Abstract
The insect fat body and the adipose tissue of vertebrates store fatty acids (FA) as triacylglycerols (TG). However, the fat body of most insects has the unique ability to rapidly produce and secrete large amounts of diacylglycerol (DG). Monoacylglycerol acyltransferase (MGAT), which catalyzes the synthesis of DG from MG, and a diacylglycerol acyltransferase (DGAT), which catalyzes the synthesis of TG from DG, are key enzymes in the metabolism of neutral glycerides. However, very little is known about these acyltransferases in insects. In the present study we have cloned two predicted MGATs and a DGAT from M. sexta and compared their sequences with predicted MGAT and DGAT homologues from a number of insect species. The comparison suggested that insects may only have a single DGAT gene, DGAT1. The apparent absence of a DGAT2 gene in insects would represent a major difference with vertebrates, which contain DGAT1 and DGAT2 genes. Insects seem to have a single MGAT gene which is similar to the MGAT2 of vertebrates. A number of conserved phosphorylation sites of potential physiological significance were identified among insect proteins and among insect and vertebrate proteins. DGAT1 and MGAT are expressed in fat body, midgut and ovaries. The relative rates of utilization of FAs for the synthesis of DG and TG correlated with the relative expression levels of MGAT and DGAT suggesting that regulation of the expression levels of these acyltransferases could be determining whether the fat body secretes DG or stores fatty acids as TG. The expression patterns of the acyltransferases suggest a role of the monoacylglycerol pathway in the production and mobilization of DG in M. sexta fat body.
1. Introduction
Insects, as most animals, store large amounts of energy in the form of triacylglycerols (TG). The main tissue for TG storage in insects is the fat body (Arrese and Soulages, 2010). TG are stored in cytosolic organelles called lipid droplets that besides serving as reservoirs of fatty acids (FA) play a role in the regulation of the metabolism of TG (Brasaemle, 2007). Utilization of the stored FA requires hydrolysis of TG by lipases. The lipolytic action releases FA that can thus be further processed by fat body adipocytes to produce acetyl-CoA or other lipids, such as hydrocarbons, waxes, phospholipids or, the most common product, DG. It is in the production of large quantities of DG, and the large concentration of sn-1,2-DG in hemolymph (Lok and van der Horst, 1980), that most insects show a clear difference with the metabolism of lipids in vertebrates. Fat body adipocytes not only produce massive amounts of DG, but they also have the ability to export DG to circulating lipoproteins. The main insect lipoprotein, lipophorin, is the carrier of the secreted DG and transports the FA to the sites of utilization, such as muscles and ovary (Beenakkers et al., 1985). Contrarily to human apoB containing lipoproteins, lipophorin particles pick up and deliver DG molecules without compromising the integrity of the apolipoproteins. Since the apolipoproteins are recycled (Downer and Chino, 1985), this process is known as the lipophorin shuttle system (van der Horst et al., 2002). The ability to secrete DG is not restricted to the fat body. The insect midgut also produces large amounts of DG that are secreted and transported by lipophorin to the fat body. The secretion and transport of DG are salient features of insects and represent a major difference with the mechanisms of FA mobilization and transport in vertebrates. The adipose tissue of vertebrates releases free fatty acids, FFA, which bind to albumin and are transported to other tissues through blood. Liver and intestine of vertebrates release TG into circulation, but this process requires the concomitant synthesis of apolipoprotein B and the intracellular assembly of lipoproteins. On the other hand, the insect midgut secretes DG, but does it without synthesizing a lipoprotein. The unique large production of DG that the fat body and midgut tissues of insects achieve is highly interesting. However, very little is known about the metabolic pathways and genes involved in the synthesis of glycerides in insects.
A major pathway for the synthesis of glycerides in animals is the monoacylglycerol pathway. In this pathway DG synthesis from MG and acyl-CoA is catalyzed by monoacylglycerol acyl transferase, MGAT (EC 2.3.1.22). Subsequent acylation of the DG in a reaction catalyzed by diacylglycerol transferase, DGAT (EC 2.3.1.20), produces TG. MGAT activity has been experimentally observed in fat body of several insects (Arrese et al., 1996; Hoffman and Downer, 1979; Peled and Tietz, 1974; Tietz et al., 1975). In vitro studies have shown that MGAT activity from M. sexta fat body produces sn-1,2-DG (Arrese et al., 1996). The production of DG is dependent on the activities of both MGAT, produces DG, and DGAT, consumes DG while producing TG. In spite of the importance of these genes to the production of DG and the mobilization of FA in insects very little research focusing on acyltransferases has been reported.
Using the genome sequence information of Manduca we were able to identify and clone two cDNAs that are homologues of the well characterized vertebrates' MGATs, and one cDNA from a transcript of M. sexta DGAT that is homologous to the vertebrate DGAT1. In this report we present a characterization and comparison of the acyltransferases gene products from M. sexta and the predicted acyltransferases from a number of insects whose genomes have been sequenced. A comparison of the expression levels of MGAT and DGAT-1 in larva and adult insects with the ability of the fat body to synthesize DG and TG provided insights into the regulation of DG and TG synthesis in M. sexta.
2. Methods
2.1. Insects
Manduca sexta eggs were purchased from Carolina Biological (Burlington, NC), and larvae were reared at 25°C on an artificial diet (Bell and Joachim, 1976). Adult insects were maintained at room temperature without food.
2.2. Cloning of Msex MGAT and MsexDGAT1
The sequence of scaffold00220 (GenBank JH668498) was used to design the primers to clone MsDGAT. The following primers, complementary to sequences found at the 5′-UTR and 3′-UTR, were used to obtain a full-length clone: MsDGAT-Fc: 5′-GCTGGCCGCAAAATTTTATGAAGTAAGC-3′ and MsDGAT-Rc: 5′-CGTTTCTGGAATCGTTGAATAATGAGAGG-3′. A product of 1610bp was expected.
Two sets of primers were used to clone Msex MGAT: one primer (MsMGAT-Rc) to target the 3′-UTR and two primers to target transcripts using alternative 5′UTRs (MsMGAT-352 and MsMGAT-346). The sequence information for primer design was obtained from the Manduca genome, scaffold00169 (GenBank JH668447.1). The sequences of the primers are: MsMGAT-Rc: 5′-GAGCCCCTAATCCAAAATGGTCG - 3′), MsMGAT-346: 5′-GTGACAAGCCAAGAAAATTAAATAGGCG-3′ and MsMGAT-352: 5′-GACAAAGATAGGGAGTTGCGTGC-3′. Products of 1157bp and 1173bp were expected for MsMGAT-352 and MSMGAT-346, respectively. Total RNA was isolated from the fat bodies of 5th instar larva and adult of M. sexta using Trizol reagent (Invitrogen). mRNA was reversed transcribed using oligod(T)18-primer. The resulting cDNA was used as template in PCR reactions using the primers indicated above. MsMGAT cDNA and MsDGAT were cloned from cDNA prepared from RNA isolated from adult and larval fat bodies, respectively. PCR products were cloned into the pGEM-T Easy Vector (Promega). For each case, at least two clones were sequenced in both directions. The cDNA sequences of both MsMGAT, and MsDGAT have been deposited in GenBank.
2.3. Quantitative PCR for analysis of DGAT and MGAT mRNA expression
Total RNA was extracted from a pool of atleast two dissected fat bodies, midguts, or ovaries using Trizol reagent (Invitrogen, CA). cDNA was synthesized from1.0μg of total RNA using qScript cDNA Supermix (Quanta) following manufacturer's instructions. Quantitative realtime PCR (qRT-PCR) was carried out using iTaq Universal SYBR Green SuperMix (Bio-Rad) and CFX Connect Real TimePCR Detection System (Bio-Rad) in 10μl reactions. The PCR reaction conditions were initial denaturation at 95°C for 1min, followed by 45 cycles alternating denaturation at 95°C for 2s and annealing/extension at 60°C for 45s. Expression of each transcript was normalized using the ribosomal protein S3 (rpS3, gi: 527679) as an internal control. Three independent sets of total RNA were analyzed in triplicate. Relative values are plotted as the mean ± SD.
