Abstract
Introns are removed from nuclear messenger RNA precursors through two sequential phospho-transesterification reactions in a dynamic RNA–protein complex called the spliceosome1,2. But whether splicing is catalysed by small nuclear RNAs3,4 in the spliceosome is unresolved. As the spliceosome is a metalloenzyme5-7, it is important to determine whether snRNAs coordinate catalytic metals. Here we show that yeast U6 snRNA coordinates a metal ion that is required for the catalytic activity of the spliceosome. With Mg2+, U6 snRNA with a sulphur substitution for the pro-RP or pro-SP non-bridging phosphoryl oxygen of nucleotide U80 reconstitutes a fully assembled yet catalytically inactive spliceosome. Adding a thiophilic ion such as Mn2+ allows the first transesterification reaction to occur in the U6/sU80(SP)- but not the U6/sU80(RP)-reconstituted spliceosome. Mg2+ competitively inhibits the Mn2+-rescued reaction, indicating that the metal-binding site at U6/U80 exists in the wild-type spliceosome and that the site changes its metal requirement for activity in the SP spliceosome. Thus, U6 snRNA contributes to pre-messenger RNA splicing through metal-ion coordination, which is consistent with RNA catalysis by the spliceosome.
The spliceosome forms by incorporating small nuclear ribonucleoprotein particles (snRNPs) and non-snRNP proteins onto premRNA, and it attains its catalytic conformation by undergoing a series of protein and RNA rearrangements1-3,8. Much evidence indicates that splicing is catalysed by the snRNAs in the spliceosome, although there is no definitive proof 3,4. Among the five spliceosomal snRNAs (U1, U2, U4, U5 and U6), U6 is most widely assumed to be involved in catalysis because it is highly conserved through evolution and very sensitive to chemical changes. In addition, U6 forms intramolecular and intermolecular helices that are analogous to autocatalytic group II introns3. If U6 snRNA were directly involved in catalysis, it might coordinate catalytic metals, because the spliceosome, like most RNA enzymes9, uses divalent metal ions in catalysis6,7. If a phosphoryl oxygen/Mg2+ interaction is crucial for function, a sulphur substitution for that oxygen would cause a switch in metal specificity from Mg2+ to a thiophilic ion such as Mn2+ (ref. 10). Several (RP)-phosphorothioate substitutions in yeast or nematode U6 snRNA fail to reconstitute active spliceosomes with Mg2+ (refs 11, 12). However, Mn2+ does not rescue these phosphorothioates, and it is unclear whether any of the oxygens coordinate metal13,14.
We tested synthetic U6 snRNA containing a single phosphorothioate (as a mixture of RP and SP diastereomers) in reconstituting Mg2+-dependent splicing in a U6-depleted yeast extract11,15. Many phosphorothioate-containing U6 RNAs reconstituted splicing well (our own unpublished data), except U6 with a sulphur substitution at U80 (Fig. 1, lane 5). Pure sU80(RP) diastereomer completely failed to reconstitute splicing (Fig. 1, lane 6)11. Notably, splicing also did not occur in the extract reconstituted with U6/sU80(SP) (Fig. 1, lane 7), suggesting that both 5′ non-bridging oxygens of U80 are important for splicing.
Figure 1.

U6 snRNA with a phosphorothioate substitution at U80 fails to reconstitute splicing. U6-depleted prp2 mutant extracts were reconstituted with T7 polymerase-transcribed, unmodified U6 (lanes 1 and 3) or ligated, synthetic U6 (lanes 4–8) in the absence (lane 1) or presence (lanes 2–8) of wild-type PRP2 protein. No U6 was added in lane 2 (−). Both input pre-mRNA and U6 RNA were labelled with 32P and were analysed by denaturing gels and autoradiography. Synthetic U6 used: O, no phosphorothioate substitution (lane 4); M, unpurified RP/SP phosphorothioate mixture with some unsulphurized RNA (lane 5); and the RP diastereomer (lane 6), SP (lane 7) and the unsulphurized RNA (lane 8) purified from the mixture. The intron–exon2 lariat (IVS–E2*) and exon 1 (E1) are products from the first transesterification reaction, and the intron lariat (IVS*) and ligated exon 1/exon 2 (E1–E2) are from the second reaction.
