Abstract
Tumor cells associated with therapy resistance (radioresistance and drug resistance) are likely to give rise to local recurrence and distant metastatic relapse. Recent studies revealed microRNA (miRNA)-mediated regulation of metastasis and epithelial-mesenchymal transition; however, whether specific miRNAs regulate tumor radioresistance and can be exploited as radiosensitizing agents remains unclear. Here we find that miR-205 promotes radiosensitivity and is downregulated in radioresistant subpopulations of breast cancer cells, and that loss of miR-205 is highly associated with poor distant relapse-free survival in breast cancer patients. Notably, therapeutic delivery of miR-205 mimics via nanoliposomes can sensitize the tumor to radiation in a xenograft model. Mechanistically, radiation suppresses miR-205 expression through ataxia telangiectasia mutated (ATM) and zinc finger E-box binding homeobox 1 (ZEB1). Moreover, miR-205 inhibits DNA damage repair by targeting ZEB1 and the ubiquitin-conjugating enzyme Ubc13. These findings identify miR-205 as a radiosensitizing miRNA and reveal a new therapeutic strategy for radioresistant tumors.
Introduction
Radiotherapy is one of the major modalities of cancer treatment. A main reason of failure in radiation treatment is intrinsic and therapy-induced radioresistant tumor cells, which display enhanced DNA repair ability1. Combining chemotherapy with radiation improves outcomes but often increases toxicity2. To overcome this problem, it is necessary to elucidate the mechanisms of resistance to radiotherapy and develop new radiosensitizers.
A growing number of studies have demonstrated that tumor initiation and progression can be regulated by miRNAs3-6, which are small non-coding RNAs acting as negative regulators of gene expression7. Whereas no miRNAs have been approved by the FDA as drugs, much progress is being made in developing them as therapeutic strategies8-10. For instance, locked nucleic acid-based miR-122 antagonist has shown therapeutic benefits in a phase 2 clinical trial to treat patients with hepatitis C virus infection11. Moreover, we and others have demonstrated the efficacy of miRNA-based therapeutic agents in preclinical models of cancer12-14. Notably, MRX34, a liposomal miR-34 mimic, entered phase 1 clinical trials in patients with advanced liver cancer in April 2013, representing the first miRNA-based drug used in cancer trials10.
Recently, several miRNAs have been found to regulate DNA damage response in cell culture systems15-20. However, whether specific miRNAs control tumor radioresistance in vivo and can be used as tumor radiosensitizers remains unclear. In this study, we show that miR-205 is downregulated in radioresistant subpopulations of breast cancer cells derived from ionizing radiation (IR) in an ATM- and ZEB1-dependent manner. We further demonstrate the therapeutic utility of the nanoliposome-encapsulated miR-205 mimic as a tumor radiosensitizer in a preclinical model. Mechanistically, miR-205 inhibits DNA damage repair and radiosensitizes tumor cells by targeting ZEB1 and Ubc13. These results suggest that delivery of miR-205 combined with radiotherapy may represent a new strategy for cancer treatment.
Results
miR-205 promotes radiosensitivity and is downregulated in radioresistant tumor cells
To establish a radioresistant model, we used γ-IR to select the radioresistant subpopulation (designated as SUM159-P2 cells) from the parental SUM159 human breast cancer cell line (Fig. 1a). Irradiation is known to cause DNA double-strand breaks (DSBs), which lead to the formation of γH2AX foci, and the persistent presence of γH2AX foci indicates delayed repair and is associated with radiosensitivity21-23. At 24 hours after irradiation, γH2AX foci remained in the parental SUM159 (SUM159-P0) cells but disappeared in SUM159-P2 cells (Supplementary Fig. 1a), suggesting that this radioresistant subline has enhanced ability to clear DNA breaks.
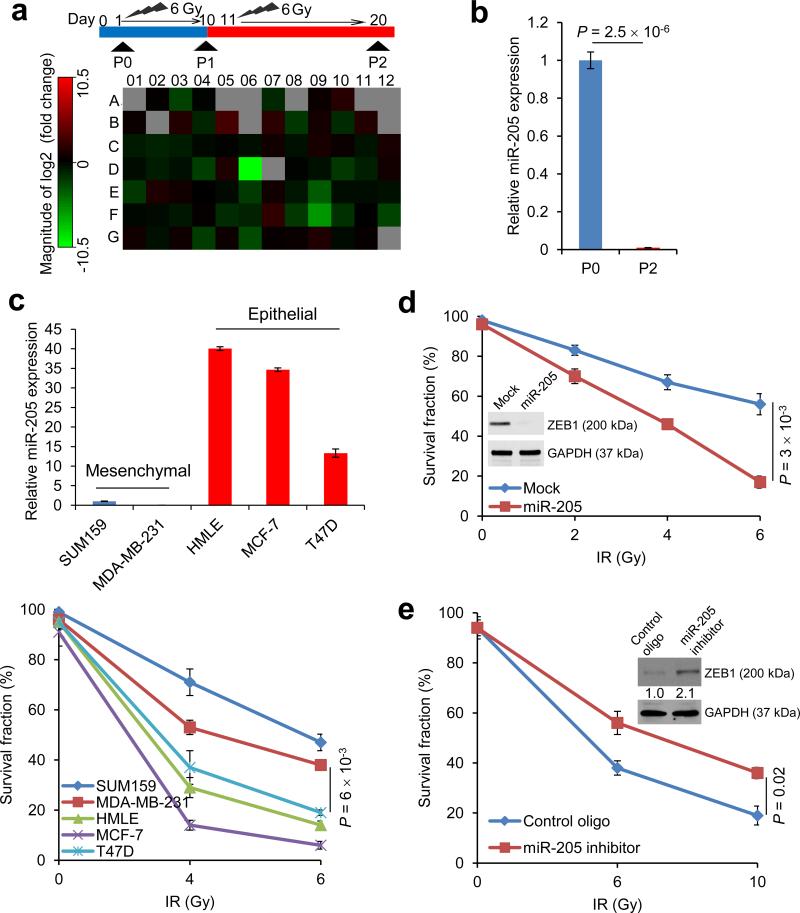
Figure 1. miR-205 increases radiosensitivity and is downregulated in radioresistant breast cancer cells.
(a) Top: schematic representation of the generation of a radioresistant subline (SUM159-P2) from the parental SUM159 cells (SUM159-P0). Bottom: miRNA expression profiling of SUM159-P2 cells relative to SUM159-P0 cells using a qPCR-based miRNA array. The expression values are shown in Supplementary Table 1.
(b) TaqMan qPCR analysis of miR-205 in SUM159-P0 and SUM159-P2 cells. n = 3 samples per group.
(c) qPCR of miR-205 (top) and clonogenic survival assays (bottom) of mesenchymal-like and epithelial-like breast cancer cell lines. n = 3 samples per group.
(d) Clonogenic survival assays of SUM159-P2 cells transduced with miR-205. n = 3 wells per group.
(e) Clonogenic survival assays of SUM159-P0 cells transfected with the miR-205 inhibitor. n = 3 wells per group. Inset in d and e: immunoblotting of ZEB1 and GAPDH.
Data in b, c, d and e are the mean of biological replicates from a representative experiment, and error bars indicate s.e.m. Statistical significance was determined by a two-tailed, unpaired Student's t-test. The experiments were repeated 3 times.
We wanted to use an unbiased approach to identify miRNAs that regulate radiosensitivity. To this end, we performed a Human Apoptosis miRNA PCR Array analysis to identify miRNAs deregulated in SUM159-P2 cells. Although most of the miRNAs on this array showed no substantial difference (fold change < 2), miR-205 stood out as a miRNA that was dramatically downregulated in SUM159-P2 cells compared with the parental SUM159 cells (fold change = 1439, Fig. 1a and Supplementary Table 1). Individual TaqMan qPCR assays further confirmed this array result (Fig. 1b).
