Abstract
Background
Current available malaria diagnostic methods each have some limitations to meet the need for real-time and large-scale screening of asymptomatic and low density malaria infection at community level. It was proposed that malaria parasite-specific low molecular-weight metabolites could be used as biomarkers for the development of a malaria diagnostic tool aimed to address this diagnostic challenge. In this study, high resolution metabolomics (HRM) was employed to identify malaria parasite-specific metabolites in Plasmodium falciparum in vitro culture samples.
Methods
Supernatants were collected at 12 hours interval from 3% haematocrit in vitro 48-hour time-course asynchronized culture system of P. falciparum. Liquid chromatography coupled with high resolution mass spectrometry was applied to discover potential parasite-specific metabolites in the cell culture supernatant. A metabolome-wide association study was performed to extract metabolites using Manhattan plot with false discovery rate (FDR) and hierarchical cluster analysis. The significant metabolites based on FDR cutoff were annotated using Metlin database. Standard curves were created using corresponding chemical compounds to accurately quantify potential Plasmodium-specific metabolites in culture supernatants.
Results
The number of significant metabolite features was 1025 in the supernatant of the Plasmodium infected culture based on Manhattan plot with FDR q=0.05. A two way hierarchical cluster analysis showed a clear segregation of the metabolic profile of parasite infected supernatant from non-infected supernatant at four time points during the 48 hour culture. Among the 1025 annotated metabolites, the intensities of four molecules were significantly increased with culture time suggesting a positive association between the quantity of these molecules and level of parasitaemia: i) 3-methylindole, a mosquito attractant, ii) succinylacetone, a haem biosynthesis inhibitor, iii) S-methyl-L-thiocitrulline, a nitric oxide synthase inhibitor, and iv) O-arachidonoyl glycidol, a fatty acid amide hydrolase inhibitor, The highest concentrations of 3-methylindole and succinylacetone were 178 ± 18.7 pmoles at 36 hours and 157±30.5 pmoles at 48 hours respectively in parasite infected supernatant.
Conclusion
HRM with bioinformatics identified four potential parasite-specific metabolite biomarkers using in vitro culture supernatants. Further study in malaria infected human is needed to determine presence of the molecules and its relationship with parasite densities.
Electronic supplementary material
The online version of this article (doi:10.1186/s12936-015-0651-1) contains supplementary material, which is available to authorized users.
Keywords: Malaria, Plasmodium falciparum, Mass spectrometry, High-resolution metabolomics (HRM), Biomarkers, Diagnostic tool
Background
Among four species of human malaria parasites, Plasmodium falciparum is responsible for most malaria-attributed morbidity and mortality. Over the past decade, successful scale-up of malaria control has resulted in substantial reductions in malaria cases and deaths [1,2]. As malaria transmission decreases due to control efforts, the epidemiology of malaria may change; that is, an increasing proportion of infections at the community level may be asymptomatic and of low parasite density [3,4]. Current malaria diagnostic tools include: 1) parasite detection by microscopic examination of blood smears, 2) antigen-based rapid diagnostic tests (RDTs), and 3) sensitive DNA-based assays. All these diagnostic methods require blood sampling by finger-prick and their implementation has been limited by either their labour/time intensive nature and requirement for specialized training and skills (microscopic method), moderate sensitivity (RDTs, microscopy), or high cost of sample preparation and supporting infrastructure needed (DNA-based methods). For programmes aiming to reduce transmission by further decreasing the parasite reservoir in humans through large scale screening approaches to detect and then to radically cure asymptomatic low-density malaria infections in real time, a field-deployable non-invasive, sensitive, low-cost, simple diagnostic tool would be very useful at the community level. Currently, available diagnostic tools cannot meet this challenge. Therefore, it was proposed to identify malaria parasite-specific low molecular-weight metabolites that could potentially be used for future development of such diagnostic tools.
As the first step for proof of concept, this study was designed to identify parasite specific low molecular-weight metabolites in an in vitro 48-hour time-course culture system of P. falciparum using high-resolution metabolomics (HRM) [5-7]. Earlier metabolomic studies on malaria have mainly focused on metabolic pathways and enzymes for the development of therapeutic strategies and interpretation of malaria pathogenesis. For example, using metabolomics with LC-MS/MS, Olszewski et al. identified the potential mechanism of cerebral malaria pathogenesis which was associated with the depletion of arginine [8]. More recently, NMR techniques in malaria metabolite profiling has been applied to identify biomarkers for infections. Tritten et al. identified two urinary metabolites in Plasmodium berghei infected mice, while a study by Sengupta et al. suggested urinary ornithine seems to have the potential as biomarkers of Plasmodium vivax malaria [9,10]. Although the two studies were conducted in rodent malaria and P. vivax malaria respectively, their analytical approaches could be used in mining for biomarkers of P. falciparum malaria infection. A study by Teng et al. showed strain-specific differences in a range of metabolites in erythrocytes infected with P. falciparum from in vitro culture and further highlighted the variation in levels of choline and phosphocholine among the strains [11]. In addition, Sana et al. investigated global mass spectrometry-based metabolomic profiling between in vitro P. falciparum infected and uninfected erythrocytes [12]. They demonstrated the alteration of metabolic profiling including the glycolysis pathway and tricarboxylic acid (TCA) cycle elucidating the mechanism of host-parasite interactions.