The following primers were used to evaluate gene expression: MsMGAT2-F 5′- GACTACTTCCCTATCACACTCGTCA-3′;MsMGAT2-R: 5′-CAGGGAATAGTTTGGG-GAAGTTTGC-3′ (148bp product); MsDGAT1-F: 5′-CTTGTTGGCCCCTACTCTATGTTACGA-3′; MsDGAT1-R : 5′-GTCACTGAGGGTATCATCCATTGCTG-3′ (150bp product); RpS3 F 5′- TACAAACTCATTGGAGGTCTGGCCGT; RpS3-R: ACGAACTTCATGGACTTGGCTCTC. The levels of MGAT mRNA determined represent the sum of both isoforms (MGAT346 and MGAT352). The primers designed do not distinguish between the MGAT isoforms.
2.4. Properties of Predicted Proteins and Phylogeny
The number and location of transmembrane helices (TMH) was predicted using the tools from the Center for Biological Sequence Analysis, Technical University of Denmark (www.cbs.dtu.dk/services/TMHMM). Phosphorylation sites were located using the publicly available search programs: Scansite at http://scansite.mit.edu and NetPhos at http://www.cbs.dtu.dk/services/NetPhos/. Protein sequences were aligned using the Alignment Explorer/Muscle (Edgar, 2004) implemented in Molecular Evolutionary Genetics Analysis (MEGA version 5.10) (Tamura et al., 2011). The default presets for gap penalties and iterations were used for aligning the sequences.
2.5. Incorporation of oleic acid in acylglycerols
Radiolabel oleic acid ([9,10(n)-3H]oleic acid, 5 μCi/insect)was administered to larval and adult insects by feeding (5th instar, day 2) or injecting (adult, day 2) as previously described (Arrese and Wells, 1997). Hemolymph and fat body were collected 4h after administering the fatty acid and analyzed individually (n=4) to determine lipid composition, total radioactivity and the distribution of radioactivity among the lipid classes (Arrese and Wells, 1997). Data were expressed as percentage of total radioactivity.
3. Results
3.1. Msex DGAT-1 gene
The candidate gene for DGAT-1 (Msex2.08486, scaffold000220) was predicted on the basis of its similarity to the best characterized human (NP_034176.1) and mouse (NP_036211.2) DGAT1 protein sequences. Using fat body cDNA and primers to target the UTRs we were able to obtain a single full-length clone of 1583bp encoding a protein of 483 amino acid residues (KF800701 and Fig. 1 Appendix). The predicted MsexDGAT1 protein sequence is 44% identical (73% similarity) to the sequences of the characterized human and mouse DGAT1 proteins, Fig. 1. Because mouse and human DGAT1 are nearly identical, in Fig. 1 we only used the sequence of the human protein for comparison with MsexDGAT. The length of MsexDGAT (483aa) is similar to that of vertebrates' DGAT1 (488aa for human). Two conserved regions that have been mentioned as potential acyl-CoA and DG binding sites are also present in MsexDGAT (Fig. 1). These two conserved motifs are characterized by the sequences FYXDWWN, for acyl-CoA binding (Yen et al., 2008), and HXXXXRHXXXP, for DG binding (Xu et al., 2008). Moreover, as shown in the Fig. 2 MsexDGAT contains the same number, 9, and location of predicted transmembrane helices (TMH) as the vertebrates' DGAT1. The C-terminal region of MsexDGAT1 (255-483) contains five TMH that constitute the conserved MBOAT domain (pfam03062) found in a number of acyltransferases (Hofmann, 2000). The isoelectric point (9.34) and mass (57,152 Da) of MsexDGAT are in agreement with the values of vertebrates DGAT1 and also of other predicted insect DGAT1 (Table 1-Appendix).
Figure 1. Sequence alignment of Human DGAT1 (NP_036211.2) and MsexDGAT1 (KF800701).

Underlined are two regions that have been proposed as potential acyl-CoA and DG binding sites. These two conserved motifs are characterized by the sequences FYXDWWN, for acyl-CoA binding (Yen et al. 2008) and HXXXXRHXXXP, for DG binding (Xu J et al 2008).
Figure 2. Features of MsexDGAT1 gene and protein structure.
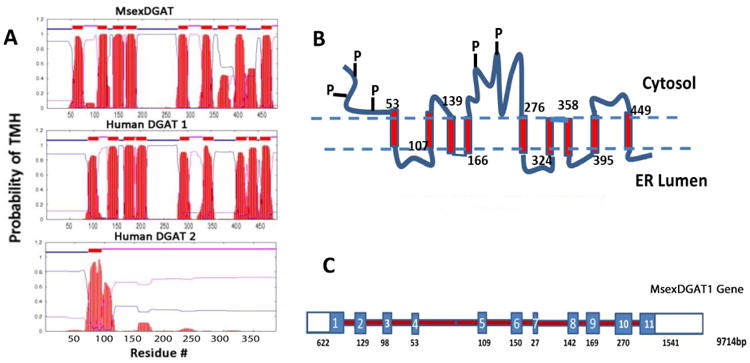
A) The number and location of transmembrane helices (TMH) was predicted using the TMHMM program (www.cbs.dtu.dk/services/TMHMM) from the Center for Biological Sequence Analysis. Sequence IDs: MsexDGAT1, KF800701; Human DGAT1, NP_036211.2; Human DGAT2, NP_115953.2. B) Structural sketch of MsexDGAT1 showing transmembrane helices, predicted regions exposed to cytosolic and lumen milieu and conserved sequence regions containing possible phosphorylation sites. Predicted TMH are indicated with rectangles and the numbers indicate the position of the first amino acid residue of each TMH. The N-terminal region (1-53) and the central region (189-275) contain conserved phosphorylation sites; C) Gene structure. The structure is based on the Manduca genome (Msex2.08486). The gene has 11 exons, indicated with rectangles, which altogether would encode a protein of 492 amino acid residues. The clone obtained and sequenced in this study encodes a protein of 483 residues that originates from a transcript that excludes the short exon 7. The total length of the gene and the lengths of the exons are indicated below the gene sketch. Introns are not drawn at scale.
A comparison of MsexDGAT1 with the predicted homologs from eight different insects (Fig. 2A, Appendix) shows that the protein is highly conserved (42.4% overall identity and 69% similarity). The conservation extends to the location and number of TMHs (not shown). As expected, if the comparison is limited to species of Lepidoptera (Bombyx mori, Danus plexipus, Heliconius melpomene and Manduca sexta) the similarity of the sequences increases to 89% (Fig. 2B, Appendix) and the identity to 78-82% (Table 1-Appendix).
Several consensus phosphorylation motifs are present in the N-terminal of MsexDGAT1. Some of these sites are 100% conserved among insect DGAT1 and some are also conserved in mouse and human DGAT1s (Fig. 3). The serine residues of MsexDGAT located at positions 17, RRAQSVTRA, 34, KVRRSQLDK, and 46, KPRDSLFSW, are probable phosphorylation sites clustering in the N-terminal of DGAT1, before the beginning of the transmembrane regions. The PKA phosphorylation motifs centered on serine residues 17 and 46 are also conserved in human and mouse DGAT1. Another pair of conserved consensus PKA phosphorylation sites, S187and S212, is found in the region that separates the two bundles of TMH present in DGAT1 (Fig. 2).
Figure 3. Conserved Consensus Phosphorylation Sites Among DGAT1.

Alignments were produced with Clustal Omega. The sequences identifiers and abbreviations used are: Hs: Homo sapiens, NP_036211.2; Mm: Mus musculus, NP_034176.1; Dp: Danus plexipus, EHJ74688.1; Hm: Heliconius melpomene, HMEL011652-PA; Msex: Manduca sexta; KF800701; Bm: Bombyx mori, XP_004927020.1; Tc: Tribolium castaneum; XP_975142.1; Dm: Drosophila melanogaster; NP_609813.1; Ag: Anopheles gambiae; XP_317656.3; Aea: Aedes aegypti, XP_001658299.1; Bt: Bombus terrestris XP_003400037.1. The indicated amino acid position numbers are based on the Msex sequence.
It is also important to note that some of the insect proteins referred to as DGAT1 in the present study have been annotated as sterol o-acyltransferases. This is the case for Danus plexipus, gi357623602, Aedes aegypti, gi157115823, and Tribolium castaneum, gi91083363. However, our study suggested that they are homologues of vertebrates' DGAT1.