Recently, cancer stem cells (CSCs), which are defined operationally as tumor-initiating cells, have been found to promote radioresistance through activation of DNA damage response24-27. Moreover, a trans-differentiation process, termed epithelial-mesenchymal transition (EMT), has been shown to generate cells with properties of stem cells or CSCs28. Interestingly, miR-205 can suppress EMT by targeting the EMT-inducing transcription factor ZEB129. We therefore reasoned that this miRNA may be negatively associated with radioresistance. Indeed, when compared with epithelial-like breast cancer cell lines, mesenchymal-like breast cancer cell lines exhibited much lower levels of miR-205 and higher clonogenic survival ability after irradiation (Fig. 1c). Next, we performed gain-of-function and loss-of-function analyses of miR-205.Overexpression of miR-205 in SUM159-P2 cells downregulated ZEB1 and sensitized these cells to radiation (Fig. 1d). Conversely, transfection of SUM159-P0 cells with the miR-205 antisense inhibitor upregulated ZEB1 and conferred radioresistance on these cells (Fig. 1e). Similar effects were also observed in MDA-MB-231 breast cancer cells (Supplementary Fig. 1b). In addition, overexpression of miR-205 rendered the A549 human lung cancer cells and the U2OS human osteosarcoma cells more radiosensitive (Supplementary Fig. 1c, d). Thus, the observed effect of miR-205 is not limited to the SUM159 breast cancer cell line.
Therapeutic delivery of nanoliposomal miR-205 mimics sensitizes tumors to radiation
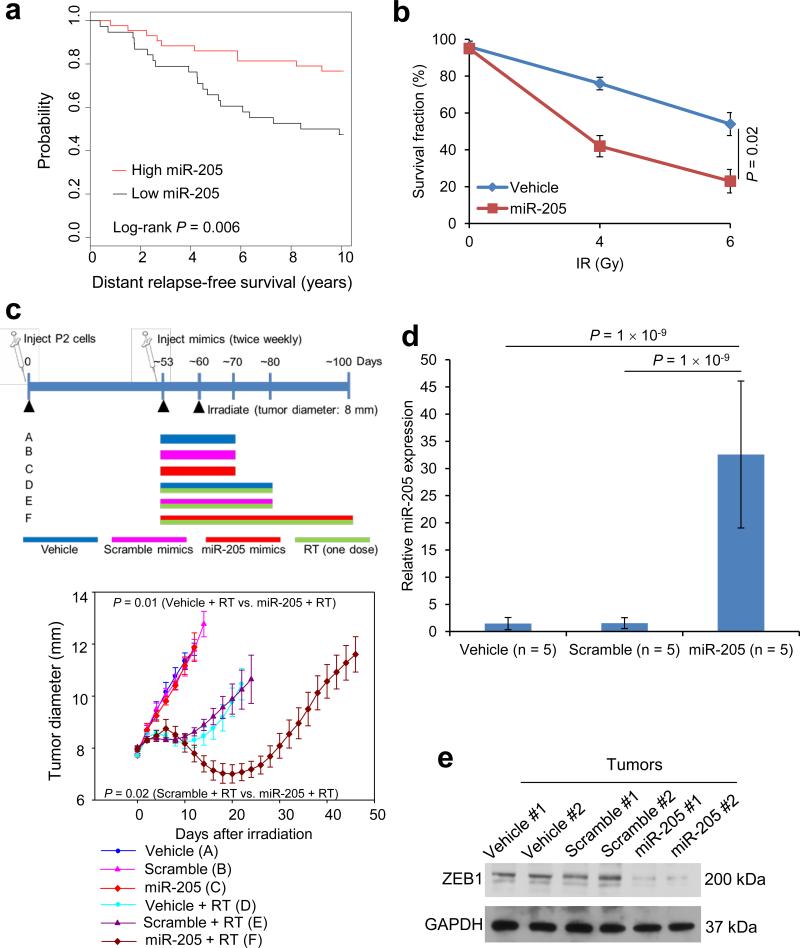
Cancer cells associated with therapy resistance (radioresistance and drug resistance) are likely to be a source for tumor recurrence and distant relapse (metastasis)30. To investigate the clinical relevance of miR-205 expression levels, we performed a retrospective analysis on a cohort of human breast cancer patients (Oxford collection) in which miRNA profiling was obtained from 207 tumor samples; 84% of these patients received radiotherapy31, 32. This analysis revealed that patients with low miR-205 expression levels (defined as the bottom 20%) in their tumors had much worse distant relapse-free survival than those with high miR-205 expression levels (defined as the top 20%; P = 0.006, Fig. 2a).
Figure 2. Therapeutic effect of miR-205 mimics in mice.
(a) Kaplan-Meier curves showing the distant relapse-free survival of patients with high (n = 43) or low (n = 38) expression of miR-205 in their breast tumors. The P value was determined by a log-rank test.
(b) Clonogenic survival assays of SUM159-P2 cells incubated with 100 nM DOPC-encapsulated miR-205 mimics. n = 3 wells per group.
(c) Top: overview of the experimental scheme. Bottom: tumor size of mice bearing SUM159-P2 xenografts. Mice were treated with the vehicle or DOPC-encapsulated miR-205 mimics or scramble mimics. Tumors were locally irradiated with a 15-Gy single dose (RT). n = 5 mice per group. General linear model multivariate analysis was performed to determine statistical significance.
(d) qPCR of miR-205 in tumor tissues from the mice described in c. n = 5 mice per group.
(e) Immunoblotting of ZEB1 and GAPDH in tumor lysates.
Data in b, c and d are the mean of biological replicates from a representative experiment, and error bars indicate s.e.m. Statistical significance in b and d was determined by a two-tailed, unpaired Student's t-test. The experiments were repeated 3 times.
To determine the therapeutic potential of miR-205 as a tumor radiosensitizer, we incorporated miR-205 mimics or the scramble mimics into nanoparticles, the neutral liposome 1,2-dioleoyl-sn-glycero-3-phosphatidylcholine (DOPC), which has been shown to remain in various organs of injected animals for at least 48 hours with no detectable toxicity33. Notably, DOPC-mediated delivery of specific siRNA or miRNA oligonucleotides injected twice weekly at a dose of 150-200 μg kg−1 body weight yielded efficient in vivo delivery and significant anti-tumor effects34-36. We incubated SUM159-P2 cells with 100 nM DOPC-encapsulated miR-205 mimics for 48 hours and assessed the effect on radioresistance in vitro. When compared with the vehicle control, cells treated with miR-205 mimics displayed a 57% reduction in clonogenic survival (Fig. 2b and Supplementary Fig. 2a, b).
Subsequently, we performed therapeutic studies in mice bearing SUM159-P2 xenograft tumors. We administered miR-205 mimics subcutaneously near the tumor, because: i) in a phase 2 clinical trial to treat patients with hepatitis C virus infection, subcutaneous injection of the anti-miR-122 locked nucleic acid (miravirsen, Santaris Pharma A/S) provided robust dose-dependent antiviral acitivity11; and ii) in mice implanted with PC-3 prostate cancer cells, subcutaneous injection of liposomal siRNA targeting BCL2 around the tumor strongly inhibited tumor growth37. When the SUM159-P2 tumor reached 8 mm in diameter, we locally irradiated the tumor with a 15-Gy single dose; one week before radiation treatment, we started the twice-weekly treatment with PBS, the scramble mimics or miR-205 mimics (250 μg kg−1) until the tumor burden or body condition reached the euthanasia criteria (Fig. 2c). Whereas radiation treatment alone led to a short initial response, treatment with miR-205 mimics alone (without tumor irradiation) had no effect on tumor size (Fig. 2c). In contrast, in mice with irradiated tumors, subcutaneous injection of nanoliposomal miR-205 mimics close to the tumor, which increased miR-205 levels in the tumor by approximately 21-fold (Fig. 2d), significantly delayed tumor growth compared with the vehicle control (P = 0.01) or scramble mimics (P = 0.02) (Fig. 2c).
