Abstract
Simplified lipid mixtures are often used to model the complex behavior of the cell plasma membrane. Indeed, as few as four components — a high-melting lipid, a nandomain-inducing low-melting lipid, a macrodomain-inducing low-melting lipid and cholesterol (chol) — can give rise to a wide range of domain sizes and patterns that are highly sensitive to lipid compositions. Though these systems are studied extensively with experiments, the molecular-level details governing their phase behavior are not yet known. We address this issue by using molecular dynamics simulations to analyze how phase separation evolves in a four-component system as it transitions from small domains to large domains. To do so, we fix concentrations of the high-melting lipid 16:0,16:0-phosphatidylcholine (DPPC) and chol, and incrementally replace the nanodomain-inducing low-melting lipid 16:0,18:2-PC (PUPC) by the macrodomain-inducing low-melting lipid 18:2,18:2-PC (DUPC). Coarse-grained simulations of this four-component system reveal that lipid demixing increases as the amount of DUPC increases. Additionally, we find that domain size and interleaflet alignment change sharply over a narrow range of replacement of PUPC by DUPC, indicating that intraleaflet and interleaflet behaviors are coupled. Corresponding united atom simulations show that only lipids within ~ 2 nm of the phase interface are significantly perturbed regardless of domain composition or size. Thus, whereas the fraction of interface-perturbed lipids is negligible for large domains, it is significant for smaller ones. Together, these results reveal characteristic traits of bilayer thermodynamic behavior in four-component mixtures, and provide a baseline for investigation of the effects of proteins and other lipids on membrane phase properties.
Keywords: Phase separation, Lipid bilayer, Nanodomains, Macrodomains, Molecular Dynamics
1. INTRODUCTION
The cell plasma membrane (PM) can exhibit nanoscopic mixing heterogeneities, or “rafts”,1,2 thought to be involved in membrane trafficking, signaling, protein sequestration and virus budding.3,4 Rafts are envisioned as distinct from the surrounding membrane, being enriched in high-melting (high-Tm) lipids and cholesterol (chol).2 Although the mechanism that produces rafts is unclear in living cells, chemically defined mixtures that model the composition of the PM outer leaflet can give rise to raft-like coexistence of distinct phases.5 Of particular biological significance is the coexistence of a liquid-ordered (Lo) phase, with fast translational diffusion and high chain order, and a liquid-disordered (Ld) phase, with fast diffusion and low chain order. Like rafts in living cells, the Lo phase in model mixtures is enriched in high-Tm lipids and chol, while the Ld phase is enriched in low-Tm lipids.5 Importantly, the properties, sizes, and even morphology of these two phases can vary greatly depending on lipid composition5,6 and bilayer curvature.7–10 Since cells can alter their membrane composition and shape, the complex functionality of a live cell's PM might be controlled in part by the same principles that influence Lo + Ld phase separation of model membranes.
Coexistence of Lo and Ld phases in lipid mixtures requires a minimum of three components: a high-Tm lipid (e.g. 18:0,18:0-phosphatidylcholine (DSPC), 16:0,16:0-PC (DPPC), or sphingomyelin), a low-Tm lipid (e.g. 16:0,18:1-PC (POPC) or 18:1,18:1-PC (DOPC)) and chol.5 Ternary model membranes containing three such components exhibit either nanoscopic Lo + Ld phase domains (“Type I” mixtures) or macroscopic Lo + Ld phase domains (“Type II” mixtures).5 The type of mixture formed is strongly influenced by the nature of the low-Tm lipid. For example, DSPC/POPC/chol is a Type I mixture11 whereas DSPC/DOPC/chol is a Type II mixture.12 Four-component systems can exhibit a natural progression between Type I and Type II mixtures, providing a complex but biologically relevant model membrane mixture.
In a revealing quaternary mixture experiment, the fractions of DSPC and chol are kept fixed while the nanodomain-inducing POPC is replaced by the macrodomain-inducing DOPC.6,13 Using a replacement ratio defined as ρ = [DOPC]/[POPC + DOPC], a specific composition of DSPC/[POPC + DOPC]/chol is chosen such that it lies within the Lo + Ld coexistence regions at both ρ = 0 (nanodomains) and ρ = 1 (macrodomains). In giant unilamellar vesicles (GUVs), compositional variation (a “trajectory”) along ρ reveals stable macroscopic phase morphologies, termed “modulated phases”, not seen in ternary systems.6,13 Modulated phases have a characteristic size-scale and a variety of interesting morphologies including stripes, honeycomb and broken-up domains.6,13 The ability to exert fine control over domain size and morphology through small changes in composition is a most useful capability of quaternary mixtures.
At a coarse level, Monte Carlo (MC) simulations and experiments have shown that the surprising range of phase morphologies observed in four-component GUVs can be explained by a competition between bending energies and line tension, the two-dimensional analog of surface tension.13,14 The membrane curvature of GUVs makes large patches of the stiff Lo phase unfavorable compared with large domains of the pliant Ld phase; competing line tension favors the coalescence of small domains to minimize interfacial energy. In MC simulations, high line tension that occurs at high ρ produces macroscopic domains, whereas low line tension that occurs at low ρ allows domains to break apart. Intermediate line tensions that occur at intermediate ρ yield energies comparable to bending energies, and the competition between the two produces modulated phases.13,14 While little is known about bending energies along a ρ-trajectory, that line tension increases with ρ is supported by both direct measurements of thermal fluctuations of domains,15 and calculations combining theoretical, simulation and experimental results.16 Together, MC simulations and experiments suggest that line tension plays a dominant role in determining domain size and phase morphology in four-component systems.
Molecular Dynamics (MD) simulations offer the unique capability to study the proposed or observed larger-scale behavior of quaternary mixtures with atomistic resolution. However, to our knowledge, there has yet to be a systematic MD study of such mixtures undergoing a “nanoto-macro” transition. And until recently, only two main ways existed to simulate phases in three- and four-component systems using MD: (1) predetermining initial lateral organization of atomistic lipids, or (2) coarse-graining (CG) lipids. In (1), rather than waiting for phase separation to occur over an unattainably long computational time, the lateral organization of atomistic lipids is pre-defined to be phase-separated.17–20 This method enables high resolution and accurate analysis of the phases, at the cost of accuracy of equilibrium lateral organization. In (2), simplifications are made to the atomistic lipid and water models such that a lipid is represented by ~ 10 beads rather than ~ 100 atoms, as in the Martini model.21 CG lipid mixtures can phase separate in a reasonable amount of computational time, at the cost of atomic resolution.22–26 Neither (1) nor (2) alone is sufficient to fully understand quaternary systems, in which large-scale phase morphologies change, but also atomistic behavior of lipids is important. For phase separation studies, bridging the gap between the long timescale, low-resolution CG simulations and the short timescale, high-resolution atomistic simulations is now possible27 with the advent of methods to convert CG models to atomistic representations.28–30
In this report, we combine the capabilities of CG and united atom (UA) simulations to investigate the phase behavior along a ρ-trajectory in a four-component lipid mixture. Our system contains chol, the high-Tm lipid DPPC (4:0,4:0-PC in CG and 16:0,16:0-PC in UA), and the two low-Tm lipids PUPC (4:0,4:2-PC in CG and 16:0,18:2-PC in UA) and DUPC (4:2,4:2-PC in CG and 18:2,18:2-PC in UA). These lipids were chosen because, in the CG simulations they reflect experimentally observed four-component phase behavior: PUPC, analogous to POPC, promotes clusters or small domains when mixed with DPPC and chol, whereas DUPC, analogous to DOPC, promotes large-scale phase separation. In the simulations, we fix the overall composition such that DPPC/[PUPC + DUPC]/chol ≈ 0.4/0.4/0.2. We then vary the relative fraction ρ = [DUPC]/[PUPC + DUPC] from ρ = 0 (small domains) to ρ = 1 (large domains) in ~ 0.1 increments. We first run these CG systems to equilibrium to study how domain composition, size, and alignment change over a ρ-trajectory. Then, using the program Backward,29 we convert the equilibrated CG structures to UA and run them further to measure phase properties at higher resolution, and to describe the phase interface along the ρ-trajectory. By providing a systematic analysis of the phase behavior of four-component lipid-only mixtures, this study can be used as a baseline to measure and understand how the addition of proteins affects the morphology, onset of phase separation and general phase properties of similar quaternary systems.
2. COMPUTATIONAL METHODS
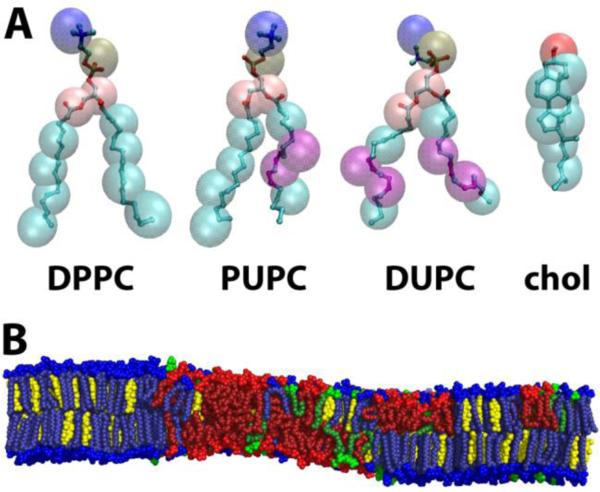
We studied lipid mixtures containing DPPC, PUPC, DUPC and chol as a model four-component system, shown in Figure 1A. The bilayer patches were square with the plane of the bilayer in the xy plane and the z-axis normal to the bilayer. Each patch contained 4,608 lipids, with a total composition of DPPC/[PUPC + DUPC]/chol ≈ 0.4/0.4/0.2. Eleven different ρ conditions were simulated at this composition, from ρ = 0 to ρ = 1 in increments of ρ ~ 0.1. Each of the 11 simulation sets was comprised of one CG simulation and one corresponding UA simulation. Figure 1B shows a snapshot of an equilibrated UA bilayer for ρ ~ 0.8.