Two DGAT genes are found in plants, vertebrates and fungi, DGAT1 and DGAT2 (Liu et al., 2012). These genes encode polypeptides that share a low degree of sequence homology and have a large difference in polypeptide length. MsexDGAT1 is clearly different from human DGAT2 (10% identity). Moreover, DGAT1 is a larger polypeptide (∼480-490aa) than DGAT2 (388aa). The structure of DGAT2 is also different. A simple prediction of TMH shows that DGAT2 has only 1 or 2 TMH, whereas DGAT1 is characterized by the presence of 9 TMH (Fig. 2). In fact, a search of the Manduca genome rendered no matches that could be confidently considered DGAT2 homologues.
The Manduca genome indicates that MsexDGAT1 gene has a length of ∼9714bps and consists of 11 coding exons evenly distributed (Fig. 2). If all exons were used it would lead to a protein of 492 amino acid residues. We were not able to clone the cDNA corresponding to a DGAT of such length, suggesting that it may not be expressed in fat body. The cDNA cloned, which encodes a 483 amino acid protein, excludes the use of exon 7.
The MBOAT domain, common to all DGAT1 proteins, is encoded by the last three exons (9-11) of the MsexDGAT gene. A very similar gene structure is found in other Lepidoptera (not shown) such as Heliconius melpomene (12 exons) and Bombyx mori (11 exons). The size of MsexDGAT1 gene (9714bp) is similar to the sizes of human (10,080bp) and murine (9543bps) DGAT1 genes (not shown). However, the number of exons in DGAT1 from vertebrates is higher (16-17) and they cluster towards the 3′end.
3.2. MGAT gene
Blast searches of nucleotide and predicted protein sequences from the Manduca genome, using as queries the sequences of the characterized vertebrates MGATs, indicated the presence of a single MGAT gene (Msex2.07183) in M. sexta. Two likely MGAT transcripts coding for proteins of 346 and 352 amino acids were predicted. The only difference between these two predicted proteins resides in the first nine N-terminal residues. The transcripts shared identical 3′-UTR sequences but differed at the 5′-ends (UTR and 1st coding exon). To confirm the occurrence of these two transcripts we designed primers targeting the 5′-UTRs of the two isoforms and a primer targeting the common 3′-UTRs. PCR of fat body cDNA showed that both isoforms were expressed. Both full length transcripts were cloned and sequenced (GenBank accession #: KF800699 and KF800700 for MsexMGAT_346 and 352, respectively). A comparison of the sequences obtained with the sequences of the well characterized human and mouse acyltransferases indicated that MsexMGATs are homologs of the vertebrates MGAT2. Fig. 4 shows an alignment of the deduced MsexMGAT protein sequences with those corresponding to human and mouse MGAT2. The MsexMGAT sequences were compared with several similar proteins from insects and vertebrates. As shown in the Fig. 3-Appendix and Table 2-Appendix, nearly all insect proteins homologous to MsexMGAT are more similar to human or mouse MGAT2 than to human or mouse DGAT2, MGAT1 or MGAT3. The single exception found was the MGAT from the ant Harpegnathos saltator that is more similar to the vertebrates MGAT1 (Table 2-Appendix). Prediction of the TMH of MsexMGAT also shows similarities with human MGAT2 and some differences with human DGAT2 (Fig. 5). In addition to the similarity in size, sequence and TMH regions, the predicted insect MGATs and the human and mouse MGAT1 and MGAT2 share a common PKA phosphorylation site near the N-terminal. This predicted phosphorylation site was marked in Fig.4 and in Fig 3-Appendix. There are no reports on the possible role of this site in the regulation of MGAT activity in vertebrates. The fact that it is also conserved in insects further suggests that it could play a role in the regulation of the synthesis of glycerides. This putative phosphorylation site is not found in vertebrate DGAT2. As it was the case for DGAT1, it is important to note that some of the insect MGAT like proteins in Table 2-Appendix were annotated as DGAT or DGAT2. This is the case for at least EHJ64327.1 (Danus plexipus) and XP 001653515.1 (Aedes aegypti). Clearly these proteins are more similar to MGATs than to DGAT2. Human and mouse DGAT2 are distinguished from all MGATs by their longer non-matching N-terminal sequences.
Figure 4. Protein sequence alignment of Human and mouse MGAT2 and MsexMGATs.
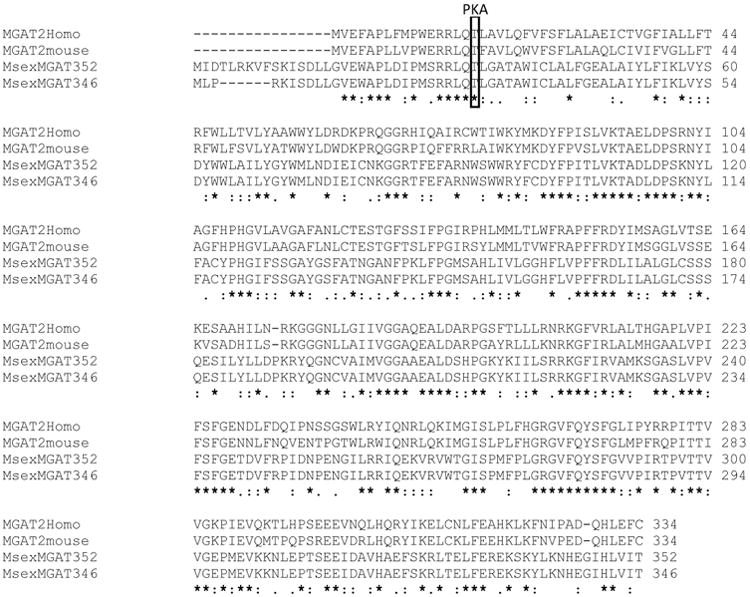
Alignments were produced with Clustal Omega. Sequence IDs: Human MGAT2: NP_036211.2; Mouse MGAT2: Q80W94, MsexMGAT 352: KF800700, MsexMGAT 346: KF800699. The marked threonine residue, represents a predicted PKA phosphorylation site that is conserved in both MGAT1 and MGAT2 of vertebrates and in the predicted insect MGATs (see also Fig. 3-Appendix).
Figure 5. Features of MsexMGAT gene and protein structure.
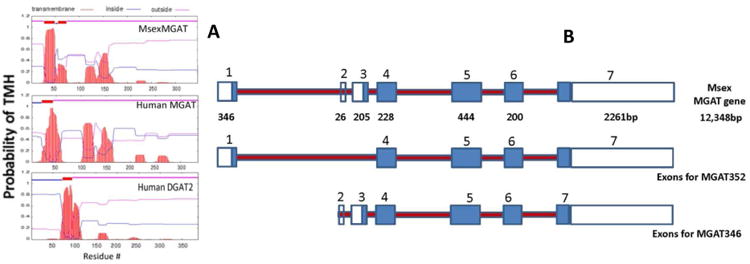
A) The number and location of transmembrane helices (TMH) was predicted using the TMHMM program, www.cbs.dtu.dk/services/TMHMM, from the Center for Biological Sequence Analysis. Sequence IDs: MsexMGAT KF800700, Human MGAT2 NP_036211.2 and Human DGAT2 NP_115953.2; B) Msex MGAT gene structure and predicted exon usage for MsexMGAT346 and MsexMGAT352. Sketch based on the Manduca genome (Msex2.07183).
The two isoforms of MsexMGAT cloned appear to derive from a single gene. The structure of the gene indicates the presence of seven exons. The shorter form, MGAT346, is the product of exons 2-7, whereas MsexMGAT352 is the splicing product containing exons 1 and 4-7 (Fig. 5).
3.3. Phylogeny of insect DGAT1 and MGAT proteins
Previous phylogenetic studies of acyltransferases have shown that DGAT1 and DGAT2 share little sequence homology and actually fall into different phylogenetic families. DGAT1 belongs to the family of MBOAT proteins, whereas DGAT2 and MGATs cluster in a group named the DGAT2 family (Cases et al., 2001; Lardizabal et al., 2001). In this report we have extended the phylogenetic study of neutral acyltranserases by analyzing the recently available DGAT and MGAT protein sequences deduced for M. sexta and homologous proteins from several other insect species. A phylogenetic tree comparing insect MGAT and DGAT1 proteins and their relationships with the well characterized human and murine proteins is shown in the Fig. 6. As expected from previous studies (Cases et al., 2001; Lardizabal et al., 2001), human MGAT and DGAT2 sequences grouped together into the so called human DGAT2 family of acyltransferases. The DGAT2 family included all predicted insect MGAT proteins. Sub-clades distinguishing MGATs from different insect orders are clearly distinguished at least among Lepidoptera, Hymenoptera and Diptera. Dipteran MGATs, however, show divergence between flies and mosquitoes. On the other hand, all predicted insect DGAT1 proteins grouped together with vertebrates DGAT1 in a clade (MBOAT family) clearly separated from the DGAT2 family. This is consistent with the notion that MBOAT and DGAT2 families originated from different prokaryote ancestors (Turchetto-Zolet et al., 2011). Moreover, as observed for insect MGATs, the phylogenetic tree of DGAT1 also shows a clear separation of sub-clades according to the insect orders.
