Abstract
Background
Thiopurine S-methyltransferase (TPMT) is a cytosolic enzyme that catalyses the S-methylation of 6-mercaptopurine and azathioprine. Low activity phenotypes are correlated with polymorphism in the TPMT gene. Patients with low or undetectable TMPT activity could develop severe myelosuppression when they are treated with standard doses of thiopurine drugs. Since ethnic differences in the TPMT gene polymorphism have been demonstrated worldwide, assessing it in the Libyan population is worthwhile.
Methods
We investigated TPMT gene polymorphism in a total of 246 Libyan healthy adult blood donors from three different Libyan regions (Tripoli, Yefren, and Tawargha) and 50 children with acute lymphoblastic leukaemia (ALL). We used polymerase chain reaction restriction length polymorphism (PCR-RFLP) and allele-specific PCR-based assays to analyse the TPMT gene for the variants *2 c.238 G>C, *3A (c.460 G>A and c.719 A>G), *3B (c.460 G>A), and *3C (c.719 A>G).
Results
Our results show that the TPMT variants associated with low enzymatic activity were detected in 3.25% (8 in 246) of adult Libyan individuals and the frequency of total mutant alleles was 1.63%. Heterozygous genotypes were TPMT*3A in three subjects (0.61%) and TPMT*3C in five subjects (1.02%). No TPMT*2 and TPMT*3B allelic variants and no homozygous or compound heterozygous mutant alleles were detected. The normal allele (wild-type) was found in 98.4% of the adult individuals studied. No mutant alleles were detected among the 50 children who had ALL.
Conclusions
We report on the presence of the TPMT*3C and *3A mutant alleles in the Libyan population. Therefore, monitoring the patients to be treated with doses of thiopurine drugs for TPMT variants is worthwhile to avoid the development of severe myelosuppression.
Keywords: TPMT gene, thiopurine drugs, polymorphisms, acute lymphoblastic leukaemia
Thiopurine S-methyltransferase (TPMT, EC2. 1.1.67) is a cytoplasmic enzyme that preferentially catalyses the S-methylation of aromatic and heterocyclic sulfhydryl compounds. This enzyme is responsible for the inactivation of antimetabolite thiopurine drugs, such as 6-mercaptopurine, 6-thioguanine, and azathioprine (1). TPMT is expressed in many cells, with the highest levels in the liver and the lowest levels in the brain and lungs (2, 3). Thiopurine drugs have been widely used in the treatment of leukaemia, autoimmune disorders, and organ transplants. However, these drugs, like many cytotoxic agents, have a relatively narrow therapeutic index, with the potential for life-threatening drug-induced toxicity, primarily myelosuppression (4, 5).
Individual differences in thiopurine drug metabolism, response, and toxicity in humans have been correlated with polymorphism of the TPMT gene. The TPMT gene is localised to chromosome 6p22.3 and is encoded by a 34-kb gene consisting of ten exons and nine introns with a cDNA of ~3,000 bp and an open reading frame of 735 bp that encodes for a 245-amino acid peptide with a molecular mass of ~35 kDa (6). TPMT activity is inherited as an autosomal co-dominant trait with large inherited variations in human tissue TPMT enzyme activity ranging from high to virtually undetectable levels of activity (1, 7).
Approximately 0.5% of Caucasians exhibit complete TPMT deficiency correlating with two mutated TPMT alleles (homozygote or compound heterozygote), and about 11% of the population reveal intermediate activity with the presence of one mutant allele (heterozygote) (8).
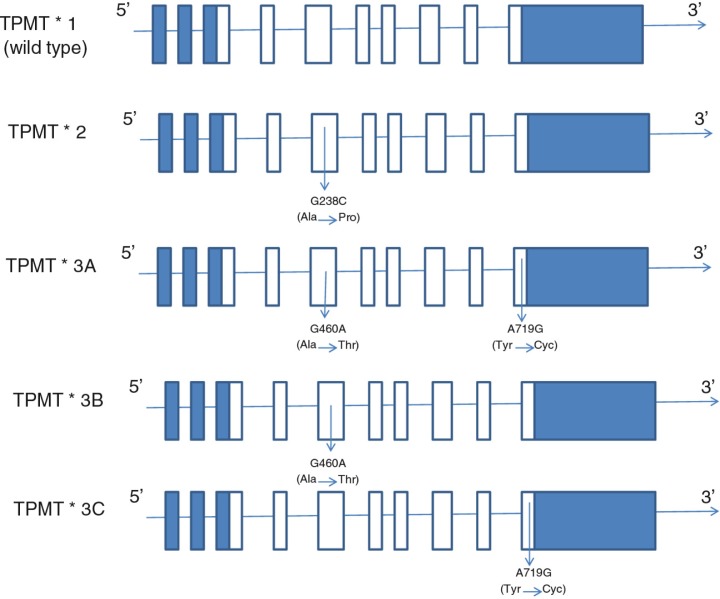
Altered TPMT activity predominantly results from single nucleotide polymorphisms. The wild-type allele is designated as TPMT*1 and three alleles designated as TPMT*2 (c.238 G>C), TPMT*3A (c.460 G>A & c.719 A>G) (9), and TPMT*3C (c.719 A>G) (Fig. 1) account for about 95% of intermediate or low enzyme activity cases (11–15). All these three alleles are associated with lower enzyme activity due to increased rates of proteolysis of the mutant proteins (16). Because of the clinical importance of TPMT pharmacogenetics, prospective determination of TPMT activity in red blood cells (RBC) is emerging as a standard clinical test prior to therapy (17), where identification of the TPMT mutant alleles allows physicians to tailor the dosage of the thiopurine drugs to the genotype of the patient or to use alternatives, improving therapeutic outcome. To date, more than 28 variant alleles of the TPMT gene have been reported. There is a large interindividual variability in the activity of TPMT. Caucasians show a trimodal distribution, with 89–94% possessing high enzymatic activity, 6–11% intermediate activity due to heterozygosity at the TPMT locus and 0.33% low activity (18). There appear to be some gender differences in TPMT activity (19–21).
Fig. 1.
The wild type and the main common mutant alleles. TPMT*1 is the most common allele (wild type), while TPMT*3A is the most common variant allele in Caucasians and TPMT*3C is the most common variant allele in East Asians. Open rectangles represent open reading frames (ORF) while the blue rectangles represent 5′- and 3′-untranslated regions (UTR). Adapted from (10).