We investigated whether U6/sU80 snRNA blocked spliceosome maturation or Mg2+-dependent transesterification. The reactions were carried out with or without the yeast PRP2 ATPase, which is required for the final ATP-dependent spliceosomal rearrangements before the first transesterification reaction16. Reconstituted spliceosomes were isolated from glycerol gradients and analysed on a non-denaturing gel (Fig. 2a). In the absence of PRP2, unmodified U6, U6/sU80(RP) and U6/sU80(SP) reconstituted a pre-catalytic SS2 spliceosome16 (Fig. 2a, lanes 1, 3 and 5), which was equivalent to mammalian spliceosome B2 (ref. 1). In the presence of PRP2, the post-PRP2 spliceosomes were formed (Fig. 2a, lanes 2, 4 and 6), which were equivalent to mammalian spliceosomes C1 and C2 (ref. 1). These spliceosome complexes were not detected in U6-depleted extracts without reconstitution (data not shown). Thus, both of the U6/sU80 RNAs support spliceosome maturation but not catalysis.
Figure 2.
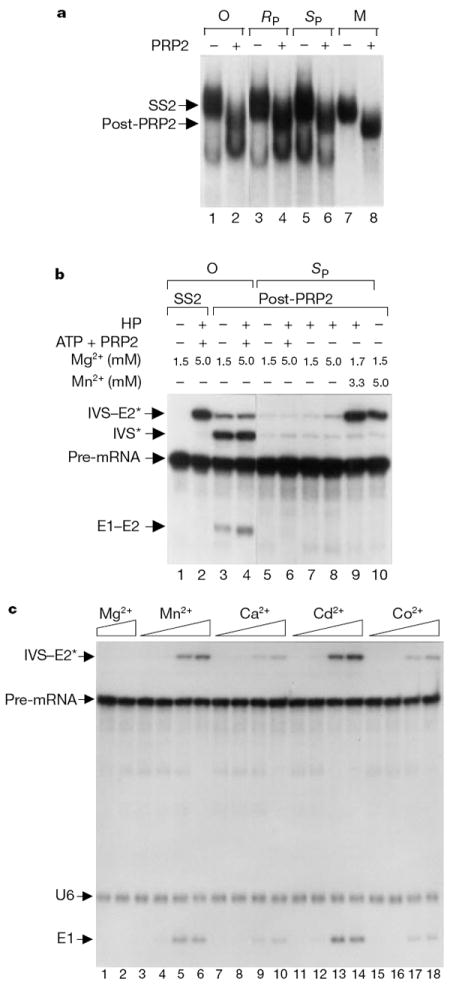
Spliceosome maturation is not affected by phosphorothioate substitution at U80 of U6 but splicing only occurs in the SP diastereomer with a thiophilic metal ion. a, U6/sU80 forms mature spliceosome. Extracts were reconstituted with unmodified U6 (lanes 1 and 2) or U6/sU80 (RP, lanes 3–4; SP, lanes 5–6), in the absence (lanes 1, 3 and 5) or presence (lanes 2, 4 and 6) of PRP2. The reaction mixture was sedimented through a glycerol gradient and the fraction containing the spliceosome was analysed on a native gel. The pre-catalytic and post-PRP2 spliceosomal markers (M; lanes 7 and 8) were generated as described16. b, The first transesterification reaction occurs in U6/sU80(SP)-containing post-PRP2 spliceosome in the presence of Mn2+. Spliceosomes isolated from glycerol gradients were incubated in buffer containing combinations of Mg2+, Mn2+, ATP, PRP2 and HP (a protein factor that facilitates splicing in purified spliceosomes16). c, Other thiophilic divalent metal ions can also rescue the transesterification reaction. The post-PRP2 U6/sU80(SP) spliceosome was incubated with metal ions without the addition of ATP, PRP2 or HP. Mg2+ was added as the sole divalent metal ion in lanes 1 (2.5 mM) and 2 (5 mM). The concentrations (mM) of the thiophilic metal (Mn2+, Ca2+, Cd2+ or Co2+) in each set were 0.02, 0.1, 0.5 and 2.5. Mg2+ was added to lanes 3–18 to make the total concentration of divalent metal ions 5 mM.