Figure 1.
Molecules used in this study. (A) CG (translucent spheres) and corresponding UA (ball-and-stick) representation of the four molecules used in this study. Purple denotes the location of double bonds. (B) Snapshot of equilibrated UA bilayer for ρ ~ 0.8, with DPPC (blue), PUPC (green), DUPC (red) and chol (yellow). Distinct regions of composition and order are observed. Molecule representations visualized in VMD version 1.9, snapshot visualized in PyMOL version 1.3.
A complete description of bilayer construction is provided in Supporting Information Section S1. All simulations were performed with GROMACS versions 4.0.5 and higher.31
2.1 Force field and molecule parameters
CG simulations used version 2.1 of the Martini force field,21,32 with all beads having the same mass. Lipid,33 chol34 and water parameters were from Martini version 2.0. PUPC parameters were not available in the Martini CG force field, and so were modeled by combining the sn-1 chain parameters of DPPC with the sn-2 chain parameters of DUPC.
UA simulations used the GROMOS 87 force field35 with added Berger Lipid parameters,36 as described in Ref. 18. DPPC,36,37 DUPC,18 and chol38 topologies were taken from existing parameterizations. UA parameters for PUPC were also not available, and were modeled by replacing the sn-2 chain parameters of DPPC with those of DUPC. The SPC model39 was used for water.
2.2 Simulation conditions
The CG simulations were performed in the NPT ensemble with 20 fs timestep. The V-rescale thermostat40 with a time constant of 1 ps and Berendsen semi-isotropic barostat41 with a time constant of 4 ps were used to maintain a temperature of 295 K and a pressure of 1 atm. Each lipid type and the solvent were independently coupled to the temperature bath. Electrostatics and van der Waals interactions were cut off at 1.2 nm, and were shifted to zero starting at 0 and 0.9 nm, respectively. Center of mass motion of the system was removed every 10 timesteps. The simulations were run for 25 μs.
Initial structures for the UA simulations were obtained by converting the final CG structures to UA representation using Backward;29 see Supporting Information Section S1. The simulations were then performed in the NPT ensemble with 2 fs timesteps. The Nosé-Hoover thermostat42,43 with a time constant of 0.5 ps and Parrinello-Rahman semi-isotropic barostat44 with a time constant of 2.0 ps were used to maintain a temperature of 300 K as in Ref. 18, and a pressure of 1 atm. Each lipid type and the solvent were independently coupled to the temperature bath. A 1.1 nm cutoff was employed for the electrostatics and van der Waals interactions. SETTLE45 was used for rigid water constraints. Particle Mesh Ewald (PME)46,47 was used for electrostatics, with a cubic interpolation order of 4 and Fourier grid spacing of 0.16. Center of mass motion of the bilayer and solvent were separately removed every 100 timesteps. The simulations were run for 200 ns.
All CG and UA simulations applied periodic boundary conditions in three dimensions, and employed the LINCS31,48 algorithm for bond constraints.
2.3 Phase determination algorithm
Determining phase patches in lipid simulations requires first deciding on a criterion for a phase. One possible choice is to use the order of lipid chains,49,50 while another is to use local composition.20 We chose to use the latter, since it is independent of lipid-specific properties: order is dependent not only on phase, but also on lipid type (see e.g. Figures S6–S9) and phase of the apposed leaflet.51 Although hidden Markov models exist to determine phases based on lipid composition,20 we chose to use a single compositional cutoff for simplicity.
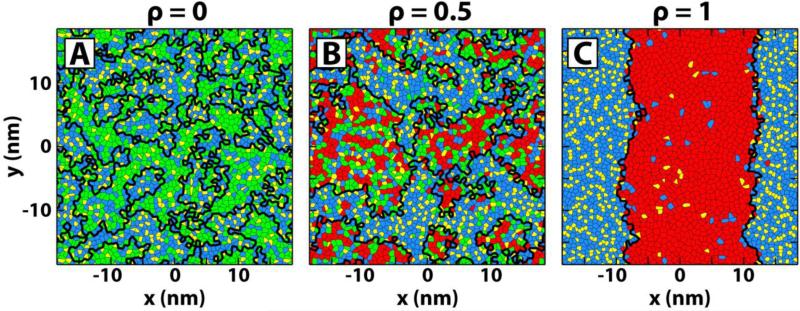
It is known from experiments that high-Tm lipids, in our case, DPPC, and chol are enriched in the Lo phase compared to the Ld phase.12 This allows use of the local concentration of DPPC and chol to determine phase domains.20 To do this, we first perform a Voronoi tessellation based on the centers of mass of each lipid within a leaflet. Next, the local environment of a particular lipid of interest (LOI) is defined to be those lipids that share a Voronoi edge with the LOI. Finally, the phase of the LOI is determined by its local concentration of DPPC + chol. If the concentration of DPPC + chol in the local environment is higher than that in the entire leaflet, the LOI is considered to be in the Lo phase; otherwise, the LOI is considered to be in the Ld phase. Continuous phase patches were determined using a connectivity matrix; patches containing fewer than 10 lipids were considered random compositional fluctuations and were included in their surrounding domain.49,50 Voronoi edges between two phases are then phase boundaries. The phase boundaries are taken to lie within the xy plane, and for calculations involving the boundaries, only those within the leaflet being analyzed are considered. Example results of the phase determination algorithm (implemented in Matlab version R2010a) are shown in Figure 2.
Figure 2.
Phase determination algorithm reveals patches of Lo and Ld phases. Voronoi tessellation of one leaflet for (A) ρ = 0, (B) ρ = 0.5 and (C) ρ = 1 with DPPC (blue), PUPC (green), DUPC (red) and chol (yellow). Coexisting phase patches, demarcated by thick black lines, increase in size and compositional difference from ρ = 0 to ρ = 1.
We note here that at low ρ in these simulations, and in ternary mixture simulations of DPPC/PUPC/chol,52,53 small clusters reveal non-ideal mixing (see e.g. web enhanced object Movie_ρTrajectoryPhaseSeparation.mpg) rather than nanoscopic phase domains. Regardless, the purpose of this work is to investigate trends in demixing as a low-Tm lipid that produces small domains is replaced by a low-Tm lipid that produces large domains. In this vein, we stick to Lo/Ld terminology for clarity and in a later section discuss the ρ value at which we believe true phase separation first appears in these simulations.
2.4 Equilibration and data analysis
Based on equilibration of CG phase interface length and UA box size (Figure S1), we determined that the CG and UA systems required 15 μs and 50 ns respectively to be sufficiently equilibrated. Consequently, data analysis for CG and UA simulations was performed only over the last 10 μs and last 150 ns of simulation time, respectively.
After equilibration, we calculated the time autocorrelations of several CG and UA properties — phase interface length, misaligned phase overlap fraction, order parameter, extent of lipid tilt and lipid tilt orientation — for the representative case of ρ = 1 (Figures S2 and S3). We chose ρ = 1 since it is expected to have the slowest correlation times due to its large domain size and high order in the Lo phase. All correlations are described in the Supporting Information Section S2, and the properties themselves are described throughout the text. Based on the correlation functions, we determined that the CG properties of interest became essentially uncorrelated by 250 ns and the UA properties became sufficiently uncorrelated by 25 ns. Thus, each 10 μs CG simulation could be split into 40 independent 250 ns traces and similarly, each 150 ns UA simulation into 6 independent 25 ns traces. For details on how averages and standard deviation error bars were calculated from these subsets, see Supporting Information Section S2.
3. RESULTS
In the following sections, CG results are in sections 3.1-3.3, UA results in 3.4-3.5. Results plotted as a function of ρ are colored from green at ρ = 0 to red at ρ = 1 unless otherwise specified.