The pattern and frequency of mutant TPMT alleles are different among various ethnic populations (22). The most prevalent TPMT mutant allele in the Caucasian and Latin American populations is TPMT*3A, which contains two nucleotide transition mutations (c. 460 G>A and c.719 A>G) in the open reading frame, leading to amino acid substitutions at codon 154 (Ala→Thr) and codon 240 (Tyr→Cys) (14, 23–26), while TPMT*3C (c.719 A>G), which includes only the codon 240 (Tyr→Cys), is predominant in East Asia and among Egyptians and African Americans (27–29).
Unlike TPMT phenotyping, genotyping is not affected by blood transfusion, less affected by pre-analytical factors, and unaffected by disease activity and drugs. Restriction digestion is a popular method because it is easy to perform and requires only basic equipment. Screening the whole TPMT gene for all 29 different alleles routinely would be technically demanding and time-consuming. Moreover, it has yet to be demonstrated whether some variant alleles result in deficient TPMT activity (14, 23–26). Since TPMT*2, TPMT*3A, and TPMT*3C make up between 60 and 95% of mutant alleles resulting in deficient TPMT activity in most populations (27–29), TPMT genotyping is usually performed only for these mutations. Consequently, patients with new mutations or from certain ethnic populations with rare TPMT variant alleles may be missed.
In this study, we used restriction fragment length polymorphism assays (RFLP-PCR) to investigate in the Libyan population the major mutant alleles of the TPMT gene: TPMT*2 (c.238 G>C), TPMT*3A (c.460 G>A and c.719 A>G), TPMT*B (c.460 G>A), and TPMT*3C (c.719 A>G) and compared the results with those published for other ethnic groups.
Materials and methods
All the chemicals used in this study were of molecular grade. The primers were purchased from Sigma-Proligo; the sequences of the primers were designed as previously described (14, 30), except primer P719NF, which we designed. The primer sequences, annealing temperatures and the sizes of polymerase chain reaction (PCR) products are listed in Table 1.
Table 1.
Primers used to detect major mutations in the TPMT gene
| Exon | Primer | Sequence (5′–3′) | Length of product (bp) | Annealing temperature (°C) |
|---|---|---|---|---|
| 5 | P2W | 5′ GTATGATTTTATGCAGGTTTG | 254 | 58 |
| 5 | P2M | 5′ GTATGATTTTATGCAGGTTTC | 254 | 58 |
| 5 | P2C | 5′ TAAATAGGAACCATCGGACAC | 254 | 58 |
| 7 | P460F | 5′ ATAACAGAGTGGGGAGGCTGC | 365 | 55 |
| 7 | P460R | 5′ CTAGAACCCAGAAAAAGTATAG | 365 | 55 |
| 10 | P719NF | 5′ GTTACTCTTTCTTGTTTCAGG | 181 | 58 |
| 10 | P719R | 5′ TCCTCAAAAACATGTCAGTGTG | 181 | 58 |
Study population
Blood samples were obtained from 246 Libyan volunteers and 50 Libyan children who had acute lymphoblastic leukaemia (ALL). Informed consent was obtained from all the participants or their guardians. The volunteers enrolled in this study were unrelated blood donors of different ages, sexes, and Libyan origins. The children were being treated at the Tripoli Medical Centre (TMC) or had been referred for evaluation because they could not tolerate chemotherapy. The TPMT genotype was determined in 126 females and 170 males. Blood samples of 1–5 ml were collected in Vacutainer tubes with EDTA as anticoagulant, transported to the laboratory of the Genetic Engineering Department at the Biotechnology Research Centre in Tripoli, and frozen at −20°C until needed for DNA extraction and subsequent PCR analysis. The blood samples from volunteers were collected from the TMC, Tripoli Central Hospital, Aljla Hospital, Yefren Hospital, and Tawargha Hospital over a period of 5 months. The study population was also subdivided into three main groups: 1) Yefren population, who are of Amazigh origin and white complexion; 2) Tawargha population, who are of African origin and black complexion; and 3) Tripoli population, which includes Libyans from different origins, including Yefren and Tawargha, and both white or black complexion. The Libyan nationality and ethnicity of all volunteers and children were confirmed by the participants’ self-declaration of race and its agreement with skin colour and officially registered identity information.
DNA extraction
The method used for DNA extraction and determination of DNA quantity and quality was according to Sambrook et al. (31). To determine that the DNA samples extracted from this procedure were suitable for PCR reaction, they were analysed by agarose gel electrophoresis.
Polymerase chain reaction
The total number of 296 DNA samples extracted were suitable for the PCR. Samples had DNA concentrations of 35–565 µg/ml. The optimum concentration for PCR is 250 µg/ml, and for this reason some DNA samples were diluted.
Three sites of known TPMT gene mutations causing TPMT deficiency (c.238 G>C, c.460 G>A, and c.719A>G) were determined according to the method described by Yates et al. (14), with minor modifications. In brief, allele-specific PCR amplification was used to detect the c.238 G>C transversion in exon 5, while PCR amplification and restriction enzyme digestion (PCR-RFLP) were used to detect the c.460 G>A and c.719 A>G mutations in exon 7 and 10, respectively.
Care was taken when PCR primers for TPMT gene analysis were designed owing to the presence of an inactive TPMT pseudogene located on chromosome 18, which has 96% homology to the TPMT gene (32). Since this inactive TPMT pseudogene is intronless, ensuring that at least one primer is complementary to the TPMT intron sequence overcomes this potential interference.
Detection of c.238 G>C (TPMT*2 Mutation)
An allele-specific PCR was used to determine whether the c.238 G>C transversion was present at the TPMT locus. The final volume for all PCR assays was 25 µl in which genomic DNA (200–250 ng) was amplified with 0.9 µl of 20 mM primer P2C (reverse) and either P2W or P2M (forward) were used in the wild-type specific or mutant-type specific reactions, respectively. The reaction included 0.125 µl of Go Taq® Flexi DNA Polymerase, 5 µl 5X Green Go Taq® Flexi Buffer (Promega), 4 µl 25 mM MgCl2 solution and 0.5 µl 10 mM dNTP. An Applied Biosystems thermocycler was used.