In contrast to the above reports, this study was designed to explore P. falciparum specific waste products, low molecular-weight metabolites, in the supernatant from the erythrocyte culture system. The rationale for profiling low molecular-weight metabolites in culture supernatants, first, was based on the hypothesis that parasite-specific small molecular wastes could be secreted into urine, saliva or sweat at high concentrations in malaria infected human and the ultimate goal is to use the small molecules as potential biomarkers for development of non-invasive and sensitive malaria diagnostic tools. This study demonstrated HRM could achieve a relatively comprehensive and quantitative analysis of Plasmodium-specific metabolites in supernatant from parasite infected culture system, and explores low molecular-weight biomarkers [6,13] associated with Plasmodium.
Methods
Parasite culture
In this study, asynchronized culture was employed. The purpose of using asynchronized culture was to capture small metabolite molecules that might be commonly released by all stages of parasites. The laboratory-adapted 3D7 strain of P. falciparum was used. The asynchronized blood stage parasites were cultured as described [14] in RPMI 1640 medium supplemented with 10% heat-inactivated O+ human serum, 1 ug/ml gentamicin, 36 uM hypoxanthine, 31 mM HEPES and 25 mM sodium bicarbonate. Four flasks of parasite culture with 3% haematocrit and 0.5% starting parasitaemia were prepared at the same time using red blood cells from different donors. At the same time, four flasks of culture without parasite inoculation but containing the same culture medium and haematocrit were also prepared. Culture materials, including supernatants and cell pellets, from all the infected and not infected flasks were collected at 12, 24, 36, and 48 hours without changing and adding culture medium. In total, 16 supernatant samples obtained from the infected flasks and the same numbers of supernatant samples from non-infected flasks were used for this study. All the samples used in this study were mycoplasma free. Parasite densities in the infected flasks were recorded as % count by blood smear reading. They increased with time, 0.83%±0.1%, 1.05%±0.06%, 1.45%±0.20% and 2.38%±0.30% (mean±SD) at 12, 24, 36 and 48 hours, respectively.
The application of C18 liquid chromatography coupled with Fourier-Transform Mass Spectrometry (FTMS)
All the samples were run in duplicate. One hundred μl aliquots of supernatant samples were treated with acetonitrile (2:1, v/v), spiked with 2.5 μl internal standard mix, and centrifuged at 14,000 × g for 5 minutes at 4°C to remove protein as described previously [15]. Then LTQ-FTMS (Thermo, hybrid linear ion trap-Fourier Transform Ion Cyclotron Resonance mass spectrometry, MA, USA) coupled with C18 liquid chromatography was run on those collected samples. HRM offers an important advantage in analysis of highly complex metabolite mixtures, such as biological extracts, because detection of mass/charge (m/z) with 5 ppm or better mass resolution and mass accuracy substantially decreases the demand for physical separation prior to detection. Detection of m/z of ions from 85 to 850 with 50,000 resolution over 10 min LC runs with data extraction using apLCMS [16] provides a minimum of 3,000 reproducible features, many with sufficient mass accuracy to allow prediction of elemental composition. A m/z feature is defined by m/z, RT (retention time), and ion intensity (integrated ion intensity for the peak). The Kyoto Encyclopedia of Genes and Genomes (KEGG) database [17] was utilized to map the features distribution on both human and Plasmodium metabolic pathways. Examination of m/z of metabolites in the KEGG [18,19] human and Plasmodium metabolomics pathways shows that less than 10% of metabolites are redundant with others in terms of elemental composition [18,19]. Identified metabolites are annotated using Metlin Mass Spectrometry Database [20,21]. Direct examination by MS/MS of selected accurate mass m/z features of human plasma showed that for many m/z, the ion dissociation patterns matched those of authentic standards with identical elution times. For such metabolites, quantification relative to stable isotope internal standards has a coefficient of variation (5 to 10%) and sensitivity (low nanomolar to picomolar), which is similar to other methods and sufficient in allowing targeted analysis of selected chemicals within the context of an information-rich non-targeted profiling of all m/z detected within biological samples.
Metabolic profiling with univariate and multivariate statistical analysis
Analyses were performed based on the results from both biological and technical replicates. Total features of culture supernatant were collected after processing mass spectral data with apLCMS. The features from duplicate LCMS analyses were averaged, log2 transformed and quantile normalized for subsequent statistical and bioinformatics analyses including univariate analysis, Manhattan plot, and false discovery rate (FDR) [22] to determine the significant metabolites between infected and non-infected cultures. Furthermore, the metabolic profiles were discriminated using Limma-hierarchical cluster analysis to separate two groups in association with metabolites. Limma is originally a package of Linear Models for Microarray to analyse the gene expression data arising from microarray or RNA-Seq technologies from Bioconductor. A core capability is the use of linear models to assess differential expression in the context of multifactor designed experiments. Limma provides the ability to make comparisons between many targets simultaneously including metabolites [23,24].