Figure 6. Phylogeny of human, mouse and insect MGAT and DGAT proteins.
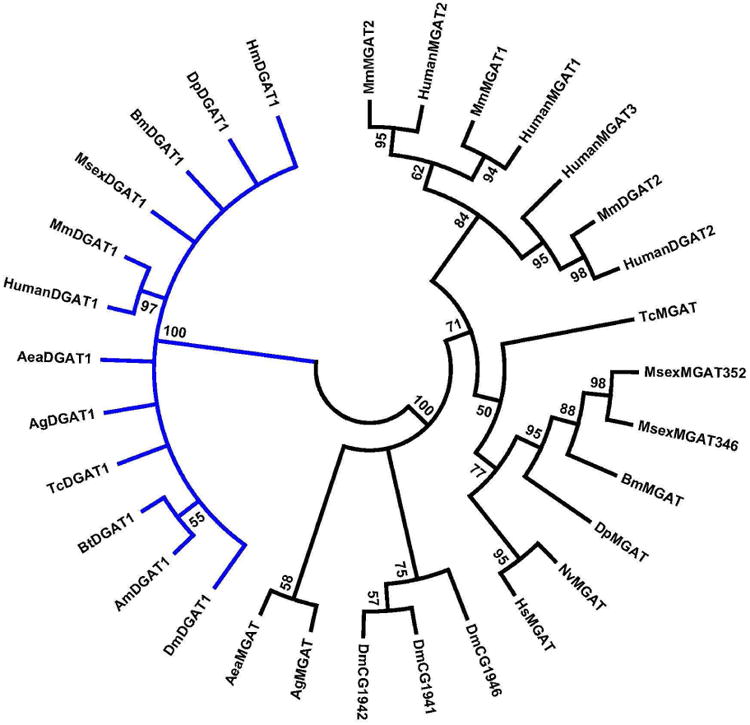
Alignments were produced with Clustal Omega and the phylogeny was inferred using the Neighbor-Joining method (Saitou and Nei, 1987). The bootstrap consensus tree inferred from 500 replicates is taken to represent the evolutionary history of the taxa analyzed (Felsenstein, 1985). Branches corresponding to partitions reproduced in less than 50% bootstrap replicates were collapsed. The evolutionary distances were computed using the JTT matrix-based method (Jones et al., 1992) and are in the units of the number of amino acid substitutions per site. The analysis involved 31 amino acid sequences. All positions containing gaps and missing data were eliminated. There were a total of 284 positions in the final dataset. Evolutionary analyses were conducted in MEGA5 (Tamura et al., 2011).
The sequence identifiers and abbreviations used for DGAT1 are: HumanDGAT1: Homo sapiens, NP_036211.2; MmDGAT1: Mus musculus, NP_034176.1; DpDGAT1: Danus plexipus; EHJ74688.1; HmDGAT1: Heliconius Melpomene, HMEL011652; MsexDGAT1: Manduca sexta; KF800701; BmDGAT1: Bombyx mori; XP_004927020.1; TcDG1: Tribolium castaneum; XP_975142.1; DmDGAT1: Drosophila melanogaster, NP_609813.1; AgDGAT1: Anopheles gambiae, XP_317656.3; AeaDG1: Aedes aegypti, XP_001658299.1; BtDGAT1: Bombus terrestris, XP_003400037.1; AmDGAT1: Apis mellifera XP_624754.2
Sequence identifiers for DGAT2 proteins: HumanDGAT2: Homo sapiens DGAT2 Isoform 1, Q96PD7; MmDGAT2: Mus musculus DGAT2, NP_080660.1.
Sequence identifiers for MGAT proteins: MsexMGAT352: Manduca sexta Predicted MGAT2 (352aa) KF800700; MsexMGAT346: Manduca sexta Predicted MGAT2 (346aa) KF800699; BmMGAT: Bombyx mori XP_004922894.1; DpMGAT: Danaus plexippus EHJ64327.1; TcMGAT: Tribolium castaneum, EEZ98240.1; NvMGAT: Nasonia vitripennis XP_001602288.1; HsMGAT: Harpegnathos saltator, EFN85928.1; HumanMGAT1: Homo sapiens MGAT1 NP_477513.2; MmMGAT1: Mus musculus MGAT1, NP_080989.2; MmMGAT2: Mus musculus MGAT2, Q80W94; HumanMGAT2: Homo sapiens MGAT2, NP_079374.2; HumanMGAT3: Homo sapiens MGAT3, NP_835470; DmCG1941: Drosophila melanogaster DGAT; DmCG1942: Drosophila melanogaster DGAT; DmCG1946: NP_610319; AgMGAT: Anopheles gambiae EDO63767.1; AeaMGAT: Aedes aegypti AAEL008878-PA.
3.4. Expression of MsexMGAT and MsexDGAT
The expression of MsexMGAT and MsexDGAT1 was 5th-instar larva, in ovaries, and in the fat bodies of feeding 5th-instar larva, wandering larva (day 1 and day 3) and adults (3rd day female). The relative levels of mRNA, as determined by qPCR, are shown in the Fig. 7. The estimated transcript levels indicate that MsexDGAT1 is highly expressed during the voracious feeding days of the last larval period and decreases ∼12-fold at the non-feeding wandering stages and between ∼3 and 7-fold in adult stages. The higher levels of DGAT1 in feeding larvae are consistent with the fact that the insect is synthesizing and storing large amounts of TG in fat body. Even though DGAT expression levels are lower in fat bodies of non-feeding states, the expression of DGAT mRNA is still significant suggesting that DGAT1 is needed even when there is no net TG synthesis. It must be noted that recycling of TG through deacylation and re-acylation reactions, an apparently futile cycle, is an active process common to the adipose tissue of vertebrates and the fat body of insects. A simple evidence of the occurrence of this process is given by the fast turnover rates of circulating non-esterified fatty acids (NEFA) in both vertebrates (Fredrickson and Gordon, 1958) and insects (Soulages et al., 1988; Soulages and Wells, 1994). Most of the NEFA, which originate from the partial or complete hydrolysis of glycerides, are not consumed. Conversely, a large fraction of the fatty acids is rapidly re-esterified before or after leaving the cells and return to the glyceride pool. This cyclic process requires, among other enzyme activities, the activity of DGAT to allow the synthesis of TG and prevent an excessive accumulation of DG and NEFA.
Figure 7. Expression of MGAT and DGAT1 mRNA.
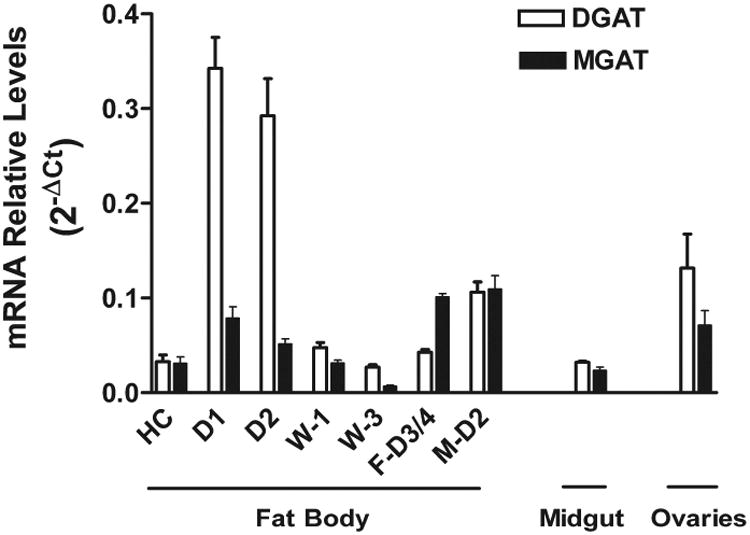
The relative levels of mRNA were determined by qPCR using as reference the ribosomal protein S3 mRNA. The levels of MGAT mRNA represent the sum of both isoforms cloned in this study. HC: headcap (4th-5th instar); D1 and D2: 5th instar feeding larva day 1 and day 2; W1 and W3: pre-pupa wandering day 1 and day 3; F-D3/4: Adult Female 3-4 days old; M-D2: Adult male day 2.