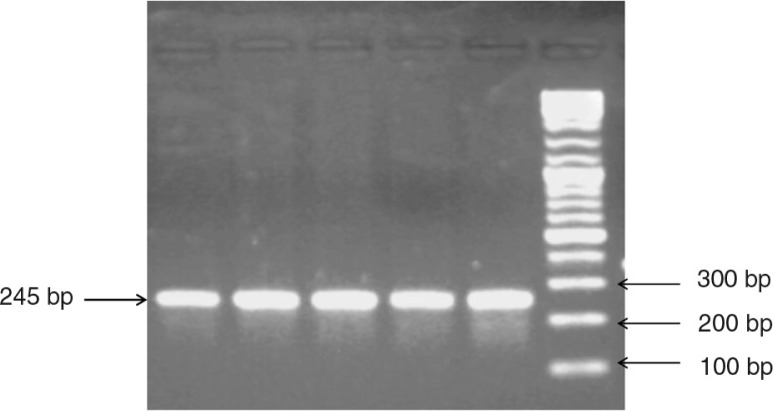
PCR amplification consisted of an initial denaturation step at 95°C for 2 min followed by 35 cycles of denaturation at 95°C for 40 s, annealing at 57°C for 30 s and extension at 72°C for 35 s. The final extension step was 72°C for 7 min. Unpurified PCR products were analysed by electrophoresis in 2.5% agarose gel and stained with ethidium bromide. A DNA fragment was amplified with P2M and P2C primers when C238 (mutant) was present, whereas a DNA fragment was amplified with P2W and P2C primers when G238 (wild-type) was present (Fig. 2).
Fig. 2.
Allele-specific PCR-based genotyping assay for the TPMT c.238 G>C mutation. PCR amplification product from wild-type exon 5 with P2W and P2C primer.
Detection of c460 G>A (TPMT*3A & TPMT*3B mutations)
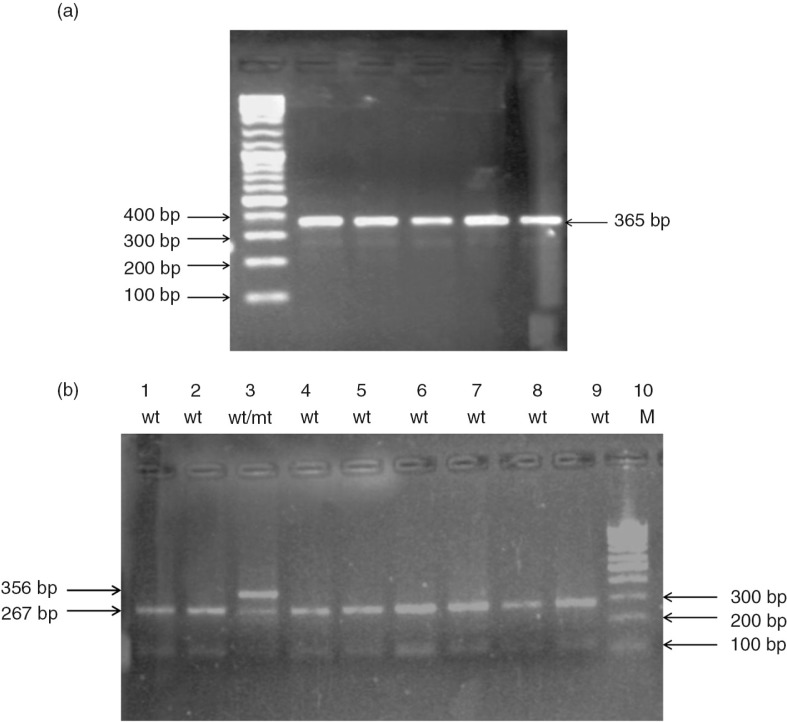
To detect the c.460 G>A mutation, a PCR assay using 0.9 µl primers (20 mM) P460F and P460R per reaction tube. The conditions were similar to those mentioned above, except that 5X Colorless Go Taq® Flexi Buffer (Promega) was used instead of 5X Green Go Taq® Flexi Buffer (Promega) and annealing was at 54°C for 30 s. The PCR product containing the fragments of 365 bp was checked in 2.5% agarose gel, and about 20 µl of PCR products were purified using the ethanol precipitation technique (31). The, 7 µl of each purified PCR product was digested with 0.3 µl of MwoI 5,000 U/ml (New England BioLabs) in 10.7 µl double-distilled H2O and 2 µl buffer 3 supplied by the manufacturer (100 mM NaCl, 50 mM Tris-HCl, 10 mM MgCl2, 1 mM dithiothreitol, pH 7.9) for 2 h at 60°C. Digested samples were electrophoresed in 2.5% agarose gel and stained with ethidium bromide. MwoI digestion of wild-type DNA yields fragments of 267 and 98 base pairs, whereas DNA containing the c.460 G>A mutation is not digested and yields an uncleaved fragment of 365 base pairs (Fig. 3).
Fig. 3.
(a) A 365-bp PCR product of exon 7 using P460F and 460R primers. (b) RFLP-PCR-based genotyping assay for TPMT c. 460 G>A. After complete digestion of the PCR product with MwoI, the mutant allele (Mut: TPMT*3B, *3A) remained as an uncleaved 365-bp fragment, while the wild allele (W: TPMT*1) yielded 267-bp and 98-bp fragments in a 2.5% agarose gel. Lane 3 shows heterozygous mutant genotype (wt/mt).