3.1 CG demixing of Lo lipids DPPC and chol, and Ld lipids PUPC and DUPC, increases along the ρ-trajectory
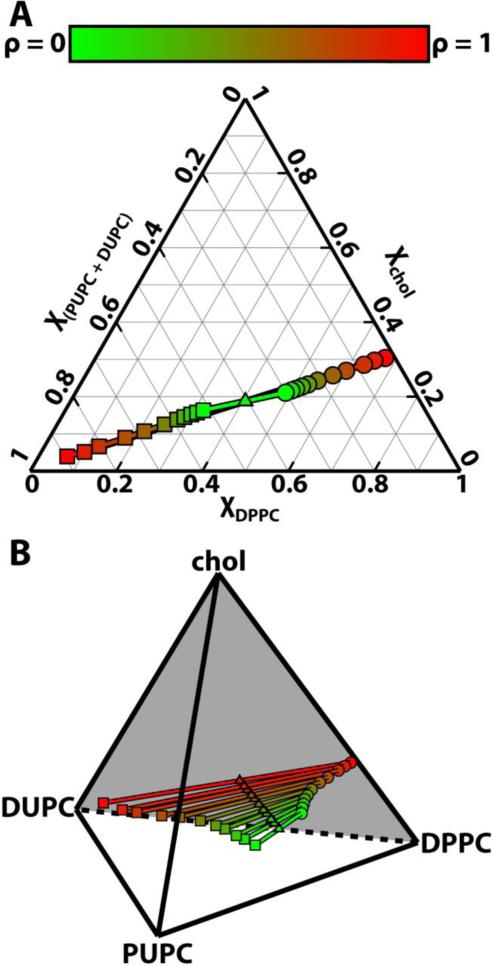
We used the phase determination algorithm from Section 2.3 on our simulation results to define regions of similar phase (Figure 2). This algorithm ensures that Ld phases are enriched in the low-Tm lipids PUPC and DUPC, while Lo phases are enriched in DPPC and chol. Once phases were determined, compositions of a given phase were calculated leaflet by leaflet based on all lipids in that phase. Figure 3 shows the coexisting Lo (circles) and Ld (squares) compositions throughout the ρ-trajectory, in terms of standard equilibrium ternary and quaternary phase diagrams at constant temperature. For each ρ value, coexisting phase compositions of Lo and Ld are connected by tielines. Triangles mark the overall composition of the bilayers simulated: DPPC/[PUPC + DUPC]/chol ≈ 0.40/0.4/0.2.
Figure 3.
Lipid demixing increases with ρ. (A) Ternary and (B) quaternary phase diagrams for DPPC/[PUPC + DUPC]/chol ≈ 0.4/0.4/0.2. Tielines connect the compositions of coexisting Lo phases (circles) and Ld phases (squares). Triangles mark the overall bilayer composition at each ρ. Results are shown from ρ = 0 (green) to ρ = 1 (red). Longer tielines at high ρ indicate increased compositional differences between coexisting phases.
The ternary and quaternary diagrams show that the compositions of coexisting Lo and Ld phases become increasingly different as ρ increases (from green to red). The differences between Lo and Ld compositions are measured by the lengths of tielines, which increase by ~ 400% from ρ = 0 to ρ = 1. Specifically, the amount of low-Tm lipid in the Lo phase decreases from 30.1 ± 0.4% at ρ = 0 to 2.3 ± 0.2 % at ρ = 1. Similarly, the amount of DPPC in the Ld phase decreases from 32.1 ± 0.3% at ρ = 0 to 6.6 ± 0.5% at ρ = 1. Thus, as DUPC replaces PUPC, the extent of demixing increases: DPPC and chol become more enriched in the Lo phase and the low-Tm lipids become more enriched in the Ld phase.
3.2 CG domain size and interleaflet domain alignment increase with ρ
The phase domains of Figure 2 show that domain size increases with ρ. To analyze the dependence of size on ρ, we calculated an intraleaflet pair correlation function, g(r), of Ld lipids. g(r) is the probability of finding two Ld lipids at a certain distance from each other in the same leaflet, normalized by the probability given a uniform random distribution:
| (1) |
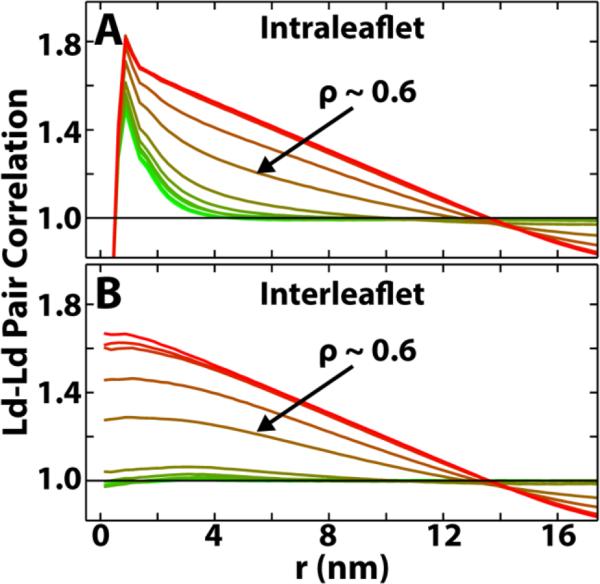
where A is the simulation area, N is the number of Ld lipids in a leaflet, are the coordinates of the lipid centers of mass, Δr = 0.25 nm is the bin size and δ is the Kronecker delta. The pair correlation functions for all ρ-trajectory simulations are shown in Figure 4A. At low ρ, pair correlations have exponential-like decays indicating only short-range correlations. At higher ρ, pair correlations decay in a much slower, linear fashion, indicating large-scale phase patches that can be limited by the finite simulation size. The observed increase in pair correlations with increasing ρ means that the characteristic size-scale of domains grows with ρ.
Figure 4.
Ld-Ld pair correlations show that both domain size and alignment increase with ρ. (A) Intraleaflet Ld-Ld pair correlation functions, g(r), plotted from ρ = 0 (green) to ρ = 1 (red). Larger correlations at higher ρ mean domain patches are larger. (B) Interleaflet Ld-Ld pair correlation functions, g1,2(r), plotted from ρ = 0 (green) to ρ = 1 (red). Larger correlations between leaflets at higher ρ mean that domain patches are more aligned between the two leaflets.
In addition to using intraleaflet pair correlations to study domain size, we also measure interleaflet correlations of phase patches to study domain alignment. Interleaflet correlations show the probability of finding two lipids in the same phase at a certain distance from each other in apposed leaflets, normalized by the probability given a uniform random distribution in the apposed leaflet. Similarly to Equation 1, we measure the interleaflet correlation, g1,2(r), of Ld lipids as:
| (2) |
where N1 and N2 are the number of Ld lipids in each of the two leaflets, and other terms are defined as in Equation 1. Interleaflet pair correlations for all ρ values are shown in Figure 4B. At low ρ, the interleaflet pair correlations remain near 1: there is no preferred alignment of phase domains between leaflets. Conversely, at high ρ, pair correlations grow significantly above 1: the phase domains in the two leaflets are more aligned.
When measuring Lo-Lo correlations with a bin size of Δr = 0.025 nm, a simulation artifact becomes apparent. It is particularly noticeable in correlations between acyl chain and chol centers of mass, as shown in Figure S4A,B. At high ρ, the Lo phase has long-range periodicity in its correlations, revealing that it is gel-like. We discuss this apparent gelation artifact in the Supplementary Information Section S3. We do not expect it to affect our simulations as it only appears in the Lo phase after phase separation has begun and is much less pronounced in the UA simulations (Figure S5). A newly developed CG cholesterol available at http://md.chem.rug.nl/cgmartini/index.php/force-field-parameters/sterols prevents this gelation in the CG simulations.
3.3 A sharp transition in CG phase morphology occurs between ρ ~ 0.5 and ρ ~ 0.8
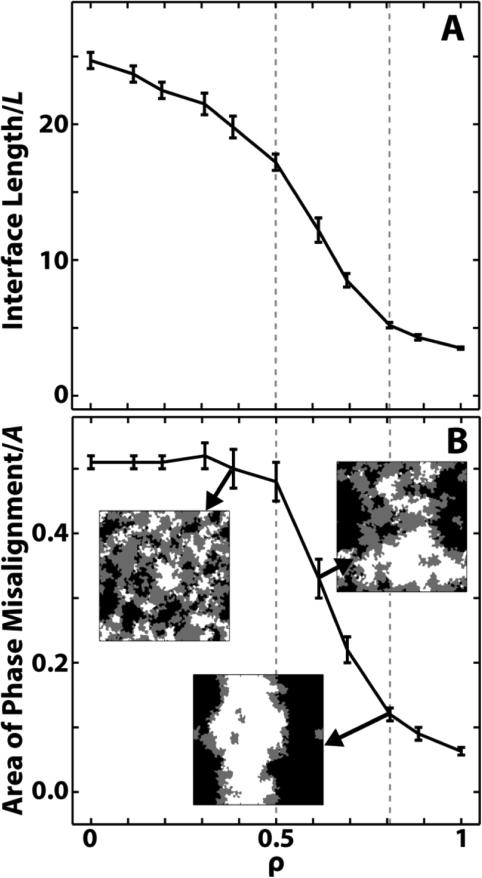
The intraleaflet pair correlation functions in Section 3.2 reveal two distinct regions of behavior: short-range correlations at low ρ and long-range correlations at high ρ. To better quantify the shift in intraleaflet phase behavior at intermediate ρ, we look at the amount of interface between Lo and Ld phases. As is often done,49,50 we normalize the interface length, in this case by the x/y box length L. We plot the normalized interface lengths as a function of ρ in Figure 5A. The normalized interface length is large, greater than 20, at low ρ and decreases smoothly until ρ ~ 0.5. A steep drop-off then starts at ρ ~ 0.6, leveling off at ρ ~ 0.8. By ρ = 1, the normalized interface length is ~ 15% that at ρ = 0.
Figure 5.
A transition in phase morphology occurs between ρ ~ 0.5 and ρ ~ 0.8 (dashed gray lines). (A) Length of the phase interface along a ρ-trajectory, normalized by the bilayer length L. A significant decrease between ρ ~ 0.5 and ρ ~ 0.8 indicates an increase in domain coalescence. (B) Area of misaligned phases along a ρ trajectory, normalized by the box area A. A significant decrease occurs between ρ ~ 0.5 and ρ ~ 0.8, indicating an increase in domain alignment. Also shown are overlayed phase plots of the two leaflets for ρ ~ 0.4, ρ ~ 0.6 and ρ ~ 0.8. Coloring: Lo across from Lo is white, Ld across from Ld is black, and Lo across from Ld is gray. Overlayed plots are 37 nm × 37 nm.