We tested whether phosphorothioate substitutions at U80 switch the metal requirement for catalysis by incubating the purified post- PRP2 spliceosomes with Mg2+ or Mn2+ (Fig. 2b). Splicing did not occur in the sU80(RP) spliceosome regardless of metal used (data not shown). No splicing occurred when the sU80(SP) spliceosome was incubated with Mg2+ as the sole divalent metal ion (Fig. 2b, lanes 5–8). However, the sU80(SP) spliceosome efficiently carried out the first transesterification reaction in the presence of Mn2+ (Fig. 2b, lanes 9 and 10). This reaction did not require additional ATP, PRP2 or the HP splicing factor, confirming that this spliceosome had been fully activated16. Other thiophilic divalent metal ions also rescued the sU80(SP) spliceosome; Cd2+ was more effective than Mn2+, whereas Ca2+ and Co2+ were less effective (Fig. 2c). Rescue occurred at submillimolar concentrations, indicating that there was specific metal interaction at the site of sulphur substitution17. Thus, these results showed that only the sU80(SP) and not sU80(RP) phosphorothioate caused a switch of metal specificity, and that the transesterification reaction could occur in the SP spliceosome once thiophilic metal occupied the sulphur-substituted site.
As purified spliceosomes are missing second-step factors16, we tested the ability of Mn2+ to rescue the second transesterification reaction in extracts (Fig. 3a). Mn2+ rescued the first but not the second transesterification reaction in U6/sU80(SP)-reconstituted extracts (Fig. 3a, lanes 14–18), whereas the U6/sU80(RP)-reconstituted extract could not be rescued (lanes 8–12). This specificity of rescue indicated that desulphurization of the phosphorothioate did not occur during the reaction. The result also suggests that the pro-SP oxygen of U80 may have a different role during the second step. The pro-SP and pro-RP oxygens, for example, may switch roles in the two spliceosomal active sites18.
Figure 3.

With U6/sU80(SP) RNA, only the first of the two transesterification reactions occurs in the presence of Mn2+, which is competitively inhibited by Mg2+. a, Mn2+ rescues the first but not the second step of splicing in U6/sU80(SP)-reconstituted reaction. Extract was reconstituted with U6 (lanes 1–6), U6/sU80(RP) (lanes 7–12), or U6/sU80(SP) (lanes 13–18) and PRP2 in the presence of Mn2+ and Mg2+. In each set of six reactions, the concentrations (mM) of Mn2+/Mg2+ were 0/2.5, 2.5/2.5, 1.25/1.25, 1.7/0.8; 2.0/0.5, 2.5/0. b, Mg2+ competitively inhibits the Mn2+-rescued reaction. Extract was reconstituted in the absence of PRP2 with U6 (lanes 1–9) or U6/sU80(SP) (lanes 10–18) and with Mg2+ (2.5 mM) as the sole divalent metal ion. Splicing was then triggered by the addition of PRP2 with 1mM Mn2+ (lanes 2–9 and 11–18) and an increasing concentration (mM) of Mg2+ (2.5, 5.0, 7.5, 10, 15, 20, 25, 30). Reactions in lanes 1 and 2 contained 2.5mM of Mg2+ and no Mn2+ (asterisk). c, The ratio of splicing efficiency between the U6/sU80(SP)- and U6-reconstituted reactions (SP/O). Splicing efficiency in each reaction (Fig. 3b) was calculated as the ratio between the products (IVS–E2*, IVS* and E1–E2) and the total (products plus pre-mRNA).
We further characterized the Mn2+-rescued site by testing whether Mg2+ could competitively inhibit the Mn2+-supported reaction in U6/sU80(SP). Pre-catalytic SS2 spliceosomes were reconstituted with unmodified U6 or U6/sU80(SP) and, without gradient purification, incubated with PRP2 and Mn2+ plus an increasing amount of Mg2+ (Fig. 3b). The ratio of splicing efficiency between the U6/sU80(SP)-reconstituted reaction and the U6-reconstituted reaction decreased as the Mg2+ concentration increased (Fig. 3c). Thus, Mg2+ could effectively occupy the Mn2+-rescued site in U6/sU80(SP) but it could not support splicing. This result indicates that sulphur substitution does not create a binding site for the rescuing metal ion that is not present in the wild-type reaction19,20. It also indicates that the metal-binding site at U80 changes its metal requirement for activity in the U6/sU80(SP)-reconstituted spliceosome.