Detection of c.719 A>G (TPMT*3A & TPMT*3C mutations)
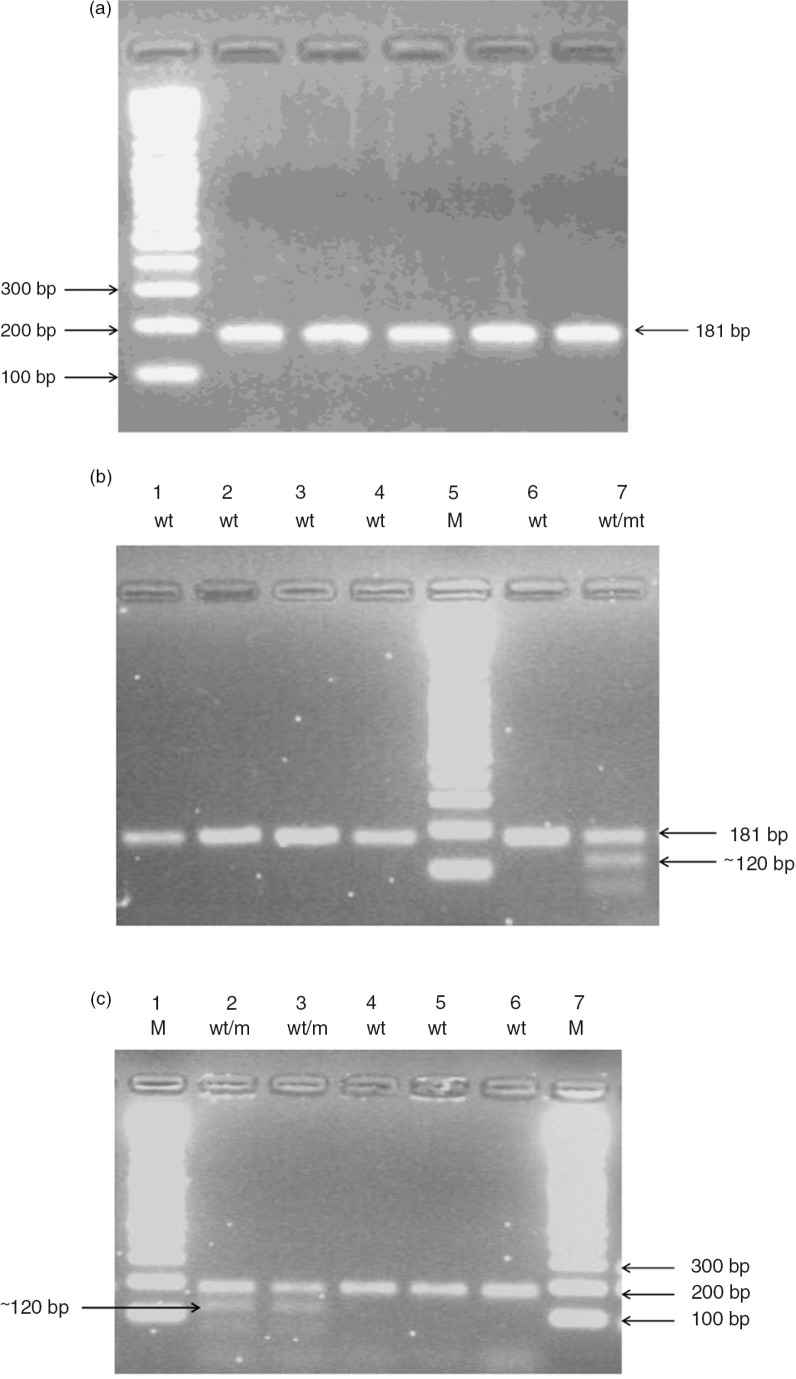
To analyse the c.719 A>G polymorphism, a 181-bp fragment containing nucleotide 719 was amplified with 0.9 µl of 20 mM primer P719NF, a new forward primer different from that previously described (14, 25), and 0.9 µl of 20 mM primer P719R under the same conditions as those used for the G460A mutation, except that annealing temperature was 57°C. The PCR products were purified as mentioned above, then 7 µl of each purified PCR product was digested with 0.2 µl of AccI 10,000 U/ml (New England BioLabs) in 10.8 µl d.d. H2O and 2 µl buffer 4 (which contained 50 mM potassium acetate, 20 mM Tris-acetate, 10 mM magnesium acetate, 1 mM DTT, pH 7.9) for 2 h at 37°C. Digested products were separated in 2.5% agarose gel. The c.719 A>G mutation introduces an AccI restriction site in the amplified fragment and yields fragments of 120 and 60 bp. Wild-type DNA yields an uncleaved fragment of 181 bp (Fig. 4).
Fig. 4.
(a) A 181-bp PCR product amplification of exon 10 using P719NF and P719R primers. (b) RFLP-PCR-based genotyping assay for TPMT c.719 A>G. Example 1: After complete digestion of the PCR product with AccI, the wild allele (wt: TPMT*1) remained as an uncleaved 181-bp fragment, while the mutant allele (m: TPMT*3C) yielded ~120 and ~60 bp fragments in a 2.5% agarose gel. Lane 7 shows heterozygous mutant genotype (wt/mt). (c) RFLP-PCR-based genotyping assay for TPMT c.719 A>G. Example 2: Lane 2 and lane 3 show heterozygous mutant genotypes (wt/mt).
The samples with one deficient allele (TPMT*1/*2, *1/*3C, *1/*3B, *1/*3A) were genotyped as heterozygous and the samples with two deficient alleles (TPMT*2/*3C, *2/*3B, *3C/*3B, *2/*3A etc.) were genotyped as homozygous. The samples that carried both the c.460 G>A and c.719A>G mutations were named TPMT *3A. Individuals with heterozygous genotypes in whom both c.460 G>A and c.719A>G mutations were detected in combination with a wild-type nucleotide in the other allele were presumed to have the TPMT*3A allele and a wild-type allele, although compound heterozygous TPMT*3B/TPMT*3C would produce the same genotyping results. However, TPMT*3B/TPMT*3C genotype is very rare worldwide.
Statistical analysis
All statistical analyses were performed using the SPSS for Windows, version 15.0 (SPSS Inc., Chicago, IL). Frequency differences of populations tested for the TPMT polymorphisms were tested by Pearson's X2 test. The level of significance (p value)<0.05 was considered statistically significant for all analyses.
Results
Genotypic analysis
The TPMT genotypes of the most prevalent mutations worldwide (TPMT*2, TPMT*3A, TPMT*3B, and TPMT*3C mutations) were determined in an unrelated Libyan population of 246 healthy adult blood donors and 50 children with ALL. Overall, there were 170 males and 126 females. Allele-specific PCR amplification was used to detect the c.238 G>C transversion in exon 5, while PCR amplification and restriction enzyme digestion (PCR-RFLP) were used to detect the c.460 G>A and c.719 A>G mutations in exon 7 and 10, respectively (Figs. 3 and 4). Mutant TPMT alleles were found in 3.25% (eight of 246) of the adult Libyan individuals: five males and three females.
TPMT*3C heterozygous allele was present in five subjects, with an allele frequency of 1.02% (Table 2). The second most common mutant allele in Libyan individuals was TPMT*3A, where three subjects were heterozygous for TPMT*3A allele, with a frequency of 0.61% making the frequency of total mutant alleles was 1.63% (Table 2).
Table 2.
Allelic frequencies of TPMT variants in a sample of 296 Libyan subjects
| Allele | SNP position | Amino acid substitution | Frequency (%) adult populationa | Frequency (%) children with ALLb |
|---|---|---|---|---|
| TPMT*1 | Wild type | 98.37 | 100 | |
| TPMT*2 | 238G>C | Ala 80 Pro | 0 | 0 |
| TPMT*3A | 460G>A 719A>G | Ala 154 Thr Tyr 240 Cyc |
0.61 | 0.0 |
| TPMT*3B | 460G>A | Ala 154 Thr | 0 | 0 |
| TPMT*3C | 719A>G | Tyr 240 Cyc | 1.02 | 0.0 |
| Total | 100 | 100 |
No. of alleles=492
No. of alleles=100; ALL: acute lymphoblastic leukaemia.
TPMT*2 (carrying c.238 G>C polymorphism) and TPMT*3B (carrying single nucleotide c.460 G>A polymorphism alone) were not found in any of the Libyan individuals. In addition, homozygous or compound heterozygous mutant alleles were not detected in any of the Libyan individuals studied. Table 2 summarises all mutant alleles found in the Libyan population.