Changes in normalized interface length from one simulation to the next can arise from changes in either phase area fractions or domain size. To determine which factor was governing our results, we measured phase area fractions. Over the entire ρ-trajectory, normalized interface length decreases significantly, whereas the area fraction of Lo decreases from 48.3 +/− 0.4% to 44.3 +/− 0.2%, and the area fraction of Ld increases from 51.7 +/− 0.4% to 55.7 +/− 0.2 %. Since only small changes in phase area fractions occur, the decrease in interface length that occurs for 0.5 ≤ ρ ≤ 0.8 must be due to increasing intraleaflet domain coalescence. This is also evident in Movie_ρTrajectoryPhaseSeparation.mpg, which shows the evolution of the Lo and Ld phase domains for all ρ over time.
A change in phase behavior at intermediate ρ is also apparent in the interleaflet pair correlations: domains are uncorrelated at low ρ but are highly correlated at high ρ. A straightforward way to quantify this change in domain alignment is by measuring area fraction of domain overlap.25,49,54 In this case, we choose to measure the area of misaligned domains, i.e. Lo across from Ld,25 normalized by the simulation area A. Since the total area fraction of each phase is ~ 50%, in the three extreme cases of perfect domain anti-alignment, perfect domain alignment, and completely random domain alignment, the area fractions of misalignment would be ~ 1, ~ 0 and ~ 0.5 respectively.
Figure 5B shows snapshots of overlayed leaflets for ρ ~ 0.4, ρ ~ 0.6 and ρ ~ 0.8 (with white corresponding to aligned Lo phases, black to aligned Ld phases, and gray to misaligned phases), together with results of the misaligned fraction calculations. For ρ ≤ 0.5, the area fraction of misaligned domains is ~ 0.5, showing domains are uncorrelated. For ρ ≥ 0.8, misalignment drops to less than 0.15. The transition between these two extremes occurs over the relatively small ρ range between 0.5 and 0.8. This is similar to the transition region observed in Figure 5A, supporting the finding that a switch in phase behavior occurs between ρ ~ 0.5 and ρ ~ 0.8. This is evident in Movie_ρTrajectoryPhaseOverlap.mpg, which shows the overlayed leaflets for all ρ over time.
3.4 UA order parameter perturbations extend ~ 2 nm into each phase and are uniform along a chain
After running the converted UA simulations, we measure the order of UA lipid chains using the carbon deuterium order parameter:
| (3) |
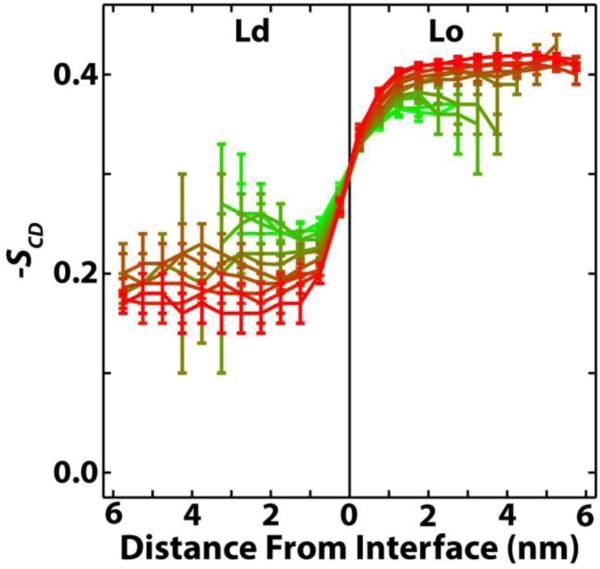
where α is the angle between the calculated CH bond and the bilayer normal, taken to be along the z-axis. Figure 6 shows −SCD averaged over all methylenes in the hydrocarbon chains of DPPC as a function of distance from the phase interface. As expected, equilibrium DPPC order in the Ld phase is substantially lower than equilibrium DPPC order in the Lo phase. As ρ increases, the Lo phase becomes more ordered and the Ld phase becomes less ordered. The changes in equilibrium order coincide with an increase (decrease) of chol in the Lo (Ld) phase as ρ increases (Figure 3), consistent with the so-called ordering effect of cholesterol.55 Whatever the differences in equilibrium order along the ρ-trajectory, DPPCs in each phase always reach bulk values within ~ 2 nm of the interface. The same is true for PUPC and DUPC (Figure S6).
Figure 6.
DPPC order parameter −SCD is only perturbed ~ 2 nm from the interface into each phase. −SCD is averaged over all methylenes in both chains of DPPC, as a function of distance from the phase interface. Results shown from ρ = 0 (green) to ρ = 1 (red). DPPC order in both phases reaches equilibrium within ~ 2 nm of the phase interface, regardless of ρ.
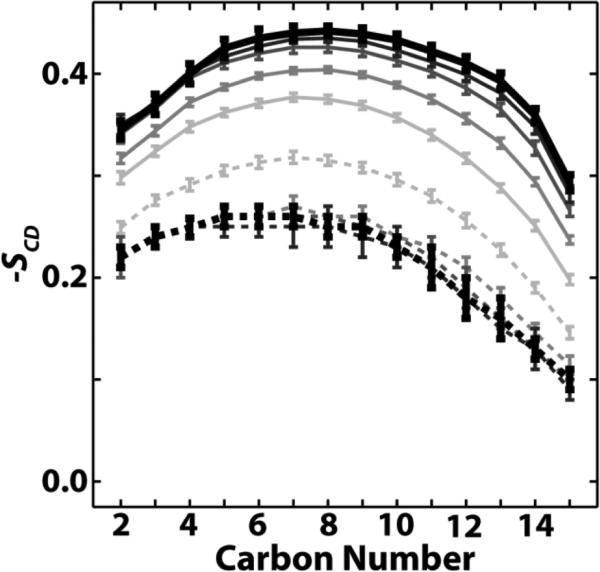
The perturbed lipids near the interface adjust to the different phase equilibria through nearly uniform changes along their acyl chains, despite there being significantly more double bonds in the Ld phase than in the Lo phase. In Figure 7, −SCD is plotted for all carbons in the DPPC sn-1 chain as a function of distance to the interface for ρ = 0.5. No part of the DPPC acyl chains is significantly more perturbed than any other. Again, changes in order do not extend far from the interface. Similar behavior is observed for the sn-2 chain of DPPC, and the sn-1 and sn-2 chains of PUPC and DUPC (Figure S7). The uniform change in order of the high-Tm lipid agrees with a previous study of a sphingomyelin/chol-enriched Lo phase surrounded by a DOPC-enriched Ld phase; whether the same was true for the low-Tm lipid is less clear.19
Figure 7.
DPPC carbons are nearly uniformly perturbed regardless of phase or distance from the phase interface. Order parameters are plotted for DPPC sn-1 carbons for Ld lipids (dashed) and Lo lipids (solid), with increasing darkness indicating increasing distance from the interface in increments of 0.5 nm. Thicker black curves are averaged over all DPPCs greater than or equal to 2 nm from the interface. Results shown for ρ = 0.5.
For corresponding CG order plots, see Figures S8 and S9.
3.5 UA lipid phase determines the extent and orientation of lipid tilt
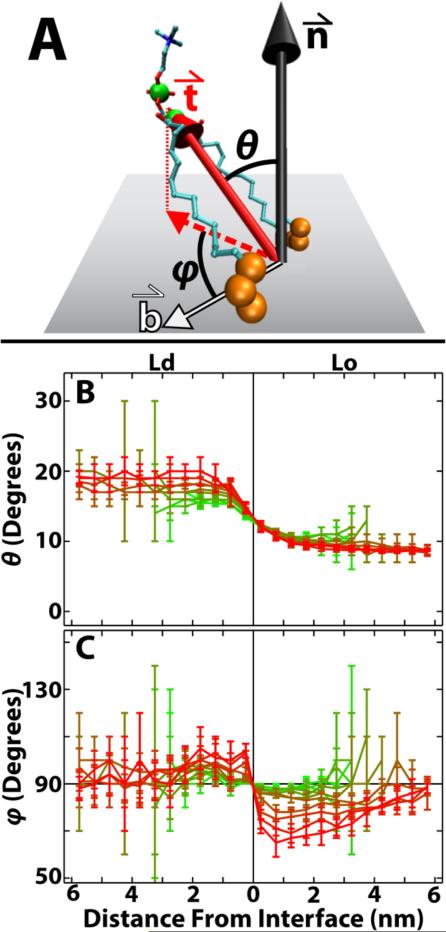
To measure lipid tilts, we assume the bilayer lies in the xy plane, and take the normal to be along the z-axis. We then define the tilt of a lipid (Figure 8A) as in Ref. 56: the tilt vector () of a PC lipid joins the combined center of mass of the last three carbons of the acyl chains to the combined center of mass of the CD atom and phosphorus of the headgroup. The polar angle (θ) of with the bilayer normal () indicates the extent of lipid tilt.
Figure 8.