Pathway analysis with KEGG
The database of Kyoto Encyclopedia of Genes and Genomes (KEGG) was utilized to map the features distribution on both human and plasmodium metabolic pathways. Detected m/z features matching known human and Plasmodium intermediary metabolites are shown in black dots in pathway maps; most human and known Plasmodium metabolic pathways are represented.
Quantification of 3-methylindole and succinylacetone
3-methylindole (M51458) and succinylacetone (D1415) were purchased from Sigma-Aldrich (MO, USA). A standard curve was made by known concentrations (0.1-0.5nmole) of the reagent grade compounds in cell culture media. Areas from the samples were plotted against concentrations and a standard curve with an r2>0.99. 3-methylindole and succinylacetone in supernatants from parasite culture were treated using 100 μl in microcentrifuge tubes. These tubes were centrifuged at 14,000× g at 4 degrees Celsius for 5 minutes. 10 μl of the supernatant was injected in mass spectrometry and resulting areas were noted. Concentrations of the compounds were calculated using the standard curve.
Results
Metabolome-wide association study (MWAS)
MWAS was used to identify changes in supernatants from non-infected and Plasmodium-infected cultures at all points (12, 24, 36, and 48 hrs). A Manhattan plot (Figure 1) combines a statistical test (e.g., p-value, ANOVA) with the magnitude of change and enables visual identification of statistically significant data-points (metabolites) that display large-magnitude changes. Multiple testing corrections like FDR adjusts p-values (q-values) derived from multiple statistical tests to correct for the occurrence of false positives. The Y axis represents the -log10 of the raw p-value comparing supernatants of culture system between non-infected and Plasmodium-infected red blood cells. The X axis indicated m/z ranging 85-850 m/z. The dotted line was shown as the FDR significant level, therefore, any m/z above this line were significantly different between two groups at FDR q=0.05. The total number of significant features was 1025 out of the 3270 detected.
Figure 1.
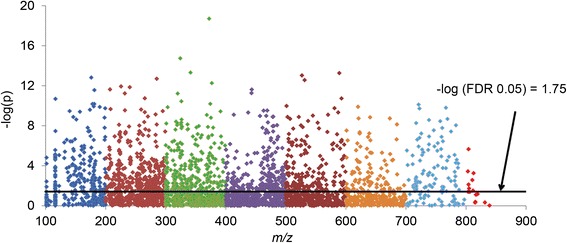
Metabolome-wide association study (MWAS). This is the snapshot of metabolites between supernatants from Plasmodium, non-infected and infected cultures at all time points using Manhattan plot on 3270 features. The features from duplicate run were averaged, log2 transformed, and quantile normalized to find out the significant features using false discovery rate (FDR). The dotted line represents FDR q=0.05. The metabolites over this line were the significant metabolites (n=1025) between two groups.
Mapping significant features through Kyoto Encyclopedia of Genes and Genomes (KEGG) human and plasmodium metabolic pathways
HRM platform provides precise metabolic phenotypes to determine the possible pathways altered by P. falciparum. The schematic representation in Additional files 1 and 2 shows that FDR significant 1025 features covering malaria parasite waste chemicals and human metabolites from supernatants were mapped through KEGG both human and Plasmodium metabolic pathways. Results show that these metabolites matched 439 metabolic compounds in both human and Plasmodium metabolic pathways. The remaining 586 of 1025 chemicals which were not matched, however, might be either waste products of the parasite or could be utilized by unidentified Plasmodium pathways.
Two way hierarchical cluster analysis (HCA) on FDR significant features in supernatant between non-infected and infected cultures
Two way HCA was performed on combined sample classification with metabolites clustering to identify which metabolites are the most important for sample grouping. In this study, HCA was performed using 1025 metabolites at FDR q=0.05, which were the key components to separate two groups using all four time points (Figure 2). HCA determines similarity measures using Euclidean distance and Pearson linear correlation. The top panel showed that two main clusters separating supernatant of non-infected from infected cultures. The sample name was listed at the bottom panel. The right panel included 1025 metabolites which contributed to discrimination of samples according to malaria infection. In addition, Figure 3 shows a broad increasing trend in significant features at FDR q=0.05 during 48 hours.
Figure 2.