Consistent with increased miR-205 levels, ZEB1 protein in the tumor was markedly downregulated by treatment with miR-205 mimics (Fig. 2e). Histopathological examination revealed massively increased necrosis in tumors treated with both miR-205 mimics and radiation (Supplementary Fig. 2c), whereas the liver displayed no necrosis, steatosis or fibrosis in any group; similarly, no pathological changes were found in the kidney (Supplementary Fig. 2d). Taken together, these results indicate that nanoparticle-mediated delivery of miR-205 mimics can sensitize tumors to localized irradiation but does not affect non-irradiated tumors or other organs.
miR-205 inhibits DNA damage repair and radioresistance by targeting ZEB1 and Ubc13
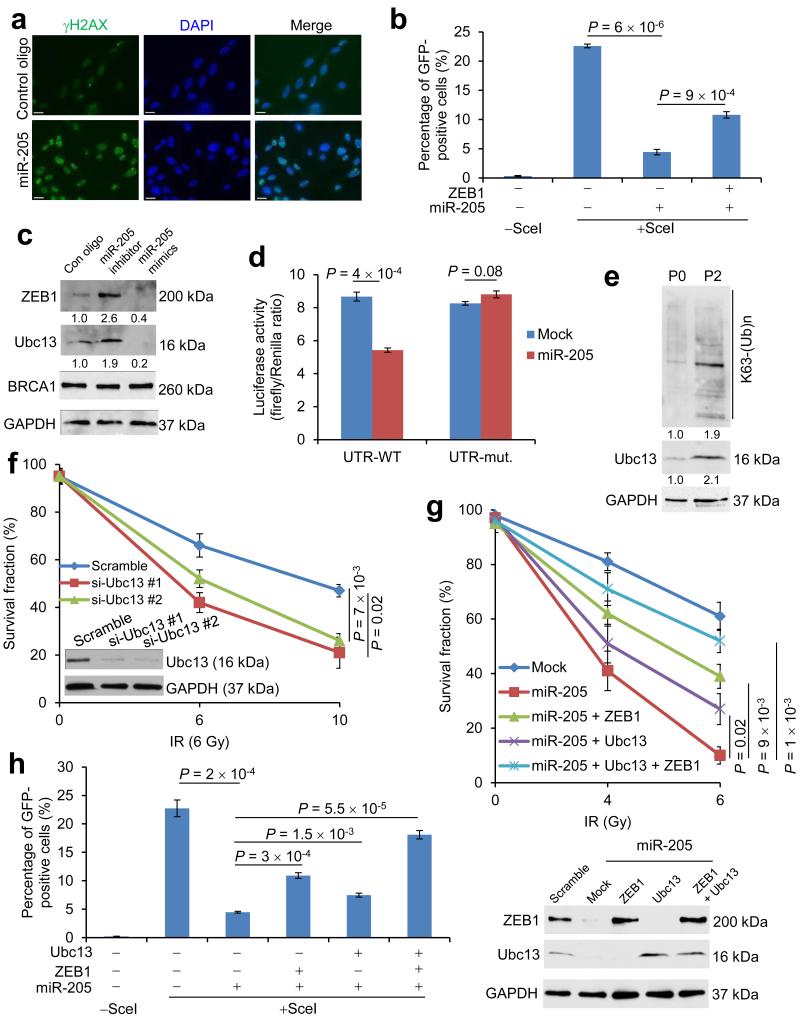
We attempted to understand how miR-205 enhances radiosensitivity. After IR treatment, γH2AX foci persist longer in radiosensitive cells than in radioresistant cells23. In miR-205-overexpressing SUM159-P2 cells but not in cells infected with a scramble control, we observed persistent presence of γH2AX foci 24 hours after exposure to 6-Gy IR (Fig. 3a), indicating that these miR-205-overexpressing cells were less able to repair DNA lesions. A critical pathway involved in DSB repair is the homologous recombination (HR) pathway38. We used a U2OS stable clone with chromosomal integration of an HR repair reporter containing two differentially mutated GFP genes (SceGFP and iGFP) oriented as direct repeats (DR-GFP); I-SceI endonuclease can generate a site-specific DSB in the SceGFP coding region, and if this DSB is repaired by HR, the expression of GFP is restored and can be quantitated by flow cytometry39, 40. We observed that upon I-SceI expression, miR-205-overexpressing U2OS cells exhibited an approximately 80% decrease in the percentage of GFP-positive cells, which indicated defective HR repair (Fig. 3b). Recently, we found that ZEB1 promotes radioresistance and HR repair through CHK141. In the present study, re-expression of ZEB1 in miR-205-overexpressing U2OS cells partially (~30%) restored HR-based repair (Fig. 3b and Supplementary Fig. 3a), suggesting that ZEB1 mediates in part the effect of miR-205 on HR repair, but other functional targets of miR-205 may also be involved.
Figure 3. miR-205 regulates DNA damage repair and radiosensitivity through ZEB1 and Ubc13.
(a) γH2AX and DAPI staining of SUM159-P2 cells transfected with miR-205 or scramble control oligonucleotides, 24 hours after 6-Gy IR. Scale bar: 20 μm.
(b) HR repair assays of U2OS_DR-GFP cells transfected with miR-205 alone or in combination with ZEB1. n = 3 wells per group.
(c) Immunoblotting of ZEB1, Ubc13, BRCA1 and GAPDH in SUM159-P0 cells transfected with miR-205 mimics or the miR-205 inhibitor.
(d) Luciferase activity of the wild-type (UTR-WT) or mutant (UTR-mut.) Ubc13 3′ UTR reporter gene in 293T cells transfected with the miR-205-expressing or empty vector. n = 3 wells per group.
(e) Immunoblotting of K63-linked polyubiquitin chains, Ubc13 and GAPDH in SUM159-P0 and SUM159-P2 cells.
(f) Clonogenic survival assays of SUM159-P2 cells transfected with Ubc13 siRNA. n = 3 wells per group.
(g) Top: clonogenic survival assays of miR-205-transduced SUM159-P2 cells with or without ectopic expression of ZEB1 and Ubc13. Bottom: immunoblotting of ZEB1, Ubc13 and GAPDH. n = 3 wells per group.
(h) HR repair assays of U2OS_DR-GFP cells transfected with miR-205 alone or in combination with ZEB1 or Ubc13, or both. n = 3 wells per group.
Data in b, d, f, g and h are the mean of biological replicates from a representative experiment, and error bars indicate s.e.m. Statistical significance was determined by a two-tailed, unpaired Student's t-test. The experiments were repeated 3 times.
Among the targets predicted by both the TargetScan42 and PicTar43 programs for miR-205, Ubc13 and BRCA1 play essential roles in HR repair44, 45. In SUM159-P0 cells, transfection of the miR-205 antisense inhibitor increased ZEB1 and Ubc13 protein levels, and transfection of miR-205 mimics downregulated ZEB1 and Ubc13, whereas BRCA1 protein level was not altered by either transfection (Fig. 3c). Similar effects were also observed in MDA-MB-231 cells (Supplementary Fig. 3b). Moreover, the activity of a luciferase reporter fused to a wild-type Ubc13 3′ untranslated region (UTR), but not that of a reporter fused to a mutant Ubc13 3′ UTR with the mutated miR-205-binding site (Supplementary Fig. 3c), was significantly reduced by overexpression of miR-205 (Fig. 3d), which confirmed Ubc13 as a direct target of this miRNA.