Among the 50 cases of children who had ALL (majority from Tripoli region; data not shown), no mutant TPMT alleles were detected (Table 2).
Among the 246 adult Libyan subjects, 154 were from Tripoli population, 54 were from the Yefren population, and 38 were from the Tawargha population. In the Tripoli population, three were heterozygous for TPMT*3C allele, with an allele frequency of 0.97%, and two were heterozygous for TPMT*3A allele, with allele frequency of 0.65%; the frequency of total mutant alleles was 1.62%. In the Yefren population, only one subject was heterozygous for the TPMT*3C allele, with an allele frequency of 0.93%, and no other mutant alleles were detected. In the Tawargha population, one subject was heterozygous for the TPMT*3C allele and one subject was heterozygous for the TPMT*3A allele, with an allele frequency of 1.32% for each allele; the frequency of total mutant alleles was 2.64% (Table 3). Although there are distribution differences in the TPMT alleles, being higher in Tawargha (2.64%), lower in Yefren (0.94%) and intermediate in the Tripoli region (1.62%), no significant relationship was found between the distribution of the TPMT alleles and region of origin either among males or females.
Table 3.
Frequencies of TPMT variant alleles (%) in different adult Libyan populations according to the region of origin
| %±S.D. | ||||||
|---|---|---|---|---|---|---|
|
| ||||||
| Gender | Region | N | *2 | *3A | *3C | P value |
| Females | Tawargha | 46 | 0.0 | 2.17±2.15 | 0 | |
| Yefren | 66 | 0.0 | 0.0 | 0 | ||
| Tripoli | 92 | 0.0 | 0 | 2.17±2.15 | ||
| Total | 204 | 0 | 0.49±0.49 | 0.98±0.69 | 0.206 | |
| Males | Tawargha | 30 | 0 | 0 | 3.33±3.28 | |
| Yefren | 42 | 0 | 0 | 2.38±2.35 | ||
| Tripoli | 216 | 0 | 0.92±0.65 | 0.46±0.46 | ||
| Total | 288 | 0 | 0.69±0.49 | 1.04±0.60 | 0.460 | |
N=No. of alleles, S.D.=standard deviation. The level of significance was set at <0.05.
Discussion
One of the best examples of the application of pharmacogenetics to clinical practice is the genetic polymorphism of the thiopurine S-methyl transferase (TPMT). Treating TPMT-deficient patients with standard doses of mercaptopurine (6MP), thioguanine or azathioprine can be fatal (33), but such patients can be successfully treated, without severe toxicity, if the dose is properly adjusted (4, 34). A high degree of concordance has been demonstrated between TPMT genotype and phenotype in Caucasians (13, 35) and the presence of mutant alleles are predictive of the phenotype, in which heterozygous patients have intermediate activity and homozygous patients have low activity, although variability can be seen between these groups (1, 22, 36). To date, of the two strategies used (genotyping and phenotyping) to identify TPMT-deficient and heterozygous patients, TPMT genotype can be easily performed.
TPMT activity is known to exhibit genetic polymorphism in most populations studied to date, and the genetic basis for this inherited trait has been elucidated in most ethnic groups and races worldwide (Table 4). The present study is the first to elucidate the genetic basis for this inherited trait in the Libyan population. The current study has illustrated that the overall frequency of TPMT alleles was 1.63% in the Libyan population. That is a relatively low distribution for this polymorphic gene in Libyans compared with many ethnic groups such as European, American and Latin American, Caucasians, as well as black Africans and African Americans. However, this proportion is higher than detected in South-East Asian, Malay, and Egyptian populations. TPMT*3C and TPMT*3A alleles are the most common mutant alleles in the Libyan population, with an allele frequency of 1.02 and 0.61%, respectively. On the other hand, TPMT*3B and TPMT*2 were not detected in Libyans. TPMT*2 was found to be the most prevalent mutation among Brazilians, Turks and Iranians, whereas TPMT*3A seems the most common variant in American and European Caucasians. The TPMT*3C allele is the only mutation found in Japanese, Thais and South-East Asians (see Table 4).
Table 4.
Published frequencies of TPMT variant alleles in different ethnic groups
| Population | N | *2 | *3A | *3C | References |
|---|---|---|---|---|---|
| Caucasian American | 564 | 0.2 | 3.2 | 0.2 | (27) |
| African American | 496 | 0.4 | 0.8 | 2.4 | (27) |
| Italian | 412 | 0.4 | 3.9 | 0.9 | (35) |
| Norwegian | 132 | _ | 3.4 | 0.3 | (37) |
| Saami Norwegian | 388 | _ | 0.0 | 3.3 | (37) |
| Bulgarian | 626 | 0.16 | 2.24 | 0.16 | (20) |
| Swedish | 1,600 | 0.06 | 3.75 | 0.44 | (38) |
| German Caucasian | 2,428 | 0.2 | 4.4 | 0.4 | (8) |
| French Caucasian | 608 | 0.7 | 3.0 | 0.4 | (39) |
| Chinese | 400 | 0.0 | 0.0 | 3.0 | (40) |
| Indian | 400 | _ | 0.0 | 2.3 | (40) |
| Malay | 400 | _ | 0.5 | 0.8 | (40) |
| Japanese | 302 | 0.0 | 0.0 | 1.6 | (41) |
| South-east Asian | 698 | 0.0 | 0.0 | 1.0 | (42) |
| Tibetan | 100 | 0.0 | 0.0 | 1.0 | (43) |
| Thai | 400 | 0.0 | 0.0 | 9.0 | (44) |
| Mexican | 218 | 0.9 | 3.2 | 1.4 | (25) |
| Brazilian | 408 | 2.2 | 1.5 | 1.0 | (45) |
| Colombian | 280 | 0.4 | 3.6 | 0.0 | (24) |
| Argentinean | 294 | 0.7 | 3.1 | 0.0 | (46) |
| Bolivian | 230 | 0.0 | 6.52 | 0.0 | (43) |
| Chilean | 420 | 0.24 | 2.86 | 0.71 | (26) |
| Turkish | 296 | 2.0 | 1.0 | 1.4 | (47) |
| Iranian | 254 | 3.93 | 0.87 | 1.57 | (48) |
| Ghanaian | 434 | 0.0 | 0.0 | 7.6 | (49) |
| Kenyan | 202 | 0.0 | 0.0 | 5.4 | (23) |
| Egyptian | 400 | _ | 0.0 | 0.3 | (29) |
| Libyan | 492 | 0.0 | 0.61 | 1.02 | Present study |
N=No. of alleles.