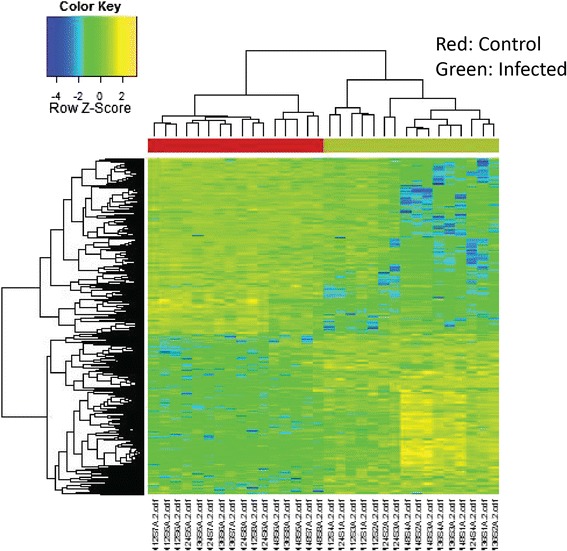
Two way hierarchical cluster analysis (HCA) on FDR significant features of supernatants between non-infected and infected cultures. HCA was performed using the 1025 metabolites at FDR q=0.05. The analysis utilized the samples from all time points to separate two groups in top bar (red clusters for control samples, green clusters for infected samples).
Figure 3.
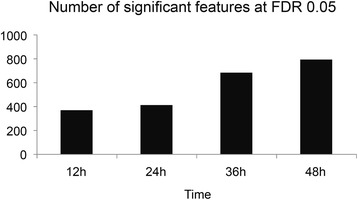
The number of significant features using FDR q=0.05 at each time point during 48 hours’ culture.
Decrease in arginine and isoleucine in the agreement with others’ results
In order to validate methodology and analytical approach used for the identification of candidate biomarker molecules, the intensity of arginine and isoleucine known to be consumed and critical for malaria parasite growth were analysed. Amino acids are the building blocks of proteins. As parasites grow within host red blood cells, they utilize large quantities of amino acids. The malaria parasite derives most of its amino acid requirements from the proteolysed haemoglobin in the host blood cells [25]. In Figure 4A, arginine decreased to zero in a time dependent fashion during 48 hours. Most importantly, hemoglobin lacks one important amino acid, isoleucine, and the parasite, therefore, has to source this from culture system. In Figure 4B, isoleucine was reduced significantly in supernatants after the 48 hour incubation period.
Figure 4.
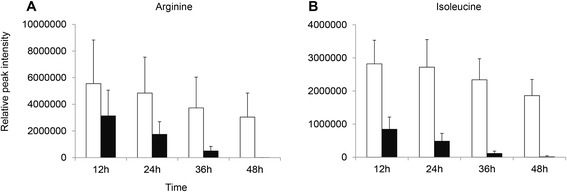
Decrease of arginine and isoleucine concentration during 48 hours’ culture. Figure 4 A: Arginine, and Figure 4 B: Isoleucine. White bars indicated supernatants from Plasmodium non-infected culture and black bar represented as supernatants from Plasmodium infected culture.
Identification of the potential biomarkers increased with culture time
Among the remaining 586 of 1025 significant metabolites (FDR q=0.05) determined by FDR, the ion intensities of several metabolites were increased with culture time in infected culture supernatants but not in non-infected culture supernatants, suggesting a positive association between the quantity of these molecules released and level of parasitaemia. These metabolites were: i) 3-methylindole (Figure 5A), ii) succinylacetone (Figure 5B), iii) S-methyl-L-thiocitrulline (Figure 5C), and iv) O-arachidonoyl glycidol (Figure 5D). The compound 3-methylindole has been shown to stimulate an odorant receptor to attract malaria mosquito vector [26] while succinylacetone has been identified as an inhibitor of haem biosynthesis [27,28]. S-methyl-L-thiocitrulline has been identified as a potent nitric oxide synthase (NOS) inhibitor to reduce nitric oxide production and endothelial dysfunction [29]. On the other hand, O-arachidonoyl glycidol was reported to be an inhibitor of fatty acid amide hydrolase [30,31]. In addition, a similar pattern was observed for phosphocholine (Additional file 3), the molecule that was reported in previous study using infected erythrocytes [11] and is involved in the phosphobase methylation for phosphatidylcholine production [32]. The observed increase in phosphocholine with culture time further validated our methodology and analytical approach for identifying the four molecules reported above.
Figure 5.
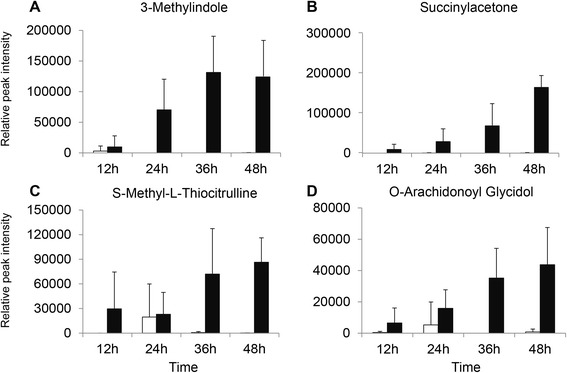
Increase in concentrations of four molecules during 48 hours’ culture. Figure 5 A: 3-methylindole, Figure 5 B: succinylacetone, Figure 5 C: S-methyl-L-thiocitrulline, and Figure 5 D: O-arachidonoyl glycidol. White bars indicated supernatants from Plasmodium non-infected culture and black bar represented as supernatants from Plasmodium infected culture.