Similar to the expression pattern of ZEB141, Ubc13 protein was significantly upregulated in SUM159-P2 cells relative to the parental SUM159 cells (Fig. 3e). Consistent with the previous finding that Ubc13 promotes HR-mediated DSB repair by conjugating lysine 63 (K63)-linked ubiquitin chains to a number of target proteins including H2AX and PCNA44, 46, 47, K63-linked poly-ubiquitination was enriched in SUM159-P2 cells (Fig. 3e).
We examined whether Ubc13 is a functional target of miR-205 in addition to ZEB1. Two independent Ubc13 siRNAs both increased the radiosensitivity of SUM159-P2 cells (Fig. 3f), which recapitulated the effect of miR-205 (Fig. 1d). Next, we re-expressed ZEB1 and Ubc13, singly or in combination, in miR-205-overexpressing SUM159-P2 or U2OS cells. Either ZEB1 or Ubc13 alone partially restored radioresistance and HR repair, and co-expression of ZEB1 and Ubc13 led to a nearly complete rescue (Fig. 3g, h and Supplementary Fig. 3d). Collectively, these results suggest that miR-205 inhibits HR-mediated DNA damage repair and radioresistance by targeting ZEB1 and Ubc13.
Radiation suppresses miR-205 expression through ATM and ZEB1
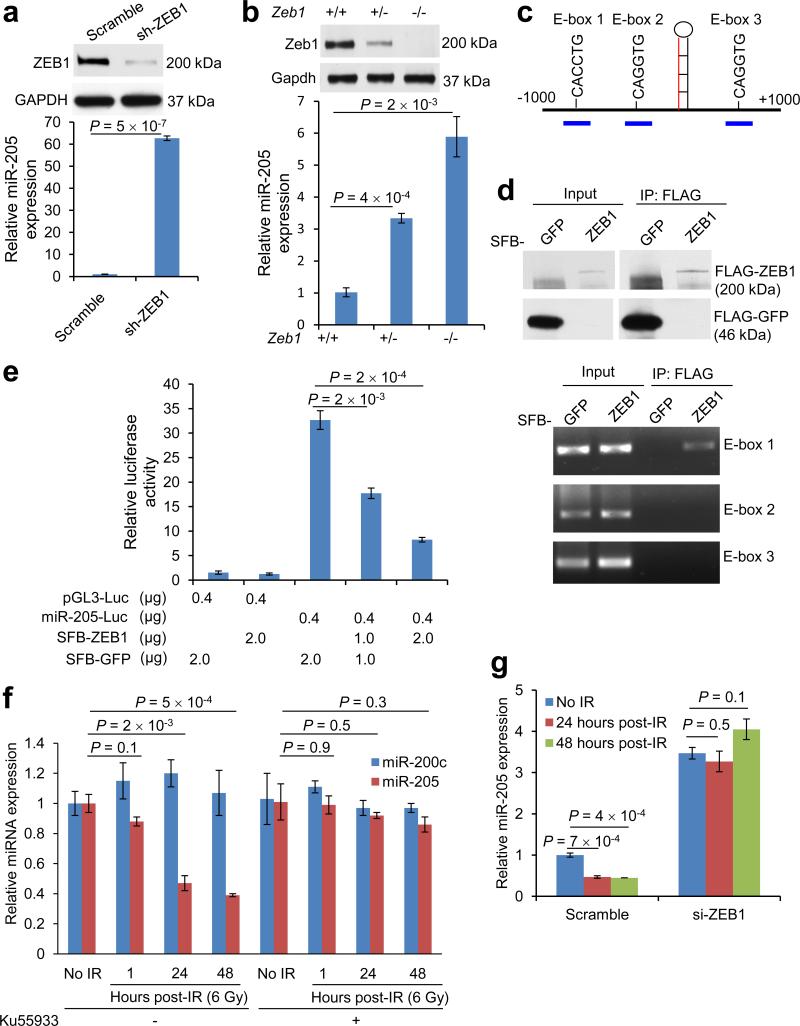
We sought to determine the mechanism by which miR-205 is downregulated in SUM159-P2 cells. Recent studies have revealed a feedback loop whereby the miR-200 family of miRNAs directly targets ZEB129 and ZEB1 represses the transcription of miR-200 genes48. We asked whether ZEB1 is also a transcriptional repressor of miR-205. Indeed, knockdown of ZEB1 in SUM159-P2 cells increased miR-205 expression levels by 63-fold (Fig. 4a). Moreover, Zeb1-deficient mouse embryonic fibroblasts (MEFs) exhibited prominent upregulation of miR-205 (Fig. 4b).
Figure 4. Radiation downregulates miR-205 expression through ZEB1 and ATM.
(a) Immunoblotting of ZEB1 and GAPDH (top) and qPCR of miR-205 (bottom) in SUM159-P2 cells transduced with ZEB1 shRNA. n = 3 samples per group.
(b) Immunoblotting of Zeb1 and Gapdh (top) and qPCR of miR-205 (bottom) in Zeb1+/+, Zeb1+/− and Zeb1−/- MEFs. n = 3 samples per group.
(c) Human mir-205 genomic locus. The three short blue lines represent PCR amplicons specific for three E-box elements.
(d) Top: 293T cells were transfected with SFB-GFP or SFB-ZEB1, immunoprecipitated with the FLAG antibody and immunoblotted with the FLAG antibody. Bottom: ChIP assays of 293T cells transfected with SFB-GFP or SFB-ZEB1. PCR was performed with primers specific for E-box elements 1, 2 and 3.
(e) Activity of a luciferase reporter fused to the putative human mir-205 promoter in 293T cells transfected with SFB-GFP or SFB-ZEB1. n = 3 wells per group.
(f) qPCR of miR-205 and miR-200c in irradiated SUM159-P0 cells with or without pretreatment with the ATM inhibitor Ku55933 (10 μM, 1 hour). n = 3 samples per group.
(g) qPCR of miR-205 in SUM159-P0 cells transfected with ZEB1 siRNA or scramble control oligonucleotides. Cells were treated with 6-Gy IR. n = 3 samples per group.
Data in a, b, e, f and g are the mean of biological replicates from a representative experiment, and error bars indicate s.e.m. Statistical significance was determined by a two-tailed, unpaired Student's t-test. The experiments were repeated 3 times.
ZEB1 is known to bind to E-box elements and act as a transcriptional repressor through recruitment of co-repressors CTBP1 and CTBP249, 50. We examined the genomic sequences flanking the human miR-205 stem-loop (pre-miR-205) and identified three E-boxes, at −417 bp (E-box 1), −186 bp (E-box 2) and +550 bp (E-box 3) (Fig. 4c). We designed PCR amplicons to assay for the presence of these three putative binding sites in chromatin immunoprecipitates. This experiment revealed that either overexpressed (Fig. 4d) or endogenous (Supplementary Fig. 4a) ZEB1 protein bound to E-box 1 located 417 bp upstream of the start site of pre-miR-205, but not to E-box 2 or E-box 3 (Fig. 4d). Moreover, SUM159-P2 exhibited increased occupancy of E-box 1 by endogenous ZEB1 than SUM159-P0 cells (Supplementary Fig. 4a). In addition, luciferase reporter assays demonstrated that overexpression of ZEB1 significantly repressed the activity of the putative mir-205 promoter (Fig. 4e). To confirm that endogenous ZEB1 represses miR-205 expression, we compared the reporter activity in SUM159-P0 versus SUM159-P2 cells (Supplementary Fig. 4b), in MDA-MB-231 versus MCF-7 cells (Supplementary Fig. 4c), and in SUM159-P2 (Supplementary Fig. 4d) or MDA-MB-231 (Supplementary Fig. 4e) cells with or without knockdown of ZEB1. In all cases, the reporter activity inversely correlated with the expression level of endogenous ZEB1 (Supplementary Fig. 4b-e).