Although this study was not designed to determine differences in allele frequency between the different Libyan populations, determining the presence of polymorphisms in a population must take into consideration the ethnic and racial mixture. Thus, it is of the greatest importance to establish the genetic basis for the TPMT polymorphism in various populations, including Libyans. Therefore, three Libyan populations (regions) were chosen according to racial and genetic mixing: the Tripoli population, among whom ethnic and racial mixture is at the highest level, and the Yefren and Tawargha populations, among whom ethnic and racial mixture is at the lowest level.
In more detail, the highest mutant alleles were found in the Tawargha population (2.64%), with equal distribution for two mutant alleles, TPMT*3A and TPMT*3C, each at 1.32%. This differs from the findings among black Africans such as the Ghanaian and Kenyan populations, among whom only TPMT*3C was detected; since the Tawargha population are African in origin and have a black complexion, this might suggest some genetic mixing with other white Caucasian populations. Surprisingly, the only mutant variant detected in the Yefren population (Amazigh origins and white complexion) was TPMT*3C, with an allele frequency of 0.93%, and no TPMT*3A or TPMT*2 alleles were detected, which are most common in European Caucasians. However, the sample sizes for the Tawargha and Yefren populations were relatively small, and that is why the result for the distribution of the TPMT alleles in these populations cannot be taken as decisive. Meanwhile, the frequency of mutant alleles in the Tripoli population, which is heterogeneous, was 1.62%, with two mutant alleles, TPMT*3A (0.65%) and TPMT*3C (0.97%), and this might be expected owing to the diversity of the Tripoli population and the very rare distribution of the TPMT*2 mutant allele in most ethnic groups. Thus, TPMT*2 was not detected in Libyans. This mutation has been found only in Caucasians and is thought to represent a more recent allele. In this study, only four mutant alleles, TPMT*2 and *3A, *3B and *3C, were genotyped in Libyans. We inferred that the samples in which these mutant alleles were not detected had the wild-type TPMT*1. However, it remains a possibility that the presence of other rare mutant alleles was not detected in this study.
Regarding the analysis of the children who had ALL, their records show that nine out of the 50 children had moderate to severe hematotoxicity from 6MP treatments, but we were surprised that we did not observe any of the major TPMT alleles among them (data not shown). Although we collected the data about the drug therapy, including the duration of 6MP therapy, maintenance therapy of 6 MP and hematotoxicity, the absence of any polymorphism prevents further analyses of the data (data not shown). This might point to presence of population specific TPMT alleles that need to be identified in further studies using more robust techniques, such as sequencing.
Conclusions
In summary, in this study we determined, for the first time, the frequency of known major TPMT mutant alleles in a Libyan sample which represented a heterogeneous population, reinforcing the importance of taking ethnicity into account when studying polymorphisms. Therefore, three Libyan populations (regions) were chosen: the Tawargha population, in whom the highest total number of mutant alleles was found (2.64%), with equal distribution of two mutant alleles (TPMT*3A 1.32% and TPMT*3C 1.32%); the Yefren population, in whom the only mutant variant detected was TPMT*3C, with an allele frequency of 0.93%; the Tripoli population, in whom the frequency of total mutant alleles was 1.62%, with two mutant alleles, TPMT*3A 0.65% and TPMT*3C 0.97%.
Acknowledgements
Thanks to Genetic Engineering Department at Biotechnology Research Center, Tripoli, Libya, for supplying genetic analysis facilities.
Conflict of interest and funding
The authors state that no conflict of interest exists.
References
- 1.McLeod HL, Siva C. The thiopurine S-methyltransferase gene locus – implications for clinical pharmacogenomics. Pharmacogenomics. 2002;3:89–98. doi: 10.1517/14622416.3.1.89. [DOI] [PubMed] [Google Scholar]
- 2.Pacifici GM, Romiti P, Giuliani L, Rane A. Thiopurine methyltransferase in humans: development and tissue distribution. Dev Pharmacol Ther. 1991;17:16–23. doi: 10.1159/000457495. [DOI] [PubMed] [Google Scholar]
- 3.Szumlanski CL, Honchel R, Scott MC, Weinshilboum RM. Human liver thiopurine methyltransferase pharmacogenetics: biochemical properties, liver-erythrocyte correlation and presence of isozymes. Pharmacogenetics. 1992;2:148–59. [PubMed] [Google Scholar]
- 4.Evans WE, Horner M, Chu YQ, Kalwinsky D, Roberts WM. Altered mercaptopurine metabolism, toxic effects, and dosage requirement in a thiopurine methyltransferase-deficient child with acute lymphocytic leukemia. J. Pediatr. 1991;119:985–9. doi: 10.1016/s0022-3476(05)83063-x. [DOI] [PubMed] [Google Scholar]
- 5.Lennard L. The clinical pharmacology of 6-mercaptopurine. Eur. J. Clin. Pharmacol. 1992;43:329–39. doi: 10.1007/BF02220605. [DOI] [PubMed] [Google Scholar]
- 6.Honchel R, Aksoy IA, Szumlanski C, Wood TC, Otterness DM, Wieben ED, et al. Human thiopurine methyltransferase: molecular cloning and expression of T84 colon carcinoma cell cDNA. Mol. Pharmacol. 1993;43:878–87. [PubMed] [Google Scholar]
- 7.Weinshilboum R. Thiopurine pharmacogenetics: clinical and molecular studies of thiopurine methyltransferase. Drug Metab Dispos. 2001;29:601–5. [PubMed] [Google Scholar]