The quantification of 3-methylindole and succinylacetone
The production of 3-methylindole and succinylacetone were measured at each time point during 48 hours in the 50 μl of supernatants from 3% haematocrit cultures. At 36 hours, the amount of 3-methylindole was highest and the concentration was 178±18.7 pmoles (Figure 6A). The generation of succinylacetone increased over the time. The amounts were 2±2 pmoles at 12 hours culture, with the highest reading at 157±30.5 pmoles at 48 hours culture (Figure 6B). Supporting information can be seen with the MS/MS data of these compounds found in Additional files 4, 5, 6 and 7.
Figure 6.
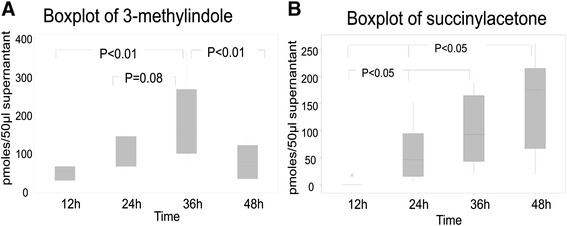
The quantification of 3-methylindole and succinylacetone at each time point during 48 hours’ culture. Figure 6 A: Boxplot of 3-methylindole, and Figure 6 B: Boxplot of succinylacetone.
Discussion
The primary objective of this study was to explore P. falciparum-specific low molecular-weight metabolites that can be used as biomarkers for future development of non-invasive malaria diagnostic tools. Using the supernatant samples from in vitro erythrocytic stage asynchronized cultures, four molecules, 3-methylindole, succinylacetone, S-methyl-L-thiocitrulline and O-arachidonoyl glycidol, were identified as potential biomarkers.
Use of HRM is a uniquely good approach to identify putative biomarkers for a complex disease like malaria since Plasmodium parasites divert nutrients toward proliferating parasite cells while the host cells try to maintain homeostasis and deal with metabolic changes during the parasites’ intraerythrocytic life cycle [33,34]. This study demonstrated the strength of HRM in measuring P. falciparum specific waste products and toxins which were expected to increase in concentration during the infection. Therefore, this approach allows for the identification of potential biomarkers associated with Plasmodium-specific products. Previous studies also showed that the analytic capabilities of HRM could measure the relative levels of all metabolites simultaneously in in vitro and in vivo malaria infection systems [12]. Furthermore, using the outcomes of HRM, KEGG mapping was performed in this study. Significant features (n=1025) were identified in both human and Plasmodium metabolic pathways to distinguish which metabolic compounds are being utilized by both. Surprisingly, 439 metabolites were found to be used in both human and plasmodium metabolic pathways. However, despite the similarities in this large number of metabolites, the pathways in which these metabolites are mapped are likely to be different due to the absence of certain metabolic pathways in plasmodium compared to human metabolic pathways. Meanwhile, of the 586 unmatched features, four (4) were found to be potential biomarkers from the parasite during the erythrocytic stage culture system. This was based on the fact that the ion intensities of the four molecules were increased with culture time, suggesting a positive association between relative quantity of these molecules and level of parasitaemia. In addition, 3-methylindole and succinylacetone molecules in the supernatant samples from 3% haematocrit 48 hour time-course cultures were further quantified. The concentration of succinylacetone peaked at 48 hours compared to 36 hours for 3-methylindole. As 3-methylindole is volatile molecule, it is possible that 3-methylindole was evaporated due to extended culture to 48 hours. Importantly, 3-methylindole has been shown to stimulate an odorant receptor to attract malaria mosquito vector [26], potentially playing a role in enhancing the probability of transmission for the parasite. Practically, 3-methylindole could be used in traps for mosquito research purposes. Although 3-methylindole is found in large intestine of humans via ingestion of tryptophan, this molecule is well-known as a highly selective pulmonary toxicant for ruminants [35]. A study showed that metabolism and bioactivation of 3-methylindole in human is mediated by human liver microsomes [36]. In spite of the fact that this molecule is found in human gastrointestinal system, it is probable that the elevation of this molecule due to malaria infection in human could still be used as a potential biomarker when urine or sweat samples are tested.
Another potential biomarker identified was succinylacetone which is known to inhibit haem biosynthesis (delta-aminolevulinate dehydrolase inhibitor) [37]. Succinylacetone either makes the haem synthesizing system nonfunctional or decreases its functionality [37]. Although this molecule has been shown to be a biomarker for Tyrosinaemia Type 1 (Tyr I), the genetic disorder is rare and worldwide prevalence is very low. Therefore, utilizing this compound as a potential biomarker in malaria-endemic areas could account for the malaria infection rather than the Tyr I. The production range of succinylacetone suggests it may be useful as a basis for developing a biosensor for non-invasive diagnosis of malaria. Two other potential metabolites were also identified in this study: S-methyl-L-thiocitrulline is a potent NOS inhibitor to reduce nitric oxide production and endothelial dysfunction [38] while O-arachidonoyl glycidol was reported to be an inhibitor of fatty acid amide hydrolase [30]. The increase in ion intensities during culture period for these two molecules indicated a potential as biomarkers for malaria diagnosis. Currently, a metabolomics study using urine, saliva, sweat and blood samples collected from P. falciparum infected people from Africa is underway. The ongoing study will provide information on concentration and stability of the molecules in malaria infected people and their association with parasite densities.