ATM kinase is a key sensor of DNA damage especially DSBs51. We previously found that ATM-mediated phosphorylation of ZEB1 at serine 585 is crucial for IR-induced stabilization of ZEB1 protein41. To determine whether IR regulates miR-205 expression through ATM and ZEB1, we irradiated SUM159-P0 cells that had been treated with an ATM kinase inhibitor (Ku55933) or transfected with ZEB1 siRNA. We found that miR-205, but not miR-200c, was significantly downregulated 24 hours after irradiation, which was accompanied by upregulation of ZEB1 (Fig. 4f and Supplementary Fig. 4f). Notably, either inhibition of ATM (Fig. 4f) or depletion of ZEB1 (Fig. 4g) completely abolished the downregulation of miR-205 by IR.
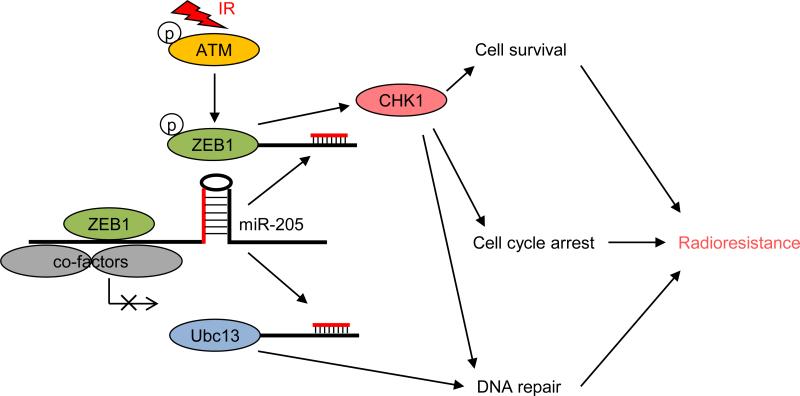
Taken together with our previous findings41, these results support a model in which ATM phosphorylates and stabilizes ZEB1 upon irradiation, and ZEB1 in turn represses its own negative regulator, miR-205, leading to further upregulation of ZEB1, increased Ubc13 levels and enhanced DNA repair and radioresistance (Fig. 5). Importantly, our study identified miR-205 mimics as a candidate radiosensitizing agent.
Figure 5.
The working model of regulation of radiosensitivity and DNA damage repair by miR-205.
Discussion
Constitutive activation of DNA damage response components has been observed in radioresistant breast tumors1. To improve the outcomes in these cases, many groups are dedicated to identifying the agents that target DNA damage response pathways which mediate radioresistance. Although numerous proteins are known to regulate the response to radiation, few of them can be targeted therapeutically. This is where miRNAs could offer new possibilities. The ability of miRNAs to act as negative gene regulators allows them to modulate signaling pathways that regulate multiple cellular processes, including the response to radiation. Consequently, they have the potential to be used as either radiosensitizers or radioprotectors.
Using a radioresistant model and an unbiased miRNA expression profiling approach, we identified miR-205 as the top downregulated miRNA in radioresistant breast cancer cells. Interestingly, miR-205 and ZEB1 negatively regulate each other and play opposing roles in regulating DNA damage response and tumor radiosensitivity. In addition to downregulating miR-205, ZEB1 has been shown to repress miR-200c expression48, and indeed, we observed upregulation of both miR-205 (Fig. 4b) and miR-200c (data not shown) in Zeb1 knockout MEFs. However, only miR-205, but not miR-200c, was downregulated by IR (Fig. 4f). One possibility is that ZEB1 may recruit a specific co-repressor to the mir-205 promoter but not to the mir-200c promoter in this context. In fact, it has been shown that ZEB1 recruits distinct transcriptional cofactors in different contexts. For example, Postigo et al52 reported that ZEB1 normally recruits the co-repressor CtBP1 but not co-activators p300 and P/CAF; however, in response to TGFβ, recruitment of p300-P/CAF leads to the displacement of CtBP1 from ZEB1, which in turn promotes the formation of a p300-SMAD transcriptional complex and activates TGFβ-responsive genes. Moreover, it should be noted that the transcription factor p6353 and the polycomb protein Mel-1854 have been shown to regulate the transcription of mir-205. Thus, ZEB1 might not be the only regulator of miR-205 expression in response to radiation.
Upregulation of ZEB1 and downregulation of miR-205 have been found in human breast tumors55-58 and other clinical cancer types59-62. Taken together with our previous finding41, our results raise the caution that radiation treatment may lead to upregulation of ZEB1, downregulation of miR-205 and therapy-induced radioresistance. Based on our data, ZEB1-targing agents, like the miR-205 mimics, represent a new class of radiosensitizing agents.
Methods
Cell culture
Mouse embryo fibroblasts (MEFs) were isolated from Zeb1-deficient embryos, genotyped and cultured in DMEM medium supplemented with 10% fetal bovine serum and 1% penicillin and streptomycin63. The 293T, MDA-MB-231, MCF-7, T47D and A549 cell lines were from ATCC and cultured under conditions specified by the manufacturer. The SUM159 cell line was from S. Ethier and cultured as described (http://www.asterand.com/Asterand/human_tissues/159PT.htm). The HMLE cell line was from R. A. Weinberg's lab stock and cultured in complete Mammary Epithelial Cell Growth Medium (MEGM from Lonza). The DR-GFP-expressing U2OS cell line was from J. Chen's lab stock and cultured in RPMI 1640 medium supplemented with 10% FBS and 1% penicillin and streptomycin.
Plasmids and shRNA
The ZEB1 expression constructs were from R. A. Weinberg. The following shRNA and ORF clones were from Open Biosystems through MD Anderson's shRNA and ORFeome Core: human ZEB1 shRNA, V3LHS-356186 (5′-AGATTTACTGTGCTGTCCT-3′); human Ubc13 (UBE2N) ORF, PLOHS_100006526. The human mir-205 genomic region, which contains the three putative ZEB1-binding sites (E-boxes 1, 2 and 3) and spans from 602 bp upstream of the start site of pre-miR-205 to 710 bp downstream of the start site of pre-miR-205, was PCR amplified from normal genomic DNA and cloned into the pGL3-Basic vector, using the following cloning primers: forward, 5′-CCCTCGAGGTTGTGGTTTTTCCTGAG-3′; reverse, 5′-CCAAGCTTCACTTAAATTTTATTTACATTTAC-3′. A Ubc13 3′ UTR fragment (500 bp) was cloned into the pMIR-REPORT vector, using the following cloning primers: forward, 5′-CTTGACTAGTATTGATACGATCATCAAGTGTGC-3′; reverse, 5′- CTAGAAGCTTGCGTCTAAATAAAGTTAGACTTGGCT-3′. The vectors used in this study are listed in Supplementary Table 2.
siRNA oligonucleotides
Two individual siRNAs that target human Ubc13 were purchased from Sigma (si-Ubc13 #1, SASI_Hs02_00326304; si-Ubc13 #2, SASI_Hs02_00326305). The on-target plus siRNA that targets human ZEB1 was purchased from Dharmacon (J-006564-10-0005). Cells were transfected with 150 nM of the indicated oligonucleotide using the Oligofectamine reagent (Invitrogen). 48 hr after siRNA transfection, cells were used for functional assays, and the remaining cells were harvested for Western blot analysis.
RNA isolation and real-time RT-PCR
Total RNA, inclusive of small RNAs, was isolated using the mirVana miRNA Isolation Kit (Ambion). Quantification of the mature form of miRNAs was performed using the TaqMan MicroRNA Assay Kit (Applied Biosystems) according to the manufacturer's instructions, and the U6 small nuclear RNA was used as an internal control. Real-time PCR and data collection were performed on a CFX96 instrument (Bio-Rad).