Lipid tilt is affected by phase and distance to interface. (A) Lipid tilt vector () connects the combined center of mass of the last three carbons on each acyl chain (large orange spheres) to the combined center of mass of the CD atom and phosphorus of the headgroup (large green spheres). The local bilayer midplane, which is assumed to lie along the xy plane, is shown as a gray rectangle. is the bilayer normal, and is the vector joining the lipid center of mass to the nearest phase interface. θ and φ are, respectively, the polar and azimuthal angles of : θ is the angle between and , and φ is the angle between (projected onto the xy plane) and . (B) θ is larger, i.e. lipids tilt more, in the Ld phase compared to the Lo phase. (C) Near the interface, lipids tilt away from the interface in Ld (φ > 90°) but towards the interface in Lo (φ < 90°). Results shown from ρ = 0 (green) to ρ = 1 (red). Molecule representation visualized in VMD version 1.9.
Figure 8B shows θ versus distance from the interface for DPPC. Throughout the ρ trajectory, DPPCs in the Ld phase tilt on average ~ 10° more than those in the Lo phase. Whereas DPPC in the Ld phase tilts more at low ρ than at high ρ, DPPC tilt in the Lo phase does not change much along the ρ-trajectory. Just as with order, tilt perturbations are gone less than 2 nm from the phase interface, regardless of ρ. Similar, but not identical, tilt behavior is seen for PUPC and DUPC, whereas chol tilts more than other lipids in the Ld phase (Figure S10). A larger chol tilt and a tendency of lipids to tilt less as chol fraction increases is consistent with previous simulations.57
While θ quantifies the magnitude of tilts, we used another angle (φ) to determine if the tilts have any preferential orientation. φ is the azimuthal angle of , defined to be the angle between (projected onto the xy plane) and the vector () connecting the lipid center of mass to the nearest phase interface (Figure 8A). Average values of φ equal to 90° indicate random orientation, while averages greater than 90° indicate that lipids tilt away from the phase interface and averages less than 90° indicate that lipids tilt toward the interface. As an example, consider a lipid in the Ld phase. If φ is less than 90°, it is tilting toward the phase interface and so tilts toward the Lo phase. Conversely, if φ is greater than 90°, it tilts away from the interface and so tilts towards the bulk Ld phase.
In Figure 8C, φ is plotted as a function of distance to the phase interface for DPPC. For large ρ, DPPCs, especially those near the interface, tilt towards the bulk Ld phase: lipids in Lo tilt towards Ld (d835df11 < 90°), and lipids in Ld tilt towards Ld (φ > 90°). This preferential orientation of tilts tends to increase with ρ. At higher ρ values, the preferential orientation persists for several nanometers in the Lo phase, but extends a much shorter distance in Ld. φ for PUPC, DUPC and chol show similar, but not identical, trends of preferential orientation (Figure S11). Though surprising, long range preferential orientation of tilts has been observed in other bilayer simulations.17
To ensure that local curvature was not dramatically altering our tilt results, we recalculated θ and φ using the local normals and the local tangent planes (data not shown). Indeed, trends in θ and the significant preferential orientation of φ at high ρ were not significantly altered using this technique.
For corresponding CG tilt plots, see Figures S12 and S13.
4. DISCUSSION
In the following sections, CG results are discussed in sections 4.1-4.2, UA results in 4.3-4.4.
4.1 Comparison of CG phase separation to experiments
The CG simulations are able to qualitatively capture four important traits of quaternary phase separation. First, both experiments58–60 and the simulations reported here show an increase in domain size as ρ increases. Second, the transition from small domains to large domains is relatively sharp in both experiments on GUVs6,60 and in simulations. Third, the simulations reveal that compositional differences between coexisting Lo and Ld phases increase with ρ which has also been observed experimentally.59 And fourth, we see that along the ρ-trajectory, increased demixing and an increased fraction of DUPC result in a predictable increase in thickness mismatch between Lo and Ld phases (SI Figures S14 and S15A). A similar increase in thickness mismatch between coexisting phases along a ρ-trajectory has been detected with small-angle neutron scattering (SANS).58
Disparities between simulation results and experiments are also revealing. Perhaps most noticeable is that the simulations do not capture the experimentally observed modulated phase patterns. This is simply because the limited size-scale and relatively flat nature of the simulations means that they are unable to sustain modulated patterns which are microns in size and likely require curvature to form.13,14 Another discrepancy, as mentioned in Section 2.3, is that the low ρ simulations are more properly termed non-ideal mixing rather than the phase domains measured experimentally. The simulations also exhibit a much larger compositional change of Lo and Ld phases along a ρ-trajectory compared to experiments,59 meaning that CG demixing is too strong. A final distinction between the simulations and experiments is that the CG Lo phase undergoes gelation at high ρ, but we emphasize this to be an artifact of the CG chol which does not occur with a newer chol model.
Some differences between experiments and the simulations may simply be due to the fact that the Martini model was parameterized to enable large size-scale and long time-scale simulations. The coarse-grained nature of the model means that while entropy may play some role in phase separation,61 it is not necessarily a dominant effect,62 and likely does not play as large a role in phase separation as it would in vitro. Similarly, the simplified treatment of electrostatics in the Martini model21,32 would affect phase behavior differently than would electrostatics in vitro and in vivo. The simplifications inherent in the Martini model are not a hindrance, but are instead essential to its success; by cutting down the computational costs, the simplifications allow for phase separation to take place in systems that would otherwise require unreasonable amounts of computational time. Thus, while our simulations involve somewhat different interactions than experimentally studied quaternary systems, we have shown that they are able to capture much of the essential experimentally observed phase behavior. Furthermore, by capturing the trends in phase separation, the CG simulations allow for conversion to a more accurate atomistic force field for more detailed analysis of phase properties.
4.2 CG intraleaflet phase separation and interleaflet domain alignment are coupled and highly dependent on the fraction of the low-Tm lipid
An abrupt transition from small, non-aligned domains to large, aligned phase patches occurs between ρ ~ 0.5 and ρ ~ 0.8 in the CG simulations. The transition window location and narrow size indicates that intraleaflet and interleaflet effects are sensitive to the fractions of PUPC and DUPC at intermediate ρ. That the intraleaflet and interleaflet transition windows are the same hints that the two effects are coupled. These observed morphological changes along the ρ-trajectory can be explained in terms of enthalpies.
Liquid-liquid phase coexistence in Martini ternary mixtures of DPPC/DUPC/chol is largely driven by enthalpy.61,62 This likely governs the quaternary system behavior as well. The unsaturated beads of DUPC and PUPC mix unfavorably with chol and with the saturated beads of DPPC.21 Since DUPC has four unsaturated beads compared to PUPC's two, DUPC is expected to segregate from DPPC and chol more readily than is PUPC. At low ρ, PUPC is the dominant low-Tm lipid and demixing is weak. When DUPC begins to outnumber PUPC at ρ > 0.5, the unfavorable interactions dominate and phase domains enlarge to minimize the interfacial penalty. This is in agreement with previous Martini studies showing that more unsaturation of the low-Tm lipid enhances demixing.52 An increase in lipid and monolayer thickness mismatch between the Lo and Ld phases as ρ increases (Figures S14 and S15A) may further drive intraleaflet phase separation53,63–65 as a way to minimize the amount of unfavorable conformational adjustments and interactions at the interface.
Coincident with the abrupt increase in domain size near ρ ~ 0.6 is the alignment of phase domains. For ρ > 0.5, the free energy contribution of aligned phases must outweigh the energetically unfavorable increasing thickness mismatch.51,66 While electrostatics, cholesterol flip-flop, interdigitation,66,67 and curvature51,54 can play a role in alignment, another influential factor is a surface tension at the bilayer midplane between the leaflet phases.68 In the Martini model, a surface tension between domains25 could arise from the unfavorable interleaflet interaction between the unsaturated beads of the Ld phase and the saturated beads and chol of the Lo phase.69 From the phase diagram, the fraction of DUPC — and in turn unsaturated beads — in the Ld phase increases with ρ, while the fraction of unsaturated beads in the Lo phase decreases with ρ. This results in a higher density of unsaturated beads near the bilayer center in the Ld phase compared to the Lo phase (Figure S15B), likely increasing the penalty for phase mismatch at higher ρ. Furthermore, the penalty for interleaflet phase mismatch grows with area of the domains.25,66 The large change in domain size near ρ ~ 0.6 is therefore expected to result in a correspondingly large increase in mismatch penalty per domain, favoring domain alignment. Since alignment only increases significantly for ρ > 0.5, it seems as though domain size, more so than just the increased surface tension between Lo and Ld phases, drives interleaflet domain alignment. This is in agreement with previous studies of bilayer mixtures that show domain alignment is greater for larger domains.25,54,61 We therefore conclude that interleaflet domain alignment in the quaternary system DPPC/PUPC/DUPC/chol is significantly affected by intraleaflet domain size, and in turn ρ.
It is interesting that the phase morphology transition window in the simulations occurs when domains reach the approximate size-scale of nanodomains measured in vitro (~ 7 nm radius for DSPC/POPC/chol).58 This can be seen in Figure 4 and Figure S4, where the intraleaflet correlations at r = 7.5 nm are much larger for ρ ≥ 0.6 compared to ρ < 0.6. As ρ ~ 0.6 is also the composition when alignment begins, it is possible that ρ ~ 0.6 marks the beginning of true phase behavior for this particular mixture and in turn most closely reflects the behavior of nanodomains measured experimentally. Larger simulations, beyond the scope of this project, could indicate if the ρ ~ 0.6 composition does indeed produce many, separate, nanodomains that are not simply limited by the simulation size.