An apparent disappearance of two amino acids, arginine and isoleucine, was observed in the 48 hr culture system. Several important host-parasite interaction studies elucidated a mechanism that isoleucine was taken by Plasmodium sp. to develop blood stage parasites [25,39]. Malaria parasite utilizes amino acids largely through the degradation of host erythrocyte haemoglobin [40]. Isoleucine is the only amino acid not present in human mature haemoglobin, resulting in utilization of this amino acid from other sources by parasites. The untargeted HRM confirmed previous finding regarding the depletion of isoleucine in culture supernatant in time dependent manner, supporting the reliability of experimental system and analytical approach in identifying the above four parasite-specific waste metabolites and toxins which were released into culture supernatant and were shown increase in concentration during 48 hr culture period.
In summary, HRM coupled with network and pathway analysis using the significant metabolites from culture supernatants of infected erythrocytes and incorporating the broader human and malaria parasite metabolomic knowledge identified four potential parasite-specific biomarkers. The findings from the current study may provide improved opportunities for innovative prevention and management programmes such as development of new malaria diagnostic tools.
Acknowledgements
This research was supported in part by the Center for Health Discovery and Well-being of Emory University, grant # NRF-2014R1A4A1007304 (Y.H.J) and grant# NRF-2014R1A1A2053787 of Korea University, and by Malaria Branch, Division of Parasitic Diseases and Malaria (DPDM), Center for Global Health, Centers for Disease Control and Prevention (CDC). DPJ is supported in part by NIH grants HL113451, ES009047, ES019776 and AG038746 and NIAID Contract HHSN272201200031C. The authors acknowledged Jae Ho Cho, Korea University in making figures. We also thank Dr. S. Patrick Kachur, Malaria Branch, DPDM, CGH, CDC for his critical review of this manuscript and valuable suggestions. The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
Additional files
Mapping significant features to Kyoto Encyclopedia of Genes andGenomes (KEGG) human metabolic pathways. This schematic representation shows the mapping of 439 matched features covering human metabolites.
Mapping significant features to Kyoto Encyclopedia of Genes and Genomes (KEGG) Plasmodium metabolic pathways. This schematic representation shows the mapping of 439 matched features covering malaria parasite metabolites.
Increase in phosphocholine concentration during 48 hours culture. White bars indicated supernatants from Plasmodium non-infected culture and black bar represented as supernatants from Plasmodium infected culture.
Spectrum of 3-methylindole itself. A) Total ion chromatography, B) MS, and C) MS/MS.
Spectrum of 3-methylindole after its addition to cell supernatant. A) Total ion chromatography, B) MS, and C) MS/MS.
Spectrum of 3-methylindole in cell supernatant. A) Total ion chromatography, B) MS, and C) MS/MS.
Spectrum of succinylacetone. A) Total ion chromatography, B) MS on itself, C) MS/MS on itself, D) MS/MS after its addition to cell supernatant, and E) MS/MS on cell supernatant without chemical addition.
Footnotes
Competing interests
The findings from the current study were filed for Emory Intellectual Property Disclosure and for CDC employee Discovery and Invention Report (EIR) respectively. Emory University was included in the CDC EIR.
Authors’ contributions
YP and DJ designed the current study and YP carried out the metabolomics experiments and comprehensive data analysis. YPS and LS conceived the concept of identification of parasite-specific low molecular-weight metabolites for future development of non-invasive, low cost, sensitive and simple malaria diagnostic tool and participated in the current study design. ETL conducted parasite cultures, and BL and KU assisted in metabolomics experiments. CM and YHJ assisted in generating additional files for this manuscript. YP, YPS and DJ wrote the paper. All authors read and approved the final manuscript.
Contributor Information
Youngja H Park, Email: yjhwang@korea.ac.kr.
Ya Ping Shi, Email: yps0@cdc.gov.
Bill Liang, Email: yliang3@emory.edu.
Carl Angelo D Medriano, Email: carlmed00@korea.ac.kr.
Young Ho Jeon, Email: yhjeon@korea.ac.kr.
Eucaris Torres, Email: hrv6@cdc.gov.
Karan Uppal, Email: kuppal2@emory.edu.
Laurence Slutsker, Email: lms5@cdc.gov.
Dean P Jones, Email: dpjones@emory.edu.
References
- 1.CDC. http://www.cdc.gov/malaria/about/index.html, Centers for Disease Control and Prevention, Atlanta, GA, USA 2014.