Data shown represent the mean of three biological replicates and each biological sample was analyzed in triplicates. The error bars represent the combined instrumental and biological standard deviations. Error bars= 2-(ΔCt +/- SD).
Midgut expresses DGAT1 at levels comparable to those of the fat body in non-feeding stages. Storage of TG in midgut, although to a lower extent than fat body, has been observed in some insects, including Manduca sexta (Canavoso and Wells, 2000). Also interesting are the relatively high levels of DGAT mRNA observed in ovaries. These levels are in agreement with the well-known fact that developing ovaries store large amounts of imported FA in the form of TG to support embryo development. Most FAs are imported in the form of DG from circulating lipophorin (Kawooya and Law, 1988; Kawooya et al., 1988). Therefore, a significant DGAT activity must be needed to convert DG into TG.
The levels of MGAT-mRNA in larval fat body and ovaries, organs that are accumulating TG, are lower than those of DGAT1. However, the MGAT levels are very similar to the DGAT levels in larval midgut, which is secreting DG, and even higher than the DGAT-mRNA level in the adult stage fat body, which is secreting large amounts of DG into the hemolymph (Fig. 7). As seen with DGAT1-mRNA levels, MGAT expression decreases significantly as the feeding larva goes through the prepupal stages (W1 and W3). However, conversely to the case of DGAT1, the levels of MsexMGAT are higher in adult insects than in the feeding 5th instar D1 and D2. This increase in MGAT and the concomitant decrease in DGAT are consistent with the fact that the fat body of adult insects is releasing DG into circulation. The increase in MGAT levels would allow a faster production of DG, whereas the 3 to 7 fold decrease in DGAT1 would slow down the rate of conversion of DG into TG favoring the secretion of DG.
The levels of MGAT and DGAT expression are expected to increase when the rates of DG and TG synthesis increase due to a net influx of FA, as occurs when the insect is feeding, or when the turnover rate of glycerides increases, for instance when remodeling of glycerides and/or reallocation of FA increases. The fact that the expression of both acyltransferases is low in the pre-pupal stages suggests low overall rates of TG and DG synthesis.
3.5. Rates of DG and TG synthesis
To study a possible correlation between the expression of MGAT and DGAT and the rates of DG and TG synthesis, we studied the relative rates of incorporation of FAs into DG and TG in fat bodies of M. sexta and compared them to the expression levels of MGAT and DGAT mRNA. To obtain a rough estimate of the rates of FA incorporation into DG and TG, radiolabeled FA were injected into the hemocele of feeding 5th instar, day 2, larva and 3-day old adults and 4h later analyzed the distribution of radiolabel among hemolymph and fat body lipids. As shown in the Table 1, most of the radiolabeled oleic acid is found in the fat body (76%) of the larva and 90% of it is associated with TG. Conversely, in adult insects the radiolabeled oleic acid is mostly found in the hemolymph (65%) and 90% of it is associated with the DG fraction. The distribution of radioactivity among fat body lipids (Table 1) also shows that a larger proportion of the FA end up associated with DG in adult insects (19%) than in larva (6%). Overall, in feeding larva the accumulation of FA in TG (68.7%) is 2.6 fold greater than in DG (26%). On the other hand, in adult insects the situation is reversed and the fraction of FA used for DG synthesis (64.9%) is 2.5-fold greater than that used for TG synthesis (25.8%). Thus, the overall relative use of FA for the synthesis of DG shows a 7-fold increase between larva and adult insects. This change suggests that the decrease in DGAT expression in adults limits the synthesis of TG allowing a greater production of DG. Fig. 8 shows that paralleling the 7-fold increase in the use of FA for DG synthesis there is a 14-fold increase in the ratio of MGAT to DGAT mRNA expression levels. This correlation suggests that the changes in the relative expression and MGAT and DGAT would play a major role defining the metabolic switch of the fat body from an organ that stores TG (in larvae) to an organ that produces and secretes DG in adult insects.
Table 1. Differences in the rates of incorporation of oleic acid into DG and TG between larval and adult stages of M. sexta.
The distribution of radiolabeled oleic acid between the glycerolipids of fat body and hemolymph was determined 4h after injecting the insects with [3H]-oleic acid. Data are expressed as percent of total radioactivity recovered in hemolymph or fat body +/- SD (n= 4 insects). Other lipids include radioactivity found in phospholipids, MG and non-esterified fatty acids.
| % of Total dpm | TG (dpm %) | DG (dpm%) | Other Lipids (dpm %) | ||
|---|---|---|---|---|---|
| Larva (5th Instar, day 2) | Hemolymph | 23.5 ± 2.5 | 1.1 ± 0.8 | 21.3 ± 0.9 | 1.1 ± 0.2 |
| Fat Body | 76.5 ± 2.5 | 67.6 ± 2.0 | 4.7 ± 0.7 | 5.5 ± 1.5 | |
| Adult (day 2) | Hemolymph | 65% ± 5.8 | 0.4 ± 0.3 | 58.2 ± 4.3 | 6.2 ± 1.2 |
| Fat Body | 35% ± 5.8 | 25.4 ± 3.3 | 6.7 ± 0.9 | 3.2 ± 0.5 | |
Figure 8. Use of Fatty Acids for the Synthesis of DG and TG; Correlation with the Expression Levels of MGAT and DGAT.
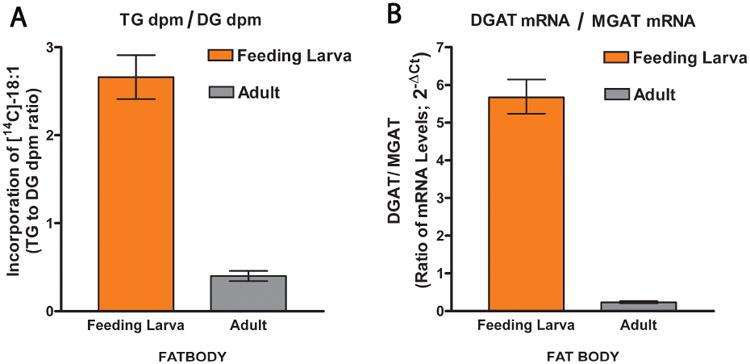
A) The ratio of radioactivity incorporated into TG and DG was determined in 5th instar feeding larvae (1st day) and in adult insects (3-day females) previously injected with radiolabeled oleic acid. The radiolabelled FA incorporated in DG includes radiolabeled DG found in both hemolymph and fat body lipids; the same applies to radiolabeled TG. Data are the mean of four biological replicates ± SD. B) The DGAT/MGAT mRNA levels ratio was determined by qPCR. Data represent the mean of three biological replicates (each of them determined in triplicate) and the error bars are 2±SD.
4. Discussion
4.1. Do insects have a DGAT2 gene?
The most common reaction for the synthesis of TG is catalyzed by DGATs using acyl-CoA and DG as substrates. DG can be used for export into the hemolymph or for the synthesis of phospholipids. Thus, the activity of DGAT is important because to a great extent its relative activity defines whether or not the tissue stores FA as TG or, in other words, the fate of DG. Two DGATs, DGAT1 and DGAT2, are found in vertebrates (Liu et al., 2012). These two enzymes catalyze the same reaction but have evolved from different ancestral precursors, and have different catalytic properties and physiological functions (Cheng et al., 2008; Yen et al., 2005).
Our study suggests that insects lack a homologue of the vertebrates DGAT2. Searches for homologues of human or murine DGAT2 in insects did not provide a convincing match. The predicted insect protein hits from blast searches using DGAT2 as query were actually more similar to MGAT2 than DGAT2 and were missing the N-terminal sequence characteristic of DGAT2. Given the fact that transgenic DGAT2-knockout mice die soon after birth (Stone et al., 2004), the apparent lack of DGAT2 in insects is somehow surprising. However, the precise reason why the DGAT2 knockout is lethal to mice is not known. A recent study has shown that adipocytes from DGAT2 knockout mice still accumulate TG to the same extent as wild type adipocytes (Harris et al., 2011); therefore, the study suggests that at least in adipose tissue DGAT1 could substitute for the lack of DGAT2. If both DGAT proteins are knocked out then the adipocytes have no TG indicating that none of the other acyltransferases present in adipocytes can substitute for the DGAT activity. Although further studies are required to confirm the absence of a second DGAT in insects, the current information suggests that DGAT1 would be the only DGAT gene present in insects.