Human apoptosis miRNA PCR array analysis
The Human Apoptosis miScript miRNA PCR Array, which profiles the expression of 84 miRNAs that regulate programmed cell death by inhibiting pro-apoptotic and anti-apoptotic gene expression, was used to compare SUM159-P2 with SUM159-P0 cells according to the manufacturer's instructions (http://www.sabiosciences.com/mirna_pcr_product/HTML/MIHS-114Z.html). Briefly, total RNA was extracted and reverse transcribed into cDNA using a miScript II Reverse Transcription Kit (Qiagen). The cDNA was combined with an RT2 SYBR Green qPCR Master Mix (Qiagen), and then equal aliquots of this mixture (25μl) were added to each well of the same PCR Array plate that contained the predispensed gene-specific primer sets. Real-time PCR and data collection were performed on a CFX96 instrument (Bio-Rad).
Luciferase reporter assay
Cells of 50% confluence in 24-well plates were transfected using Fugene 6 (Roche). The firefly luciferase reporter gene construct (400 ng) and the pRL-SV40 Renilla luciferase construct (1 ng, for normalization) were used for cotransfection, with or without 1-2 μg of the SFB-ZEB1 construct. Cell extracts were prepared 24 hours after transfection, and the luciferase activity was measured using the Dual-Luciferase Reporter Assay System (Promega).
Immunoblotting
Western blot analysis was performed with precast gradient gels (Bio-Rad) using standard methods. Briefly, cells were lysed in the RIPA buffer containing protease inhibitors (Roche) and phosphatase inhibitors (Sigma). Proteins were separated by SDS-PAGE and blotted onto a nitrocellulose membrane (Bio-Rad). Membranes were probed with the specific primary antibodies, followed by peroxidase-conjugated secondary antibodies. The bands were visualized by chemiluminescence (Denville Scientific). The following antibodies were used: antibodies to ZEB1 (1:1000, Bethyl Laboratories, A301-922A), γH2AX (1:1000, Cell Signaling Technology, 2577), K63-linkage specific polyubiquitin (1:1000, Cell signaling Technology, 12930), BRCA1 (1:1000, Bethyl Laboratories, A300-000A), Ubc13 (1:1000, Cell Signaling Technology, 4919), FLAG (1:5000, Sigma, F3165) and GAPDH (1:3000, Thermo, MA5-15738). The ImageJ program (http://rsbweb.nih.gov/ij/download.html) was used for densitometric analysis of Western blots, and the quantification results were normalized to the loading control. Uncropped images of blots are shown in Supplementary Fig. 5.
Immunofluorescence
Cells were cultured in chamber slides overnight and fixed with 3.7% formaldehyde in PBS for 20 min at 4°C, followed by permeabilization with 0.5% Triton X-100 in PBS for 30 min. Cells were then blocked for non-specific binding with 10% goat serum in PBS and Tween-20 (PBST) overnight, and incubated with the anti-γH2AX antibody (1:300, 1:1000, Millipore, 07-164) for 1 hr at 37°C, followed by incubation with Alexa Fluor 594 goat anti-mouse IgG (1:400, Invitrogen, A11005) for 1 hr at 37 °C. Cover slips were mounted on slides using anti-fade mounting medium with DAPI. Immunofluorescence images were acquired on a Zeiss Axio Observer Z1 fluorescence microscope.
Chromatin immunoprecipitation (ChIP) assay
ChIP was performed with SUM159-P0, SUM159-P2, or 293T cells transfected with SFB-GFP control or SFB-ZEB1, by using a ChIP Assay Kit (Millipore, 17-295) according to the manufacturer's instructions. After immunoprecipitation with the ZEB1 antibody (1:100, Bethyl Laboratories, A301-922A) or FLAG antibody-conjugated beads (Sigma, M8823), protein-DNA crosslinks were reversed and DNA was purified to remove the chromatin proteins and used for PCR. The PCR primers are: E-box 1, 5′-GGCCTTGGTAAGCAAGCTCAG-3′ (forward) and 5′-CTCGGTGTGGTTGGTCCTTTC-3′(reverse); E-box 2, 5′-TGGCCCCACAGACCCACCT-3′ (forward) and 5′-CATCCTCCAAGGAGGCTGCA-3′ (reverse); E-box 3, 5′-AACTGAGACATGAACTTG-3′ (forward) and 5′-AGGGTTCCTTGCTGATTG-3′ (reverse).
Clonogenic assay
Equal numbers of cells were plated in 10-cm tissue culture dishes at a clonogenic density (500 cells per dish) and irradiated by using a JL Shepherd Mark I-68A 137Cs irradiator with the indicated doses. Cells were incubated for 10-14 days. Colonies were stained with crystal violet and counted using a Gel Doc EZ Imager instrument (Bio-Rad) with the Quantity One software. The survival fraction was calculated as: (number of colonies / number of cells plated) irradiated / (number of colonies / number of cells plated) non-irradiated.
Homologous recombination (HR) repair assay
A U2OS cell clone stably expressing an HR repair reporter was described previously40. Briefly, 2 days after transfection with the indicated miRNA mimics, 1 × 106 U2OS cells expressing the HR repair reporter were electroporated with 10 μg of pCBASce, an I-SceI expression vector described previously64. Cells were harvested 2 days after electroporation and subjected to flow cytometry analysis to determine the percentage of GFP-positive cells resulting from HR-based repair of I-SceI-induced DSBs.
Tumor radiosensitivity study
Animal experiments were performed as previously described65 in accordance with a protocol approved by the Institutional Animal Care and Use Committee of MD Anderson Cancer Center, and mice were euthanized when they met the institutional euthanasia criteria for tumor size and overall health condition. When used in a power calculation, our sample size predetermination experiments indicated that 5 mice per group can identify the expected effect of miR-205 on tumor radiosensitivity (P < 0.05) with 100% power. Solitary tumor xenografts were produced in the muscle of the right hind limb of 3-month-old female athymic nude mice (NCR Nu/Nu) by inoculation of 3 × 106 SUM159-P2 cells. Mice were randomly assigned to no treatment or treatment groups. Radiation treatment (RT) was initiated when tumors grew to 8.0 mm (range: 7.7–8.2 mm) in diameter. Single doses (15 Gy once) were delivered to the tumor-bearing limb of mice using a small-animal irradiator (Co-V, Theratron 780; MDS Nordion, Ottawa, Ontario) with a cobalt Co60 source (field size, 10 × 10 cm; source axis distance, 64.9 cm), at a dose rate of 0.955 Gy min−1. During irradiation, unanesthetized mice were mechanically immobilized in a jig so that the tumor was exposed in radiation field but the animal's body was shielded from radiation exposure. Three mutually orthogonal diameters of the tumor were measured every other day with a caliper, and the mean value was calculated and used as the tumor diameter. An investigator (Li Wang) who measured tumor size was blinded to the group allocation during all animal experiments and outcome assessment. General linear model multivariate analysis was performed to determine statistical significance using the SPSS 14.0 software package.
Liposomal miRNA preparation and in vivo treatment
Nanoliposomal miRNA mimics were prepared and used as previously described36. Briefly, miRNA oligonucleotides and DOPC were mixed in the presence of excess tertiary butanol at a ratio of 1:10 (miRNA:DOPC). Before in vivo administration, the preparation was hydrated with normal 0.9% saline (200 μl per mouse) for subcutaneous injection. One week before tumors grew to 8.0 mm (range: 7.7–8.2 mm) in diameter, mice were injected with 250 μg kg−1 of DOPC-encapsulated miR-205 mimics or a scramble control. Injection was performed twice per week till the end point.
Statistical analysis
Each experiment was repeated three times or more. Unless otherwise noted, data are presented as mean ± s.e.m, and Student's t test (unpaired, two-tailed) was used to compare two groups for independent samples. The data analyzed by t-test meet normal distribution; we used an F-test to compare variances, and the variances are not significantly different. Therefore, when using an unpaired t-test, we assumed equal variance, and no samples were excluded from the analysis. P < 0.05 was considered statistically significant.