- 2.WHO . World Malaria Reports 2011. Geneva: World Health Organization; 2011. [Google Scholar]
- 3.Harris I, Sharrock WW, Bain LM, Gray KA, Bobogare A, Boaz L, et al. A large proportion of asymptomatic Plasmodium infections with low and sub-microscopic parasite densities in the low transmission setting of Temotu Province, Solomon Islands: challenges for malaria diagnostics in an elimination setting. Malar J. 2010;9:254. doi: 10.1186/1475-2875-9-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mosha JF, Sturrock HJ, Greenhouse B, Greenwood B, Sutherland CJ, Gadalla N, et al. Epidemiology of subpatent Plasmodium falciparum infection: implications for detection of hotspots with imperfect diagnostics. Malar J. 2013;12:221. doi: 10.1186/1475-2875-12-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dettmer K, Hammock BD. Metabolomics–a new exciting field within the “omics” sciences. Environ Health Perspect. 2004;112:A396–7. doi: 10.1289/ehp.112-1241997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jones DP, Park Y, Ziegler TR. Nutritional metabolomics: progress in addressing complexity in diet and health. Annu Rev Nutr. 2012;32:183–202. doi: 10.1146/annurev-nutr-072610-145159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Park YH, Lee K, Soltow QA, Strobel FH, Brigham KL, Parker RE, et al. High-performance metabolic profiling of plasma from seven mammalian species for simultaneous environmental chemical surveillance and bioeffect monitoring. Toxicology. 2012;295:47–55. doi: 10.1016/j.tox.2012.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Olszewski KL, Morrisey JM, Wilinski D, Burns JM, Vaidya AB, Rabinowitz JD, et al. Host-parasite interactions revealed by Plasmodium falciparum metabolomics. Cell Host Microbe. 2009;5:191–9. doi: 10.1016/j.chom.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tritten L, Keiser J, Godejohann M, Utzinger J, Vargas M, Beckonert O, et al. Metabolic profiling framework for discovery of candidate diagnostic markers of malaria. Sci Rep. 2013;3:2769. doi: 10.1038/srep02769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sengupta A, Ghosh S, Basant A, Malusare S, Johri P, Pathak S, et al. Global host metabolic response to Plasmodium vivax infection: a 1H NMR based urinary metabonomic study. Malar J. 2011;10:384. doi: 10.1186/1475-2875-10-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Teng R, Lehane AM, Winterberg M, Shafik SH, Summers RL, Martin RE, et al. 1H-NMR metabolite profiles of different strains of Plasmodium falciparum. Biosci Rep. 2014;34:e00150. doi: 10.1042/BSR20140134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sana TR, Gordon DB, Fischer SM, Tichy SE, Kitagawa N, Lai C, et al. Global mass spectrometry based metabolomics profiling of erythrocytes infected with Plasmodium falciparum. PLoS One. 2013;8:e60840. doi: 10.1371/journal.pone.0060840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu W, Bennett BD, Rabinowitz JD. Analytical strategies for LC-MS-based targeted metabolomics. J Chromatogr B Analyt Technol Biomed Life Sci. 2008;871:236–42. doi: 10.1016/j.jchromb.2008.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Trager W, Jensen JB. Human malaria parasites in continuous culture. Science. 1976;193:673–5. doi: 10.1126/science.781840. [DOI] [PubMed] [Google Scholar]
- 15.Johnson JM, Yu T, Strobel FH, Jones DP. A practical approach to detect unique metabolic patterns for personalized medicine. Analyst. 2010;135:2864–70. doi: 10.1039/c0an00333f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu T, Park Y, Johnson JM, Jones DP. apLCMS–adaptive processing of high-resolution LC/MS data. Bioinformatics. 2009;25:1930–6. doi: 10.1093/bioinformatics/btp291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kanehisa M, Goto S, Sato Y, Kawashima M, Furumichi M, Tanabe M: Kyoto Encyclopedia of Genes and Genomes. http://www.genome.jp/kegg Accessed December 2014.
- 18.Kanehisa M. The KEGG database. Novartis Found Symp. 2002;247:91–101. doi: 10.1002/0470857897.ch8. [DOI] [PubMed] [Google Scholar]
- 19.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith CA, O'Maille G, Want EJ, Qin C, Trauger SA, Brandon TR, et al. METLIN: a metabolite mass spectral database. Ther Drug Monit. 2005;27:747–51. doi: 10.1097/01.ftd.0000179845.53213.39. [DOI] [PubMed] [Google Scholar]
- 21.Smith CA, O’Maille G, Want EJ, Qin C, Trauger SA, Brandon TR, et al. METLIN: Metabolite and Tandem MS Database. https://metlin.scripps.edu/index.php. Accessed December 10 2014.