4.2. MsexDGAT1 characterization
The comparison of MsexDGAT1 with predicted acyltransferases from a number of insects provided a broad characterization of insect DGAT1. Several of the predicted phosphorylation sites identified were found to be conserved among insect DGAT1, and also between insect and vertebrate DGAT1. Several of these sites are located in the N-terminal region. Interestingly, deletion of the N-terminal of the mouse DGAT1 renders the protein more active (McFie et al., 2010) suggesting that phosphorylation in this protein region could play an important regulatory role in the metabolism of TG. For instance, the adipokinetic hormone, AKH, is known to activate PKA and promote activation of lipolysis (Arrese et al., 1999; Arrese et al., 2001). These events lead to the accumulation of DG in fat body and also a massive secretion of DG into the hemolymph (Arrese et al., 2001). The production of DG induced by AKH could be partly facilitated by inhibition of DGAT1 upon PKA mediated phosphorylation. Clearly, further studies on the possible role of the predicted phosphorylation sites in the function of DGAT1 are likely to contribute to the understanding of the metabolism of TG in insects. The high conservation of DGAT1 between vertebrates and insects suggests that several properties of this enzyme could also be conserved. Human DGAT1 has also been shown to have MGAT activity and catalyze the synthesis of waxes, and retinyl esters (Yen et al., 2005). This could also be the case of DGAT1 from insects. The synthesis of waxes is very important to prevent dehydration by lowering the water permeability of the cuticle and eggs (Lockey, 1988). DGAT1-knockout mice are viable and fertile even though their phenotype shows major changes in lipid metabolism and energy homeostasis (Smith et al., 2000). Thus, among other changes, mice lacking DGAT1 are leaner, more sensitive to insulin, and have dry fur apparently due to their deficiency in the synthesis of waxes (Smith et al., 2000). Relevant studies on insect DGATs are limited to one report on an ethyl methanesulfonate generated Drosophila mutant. Female flies carrying mutations in the midway gene are sterile. The midway gene was shown to encode a DGAT1 and play a major role in the accumulation of TG in ovaries (Buszczak et al., 2002). Mutations in the midway gene decrease the accumulation of TG in nurse cells and this has been suggested as the cause of early nurse cell degeneration and sterility (Buszczak et al., 2002).
This study shows that DGAT1 is expressed in the three major M. sexta organs that are known to store TG, such as ovaries (Ziegler and Van Antwerpen, 2006), midgut (Canavoso and Wells, 2000) and fat body (Arrese and Soulages, 2010). We have not studied other tissues, yet. However, DGAT1 is likely to be broadly expressed. The synthesis of pheromones in moths takes place in the pheromone gland (PG) using FAs as precursors; i.e. Bombykol is made from palmitic acid. DGAT1 should be highly expressed in the pheromone producing cells of female moths before maturity. At that time the PG shows an increase in lipid droplets due to the accumulation of TG (Fonagy et al., 2001). In this regard, a recent study has shown that the expression of a “DGAT2” transcript in PG of Bombyx mori increases during the preparation for mating (Du et al., 2012). Our study suggests that the “BmDGAT2” gene actually encodes an MGAT-2 like protein. Therefore, the expression of both MGAT and DGAT 1 would be needed for the synthesis of TG in PG.
The developmental changes in the expression of MsexDGAT1 mRNA observed in this study suggest that the expression of DGAT1 in fat body is regulated according to the physiological needs of the insect. This was exemplified by the ∼7-fold higher fat body levels of DGAT1 mRNA in feeding larvae, which is accumulating TG, than in adult insects, which are mobilizing and burning fat (Fig. 7). These changes suggest that DGAT1 could be a rate limiting enzyme in the synthesis of fat body TG. Regulation of the expression of DGAT1 is important because this enzyme catalyzes a metabolic step centered at the branch point where the metabolic fate of DG is at least temporarily defined. Conversion of DG to TG for storage reduces the availability of DG for phospholipid synthesis and for export to the hemolymph.
4.3. MsexMGATs
The two MsexMGATs cloned and all predicted insect MGAT proteins, with the exception of the Harpegnathos saltator MGAT, were more similar to the vertebrates' MGAT2 than to MGAT1 and MGAT3. The MGAT3 gene is found only in higher mammals and humans but not in rodents (Cheng et al., 2003) and, therefore, its absence in insects is not surprising. The higher similarity with MGAT2 may be related to the kinetic properties of the enzyme. MGAT2 is expressed in most tissues in vertebrates, but it is highly abundant in intestine (Cao et al., 2003), where the MG-pathway is prevalent. Therefore, it is possible that the similarity of insect MGATs with the vertebrates MGAT2 be due to the significance of the MG-pathway in the metabolism of neutral glycerides.
In contrast to MsexDGAT, the expression of MsexMGAT is up-regulated and shows its highest levels in the adult stage. The changes in MGAT expression between the feeding larval and adult stages, a 2-fold change, are not as pronounced as observed for DGAT. However, the combined regulation of the expression of MGAT and DGAT leads to major changes between the ratios of the expression levels of DGAT1/MGAT. Since up-regulation of MGAT is expected to increase the synthesis of DG whereas down-regulation of DGAT is expected to decrease the rate of TG synthesis, these simultaneous changes should lead to an increase in the net production and accumulation of DG in adult insects (TG in larvae). The fact that this is exactly what is observed, as shown by the incorporation of FA in glycerides (Fig. 8) suggests that MsexMGAT and DGAT are key enzymes defining whether the fat body stores TG or release DG into circulation.
Research Highlights.
Monoacylglycerol and diacylglycerol acyltransferases catalyze the synthesis of DG from MG and TG from DG, respectively.
In contrast to vertebrates, which have DGAT1 and DGAT2 genes, insects seem to have a DGAT1 gene only.
A higher expression of DGAT1 in fat body is observed when Manduca sexta is accumulating TG.
Relative expression levels of MGAT and DGAT appear to define whether the fat body secretes DG or stores TG
Acknowledgments
This research was supported by Oklahoma Agricultural Experiment Station (OKL02398), Oklahoma State University and by Grant Number R01GM064677 from the National Institute of General Medical Sciences. The content is solely the responsibility of the authors and does not necessarily represent the official use of the National Institute of General Medical Sciences or the National Institutes of Health.
Appendix
Figure 1. DNA and protein sequence of MsexDGAT1.

The figure shows the DNA and predicted ORF amino acid sequence of the Msex DGAT1 cloned in this study. Start and stop codons are highlighted.
Figure 2.


A) Multiple sequence alignment of insect proteins similar to Manduca DGAT1. The sequences identifiers and abbreviations used are: Msex483: Manduca sexta DGAT1 KF800701; Bm: Bombyx mori; XP_004927020.1, DGAT1-like isoform X1; Dp: Danaus plexipus; EHJ74688.1, sterol o-acyltransferase; Heliconius: Heliconius melpomene HMEL011652; Dm: Drosophila melanogaster, midway, isoform A, NP_609813.1; Ag: Anopheles gambiae, XP_317656.3; Aea: Aedes aegypti XP_001658299.1, sterol o-acyltransferase; Tc: Tribolium castaneum sterol o-acyltransferase, XP_975142.1; Bt: Bombus terrestris DGAT1-like, XP_003400037.1.
B) Multiple sequence alignment DGAT1 from Lepidoptera. Msex483: Manduca sexta DGAT1 KF800701; Bm: Bombyx mori; XP_004927020.1, Pred. DGAT1-like isoform X1; Dp: Danaus plexipus, EHJ74688.1, Pred.sterol o-acyltransferase; Heliconius: Heliconius melpomene HMEL011652.
Figure 3. Protein sequence alignment of vertebrate MGAT and DGAT2 and predicted insect MGATs.
Sequence identifiers for DGAT2 proteins: HumanDGAT2: Q96PD7; MmDGAT2: Mus musculus DGAT2, NP_080660.1.