Supplementary Material
Acknowledgements
We thank the shRNA and ORFeome Core (supported by the core grant P30CA016672 from NIH) at MD Anderson Cancer Center and Drs. Z. Gong, J. Yuan and A. Lin for reagents and technical assistance. We thank members of the Ma Laboratory for discussion. This work is supported by NIH grants R00CA138572 (to L.M.), R01CA166051 (to L.M.), R01CA181029 (to L.M.), R44GM086937 (to X.Y.) and R43GM100777 (to X.Y.), and a CPRIT Scholar Award R1004 (to L.M.). L. M. is a R. Lee Clark Fellow of The University of Texas MD Anderson Cancer Center. M.J.Y. is supported in part by NIH R01CA164346, Developmental Research Award in Leukemia SPORE CA100632, Center for Inflammation and Cancer, Center for Genetics and Genomics, Institutional Research Grant and Sister Institution Network fund of MD Anderson Cancer Center. L. W. and K. K. A. are supported by the Cancer Center Support Grant P30CA016672 and the Gilbert H. Fletcher Chair. B.G.D. and W.A.W. are supported by a Komen Foundation Grant KG101478. Y. H. is supported in part by NIH U54CA151668. We wish to dedicate this work to the memory of Dr. K. Kian Ang.
Footnotes
Author contributions
P.Z. and L.M. conceived and designed the project. P.Z. performed and analyzed most of the experiments. L.W. and K.K.A. performed tumor radiosensitivity studies. C.R.-A., G.L.-B., A.K.S. and Y.H. assisted with the nanoliposomal miRNA delivery study. Y.Y. and H.L. performed computational data analysis. B.G.D. and W.A.W. established the radioresistant subline. D.C. made some constructs. Y.S. maintained shRNA and ORF clones. M.J.Y. performed histopathological analysis. Y.L. and D.C.D. provided Zeb1-deficient MEFs. X.Y. synthesized miRNA mimics. J.C. provided DR-GFP-expressing U2OS cells. P.Z. and L.M. wrote the manuscript with input from all other authors.
Competing financial interests
X.Y. is an employee of AM Biotechnologies LLC. The remaining authors declare no competing financial interests.
Accession codes
The miRNA qPCR array data have been deposited in the Gene Expression Omnibus database under accession code GSE62511.
References
- 1.Jameel JK, Rao VS, Cawkwell L, Drew PJ. Radioresistance in carcinoma of the breast. Breast. 2004;13:452–460. doi: 10.1016/j.breast.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 2.Horsman MR, et al. Tumor radiosensitizers--current status of development of various approaches: report of an International Atomic Energy Agency meeting. Int J Radiat Oncol Biol Phys. 2006;64:551–561. doi: 10.1016/j.ijrobp.2005.09.032. [DOI] [PubMed] [Google Scholar]
- 3.Calin GA, et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.He L, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682–688. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- 6.Iorio MV, Croce CM. MicroRNAs in cancer: small molecules with a huge impact. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2009;27:5848–5856. doi: 10.1200/JCO.2009.24.0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bader AG, Brown D, Stoudemire J, Lammers P. Developing therapeutic microRNAs for cancer. Gene therapy. 2011;18:1121–1126. doi: 10.1038/gt.2011.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kasinski AL, Slack FJ. Epigenetics and genetics. MicroRNAs en route to the clinic: progress in validating and targeting microRNAs for cancer therapy. Nature reviews. Cancer. 2011;11:849–864. doi: 10.1038/nrc3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ling H, Fabbri M, Calin GA. MicroRNAs and other non-coding RNAs as targets for anticancer drug development. Nature reviews. Drug discovery. 2013;12:847–865. doi: 10.1038/nrd4140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Janssen HL, et al. Treatment of HCV infection by targeting microRNA. The New England journal of medicine. 2013;368:1685–1694. doi: 10.1056/NEJMoa1209026. [DOI] [PubMed] [Google Scholar]
- 12.Ma L, et al. Therapeutic silencing of miR-10b inhibits metastasis in a mouse mammary tumor model. Nature biotechnology. 2010;28:341–347. doi: 10.1038/nbt.1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu C, et al. The microRNA miR-34a inhibits prostate cancer stem cells and metastasis by directly repressing CD44. Nature medicine. 2011;17:211–215. doi: 10.1038/nm.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ell B, et al. Tumor-induced osteoclast miRNA changes as regulators and biomarkers of osteolytic bone metastasis. Cancer cell. 2013;24:542–556. doi: 10.1016/j.ccr.2013.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Taira N, et al. Induction of amphiregulin by p53 promotes apoptosis via control of microRNA biogenesis in response to DNA damage. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:717–722. doi: 10.1073/pnas.1313675111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun C, et al. miR-9 regulation of BRCA1 and ovarian cancer sensitivity to cisplatin and PARP inhibition. Journal of the National Cancer Institute. 2013;105:1750–1758. doi: 10.1093/jnci/djt302. [DOI] [PubMed] [Google Scholar]
- 17.Zhou Y, et al. miR-203 induces oxaliplatin resistance in colorectal cancer cells by negatively regulating ATM kinase. Molecular oncology. 2014;8:83–92. doi: 10.1016/j.molonc.2013.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martin NT, et al. ATM-dependent MiR-335 targets CtIP and modulates the DNA damage response. PLoS genetics. 2013;9:e1003505. doi: 10.1371/journal.pgen.1003505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chowdhury D, Choi YE, Brault ME. Charity begins at home: non-coding RNA functions in DNA repair. Nature reviews. Molecular cell biology. 2013;14:181–189. doi: 10.1038/nrm3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Metheetrairut C, Slack FJ. MicroRNAs in the ionizing radiation response and in radiotherapy. Current opinion in genetics & development. 2013;23:12–19. doi: 10.1016/j.gde.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Banath JP, Macphail SH, Olive PL. Radiation sensitivity, H2AX phosphorylation, and kinetics of repair of DNA strand breaks in irradiated cervical cancer cell lines. Cancer Res. 2004;64:7144–7149. doi: 10.1158/0008-5472.CAN-04-1433. [DOI] [PubMed] [Google Scholar]
- 22.Olive PL, Banath JP. Phosphorylation of histone H2AX as a measure of radiosensitivity. Int J Radiat Oncol Biol Phys. 2004;58:331–335. doi: 10.1016/j.ijrobp.2003.09.028. [DOI] [PubMed] [Google Scholar]
- 23.Taneja N, et al. Histone H2AX phosphorylation as a predictor of radiosensitivity and target for radiotherapy. J Biol Chem. 2004;279:2273–2280. doi: 10.1074/jbc.M310030200. [DOI] [PubMed] [Google Scholar]
- 24.Bao S, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 25.Baumann M, Krause M, Hill R. Exploring the role of cancer stem cells in radioresistance. Nature reviews. Cancer. 2008;8:545–554. doi: 10.1038/nrc2419. [DOI] [PubMed] [Google Scholar]
- 26.Phillips TM, McBride WH, Pajonk F. The response of CD24(−/low)/CD44+ breast cancer-initiating cells to radiation. Journal of the National Cancer Institute. 2006;98:1777–1785. doi: 10.1093/jnci/djj495. [DOI] [PubMed] [Google Scholar]
- 27.Debeb BG, Xu W, Woodward WA. Radiation resistance of breast cancer stem cells: understanding the clinical framework. J Mammary Gland Biol Neoplasia. 2009;14:11–17. doi: 10.1007/s10911-009-9114-z. [DOI] [PubMed] [Google Scholar]
- 28.Mani SA, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gregory PA, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 30.Brabletz T, Lyden D, Steeg PS, Werb Z. Roadblocks to translational advances on metastasis research. Nature medicine. 2013;19:1104–1109. doi: 10.1038/nm.3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Camps C, et al. hsa-miR-210 Is induced by hypoxia and is an independent prognostic factor in breast cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2008;14:1340–1348. doi: 10.1158/1078-0432.CCR-07-1755. [DOI] [PubMed] [Google Scholar]