- 22.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Statist Soc B. 1995;B57:289–300. [Google Scholar]
- 23.Cribbs SK, Park Y, Guidot DM, Martin GS, Brown LA, Lennox J, et al. Metabolomics of bronchoalveolar lavage differentiate healthy HIV-1-infected subjects from controls. AIDS Res Hum Retroviruses. 2014;30:579–85. doi: 10.1089/aid.2013.0198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Neujahr DC, Uppal K, Force SD, Fernandez F, Lawrence C, Pickens A, et al. Bile acid aspiration associated with lung chemical profile linked to other biomarkers of injury after lung transplantation. Am J Transplant. 2014;14:841–8. doi: 10.1111/ajt.12631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Istvan ES, Dharia NV, Bopp SE, Gluzman I, Winzeler EA, Goldberg DE. Validation of isoleucine utilization targets in Plasmodium falciparum. Proc Natl Acad Sci U S A. 2011;108:1627–32. doi: 10.1073/pnas.1011560108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu P, Leal WS. Probing insect odorant receptors with their cognate ligands: insights into structural features. Biochem Biophys Res Commun. 2013;435:477–82. doi: 10.1016/j.bbrc.2013.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ebert PS, Hess RA, Frykholm BC, Tschudy DP. Succinylacetone, a potent inhibitor of heme biosynthesis: effect on cell growth, heme content and delta-aminolevulinic acid dehydratase activity of malignant murine erythroleukemia cells. Biochem Biophys Res Commun. 1979;88:1382–90. doi: 10.1016/0006-291X(79)91133-1. [DOI] [PubMed] [Google Scholar]
- 28.Tschudy DP, Hess RA, Frykholm BC. Inhibition of delta-aminolevulinic acid dehydrase by 4,6-dioxoheptanoic acid. J Biol Chem. 1981;256:9915–23. [PubMed] [Google Scholar]
- 29.Bradshaw HB, Rimmerman N, Hu SS, Burstein S, Walker JM. Novel endogenous N-acyl glycines identification and characterization. Vitam Horm. 2009;81:191–205. doi: 10.1016/S0083-6729(09)81008-X. [DOI] [PubMed] [Google Scholar]
- 30.Bradshaw HB, Rimmerman N, Hu SS, Benton VM, Stuart JM, Masuda K, et al. The endocannabinoid anandamide is a precursor for the signaling lipid N-arachidonoyl glycine by two distinct pathways. BMC Biochem. 2009;10:14. doi: 10.1186/1471-2091-10-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McHugh D, Hu SS, Rimmerman N, Juknat A, Vogel Z, Walker JM, et al. N-arachidonoyl glycine, an abundant endogenous lipid, potently drives directed cellular migration through GPR18, the putative abnormal cannabidiol receptor. BMC Neurosci. 2010;11:44. doi: 10.1186/1471-2202-11-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saen-Oon S, Lee SG, Jez JM, Guallar V. An alternative mechanism for the methylation of phosphoethanolamine catalyzed by Plasmodium falciparum phosphoethanolamine methyltransferase. J Biol Chem. 2014;289:33815–25. doi: 10.1074/jbc.M114.611319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lakshmanan V, Rhee KY, Daily JP. Metabolomics and malaria biology. Mol Biochem Parasitol. 2011;175:104–11. doi: 10.1016/j.molbiopara.2010.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.LeRoux M, Lakshmanan V, Daily JP. Plasmodium falciparum biology: analysis of in vitro versus in vivo growth conditions. Trends Parasitol. 2009;25:474–81. doi: 10.1016/j.pt.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 35.Ruangyuttikam W. Metabolism and mechanism of toxicity of 3-methylindole in human and goat tissues. Dissertation: University of Utah, Department of Pharmacology and Toxicology; 1991. [Google Scholar]
- 36.Yan Z, Easterwood LM, Maher N, Torres R, Huebert N, Yost GS. Metabolism and bioactivation of 3-methylindole by human liver microsomes. Chem Res Toxicol. 2007;20:140–8. doi: 10.1021/tx060239e. [DOI] [PubMed] [Google Scholar]
- 37.Srivastava P, Pandey VC. Heme synthesizing enzymes of Plasmodium knowlesi: a simian malaria parasite. Exp Parasitol. 1998;88:60–3. doi: 10.1006/expr.1998.4193. [DOI] [PubMed] [Google Scholar]
- 38.Furfine ES, Harmon MF, Paith JE, Knowles RG, Salter M, Kiff RJ, et al. Potent and selective inhibition of human nitric oxide synthases. Selective inhibition of neuronal nitric oxide synthase by S-methyl-L-thiocitrulline and S-ethyl-L-thiocitrulline. J Biol Chem. 1994;269:26677–83. [PubMed] [Google Scholar]
- 39.Babbitt SE, Altenhofen L, Cobbold SA, Istvan ES, Fennell C, Doerig C, et al. Plasmodium falciparum responds to amino acid starvation by entering into a hibernatory state. Proc Natl Acad Sci U S A. 2012;109:E3278–87. doi: 10.1073/pnas.1209823109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu J, Istvan ES, Gluzman IY, Gross J, Goldberg DE. Plasmodium falciparum ensures its amino acid supply with multiple acquisition pathways and redundant proteolytic enzyme systems. Proc Natl Acad Sci U S A. 2006;103:8840–5. doi: 10.1073/pnas.0601876103. [DOI] [PMC free article] [PubMed] [Google Scholar]