We have identified, to our knowledge, the first snRNA site that contributes to catalysis in the spliceosome through metal-ion coordination. Ribozymes such as hammerhead21,22, group I intron of Tetrahymena23 and RNase P (ref. 24) have phosphoryl oxygens coordinating metal that contributes to catalysis. It will be of interest to know the exact geometry of the Mg2+ coordinated by the pro-SP oxygen of U80 with respect to the 5′ splice site, and whether this metal ion is the one that interacts with the leaving group6. U80 of the yeast U6 snRNA is situated at the bulge region of its 3′ intramolecular stem loop (3′ISL, Fig. 4). This 3′ISL of U6 and the helix Ib of U2–U6 have been proposed to be functionally analogous to the catalytically important domain 5 of group II introns, partly on the basis of (RP)-phosphorothioate interference analysis12,25. For U80 to coordinate a Mg2+ at or near the 5′ splice site, it has to fold over to the scissile phosphate. A tertiary interaction between U2/A25 and U6/G52 identified in yeast26 and in human27 might be involved in this folding. Tertiary interactions (λ–λ′) have recently been described in a group II intron between the conserved G at the fifth position of the intron and the two conserved G·C base pairs above the corresponding U80 bulge in domain 5 (ref. 28). If similar interactions also occurred in the spliceosome, they could juxtapose the U80 of U6 and the 5′ splice site. Thus, U80 could coordinate the Mg2+ that stabilizes the leaving group at the 5′ splice junction6, the putative Mg2+ that activates the nucleophilic hydroxyl at the branchpoint5, or an Mg2+ nearby (Fig. 4). In conclusion, we have elucidated a catalytic role for theU6 snRNA in pre-mRNA splicing, and provided evidence to support the notion that the spliceosome is fundamentally an RNA enzyme.
Figure 4.

A representation of RNAs and metal ions in the yeast spliceosome before the first catalytic step. Intermolecular helices between U6 and U2 (Ia and Ib), U2 and pre-mRNA at the branch site (U2/BS), U6 and pre-mRNA at the 5′ splice site (U6/5′SS), as well as the 3′ intramolecular stem-loop (3′ISL) in U6 are depicted1-3,8. The branchpoint adenine is highlighted with a star. The U80 of U6 is shaded. Three Mg2+ ions are shown as circles: one stabilizes the leaving oxygen at the 5′ scissile phosphate6 (lined); one activates the attacking oxygen5 (dotted); and one binds to the pro-SP oxygen of U80 (filled). The tertiary interaction between G52 of U6 and A25 of U2 (refs 26, 27), a putative interaction between 3′ISL of U6 and the U6/5′SS helix based on the λ-λ′ interaction in a group II intron28, and the potential relationship between the metals are linked with a line.
Methods
RNA oligonucleotides
We synthesized RNA oligonucleotides using CE (β-cyanoethyl) phosphoramidites with 2′TBDMS (t-butyl-dimethylsilyl) protecting groups (Glen Research) on an ABI 394 DNA/RNA synthesizer at the Caltech Biopolymer Center. Sulphurization of a non-bridging oxygen was performed by using 3H-1,2-benzodithiol-3-one-1,1-dioxide. After synthesis, we deprotected the preparation using triethylamine trihydrofluoride, and purified the full-length oligonucleotide from a denaturing polyacrylamide gel or through a diethylaminoethyl (DEAE) column. The oligomer containing sU80 was resolved by a C18 Beckman Ultrasphere ODS column on a Hewlett-Packard high-performance liquid chromatography (HPLC) into three peaks: first, the unmodified oligonucleotide; second, the RP diastereomer; and third, the SP diastereomer. The exact mass of the oligomers was confirmed by mass spectrometry on a Voyager Elite instrument at the Caltech Protein/Peptide Micro Analytical Laboratory (PPMAL) facility. The RP and SP diastereomers were verified by digesting 32P-labelled oligomers with snake venom phosphodiesterase, which preferentially cleaves the RP isomer29
Pre-mRNA and U6 snRNA
SP6 RNA polymerase was used to produce 32P-labelled actin pre-mRNA16. U6 snRNA was made by using T7 RNA polymerase11 or by ligating four synthetic RNA oligonucleotides15,30. Four RNA oligos, U6/1–37, U6/38–69, U6/70–83, and U6/84–112 were ligated to generate U6/U80 RNA on a DNA template, cdU6/19–100. Before ligation, three RNA oligomers, U6/38–69, U6/70–83, and U6/84–112 were pooled and phosphorylated for 60 min at 37 °C in 10 μl reaction containing 50 mM Tris pH 7.5, 10 mM MgCl2, 5 mM DTT, 60 μM ATP, 10 pmol [γ-32P]ATP, 10 units of RNasin and 10 units of T4 polynucleotide kinase. The phosphorylation reaction was terminated by heating at 65 °C for 20 min. The fourth RNA oligomer, U6/1–37, and the DNA template, cdU6/19–100, were then added and the mixture was heated at 90 °C for 1 min and cooled at room temperature. Ligations were performed at 30 °C for 4 h in 25 μl reaction containing 5,000 units of T4 DNA ligase, 20 units of RNasin, 4% of PEG and 1 × T4 DNA ligase buffer (New England Biolabs). Ligated RNA was isolated from a 7.5% polyacrylamide/urea gel.