Experimentally, the transition window from nanoscopic to macroscopic domains can vary substantially depending on the mixture used. In some systems, this transition window is narrow and low, and in other cases it is high and broad.13 Thus, there is nothing universal about the region 0.5 ≤ ρ ≤ 0.8. Instead, the transition window simply represents a regime in this particular four-component mixture where the interfacial penalty in the simulation becomes large enough to induce the formation of large domains. We expect that the location and width of such a transition window in vitro are likely affected by water entropy, electrostatics of the different lipid moieties, and other atomistic properties that are outside the scope of the Martini model. Regardless, differences between the high-Tm lipid and the low-Tm lipid are expected to strongly affect the location of the transition window in both experiments and simulations: larger differences, e.g. in thickness, unsaturation, branching, result in larger interfacial penalties and in turn, lower transition windows. Whether or not the experimentally observed intraleaflet transition window is also in general an interleaflet transition window is not currently known. In all experimental quaternary systems to date, macroscopic phase domains are aligned between the two leaflets, consistent with the simulations. However, the simulations indicate that alignment is likely lower for nanodomains than for macrodomains, which has yet to be tested experimentally.
4.3 Energy penalty of the UA phase interface is spread out over only a few lipid shells
Line tension plays a crucial role in phase separation of lipid mixtures and acts as a competing interaction producing modulated phases.14,70 The atomistic underpinnings of line tension are not well understood, though it is hypothesized to be a function of the bending moduli, tilt moduli, spontaneous curvatures and thickness mismatch of the two phases.64 Here, we determine how lipids adjust to the interface in the UA systems and thus the width of lipid layers whose perturbed energy contributes to line tension.
As shown in Figures 6 and 8B, the Ld/Lo phase interface — in terms of order and extent of lipid tilt — extends ~ 2 nm into each phase, with the largest changes occurring within 1 nm of the interface, consistent with other simulations and mesoscopic modeling of phase separated bilayers.51,71 Perhaps as a manifestation of order and tilt perturbations, we also find that lipid thicknesses of the UA PC lipids in the Lo and Ld phases reaches equilibrium within ~ 1.5 nm of the interface (Figure S16). The interfacial width of 1-2 nm in each phase is relatively constant along the ρ-trajectory despite increasing equilibrium differences in order, tilt and thickness between Lo and Ld phases. Therefore, lipids within ~ 2 nm of the phase interface must bear the brunt of the energetic cost of the interface regardless of domain size, alignment, composition or equilibrium phase properties.
Assuming an interface width of just 2 nm into each phase, a circular 7 nm radius nanodomain58 (similar to that observed at ρ ~ 0.6) would have ~ 50% of its lipids within the interfacial environment. The fraction of lipids at the interface drops off with domain size, reaching 1% at a domain radius of ~ 400 nm. These interface percentages are inherently overestimates as they do not account for the magnitude of perturbations, which are most significant within 1 nm of the interface. Nevertheless, they demonstrate that while the width of the interface might only be one or two lipids in each phase, it can make up a large fraction of small domains. This could be crucial for interpreting experiments, such as those involving electron spin resonance (ESR) and SANS, that assume homogeneity of the phases. It may also affect the partitioning of molecules, some of which may prefer the environment of the interface.72
4.4 The Ld phase alters molecule behavior deep within the Lo phase in UA simulations
One might naively assume that all phase properties change to their bulk values through an intermediate transition at the interface. Indeed, we saw that this was the case for order and the magnitude of lipid tilts, which reach bulk values within ~ 2 nm of the interface. However, an interesting property of the UA lipids, which is distinct from bulk phase properties and persists beyond the interfacial length of ~ 2 nm, is the orientation of lipid tilt with respect to the boundaries. Rather than being randomly oriented, lipids several nanometers from the phase interface preferentially tilt towards the Ld phase at high ρ. This spatially extended preferred orientation could be explained by the nature of the Ld phase, which is more disordered and fluid-like than the Lo phase. As ρ increases, the Ld phase becomes even more disordered and the Lo phase becomes even more ordered. The Ld phase can then more easily accommodate lipid tilts compared to the Lo phase, and preferential orientation of lipid tilts towards the Ld phase increases. By ρ = 1, lipids in the Lo phase as far as 6 nm from the interface significantly preferentially tilt towards the Ld phase.
There is some uncertainty as to whether the extended tilt orientation observed here represents the true behavior of lipids in vitro or is instead an artifact of the UA simulation. If the preferential orientation is an artifact, it is important for other researchers to be aware of it. However, recent findings of long-range tilt orientations in simulations of bilayers17 provide support that the effect may be real, though such correlations were only observed in asymmetric bilayers and were not detected in corresponding symmetric bilayers as in our present study. If the observed long-range interaction is not an artifact, then it may affect membrane shape,17,73 allow for transmission of information beyond nearest neighbors, and alter the behavior of molecules deep within the bulk phases. So while the interface itself may only be 3-4 nm wide in total, its presence may have more far-reaching effects.
5. CONCLUSION
We found several interesting behaviors of a four-component lipid bilayer through use of a series of CG and UA simulations that take the mixture from nanoscopic to macroscopic phase separation. The CG simulations show that domain size and interleaflet alignment are coupled, and that domain alignment is especially sensitive to the fractions of low-Tm lipids. They also indicate that compositions of the coexisting Lo and Ld phases become more distinct as DUPC replaces PUPC. The UA simulations show that the phase interface, in terms of perturbed order and extent of lipid tilt, does not extend very far into either phase. However, the interface is not negligible, and can make up a significant fraction of smaller domains.
The UA simulations also revealed two surprising aspects of lipids in the coexisting phases. First, the UA lipids were shown to adjust to the different phase orders through uniform changes in their acyl chains, despite the high fraction of double bonds in the Ld phase compared to the Lo phase. Second, the UA simulations showed a long-range preferential tilt of lipids toward the Ld phase, meaning that lipids in a phase far from the interface can be affected by a distant, coexisting phase.
Though no experiments have been performed on the DPPC/PUPC/DUPC/chol system simulated here, direct comparison of such experiments to the simulations could be very informative. Fluorescence microscopy, Förster resonance energy transfer and SANS could provide information about domain composition, size and thickness along an experimental ρ-trajectory.6,13,58,59 One could also find the ρ value where an abrupt transition in domain morphology occurs. ESR analysis of a spin-labeled lipid could be used to study equilibrium phase properties, and could provide an estimate for the fraction of phase interface by determining the amount of lipids with order between the equilibrium orders of the Lo and Ld phases. These results could all be directly compared to the results reported here in order to learn both how to improve force fields and molecular parameterizations, and in what ways the limitations of box size and coarse-graining affect simulation results. Such a comparison would have implications for the reliability of simulation properties that are not amenable to experimental measurements, such as pressure profiles and atomistic-level details of lipid behavior.
By providing a thorough analysis of a four-component mixture, CG and UA simulations reported here will be useful as a control for future studies addressing how addition of other molecules, membrane proteins in particular, affect the phase behavior in similar complex mixtures. They also form a basis for comparison to complementary experiments.
ACKNOWLEDGMENT
This work was supported in part by funding from the U.S. National Science Foundation (MCB-1410926) and the U.S. National Institutes of Health (GM105684) to G.W.F. D.G.A. was supported by the U.S. National Science Foundation Graduate Research Fellowship (DGE-114153) and by NIH Training Grant 1-T32-GM08267. This work used the Extreme Science and Engineering Discovery Environment (XSEDE), which is supported by National Science Foundation grant number ACI-1053575. Simulations were predominantly run on the Lonestar and Gordon clusters. We thank Edward Lyman, Jonathan Amazon, Milka Doktorova, Michael Weiner, Benoit Palmieri, Alan Grossfield and Huimin Chen for useful discussions.