- 8.Schaeffeler E, Fischer C, Brockmeier D, Wernet D, Moerike K, Eichelbaum M, et al. Comprehensive analysis of thiopurine S-methyltransferase phenotype-genotype correlation in a large population of German-Caucasians and identification of novel TPMT variants. Pharmacogenetics. 2004;14:407–17. doi: 10.1097/01.fpc.0000114745.08559.db. [DOI] [PubMed] [Google Scholar]
- 9.Melaouhia S, Fekih M, Garat A, Allorge D, Ferchichi H, Klouz A, et al. Allele frequency of inosine triphosphate pyrophosphatase (ITPA) and thiopurine-S-methyl transferase (TPMT) genes in the Tunisian population. Clin Res Hepatol Gastroenterol. 2012;36:178–84. doi: 10.1016/j.clinre.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 10.Schaeffeler E, Lang T, Zanger UM, Eichelbaum M, Schwab M. High-throughput genotyping of thiopurine S-methyltransferase by denaturing HPLC. Clin Chem. 2001;47:548–55. [PubMed] [Google Scholar]
- 11.Tai HL, Krynetski EY, Yates CR, Loennechen T, Fessing MY, Krynetskaia NF, et al. Thiopurine S-methyltransferase deficiency: two nucleotide transitions define the most prevalent mutant allele associated with loss of catalytic activity in Caucasians. Am J Hum Genet. 1996;58:694–702. [PMC free article] [PubMed] [Google Scholar]
- 12.Otterness D, Szumlanski C, Lennard L, Klemetsdal B, Aarbakke J, Park-Hah JO, et al. Human thiopurine methyltransferase pharmacogenetics: gene sequence polymorphisms. Clin Pharmacol Ther. 1997;62:60–73. doi: 10.1016/S0009-9236(97)90152-1. [DOI] [PubMed] [Google Scholar]
- 13.McLeod HL, Krynetski EY, Relling MV, Evans WE. Genetic polymorphism of thiopurine methyltransferase and its clinical relevance for childhood acute lymphoblastic leukemia. Leukemia. 2000;14:567–72. doi: 10.1038/sj.leu.2401723. [DOI] [PubMed] [Google Scholar]
- 14.Yates CR, Krynetski EY, Loennechen T, Fessing MY, Tai HL, Pui CH, et al. Molecular diagnosis of thiopurine S-methyltransferase deficiency: genetic basis for azathioprine and mercaptopurine intolerance. Ann Intern Med. 1997;126:608–14. doi: 10.7326/0003-4819-126-8-199704150-00003. [DOI] [PubMed] [Google Scholar]
- 15.Krynetski EY, Schuetz JD, Galpin AJ, Pui CH, Relling MV, Evans WE. A single point mutation leading to loss of catalytic activity in human thiopurine S-methyltransferase. Proc Natl Acad Sci U S A. 1995;92:949–53. doi: 10.1073/pnas.92.4.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tai HL, Fessing MY, Bonten EJ, Yanishevsky Y, d'Azzo A, Krynetski EY, et al. Enhanced proteasomal degradation of mutant human thiopurine S-methyltransferase (TPMT) in mammalian cells: mechanism for TPMT protein deficiency inherited by TPMT*2, TPMT*3A, TPMT*3B or TPMT*3C. Pharmacogenetics. 1999;9:641–50. doi: 10.1097/01213011-199910000-00011. [DOI] [PubMed] [Google Scholar]
- 17.Marshall E. Preventing toxicity with a gene test. Science. 2003;302:588–90. doi: 10.1126/science.302.5645.588. [DOI] [PubMed] [Google Scholar]
- 18.McLeod HL, Lin JS, Scott EP, Pui CH, Evans WE. Thiopurine methyltransferase activity in American white subjects and black subjects. Clin Pharmacol Ther. 1994;55:15–20. doi: 10.1038/clpt.1994.4. [DOI] [PubMed] [Google Scholar]
- 19.Jang IJ, Shin SG, Lee KH, Yim DS, Lee MS, Koo HH, et al. Erythrocyte thiopurine methyltransferase activity in a Korean population. Br J Clin Pharmacol. 1996;42:638–41. doi: 10.1111/j.1365-2125.1996.tb00122.x. [DOI] [PubMed] [Google Scholar]
- 20.Indjova D, Atanasova S, Shipkova M, Armstrong VW, Oellerich M, Svinarov D. Phenotypic and genotypic analysis of thiopurine S-methyltransferase polymorphism in the Bulgarian population. Ther Drug Monit. 2003;25:631–6. doi: 10.1097/00007691-200310000-00013. [DOI] [PubMed] [Google Scholar]
- 21.Park-Hah JO, Klemetsdal B, Lysaa R, Choi KH, Aarbakke J. Thiopurine methyltransferase activity in a Korean population sample of children. Clin Pharmacol Ther. 1996;60:68–74. doi: 10.1016/S0009-9236(96)90169-1. [DOI] [PubMed] [Google Scholar]
- 22.Krynetski EY, Evans WE. Genetic polymorphism of thiopurine S-methyltransferase: molecular mechanisms and clinical importance. Pharmacology. 2000;61:136–46. doi: 10.1159/000028394. [DOI] [PubMed] [Google Scholar]
- 23.McLeod HL, Pritchard SC, Githang'a J, Indalo A, Ameyaw MM, Powrie RH, et al. Ethnic differences in thiopurine methyltransferase pharmacogenetics: evidence for allele specificity in Caucasian and Kenyan individuals. Pharmacogenetics. 1999;9:773–6. doi: 10.1097/00008571-199912000-00012. [DOI] [PubMed] [Google Scholar]
- 24.Isaza C, Henao J, Lopez AM, Cacabelos R. Allelic variants of the thiopurine methyltransferase (TPMT) gene in the Colombian population. Methods Find Exp Clin Pharmacol. 2003;25:423–9. doi: 10.1358/mf.2003.25.6.769646. [DOI] [PubMed] [Google Scholar]
- 25.Taja-Chayeb L, Vidal-Millan S, Gutierrez O, Ostrosky-Wegman P, Duenas-Gonzalez A, Candelaria M. Thiopurine S-methyltransferase gene (TMPT) polymorphisms in a Mexican population of healthy individuals and leukemic patients. Med Oncol. 2008;25:56–62. doi: 10.1007/s12032-007-9002-6. [DOI] [PubMed] [Google Scholar]
- 26.Alvarez LL, Venegas SM, Larrondo LM, Becerra BN, Castro LA, Quera PR. [Thiopurine S-methyltransferase gene polymorphism in Chilean blood donors] Rev Med Chil. 2009;137:185–92. [PubMed] [Google Scholar]
- 27.Hon YY, Fessing MY, Pui CH, Relling MV, Krynetski EY, Evans WE. Polymorphism of the thiopurine S-methyltransferase gene in African Americans. Hum Mol Genet. 1999;8:371–6. doi: 10.1093/hmg/8.2.371. [DOI] [PubMed] [Google Scholar]
