture, Sports, Science and Technology-supported program for the strategic research foundation at private universities (grant No. S1201032), Japan.
References
- 1.Perry RJ, Watson P, Hodge JR. The nature and staging of attention dysfunction in early (minimal and mild) Alzheimer's disease: relationship to episodic and semantic memory impairment. Neuropsychologia. 2000;38:252–271. doi: 10.1016/s0028-3932(99)00079-2. [DOI] [PubMed] [Google Scholar]
- 2.Baudic S, Barba GD, Thibaudet MC, Smagghe A, Remy P, Traykov L. Executive function deficit in early Alzheimer's disease and their relations with episodic memory. Arch Clin Neuropsychol. 2006;21:15–21. doi: 10.1016/j.acn.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 3.Nagata T, Shinagawa S, Ochiai Y, Kada H, Kasahara H, Nukariya K, Nakayama K. Relationship of frontal lobe dysfunction and aberrant motor behaviors in patients with Alzheimer's disease. Int Psychogeriatr. 2010;22:463–469. doi: 10.1017/S1041610209991323. [DOI] [PubMed] [Google Scholar]
- 4.Hardy J, Allsop D. Amyloid deposition as the central event in the aetiology of Alzheimer's disease. Trends Pharmacol Sci. 1991;12:383–388. doi: 10.1016/0165-6147(91)90609-v. [DOI] [PubMed] [Google Scholar]
- 5.Braak H, Braak E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol. 1991;82:239–259. doi: 10.1007/BF00308809. [DOI] [PubMed] [Google Scholar]
- 6.Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW, Roses AD, Haines JL, Pericak-Vance MA. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer's disease in late onset families. Science. 1993;261:921–923. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- 7.Harold D, Abraham R, Hollingworth P, Sims R, Gerrish A, Hamshere ML, Pahwa JS, Moskvina V. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer's disease. Nat Genet. 2009;41:1088–1093. doi: 10.1038/ng.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bell RD, Winkler EA, Singh I, Sagare AP, Deane R, Wu Z, Holtzman DM, Betsholtz C, Armulik A, Sallstrom J, Berk BC, Zlokovic BV. Apolipoprotein E controls cerebrovascular integrity via cyclophilin A. Nature. 2012;485:512–516. doi: 10.1038/nature11087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iwata A, Nagata K, Hatsuta H, Takuma H, Bundo M, Iwamoto K, Tamaoka A, Murayama S, Saido T, Tsuji S. Altered CpG methylation in sporadic Alzheimer's disease is associated with APP and MAPT dysregulation. Hum Mol Genet. 2014;23:648–656. doi: 10.1093/hmg/ddt451. [DOI] [PubMed] [Google Scholar]
- 10.Lu H, Liu X, Deng Y, Qing H. DNA methylation, a hand behind neurodegenerative diseases. Front Aging Neurosci. 2013;5:85. doi: 10.3389/fnagi.2013.00085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maisonpierre PC, Belluscio L, Friedman B, Alderson RF, Wiegand SJ, Furth ME, Lindsay RM, Yancopoulos GD. NT-3, BDNF, and NGF in the developing rat nervous system: parallel as well as reciprocal patterns of expression. Neuron. 1990;5:501–509. doi: 10.1016/0896-6273(90)90089-x. [DOI] [PubMed] [Google Scholar]
- 12.Nagahara AH, Merrill DA, Coppola G, Tsukada S, Schroeder BE, Shaked GM, Wang L, Blesch A, Kim A, Conner JM, Rockenstein E, Chao MV, Koo EH, Geschwind D, Masliah E, Chiba AA, Tuszynski MH. Neuroprotective effects of brain-derived neurotrophic factor in rodent and primate models of Alzheimer's disease. Nat Med. 2009;15:331–337. doi: 10.1038/nm.1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Borroni B, Archetti S, Costanzi C, Grassi M, Ferrari M, Radeghieri A, Caimi L, Caltagirone C, Di Luca M, Padovani A. ITINAD Working Group. Role of BDNF Val66Met functional polymorphism in Alzheimer's disease-related depression. Neurobiol Aging. 2009;30:1406–1412. doi: 10.1016/j.neurobiolaging.2007.11.023. [DOI] [PubMed] [Google Scholar]