Splicing and spliceosome assays
Extract preparation, in vitro splicing, gradient and gel analysis of spliceosomes, and spliceosome conversion assays were done as described16. U6 reconstitution was done by cleaving endogenous U6 snRNA in heat-inactivated prp2 mutant extracts in the presence of 450nM d1 (cdU6/28–54) oligodeoxyribonucleotide and 2mM ATP11. Pre-mRNA (0.4 nM), U6 RNA (4 nM), divalent metal ions in chloride form, potassium phosphate and PEG (4.8%) were added and incubated at 23 °C for 30 min to allow reconstitution and spliceosome assembly. The PRP2 protein was added and incubated for splicing at 23 °C for 15 min. 32P-labelled RNA on gels was quantitated with a Phosphorimager.
Acknowledgments
We thank J. Abelson for U6 plasmids; D. McPheeters and D. Ryan for advice on RNA ligation procedure; and T. Nilsen, E. Sontheimer, D. McPheeters, J. Rossi, G. Edwalds- Gilbert and E. Silverman for critical reading of the manuscript. This work is supported by grants from NIH to J.T. and R.-J.L. The Molecular Dynamics phosphorimager was purchased with a core grant from NSF.
References
- 1.Sharp PA. Split genes and RNA splicing. Cell. 1994;77:805–815. doi: 10.1016/0092-8674(94)90130-9. [DOI] [PubMed] [Google Scholar]
- 2.Guthrie C. The spliceosome is a dynamic ribonucleoprotein machine. Harvey Lect. 1994;90:59–80. [PubMed] [Google Scholar]
- 3.Nilsen TW. In: RNA Structure and Function. Simons R, Grunberg-Manago M, editors. Cold Spring Harbor Laboratory Press; Cold Spring Harbor: 1998. pp. 279–307. [Google Scholar]
- 4.Collins CA, Guthrie C. The question remains: is the spliceosome a ribozyme? Nature Struct Biol. 2000;7:850–854. doi: 10.1038/79598. [DOI] [PubMed] [Google Scholar]
- 5.Steitz TA, Steitz JA. A general two-metal-ion mechanism for catalytic RNA. Proc Natl Acad Sci USA. 1993;90:6498–6502. doi: 10.1073/pnas.90.14.6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sontheimer EJ, Sun S, Piccirilli JA. Metal ion catalysis during splicing of premessenger RNA. Nature. 1997;388:801–805. doi: 10.1038/42068. [DOI] [PubMed] [Google Scholar]
- 7.Gordon PM, Sontheimer EJ, Piccirilli JA. Metal ion catalysis during the exon-ligation step of nuclear pre-mRNA splicing: extending the parallels between the spliceosome and group II introns. RNA. 2000;6:199–205. doi: 10.1017/s1355838200992069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Staley JP, Guthrie C. Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell. 1998;92:315–326. doi: 10.1016/s0092-8674(00)80925-3. [DOI] [PubMed] [Google Scholar]
- 9.Pyle AM. Ribozymes: a distinct class of metalloenzymes. Science. 1993;261:709–714. doi: 10.1126/science.7688142. [DOI] [PubMed] [Google Scholar]
- 10.Sigel RKO, Song B, Sigel H. Stabilities and structures of metal ion complexes of adenosine 5′-O-thiomonophosphate (AMPS2−) in comparison with those of its parent nucleotide (AMP2−) in aqueous solution. J Am Chem Soc. 1997;119:744–755. [Google Scholar]
- 11.Fabrizio P, Abelson J. Thiophosphates in yeast U6 snRNA specifically affect pre-mRNA splicing in vitro. Nucleic Acids Res. 1992;20:3659–3664. doi: 10.1093/nar/20.14.3659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu YT, Maroney PA, Darzynkiwicz E, Nilsen TW. U6 snRNA function in nuclear pre-mRNA splicing: a phosphorothioate interference analysis of the U6 phosphate backbone. RNA. 1995;1:46–54. [PMC free article] [PubMed] [Google Scholar]
- 13.Brautigam CA, Steitz TA. Structural principles for the inhibition of the 3′-5′ exonuclease activity of Escherichia coli DNA polymerase I by phosphorothioates. J Mol Biol. 1998;277:363–377. doi: 10.1006/jmbi.1997.1586. [DOI] [PubMed] [Google Scholar]