- 32.Buffa FM, et al. microRNA-associated progression pathways and potential therapeutic targets identified by integrated mRNA and microRNA expression profiling in breast cancer. Cancer Res. 2011;71:5635–5645. doi: 10.1158/0008-5472.CAN-11-0489. [DOI] [PubMed] [Google Scholar]
- 33.Gutierrez-Puente Y, et al. Safety, pharmacokinetics, and tissue distribution of liposomal P-ethoxy antisense oligonucleotides targeted to Bcl-2. J Pharmacol Exp Ther. 1999;291:865–869. [PubMed] [Google Scholar]
- 34.Lu C, et al. Regulation of tumor angiogenesis by EZH2. Cancer cell. 2010;18:185–197. doi: 10.1016/j.ccr.2010.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Landen CN, Jr., et al. Therapeutic EphA2 gene targeting in vivo using neutral liposomal small interfering RNA delivery. Cancer Res. 2005;65:6910–6918. doi: 10.1158/0008-5472.CAN-05-0530. [DOI] [PubMed] [Google Scholar]
- 36.Nishimura M, et al. Therapeutic synergy between microRNA and siRNA in ovarian cancer treatment. Cancer discovery. 2013;3:1302–1315. doi: 10.1158/2159-8290.CD-13-0159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yano J, et al. Antitumor activity of small interfering RNA/cationic liposome complex in mouse models of cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2004;10:7721–7726. doi: 10.1158/1078-0432.CCR-04-1049. [DOI] [PubMed] [Google Scholar]
- 38.Sung P, Klein H. Mechanism of homologous recombination: mediators and helicases take on regulatory functions. Nature reviews. Molecular cell biology. 2006;7:739–750. doi: 10.1038/nrm2008. [DOI] [PubMed] [Google Scholar]
- 39.Pierce AJ, Johnson RD, Thompson LH, Jasin M. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 1999;13:2633–2638. doi: 10.1101/gad.13.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weinstock DM, Nakanishi K, Helgadottir HR, Jasin M. Assaying double-strand break repair pathway choice in mammalian cells using a targeted endonuclease or the RAG recombinase. Methods Enzymol. 2006;409:524–540. doi: 10.1016/S0076-6879(05)09031-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang P, et al. ATM-mediated stabilization of ZEB1 promotes DNA damage response and radioresistance through CHK1. Nat Cell Biol. 2014;16:864–875. doi: 10.1038/ncb3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 43.Krek A, et al. Combinatorial microRNA target predictions. Nature genetics. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 44.Zhao GY, et al. A critical role for the ubiquitin-conjugating enzyme Ubc13 in initiating homologous recombination. Molecular cell. 2007;25:663–675. doi: 10.1016/j.molcel.2007.01.029. [DOI] [PubMed] [Google Scholar]
- 45.Moynahan ME, Chiu JW, Koller BH, Jasin M. Brca1 controls homology-directed DNA repair. Molecular cell. 1999;4:511–518. doi: 10.1016/s1097-2765(00)80202-6. [DOI] [PubMed] [Google Scholar]
- 46.Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419:135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- 47.Haracska L, Torres-Ramos CA, Johnson RE, Prakash S, Prakash L. Opposing effects of ubiquitin conjugation and SUMO modification of PCNA on replicational bypass of DNA lesions in Saccharomyces cerevisiae. Molecular and cellular biology. 2004;24:4267–4274. doi: 10.1128/MCB.24.10.4267-4274.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Burk U, et al. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO reports. 2008;9:582–589. doi: 10.1038/embor.2008.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Furusawa T, Moribe H, Kondoh H, Higashi Y. Identification of CtBP1 and CtBP2 as corepressors of zinc finger-homeodomain factor deltaEF1. Molecular and cellular biology. 1999;19:8581–8590. doi: 10.1128/mcb.19.12.8581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Postigo AA, Dean DC. ZEB represses transcription through interaction with the corepressor CtBP. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:6683–6688. doi: 10.1073/pnas.96.12.6683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shiloh Y. ATM and related protein kinases: safeguarding genome integrity. Nature reviews. Cancer. 2003;3:155–168. doi: 10.1038/nrc1011. [DOI] [PubMed] [Google Scholar]
- 52.Postigo AA, Depp JL, Taylor JJ, Kroll KL. Regulation of Smad signaling through a differential recruitment of coactivators and corepressors by ZEB proteins. The EMBO journal. 2003;22:2453–2462. doi: 10.1093/emboj/cdg226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tucci P, et al. Loss of p63 and its microRNA-205 target results in enhanced cell migration and metastasis in prostate cancer. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:15312–15317. doi: 10.1073/pnas.1110977109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee JY, et al. Loss of the polycomb protein Mel-18 enhances the epithelial mesenchymal transition by ZEB1 and ZEB2 expression through the downregulation of miR-205 in breast cancer. Oncogene. 2014;33:1325–1335. doi: 10.1038/onc.2013.53. [DOI] [PubMed] [Google Scholar]
- 55.Karihtala P, et al. Vimentin, zeb1 and Sip1 are up-regulated in triple-negative and basal-like breast cancers: association with an aggressive tumour phenotype. Breast Cancer Res Treat. 2013;138:81–90. doi: 10.1007/s10549-013-2442-0. [DOI] [PubMed] [Google Scholar]
- 56.Graham TR, et al. Reciprocal regulation of ZEB1 and AR in triple negative breast cancer cells. Breast Cancer Res Treat. 2010;123:139–147. doi: 10.1007/s10549-009-0623-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu H, Zhu S, Mo YY. Suppression of cell growth and invasion by miR-205 in breast cancer. Cell Res. 2009;19:439–448. doi: 10.1038/cr.2009.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Iorio MV, et al. microRNA-205 regulates HER3 in human breast cancer. Cancer Res. 2009;69:2195–2200. doi: 10.1158/0008-5472.CAN-08-2920. [DOI] [PubMed] [Google Scholar]
- 59.Xu Y, Brenn T, Brown ER, Doherty V, Melton DW. Differential expression of microRNAs during melanoma progression: miR-200c, miR-205 and miR-211 are downregulated in melanoma and act as tumour suppressors. Br J Cancer. 2012;106:553–561. doi: 10.1038/bjc.2011.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Verdoodt B, et al. MicroRNA-205, a novel regulator of the anti-apoptotic protein Bcl2, is downregulated in prostate cancer. Int J Oncol. 2013;43:307–314. doi: 10.3892/ijo.2013.1915. [DOI] [PubMed] [Google Scholar]
- 61.Kenney PA, et al. Novel ZEB1 expression in bladder tumorigenesis. BJU Int. 2011;107:656–663. doi: 10.1111/j.1464-410X.2010.09489.x. [DOI] [PubMed] [Google Scholar]
- 62.Spoelstra NS, et al. The transcription factor ZEB1 is aberrantly expressed in aggressive uterine cancers. Cancer Res. 2006;66:3893–3902. doi: 10.1158/0008-5472.CAN-05-2881. [DOI] [PubMed] [Google Scholar]
- 63.Liu Y, El-Naggar S, Darling DS, Higashi Y, Dean DC. Zeb1 links epithelial mesenchymal transition and cellular senescence. Development. 2008;135:579–588. doi: 10.1242/dev.007047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Richardson C, Moynahan ME, Jasin M. Double-strand break repair by interchromosomal recombination: suppression of chromosomal translocations. Genes Dev. 1998;12:3831–3842. doi: 10.1101/gad.12.24.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang L, et al. MK-4827, a PARP-1/-2 inhibitor, strongly enhances response of human lung and breast cancer xenografts to radiation. Invest New Drugs. 2012;30:2113–2120. doi: 10.1007/s10637-011-9770-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.