Sequence identifiers for MGAT proteins: HumanMGAT1: Homo sapiens MGAT1 NP_477513.2; MmMGAT1: Mus musculus MGAT1, NP_080989.2; MmMGAT2: Mus musculus MGAT2, Q80W94; HumanMGAT2: Homo sapiens MGAT2, NP_079374.2; DpMGAT: Danaus plexippus EHJ64327.1; ; MsexMG352: Manduca sexta MGAT2 (352aa) KF800700; MsexMG346: Manduca sexta MGAT2 (346aa) KF800699; BmMGAT: Bombyx mori XP_004922894.1 Dm: Drosophila melanogaster CG1941; AgMGAT: Anopheles gambiae EDO63767.1; AeaMGAT: Aedes aegypti AAEL008878-PA; NvMGAT: Nasonia vitripennis XP_001602288.1; HsMGAT: Harpegnathos saltator, EFN85928.1.
The rectangle encloses a conserved threonine residue that is a probable PKA phosphorylation site. It is only conserved among MGAT proteins.
Table 1. Identity Matrix, Masses and Isoelectric points of DGAT1.
Cells showing the % identity among Lepidoptera DGAT1 proteins are highlighted. Protein sequence identifiers: Human: Homo sapiens DGAT1, NP_036211.2; Mouse: Mus musculus, NP_034176.1; Dp: Danaus plexipus; EHJ74688.1, sterol o-acyltransferase; Hm: Heliconius melpomene gnl|BL_ORD_ID|7464 HMEL011652; Msex: Manduca sexta DGAT1 KF800701; Bm: Bombyx mori; gi512909843, XP_004927020.1, DGAT1-like isoform X1; Tc: Tribolium castaneum; XP_975142.1, sterol o-acyltransferase; Dm: Drosophila melanogaster;gi19921444, NP_609813.1 midway, isoform A; Bt: Bombus terrestris XP_003400037.1, DGAT1-like; Ag: Anopheles gambiae; gi 158297422, XP_317656.3, AGAP007843-PA; Aea: Aedes aegypti XP_001658299.1, sterol o-acyltransferase.
| Human | Mouse | Dp | Hm | Msex | Bm | Tc | Dm | Bt | Ag | Aae | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Human | 100 | 86 | 44 | 46 | 46 | 47 | 45 | 44 | 45 | 46 | 47 |
| Mouse | 86 | 100 | 43 | 45 | 46 | 46 | 45 | 44 | 45 | 46 | 46 |
| Dp | 44 | 43 | 100 | 82 | 80 | 77 | 61 | 63 | 62 | 64 | 67 |
| Hm | 46 | 45 | 82 | 100 | 78 | 79 | 61 | 60 | 61 | 63 | 63 |
| Msex | 46 | 46 | 80 | 78 | 100 | 82 | 60 | 62 | 61 | 63 | 64 |
| Bm | 47 | 46 | 77 | 79 | 82 | 100 | 58 | 60 | 57 | 61 | 63 |
| Tc | 45 | 45 | 61 | 61 | 60 | 58 | 100 | 63 | 68 | 65 | 69 |
| Dm | 44 | 44 | 63 | 60 | 62 | 60 | 63 | 100 | 65 | 68 | 70 |
| Bt | 45 | 45 | 62 | 61 | 61 | 57 | 68 | 65 | 100 | 70 | 71 |
| Ag | 46 | 46 | 64 | 63 | 63 | 61 | 65 | 68 | 70 | 100 | 85 |
| Aae | 47 | 46 | 67 | 63 | 64 | 63 | 69 | 70 | 71 | 85 | 100 |
| Mass (kDa) | 55.3 | 56.8 | 55.2 | 57.1 | 57.1 | 56.7 | 56 | 54.7 | 56.7 | 54.9 | 55.3 |
| Isoelect. Pt. | 9.41 | 9.41 | 9.53 | 9.19 | 9.34 | 9.46 | 9.29 | 9.26 | 9.47 | 9.39 | 9.71 |
Table 2. Identity matrix of insect, human and mouse proteins from the DGAT2 family.
The cells corresponding to the insect/vertebrate pairs of proteins that have the highest % identity have been highlighted. The diagonal was highlighted for clarity, only. MsexMGAT346: KF800699; MsexMGAT352: KF800700; Bm: Bombyx mori MGAT2-like XP_004922894.1; BmDGAT2: Bombyx mori bgibmga008049-ta; Dp: Danus plexipus, EHJ64327.1; Nv: Nasonia vitripennis MGAT2-like, XP_001602288.1; Hs: Harpegnathos saltator MGAT1, EFN85928.1; Tc: Tribolium castaneum MGAT2, EEZ98240.1; Ag: Anopheles gambiae, EDO63767.1; Aae: Aedes aegypti, EAT39311.1; Dm: Drosophila melanogaster CG1941, CG1942 and CG1946; Human MGAT1: NP_477513.2; Mouse MGAT1: NP_080989.2; Human MGAT2: NP_079374.2; Mouse MGAT2: Q80W94;Human DGAT2: Homo sapiens DGAT2 Isoform 1, NP_115953; Mouse DGAT2: NP_080660.1; Human MGAT3: NP_835470.
| Msex MGAT346 | Msex MGAT352 | Bm | *BmDGAT2 | *Dp | Nv | Hs | Te | Ag | *Aae | *DmCG1941 | *DmCG1942 | *DmCG1946 | Human MGAT1 | Mouse MGAT1 | Human MGAT2 | Mouse MGAT2 | Human DGAT2 | Mouse DGAT2 | Human MGAT3 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Msex MGAT346 | 100 | 99.1 | 78.3 | 78.6 | 71.6 | 49.7 | 46.2 | 43.5 | 43.5 | 42.9 | 36.8 | 39.3 | 37.4 | 41.8 | 41.8 | 44.6 | 44.3 | 36.6 | 36.6 | 37.6 |
| Msex MGAT352 | 99.1 | 100 | 76.9 | 76.7 | 71.8 | 49.7 | 46.2 | 43.5 | 43.5 | 42.9 | 36.8 | 39.3 | 37.4 | 41.8 | 41.8 | 44.6 | 44.3 | 35.7 | 35.7 | 37.6 |
| Bm | 78.3 | 76.9 | 100 | 98.0 | 70.6 | 50.6 | 46.8 | 44.6 | 46.8 | 45.6 | 39.2 | 41.1 | 38.9 | 41.5 | 42.4 | 46.7 | 45.2 | 37.4 | 37.7 | 40.3 |
| BmDGAT2 | 78.6 | 76.7 | 98.0 | 100 | 70.4 | 50.6 | 46.8 | 44.6 | 46.8 | 45.6 | 39.2 | 41.1 | 38.9 | 41.5 | 42.4 | 46.7 | 45.2 | 37.0 | 37.3 | 40.3 |
| Dp | 71.6 | 71.8 | 70.6 | 70.4 | 100 | 50.0 | 47.9 | 44.1 | 47.0 | 45.9 | 37.4 | 40.2 | 40.1 | 39.1 | 39.7 | 44.0 | 44.3 | 35.5 | 34.7 | 37.9 |
| Nv | 49.7 | 49.7 | 50.6 | 50.6 | 50.0 | 100 | 57.2 | 45.2 | 43.6 | 47.3 | 38.6 | 39.0 | 38.1 | 40.6 | 41.2 | 41.9 | 43.1 | 39.1 | 39.6 | 37.0 |
| Hs | 46.2 | 46.2 | 46.8 | 46.8 | 47.9 | 57.2 | 100 | 47.6 | 41.6 | 41.9 | 34.1 | 36.6 | 36.5 | 42.4 | 40.3 | 41.0 | 40.4 | 38.8 | 39.1 | 36.4 |
| Tc | 43.5 | 43.5 | 44.6 | 44.6 | 44.1 | 45.2 | 47.6 | 100 | 41.9 | 41.9 | 35.1 | 38.9 | 37.9 | 40.4 | 40.1 | 47.1 | 47.1 | 37.9 | 37.9 | 37.4 |
| Ag | 43.5 | 43.5 | 46.8 | 46.8 | 47.0 | 43.6 | 41.6 | 41.9 | 100 | 67.1 | 42.6 | 42.2 | 41.8 | 39.7 | 37.6 | 42.2 | 40.7 | 35.1 | 35.1 | 34.4 |
| Aae | 42.9 | 42.9 | 45.6 | 45.6 | 45.9 | 47.3 | 41.9 | 41.9 | 67.1 | 100 | 42.9 | 43.3 | 41.8 | 38.5 | 37.6 | 42.2 | 42.5 | 34.8 | 34.8 | 35.3 |
| Dm CG1941 | 36.8 | 36.8 | 39.2 | 39.2 | 37.4 | 38.6 | 34.1 | 35.1 | 42.6 | 42.9 | 100 | 72.9 | 67.1 | 34.0 | 35.2 | 36.8 | 38.0 | 33.7 | 34.0 | 36.0 |
| Dm CG1942 | 39.3 | 39.3 | 41.1 | 41.1 | 40.2 | 39.0 | 36.6 | 38.9 | 42.2 | 43.3 | 72.9 | 100 | 67.8 | 35.6 | 36.8 | 39.0 | 39.0 | 34.4 | 34.4 | 38.0 |
| Dm CG1946 | 37.4 | 37.4 | 38.9 | 38.9 | 40.1 | 38.1 | 36.5 | 37.9 | 41.8 | 41.8 | 67.1 | 67.8 | 100 | 34.6 | 36.8 | 38.1 | 38.7 | 32.5 | 32.5 | 36.1 |