- 14.Nagata T, Shinagawa S, Nukariya K, Yamada H, Nakayama K. Association between BDNF polymorphism (Val66Met) and executive function in patients with amnestic mild cognitive impairment or mild Alzheimer disease. Dement Geriatr Cogn Disord. 2012;33:266–272. doi: 10.1159/000339358. [DOI] [PubMed] [Google Scholar]
- 15.Chen ZY, Patel PD, Sant G, Meng CX, Teng KK, Hempstead BL, Lee FS. Variant brain-derived neurotrophic factor (BDNF) (Met66) alters the intracellular trafficking and activity-dependent secretion of wild-type BDNF in neurosecretory cells and cortical neurons. J Neurosci. 2004;24:4401–4411. doi: 10.1523/JNEUROSCI.0348-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rao JS, Keleshian VL, Klein S, Rapoport SI. Epigenetic modifications in frontal cortex from Alzheimer's disease and bipolar disorder patients. Transl Psychiatry. 2012;2:e132. doi: 10.1038/tp.2012.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fuchikami M, Morinobu S, Segawa M, Okamoto Y, Yamawaki S, Ozaki N, Inoue T, Kusumi I, Koyama T, Tsuchiyama K, Terao T. DNA methylation profiles of the brain-derived neurotrophic factor (BDNF) gene as a potent diagnostic biomarker in major depression. PLoS One. 2011;6:e23881. doi: 10.1371/journal.pone.0023881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim JM, Stewart R, Kang HJ, Kim SY, Kim SW, Shin IS, Park MS, Kim HR, Shin MG, Cho KH, Yoon JS. A longitudinal study of BDNF promoter methylation and genotype with poststroke depression. J Affect Disord. 2013;149:93–99. doi: 10.1016/j.jad.2013.01.008. [DOI] [PubMed] [Google Scholar]
- 19.Hock C, Heese K, Hulette C, Rosenberg C, Otten U. Region-specific neurotrophin imbalances in Alzheimer disease: decreased levels of brain-derived neurotrophin factor and increased levels of nerve growth factor in hippocampus and cortical areas. Arch Neurol. 2000;57:846–851. doi: 10.1001/archneur.57.6.846. [DOI] [PubMed] [Google Scholar]
- 20.Phillips HS, Hains JM, Armanini M, Laramee GR, Johnson SA, Winslow JW. BDNF mRNA is decreased in the hippocampus of individuals with Alzheimer's disease. Neuron. 1991;7:695–702. doi: 10.1016/0896-6273(91)90273-3. [DOI] [PubMed] [Google Scholar]
- 21.McKhann G, Drachman D, Folstein M, Katzman R, Price D, Stadlan EM. Clinical diagnosis of Alzheimer's disease: report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer's Disease. Neurology. 1984;34:939–944. doi: 10.1212/wnl.34.7.939. [DOI] [PubMed] [Google Scholar]
- 22.Reisberg B, Borenstein J, Salob SP, Ferris SH, Franssen E, Georgotas A. Behavioral symptoms in Alzheimer's disease: phenomenology and treatment. J Clin Psychiatry. 1987;48((suppl)):9–15. [PubMed] [Google Scholar]
- 23.Dubois B, Slachevsky A, Litvan I, Pillon B. The FAB: a Frontal Assessment Battery at bedside. Neurology. 2000;55:1621–1626. doi: 10.1212/wnl.55.11.1621. [DOI] [PubMed] [Google Scholar]
- 24.Folstein MF, Folstein SE, McHugh PR. ‘Mini-mental state‘. A practical method for grading the cognitive state of patients for the clinician. J Psychiatr Res. 1975;12:189–198. doi: 10.1016/0022-3956(75)90026-6. [DOI] [PubMed] [Google Scholar]
- 25.Carlberg L, Scheibelreiter J, Hassler MR, Schloegelhofer M, Schmoeger M, Ludwig B, Kasper S, Aschauer H, Egger G, Schosser A. Brain-derived neurotrophic factor (BDNF)-epigenetic regulation in unipolar and bipolar affective disorder. J Affect Disord. 2014;168:399–406. doi: 10.1016/j.jad.2014.07.022. [DOI] [PubMed] [Google Scholar]