- 14.Smith JS, Nikonowicz EP. Phosphorothioate substitution can substantially alter RNA conformation. Biochemistry. 2000;39:5642–5652. doi: 10.1021/bi992712b. [DOI] [PubMed] [Google Scholar]
- 15.Kim CH, Abelson J. Site-specific crosslinks of yeast U6 snRNA to the pre-mRNA near the 5′ splice site. RNA. 1996;2:995–1010. [PMC free article] [PubMed] [Google Scholar]
- 16.Kim SH, Lin RJ. Spliceosome activation by PRP2 ATPase prior to the first transesterification reaction of pre-mRNA splicing. Mol Cell Biol. 1996;16:6810–6819. doi: 10.1128/mcb.16.12.6810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Basu S, Strobel SA. Thiophilic metal ion rescue of phosphorothioate interference within the Tetrahymena ribozyme P4-P6 domain. RNA. 1999;5:1399–1407. doi: 10.1017/s135583829999115x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moore MJ, Sharp PA. Evidence for two active sites in the spliceosome provided by stereochemistry of pre-mRNA splicing. Nature. 1993;365:364–368. doi: 10.1038/365364a0. [DOI] [PubMed] [Google Scholar]
- 19.Piccirilli JA, Vyle JS, Caruthers MH, Cech TR. Metal ion catalysis in the Tetrahymena ribozyme reaction. Nature. 1993;361:85–88. doi: 10.1038/361085a0. [DOI] [PubMed] [Google Scholar]
- 20.Weinstein LB, Jones BC, Cosstick R, Cech TR. A second catalytic metal ion in group I ribozyme. Nature. 1997;388:805–808. doi: 10.1038/42076. [DOI] [PubMed] [Google Scholar]
- 21.Peracchi A, Beigelman L, Scott EC, Uhlenbeck OC, Herschlag D. Involvement of a specific metal ion in the transition of the hammerhead ribozyme to its catalytic conformation. J Biol Chem. 1997;272:26822–26826. doi: 10.1074/jbc.272.43.26822. [DOI] [PubMed] [Google Scholar]
- 22.Murray JB, Scott WG. Does a single metal ion bridge the A-9 and scissile phosphate groups in the catalytically active hammerhead ribozyme structure? J Mol Biol. 2000;296:33–41. doi: 10.1006/jmbi.1999.3428. [DOI] [PubMed] [Google Scholar]
- 23.Christian EL, Yarus M. Metal coordination sites that contribute to structure and catalysis in the group I intron from Tetrahymena. Biochemistry. 1993;32:4475–4480. doi: 10.1021/bi00068a001. [DOI] [PubMed] [Google Scholar]
- 24.Christian EL, Kaye NM, Harris ME. Helix P4 is a divalent metal ion binding site in the conserved core of the ribonuclease P ribozyme. RNA. 2000;6:511–519. doi: 10.1017/s1355838200000042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chanfreau G, Jacquier A. Catalytic site components common to both splicing steps of a group II intron. Science. 1994;266:1383–1387. doi: 10.1126/science.7973729. [DOI] [PubMed] [Google Scholar]
- 26.Madhani HD, Guthrie C. Randomization-selection analysis of snRNAs in vivo: evidence for a tertiary interaction in the spliceosome. Genes Dev. 1994;8:1071–1086. doi: 10.1101/gad.8.9.1071. [DOI] [PubMed] [Google Scholar]
- 27.Valadkhan S, Manley JL. A tertiary interaction detected in a human U2–U6 snRNA complex assembled in vitro resembles a genetically proven interaction in yeast. RNA. 2000;6:206–219. doi: 10.1017/s1355838200992197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boudvillain M, de Lencastre A, Pyle AM. A tertiary interaction that links active-site domains to the 5′ splice site of a group II intron. Nature. 2000;406:315–318. doi: 10.1038/35018589. [DOI] [PubMed] [Google Scholar]
- 29.Burgers PM, Eckstein F. Absolute configuration of the diastereomers of adenosine 5′-O-(1-thiotriphosphate): consequences for the stereochemistry of polymerization by DNA-dependent RNA polymerase from Escherichia coli. Proc Natl Acad Sci USA. 1978;75:4798–4800. doi: 10.1073/pnas.75.10.4798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moore MJ, Sharp PA. Site-specific modification of pre-mRNA: the 2′-hydroxyl groups at the splice sites. Science. 1992;256:992–997. doi: 10.1126/science.1589782. [DOI] [PubMed] [Google Scholar]