- 28.Collie-Duguid ES, Pritchard SC, Powrie RH, Sludden J, Collier DA, Li T, et al. The frequency and distribution of thiopurine methyltransferase alleles in Caucasian and Asian populations. Pharmacogenetics. 1999;9:37–42. doi: 10.1097/00008571-199902000-00006. [DOI] [PubMed] [Google Scholar]
- 29.Hamdy SI, Hiratsuka M, Narahara K, Endo N, El-Enany M, Moursi N, et al. Genotype and allele frequencies of TPMT, NAT2, GST, SULT1A1 and MDR-1 in the Egyptian population. Br J Clin Pharmacol. 2003;55:560–9. doi: 10.1046/j.1365-2125.2003.01786.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang JP, Guan YY, Wu JH, Xu AL, Zhou S, Huang M. Phenotyping and genotyping study of thiopurine S-methyltransferase in healthy Chinese children: a comparison of Han and Yao ethnic groups. Br J Clin Pharmacol. 2004;58:163–8. doi: 10.1111/j.1365-2125.2004.02113.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Plainview: Cold Harbor Lab. Press; 1989. [Google Scholar]
- 32.Lee D, Szumlanski C, Houtman J, Honchel R, Rojas K, Overhauser J, et al. Thiopurine methyltransferase pharmacogenetics. Cloning of human liver cDNA and a processed pseudogene on human chromosome 18q21.1. Drug Metab Dispos. 1995;23:398–405. [PubMed] [Google Scholar]
- 33.Schutz E, Gummert J, Mohr F, Oellerich M. Azathioprine-induced myelosuppression in thiopurine methyltransferase deficient heart transplant recipient. Lancet. 1993;341:436. doi: 10.1016/0140-6736(93)93028-y. [DOI] [PubMed] [Google Scholar]
- 34.McLeod HL, Miller DR, Evans WE. Azathioprine-induced myelosuppression in thiopurine methyltransferase deficient heart transplant recipient. Lancet. 1993;341:1151. doi: 10.1016/0140-6736(93)93168-z. [DOI] [PubMed] [Google Scholar]
- 35.Rossi AM, Bianchi M, Guarnieri C, Barale R, Pacifici GM. Genotype-phenotype correlation for thiopurine S-methyltransferase in healthy Italian subjects. Eur J Clin Pharmacol. 2001;57:51–4. doi: 10.1007/s002280000246. [DOI] [PubMed] [Google Scholar]
- 36.Relling MV, Hancock ML, Rivera GK, Sandlund JT, Ribeiro RC, Krynetski EY, et al. Mercaptopurine therapy intolerance and heterozygosity at the thiopurine S-methyltransferase gene locus. J Natl Cancer Inst. 1999;91:2001–8. doi: 10.1093/jnci/91.23.2001. [DOI] [PubMed] [Google Scholar]
- 37.Loennechen T, Utsi E, Hartz I, Lysaa R, Kildalsen H, Aarbakke J. Detection of one single mutation predicts thiopurine S-methyltransferase activity in a population of Saami in northern Norway. Clin Pharmacol Ther. 2001;70:183–8. doi: 10.1067/mcp.2001.117445. [DOI] [PubMed] [Google Scholar]
- 38.Haglund S, Lindqvist M, Almer S, Peterson C, Taipalensuu J. Pyrosequencing of TPMT alleles in a general Swedish population and in patients with inflammatory bowel disease. Clin Chem. 2004;50:288–95. doi: 10.1373/clinchem.2003.023846. [DOI] [PubMed] [Google Scholar]
- 39.Ganiere-Monteil C, Medard Y, Lejus C, Bruneau B, Pineau A, Fenneteau O, et al. Phenotype and genotype for thiopurine methyltransferase activity in the French Caucasian population: impact of age. Eur J Clin Pharmacol. 2004;60:89–96. doi: 10.1007/s00228-004-0732-5. [DOI] [PubMed] [Google Scholar]
- 40.Kham SK, Tan PL, Tay AH, Heng CK, Yeoh AE, Quah TC. Thiopurine methyltransferase polymorphisms in a multiracial Asian population and children with acute lymphoblastic leukemia. J Pediatr Hematol Oncol. 2002;24:353–9. doi: 10.1097/00043426-200206000-00006. [DOI] [PubMed] [Google Scholar]
- 41.Kumagai K, Hiyama K, Ishioka S, Sato H, Yamanishi Y, McLeod HL, et al. Allelotype frequency of the thiopurine methyltransferase (TPMT) gene in Japanese. Pharmacogenetics. 2001;11:275–8. doi: 10.1097/00008571-200104000-00012. [DOI] [PubMed] [Google Scholar]
- 42.Chang JG, Lee LS, Chen CM, Shih MC, Wu MC, Tsai FJ, et al. Molecular analysis of thiopurine S-methyltransferase alleles in South-east Asian populations. Pharmacogenetics. 2002;12:191–5. doi: 10.1097/00008571-200204000-00003. [DOI] [PubMed] [Google Scholar]
- 43.Lu HF, Shih MC, Hsueh SC, Chen CM, Chang JY, Chang JG. Molecular analysis of the thiopurine S-methyltransferase alleles in Bolivians and Tibetans. J Clin Pharm Ther. 2005;30:491–6. doi: 10.1111/j.1365-2710.2005.00640_1.x. [DOI] [PubMed] [Google Scholar]
- 44.Srimartpirom S, Tassaneeyakul W, Kukongviriyapan V, Tassaneeyakul W. Thiopurine S-methyltransferase genetic polymorphism in the Thai population. Br J Clin Pharmacol. 2004;58:66–70. doi: 10.1111/j.1365-2125.2004.02112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Boson WL, Romano-Silva MA, Correa H, Falcao RP, Teixeira-Vidigal PV, De Marco L. Thiopurine methyltransferase polymorphisms in a Brazilian population. Pharmacogenomics J. 2003;3:178–82. doi: 10.1038/sj.tpj.6500175. [DOI] [PubMed] [Google Scholar]
- 46.Larovere LE, de Kremer RD, Lambooy LH, De Abreu RA. Genetic polymorphism of thiopurine S-methyltransferase in Argentina. Ann Clin Biochem. 2003;40:388–93. doi: 10.1258/000456303766477039. [DOI] [PubMed] [Google Scholar]
- 47.Sayitoglu MA, Yildiz I, Hatirnaz O, Ozbek U. Common cytochrome p4503 (CYP3A4 and CYP3A5) and thiopurine S-methyl transferase (TPMT) polymorphisms in Turkish population. Turk J Med Sci. 2006;36:11–5. [Google Scholar]
- 48.Azad M, Kaviani S, Soleimani M, Noruzinia M, Hajfathali A. Common polymorphism's analysis of thiopurine S-methyltransferase (TPMT) in Iranian population. Yakhteh Med J. 2009;11:311–6. [Google Scholar]
- 49.Ameyaw MM, Collie-Duguid ES, Powrie RH, Ofori-Adjei D, McLeod HL. Thiopurine methyltransferase alleles in British and Ghanaian populations. Hum Mol Genet. 1999;8:367–70. doi: 10.1093/hmg/8.2.367. [DOI] [PubMed] [Google Scholar]