Footnotes
Supporting Information Available: Description of bilayer construction and data acquisition; analysis of cholesterol artifact, order parameter, lipid tilts, lipid thickness and unsaturation densities; 16 Figures; 2 Movies. This information is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Simons K, Sampaio JL. Membrane Organization and Lipid Rafts. Cold Spring Harbor Perspect. Biol. 2011;3:1–17. doi: 10.1101/cshperspect.a004697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lingwood D, Simons K. Lipid Rafts as a Membrane-Organizing Principle. Science. 2010;327:46–50. doi: 10.1126/science.1174621. [DOI] [PubMed] [Google Scholar]
- 3.Rajendran L, Simons K. Lipid Rafts and Membrane Dynamics. J. Cell Sci. 2005;118:1099–1102. doi: 10.1242/jcs.01681. [DOI] [PubMed] [Google Scholar]
- 4.Simons K, Gerl MJ. Revitalizing Membrane Rafts: New Tools and Insights. Nat. Rev. Mol. Cell Biol. 2010;11:688–699. doi: 10.1038/nrm2977. [DOI] [PubMed] [Google Scholar]
- 5.Feigenson GW. Phase Diagrams and Lipid Domains in Multicomponent Lipid Bilayer Mixtures. Biochim. Biophys. Acta. 2009;1788:47–52. doi: 10.1016/j.bbamem.2008.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Konyakhina TM, Goh SL, Amazon J, Heberle FA, Wu J, Feigenson GW. Control of a Nanoscopic-to-Macroscopic Transition: Modulated Phases in Four-Component DSPC/DOPC/POPC/Chol Giant Unilamellar Vesicles. Biophys. J. 2011;101:L8–10. doi: 10.1016/j.bpj.2011.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heinrich M, Tian A, Esposito C, Baumgart T. Dynamic Sorting of Lipids and Proteins in Membrane Tubes with a Moving Phase Boundary. Proc. Natl. Acad. Sci. U. S. A. 2010;107:7208–7213. doi: 10.1073/pnas.0913997107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Amazon JJ, Feigenson GW. Lattice Simulations of Phase Morphology on Lipid Bilayers: Renormalization, Membrane Shape, and Electrostatic Dipole Interactions. Phys. Rev. E. 2014;89:022702. doi: 10.1103/PhysRevE.89.022702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu J, Weikl T, Lipowsky R. Vesicles with Multiple Membrane Domains. Soft Matter. 2011;7:6092–6102. [Google Scholar]
- 10.Parthasarathy R, Yu C, Groves JT. Curvature-Modulated Phase Separation in Lipid Bilayer Membranes. Langmuir. 2006;22:5095–5099. doi: 10.1021/la060390o. [DOI] [PubMed] [Google Scholar]
- 11.Heberle FA, Wu J, Goh SL, Petruzielo RS, Feigenson GW. Comparison of Three Ternary Lipid Bilayer Mixtures: FRET and ESR Reveal Nanodomains. Biophys. J. 2010;99:3309–3318. doi: 10.1016/j.bpj.2010.09.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao J, Wu J, Heberle FA, Mills TT, Klawitter P, Huang G, Costanza G, Feigenson GW. Phase Studies of Model Biomembranes: Complex Behavior of DSPC/DOPC/cholesterol. Biochim. Biophys. Acta. 2007;1768:2764–2776. doi: 10.1016/j.bbamem.2007.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goh SL, Amazon JJ, Feigenson GW. Toward a Better Raft Model: Modulated Phases in the Four-Component Bilayer, DSPC/DOPC/POPC/CHOL. Biophys. J. 2013;104:853–862. doi: 10.1016/j.bpj.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Amazon JJ, Goh SL, Feigenson GW. Competition between Line Tension and Curvature Stabilizes Modulated Phase Patterns on the Surface of Giant Unilamellar Vesicles: A Simulation Study. Phys. Rev. E. 2013;87:022708. doi: 10.1103/PhysRevE.87.022708. [DOI] [PubMed] [Google Scholar]
- 15.Hassan-Zadeh E, Baykal-Caglar E, Alwarawrah M, Huang J. Complex Roles of Hybrid Lipids in the Composition, Order, and Size of Lipid Membrane Domains. Langmuir. 2014;30:1361–1369. doi: 10.1021/la4044733. [DOI] [PubMed] [Google Scholar]
- 16.Kollmitzer B, Heftberger P, Rappolt M, Pabst G. Monolayer Spontaneous Curvature of Raft-Forming Membrane Lipids. Soft Matter. 2013;9:10877–10884. doi: 10.1039/C3SM51829A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Polley A, Vemparala S, Rao M. Atomistic Simulations of a Multicomponent Asymmetric Lipid Bilayer. J. Phys. Chem. B. 2012;116:13403–13410. doi: 10.1021/jp3032868. [DOI] [PubMed] [Google Scholar]
- 18.Hakobyan D, Heuer A. Phase Separation in a Lipid/cholesterol System: Comparison of Coarse-Grained and United-Atom Simulations. J. Phys. Chem. B. 2013;117:3841–3851. doi: 10.1021/jp312245y. [DOI] [PubMed] [Google Scholar]
- 19.Pandit SA, Vasudevan S, Chiu SW, Mashl RJ, Jakobsson E, Scott HL. Sphingomyelin-Cholesterol Domains in Phospholipid Membranes: Atomistic Simulation. Biophys. J. 2004;87:1092–1100. doi: 10.1529/biophysj.104.041939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sodt AJ, Sandar ML, Gawrisch K, Pastor RW, Lyman E. The Molecular Structure of the Liquid-Ordered Phase of Lipid Bilayers. J. Am. Chem. Soc. 2014;136:725–732. doi: 10.1021/ja4105667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marrink SJ, Risselada HJ, Yefimov S, Tieleman DP, de Vries AH. The MARTINI Force Field: Coarse Grained Model for Biomolecular Simulations. J. Phys. Chem. B. 2007;111:7812–7824. doi: 10.1021/jp071097f. [DOI] [PubMed] [Google Scholar]
- 22.Bennett WFD, Tieleman DP. Computer Simulations of Lipid Membrane Domains. Biochim. Biophys. Acta. 2013;1828:1765–1776. doi: 10.1016/j.bbamem.2013.03.004. [DOI] [PubMed] [Google Scholar]
- 23.Meinhardt S, Vink RLC, Schmid F. Monolayer Curvature Stabilizes Nanoscale Raft Domains in Mixed Lipid Bilayers. Proc. Natl. Acad. Sci. U. S. A. 2013;110:4476–4481. doi: 10.1073/pnas.1221075110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stevens MJ. Complementary Matching in Domain Formation within Lipid Bilayers. J. Am. Chem. Soc. 2005;127:15330–15331. doi: 10.1021/ja043611q. [DOI] [PubMed] [Google Scholar]
- 25.Risselada HJ, Marrink SJ. The Molecular Face of Lipid Rafts in Model Membranes. Proc. Natl. Acad. Sci. U. S. A. 2008;105:17367–17372. doi: 10.1073/pnas.0807527105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ingólfsson HI, Melo MN, van Eerden FJ, Arnarez C, López CA, Wassenaar TA, Periole X, De Vries AH, Tieleman DP, Marrink SJ. Lipid Organization of the Plasma Membrane Lipid Organization of the Plasma Membrane. J. Am. Chem. Soc. 2014 doi: 10.1021/ja507832e. [DOI] [PubMed] [Google Scholar]
- 27.Schäfer LV, de Jong DH, Holt A, Rzepiela AJ, de Vries AH, Poolman B, Killian JA, Marrink SJ. Lipid Packing Drives the Segregation of Transmembrane Helices into Disordered Lipid Domains in Model Membranes. Proc. Natl. Acad. Sci. U. S. A. 2011;108:1343–1348. doi: 10.1073/pnas.1009362108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rzepiela AJ, Schäfer LV, Goga N, Risselada HJ, de Vries AH, Marrink SJ. Reconstruction of Atomistic Details from Coarse-Grained Structures. J. Comput. Chem. 2010;31:1333–1343. doi: 10.1002/jcc.21415. [DOI] [PubMed] [Google Scholar]
- 29.Wassenaar TA, Pluhackova K, Böckmann RA, Marrink SJ, Tieleman DP. Going Backward: A Flexible Geometric Approach to Reverse Transformation from Coarse Grained to Atomistic Models. J. Chem. Theory Comput. 2013;10:676–690. doi: 10.1021/ct400617g. [DOI] [PubMed] [Google Scholar]
- 30.Stansfeld PJ, Sansom MSP. From Coarse Grained to Atomistic: A Serial Multiscale Approach to Membrane Protein Simulations. J. Chem. Theory Comput. 2011;7:1157–1166. doi: 10.1021/ct100569y. [DOI] [PubMed] [Google Scholar]
- 31.Hess B, Kutzner C, van der Spoel D, Lindahl E. GROMACS 4: Algorithms for Highly Efficient, Load-Balanced, and Scalable Molecular Simulation. J. Chem. Theory Comput. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 32.Monticelli L, Kandasamy SK, Periole X, Larson RG, Tieleman DP, Marrink S-J. The MARTINI Coarse-Grained Force Field: Extension to Proteins. J. Chem. Theory Comput. 2008;4:819–834. doi: 10.1021/ct700324x. [DOI] [PubMed] [Google Scholar]
- 33.Marrink SJ, de Vries AH, Mark AE. Coarse Grained Model for Semiquantitative Lipid Simulations. J. Phys. Chem. B. 2004;108:750–760. [Google Scholar]
- 34.Marrink SJ, de Vries AH, Harroun TA, Katsaras J, Wassall SR. Cholesterol Shows Preference for the Interior of Polyunsaturated Lipid Membranes. J. Am. Chem. Soc. 2008;130:10–11. doi: 10.1021/ja076641c. [DOI] [PubMed] [Google Scholar]
- 35.Van Gunsteren WF, Berendsen HJC. Groningen Molecular Simulation (GROMOS) Library Manual. Biomos; Groningen, The Netherlands: 1987. [Google Scholar]