- 26.Song Y, Miyaki K, Suzuki T, Sasaki Y, Tsutsumi A, Kawakami N, Shimazu A, Takahashi M, Inoue A, Kan C, Kurioka S, Shimbo T. Altered DNA methylation status of human brain derived neurotrophis factor gene could be useful as biomarker of depression. Am J Med Genet B Neuropsychiatr Genet. 2014;165B:357–364. doi: 10.1002/ajmg.b.32238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fuchikami M, Yamamoto S, Morinobu S, Takei S, Yamawaki S. Epigenetic regulation of BDNF gene in response to stress. Psychiatry Investig. 2010;7:251–256. doi: 10.4306/pi.2010.7.4.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Keleshian VL, Modi HR, Rapoport SI, Rao JS. Aging is associated with altered inflammatory, arachidonic acid cascade, and synaptic markers, influenced by epigenetic modifications, in the human frontal cortex. J Neurochem. 2013;125:63–73. doi: 10.1111/jnc.12153. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 29.Wang BY, Zhong Y, Zhao Z, Miao Y. Epigenetic suppression of hippocampal BDNF mediates the memory deficiency induced by amyloid fibrils. Pharmacol Biochem Behav. 2014;126C:83–89. doi: 10.1016/j.pbb.2014.09.009. [DOI] [PubMed] [Google Scholar]
- 30.Bie B, Wu J, Yang H, Xu JJ, Brown DL, Naguib M. Epigenetic suppression of neuroligin 1 underlies amyloid-induced memory deficiency. Nat Neurosci. 2014;17:223–231. doi: 10.1038/nn.3618. [DOI] [PubMed] [Google Scholar]
- 31.Taher N, McKenzie C, Garrett R, Baker M, Fox N, Isaacs GD. Amyloid-β alters the DNA methylation status of cell-fate genes in an Alzheimer's disease model. J Alzheimers Dis. 2014;38:831–844. doi: 10.3233/JAD-131061. [DOI] [PubMed] [Google Scholar]
- 32.Lubin FD, Roth TL, Sweatt JD. Epigenetic regulation of BDNF gene transcription in the consolidation of fear memory. J Neurosci. 2008;28:10576–10586. doi: 10.1523/JNEUROSCI.1786-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nagata T, Kobayashi N, Shinagawa S, Yamada H, Kondo K, Nakayama K. Plasma BDNF levels are correlated with aggressiveness in patients with amnestic mild cognitive impairment or Alzheimer disease. J Neural Transm. 2014;121:433–441. doi: 10.1007/s00702-013-1121-y. [DOI] [PubMed] [Google Scholar]
- 34.Karege F, Bondolfi G, Gervasoni N, Schwald M, Aubry JM, Bertschy G. Low brain-derived neurotrophic factor (BDNF) levels in serum of depressed patients probably results from lowered platelet BDNF release unrelated to platelet reactivity. Biol Psychiatry. 2005;57:1068–1072. doi: 10.1016/j.biopsych.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 35.Dell'Osso B, D'Addario C, Carlotta Palazzo M, Benatti B, Camuri G, Galimberti D, Fenoglio C, Scarpini E, Di Francesco A, Maccarrone M, Altamura AC. Epigenetic modulation of BDNF gene: differences in DNA methylation between unipolar and bipolar patients. J Affect Disord. 2014;166:330–333. doi: 10.1016/j.jad.2014.05.020. [DOI] [PubMed] [Google Scholar]
- 36.Leyhe T, Stransky E, Eschweiler GW, Buchkremer G, Laske C. Increase of BDNF serum concentration during donepezil treatment of patients with early Alzheimer's disease. Eur Arch Psychiatry Clin Neurosci. 2008;258:124–128. doi: 10.1007/s00406-007-0764-9. [DOI] [PubMed] [Google Scholar]
- 37.Lopez JP, Mamdani F, Labonte B, Beaulieu MM, Yang JP, Berlim MT, Ernst C, Turecki G. Epigenetic regulation of BDNF expression according to antidepressant response. Mol Psychiatry. 2013;18:398–399. doi: 10.1038/mp.2012.38. [DOI] [PMC free article] [PubMed] [Google Scholar]