- 36.Berger O, Edholm O, Jähnig F. Molecular Dynamics Simulations of a Fluid Bilayer of Dipalmitoylphosphatidylcholine at Full Hydration, Constant Pressure, and Constant Temperature. Biophys. J. 1997;72:2002–2013. doi: 10.1016/S0006-3495(97)78845-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tieleman DP, Berendsen HJC. Molecular Dynamics Simulations of a Fully Hydrated Dipalmitoylphosphatidylcholine Bilayer with Different Macroscopic Boundary Conditions and Parameters. J. Chem. Phys. 1996;105:4871–4880. [Google Scholar]
- 38.Höltje M, Förster T, Brandt B, Engels T, von Rybinski W, Höltje H-D. Molecular Dynamics Simulations of Stratum Corneum Lipid Models: Fatty Acids and Cholesterol. Biochim. Biophys. Acta. 2001;1511:156–167. doi: 10.1016/s0005-2736(01)00270-x. [DOI] [PubMed] [Google Scholar]
- 39.Berendsen HJC, Postma JPM, van Gunsteren WF, Hermans J. Interaction Models for Water in Relation to Protein Hydration. In: Pullman B, editor. Intermolecular Forces. D. Reidel Publishing Company; Dordrecht, The Netherlands: 1981. pp. 331–342. [Google Scholar]
- 40.Bussi G, Donadio D, Parrinello M. Canonical Sampling through Velocity Rescaling. J. Chem. Phys. 2007;126:014101. doi: 10.1063/1.2408420. [DOI] [PubMed] [Google Scholar]
- 41.Berendsen HJC, Postma JPM, van Gunsteren WF, DiNola A, Haak JR. Molecular Dynamics with Coupling to an External Bath. J. Chem. Phys. 1984;81:3684–3690. [Google Scholar]
- 42.Nosé S. A Molecular Dynamics Method for Simulations in the Canonical Ensemble. Mol. Phys. 1984;55:255–268. [Google Scholar]
- 43.Hoover WG. Canonical Dynamics: Equilibrium Phase-Space Distributions. Phys. Rev. A. 1985;31:1695–1697. doi: 10.1103/physreva.31.1695. [DOI] [PubMed] [Google Scholar]
- 44.Parrinello M, Rahman A. Polymorphic Transitions in Single Crystals: A New Molecular Dynamics Method. J. Appl. Phys. 1981;52:7182–7190. [Google Scholar]
- 45.Miyamoto S, Kollman PA. Settle: An Analytical Version of the SHAKE and RATTLE Algorithm for Rigid Water Models. J. Comput. Chem. 1992;13:952–962. [Google Scholar]
- 46.Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG. A Smooth Particle Mesh Ewald Method. J. Chem. Phys. 1995;103:8577–8593. [Google Scholar]
- 47.Darden T, York D, Pedersen L. Particle Mesh Ewald: An Nlog(N) Method for Ewald Sums in Large Systems. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- 48.Hess B, Bekker H, Berendsen HJC, Fraaije JGEM. LINCS: A Linear Constraint Solver for Molecular Simulations. J. Comput. Chem. 1997;18:1463–1472. [Google Scholar]
- 49.Baoukina S, Mendez-Villuendas E, Bennett WFD, Tieleman DP. Computer Simulations of the Phase Separation in Model Membranes. Faraday Discuss. 2013;161:63–75. doi: 10.1039/c2fd20117h. [DOI] [PubMed] [Google Scholar]
- 50.Baoukina S, Mendez-Villuendas E, Tieleman DP. Molecular View of Phase Coexistence in Lipid Monolayers. J. Am. Chem. Soc. 2012;134:17543–17553. doi: 10.1021/ja304792p. [DOI] [PubMed] [Google Scholar]
- 51.Perlmutter JD, Sachs JN. Interleaflet Interaction and Asymmetry in Phase Separated Lipid Bilayers: Molecular Dynamics Simulations. J. Am. Chem. Soc. 2011;133:6563–6577. doi: 10.1021/ja106626r. [DOI] [PubMed] [Google Scholar]
- 52.Rosetti C, Pastorino C. Comparison of Ternary Bilayer Mixtures with Asymmetric or Symmetric Unsaturated Phosphatidylcholine Lipids by Coarse Grained Molecular Dynamics Simulations. J. Phys. Chem. B. 2012;116:3525–3537. doi: 10.1021/jp212406u. [DOI] [PubMed] [Google Scholar]
- 53.Domański J, Marrink SJ, Schäfer LV. Transmembrane Helices Can Induce Domain Formation in Crowded Model Membranes. Biochim. Biophys. Acta. 2012;1818:984–994. doi: 10.1016/j.bbamem.2011.08.021. [DOI] [PubMed] [Google Scholar]
- 54.Pantano DA, Moore PB, Klein ML, Discher DE. Raft Registration across Bilayers in a Molecularly Detailed Model. Soft Matter. 2011;7:8182–8191. [Google Scholar]
- 55.Róg T, Pasenkiewicz-Gierula M, Vattulainen I, Karttunen M. Ordering Effects of Cholesterol and Its Analogues. Biochim. Biophys. Acta. 2009;1788:97–121. doi: 10.1016/j.bbamem.2008.08.022. [DOI] [PubMed] [Google Scholar]
- 56.Khelashvili G, Kollmitzer B, Heftberger P, Pabst G, Harries D. Calculating the Bending Modulus for Multicomponent Lipid Membranes in Different Thermodynamic Phases. J. Chem. Theory Comput. 2013;9:3866–3871. doi: 10.1021/ct400492e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hofsäß C, Lindahl E, Edholm O. Molecular Dynamics Simulations of Phospholipid Bilayers with Cholesterol. Biophys. J. 2003;84:2192–2206. doi: 10.1016/S0006-3495(03)75025-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Heberle FA, Petruzielo RS, Pan J, Drazba P, Kučerka N, Standaert RF, Feigenson GW, Katsaras J. Bilayer Thickness Mismatch Controls Domain Size in Model Membranes. J. Am. Chem. Soc. 2013;135:6853–6859. doi: 10.1021/ja3113615. [DOI] [PubMed] [Google Scholar]
- 59.Konyakhina TM, Wu J, Mastroianni JD, Heberle FA, Feigenson GW. Phase Diagram of a 4-Component Lipid Mixture: DSPC/DOPC/POPC/chol. Biochim. Biophys. Acta. 2013;1828:2204–2214. doi: 10.1016/j.bbamem.2013.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Heberle FA, Doktorova M, Goh SL, Standaert RF, Katsaras J, Feigenson GW. Hybrid and Nonhybrid Lipids Exert Common E Ff Ects on Membrane Raft Size and Morphology. J. Am. Chem. Soc. 2013;135:14932–14935. doi: 10.1021/ja407624c. [DOI] [PubMed] [Google Scholar]
- 61.Hakobyan D, Heuer A. Key Molecular Requirements for Raft Formation in Lipid/cholesterol Membranes. PLoS One. 2014;9:e87369. doi: 10.1371/journal.pone.0087369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Davis RS, Sunil Kumar PB, Sperotto MM, Laradji M. Predictions of Phase Separation in Three-Component Lipid Membranes by the MARTINI Force Field. J. Phys. Chem. B. 2013;117:4072–4080. doi: 10.1021/jp4000686. [DOI] [PubMed] [Google Scholar]
- 63.García-Sáez AJ, Chiantia S, Schwille P. Effect of Line Tension on the Lateral Organization of Lipid Membranes. J. Biol. Chem. 2007;282:33537–33544. doi: 10.1074/jbc.M706162200. [DOI] [PubMed] [Google Scholar]
- 64.Kuzmin PI, Akimov SA, Chizmadzhev YA, Zimmerberg J, Cohen FS. Line Tension and Interaction Energies of Membrane Rafts Calculated from Lipid Splay and Tilt. Biophys. J. 2005;88:1120–1133. doi: 10.1529/biophysj.104.048223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Muddana HS, Chiang HH, Butler PJ. Tuning Membrane Phase Separation Using Nonlipid Amphiphiles. Biophys. J. 2012;102:489–497. doi: 10.1016/j.bpj.2011.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.May S. Trans-Monolayer Coupling of Fluid Domains in Lipid Bilayers. Soft Matter. 2009;5:3148–3156. [Google Scholar]
- 67.Chiantia S, London E. Acyl Chain Length and Saturation Modulate Interleaflet Coupling in Asymmetric Bilayers: Effects on Dynamics and Structural Order. Biophys. J. 2012;103:2311–2319. doi: 10.1016/j.bpj.2012.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Collins MD. Interleaflet Coupling Mechanisms in Bilayers of Lipids and Cholesterol. Biophys. J. 2008;94:L32–4. doi: 10.1529/biophysj.107.124362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rosetti C, Pastorino C. Polyunsaturated and Saturated Phospholipids in Mixed Bilayers: A Study from the Molecular Scale to the Lateral Lipid Organization. J. Phys. Chem. B. 2010;115:1002–1013. doi: 10.1021/jp1082888. [DOI] [PubMed] [Google Scholar]
- 70.Seul M, Andelman D. Domain Shapes and Patterns: The Phenomenology of Modulated Phases. Science. 1995;267:476–483. doi: 10.1126/science.267.5197.476. [DOI] [PubMed] [Google Scholar]
- 71.Shi Q, Voth GA. Multi-Scale Modeling of Phase Separation in Mixed Lipid Bilayers. Biophys. J. 2005;89:2385–2394. doi: 10.1529/biophysj.105.063784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Palmieri B, Yamamoto T, Brewster RC, Safran SA. Line Active Molecules Promote Inhomogeneous Structures in Membranes: Theory, Simulations and Experiments. Adv. Colloid Interface Sci. 2014;208:58–65. doi: 10.1016/j.cis.2014.02.007. [DOI] [PubMed] [Google Scholar]
- 73.Sarasij RC, Mayor S, Rao M. Chirality-Induced Budding: A Raft-Mediated Mechanism for Endocytosis and Morphology of Caveolae? Biophys. J. 2007;92:3140–3158. doi: 10.1529/biophysj.106.085662. [DOI] [PMC free article] [PubMed] [Google Scholar]