| Human MGAT1 | 41.8 | 41.8 | 41.5 | 41.5 | 39.1 | 40.6 | 42.4 | 40.4 | 39.7 | 38.5 | 34.0 | 35.6 | 34.6 | 100 | 72.5 | 53.0 | 52.1 | 48.7 | 48.1 | 43.2 |
| Mouse MGAT1 | 41.8 | 41.8 | 42.4 | 42.4 | 39.7 | 41.2 | 40.3 | 40.1 | 37.6 | 37.6 | 35.2 | 36.8 | 36.8 | 72.5 | 100 | 53.0 | 52.7 | 45.7 | 45.1 | 40.8 |
| Human MGAT2 | 44.6 | 44.6 | 46.7 | 46.7 | 44.0 | 41.9 | 41.0 | 47.1 | 42.2 | 42.2 | 36.8 | 39.0 | 38.1 | 53.0 | 53.0 | 100 | 81.1 | 45.8 | 45.5 | 46.4 |
| Mouse MGAT2 | 44.3 | 44.3 | 45.2 | 45.2 | 44.3 | 43.1 | 40.4 | 47.1 | 40.7 | 42.5 | 38.0 | 39.0 | 38.7 | 52.1 | 52.7 | 81.1 | 100 | 46.4 | 46.7 | 46.39 |
| Human DGAT2 | 36.6 | 35.7 | 37.4 | 37.0 | 35.5 | 39.1 | 38.8 | 37.9 | 35.1 | 34.8 | 33.7 | 34.4 | 32.5 | 48.7 | 45.7 | 45.8 | 46.4 | 100 | 95.1 | 49.9 |
| Mouse DGAT2 | 36.6 | 35.7 | 37.7 | 37.3 | 34.7 | 39.6 | 39.1 | 37.9 | 35.1 | 34.8 | 34.0 | 34.4 | 32.5 | 48.1 | 45.1 | 45.5 | 46.7 | 95.1 | 100 | 50.2 |
| Human MGAT3 | 37.6 | 37.6 | 40.3 | 40.3 | 37.9 | 37.0 | 36.4 | 37.4 | 34.4 | 35.3 | 36.0 | 38.0 | 36.1 | 43.2 | 40.8 | 46.4 | 46.4 | 49.9 | 50.2 | 100 |
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arrese EL, Flowers MT, Gazard JL, Wells MA. Calcium and cAMP are second messengers in the adipokinetic hormone-induced lipolysis of triacylglycerols in Manduca sexta fat body. Journal of lipid research. 1999;40:556–564. [PubMed] [Google Scholar]
- Arrese EL, Gazard JL, Flowers MT, Soulages JL, Wells MA. Diacylglycerol transport in the insect fat body:evidence of involvement of lipid droplets and the cytosolic fraction. J Lipid Res. 2001;42:225–234. [PubMed] [Google Scholar]
- Arrese EL, Rojas-Rivas BI, Wells MA. Synthesis of sn-1,2-diacylglycerols by monoacylglycerol acyltransferase from Manduca sexta fat body. Archives of insect biochemistry and physiology. 1996;31:325–335. doi: 10.1002/(SICI)1520-6327(1996)31:3<325::AID-ARCH7>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Arrese EL, Soulages JL. Insect fat body: energy, metabolism, and regulation. Annual review of entomology. 2010;55:207–225. doi: 10.1146/annurev-ento-112408-085356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrese EL, Wells MA. Adipokinetic hormone-induced lipolysis in the fat body of an insect, Manduca sexta: synthesis of sn-1,2-diacylglycerols. Journal of lipid research. 1997;38:68–76. [PubMed] [Google Scholar]
- Beenakkers AM, Van der Horst DJ, Van Marrewijk WJ. Insect lipids and lipoproteins, and their role in physiological processes. Prog Lipid Res. 1985;24:19–67. doi: 10.1016/0163-7827(85)90007-4. [DOI] [PubMed] [Google Scholar]
- Bell RA, Joachim FG. Techniques for rearing laboratory colonies of tobacco hornworms and pink bollworms. Annals of the Entomological Society of America. 1976;69:365–373. [Google Scholar]
- Brasaemle DL. Thematic review series: adipocyte biology. The perilipin family of structural lipid droplet proteins: stabilization of lipid droplets and control of lipolysis. Journal of lipid research. 2007;48:2547–2559. doi: 10.1194/jlr.R700014-JLR200. [DOI] [PubMed] [Google Scholar]
- Buszczak M, Lu X, Segraves WA, Chang TY, Cooley L. Mutations in the midway gene disrupt a Drosophila acyl coenzyme A: diacylglycerol acyltransferase. Genetics. 2002;160:1511–1518. doi: 10.1093/genetics/160.4.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canavoso LE, Wells MA. Metabolic pathways for diacylglycerol biosynthesis and release in the midgut of larval Manduca sexta. Insect biochemistry and molecular biology. 2000;30:1173–1180. doi: 10.1016/s0965-1748(00)00094-1. [DOI] [PubMed] [Google Scholar]
- Cao J, Lockwood J, Burn P, Shi Y. Cloning and functional characterization of a mouse intestinal acyl-CoA:monoacylglycerol acyltransferase, MGAT2. The Journal of biological chemistry. 2003;278:13860–13866. doi: 10.1074/jbc.M300139200. [DOI] [PubMed] [Google Scholar]
- Cases S, Stone SJ, Zhou P, Yen E, Tow B, Lardizabal KD, Voelker T, Farese RV., Jr Cloning of DGAT2, a second mammalian diacylglycerol acyltransferase, and related family members. The Journal of biological chemistry. 2001;276:38870–38876. doi: 10.1074/jbc.M106219200. [DOI] [PubMed] [Google Scholar]
- Cheng D, Iqbal J, Devenny J, Chu CH, Chen L, Dong J, Seethala R, Keim WJ, Azzara AV, Lawrence RM, Pelleymounter MA, Hussain MM. Acylation of acylglycerols by acyl coenzyme A:diacylglycerol acyltransferase 1 (DGAT1). Functional importance of DGAT1 in the intestinal fat absorption. The Journal of biological chemistry. 2008;283:29802–29811. doi: 10.1074/jbc.M800494200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng D, Nelson TC, Chen J, Walker SG, Wardwell-Swanson J, Meegalla R, Taub R, Billheimer JT, Ramaker M, Feder JN. Identification of acyl coenzyme A:monoacylglycerol acyltransferase 3, an intestinal specific enzyme implicated in dietary fat absorption. The Journal of biological chemistry. 2003;278:13611–13614. doi: 10.1074/jbc.C300042200. [DOI] [PubMed] [Google Scholar]
- Downer RGH, Chino H. Turnover of protein and diacylglycerol components of lipophorin in insect haemolymph. Insect Biochemistry. 1985;15:627–630. [Google Scholar]
- Du M, Zhang S, Zhu B, Yin X, An S. Identification of a diacylglycerol acyltransferase 2 gene involved in pheromone biosynthesis activating neuropeptide stimulated pheromone production in Bombyx mori. J Insect Physiol. 2012;58:699–703. doi: 10.1016/j.jinsphys.2012.02.002. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic acids research. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. Confidence-Limits on Phylogenies - an Approach Using the Bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Fonagy A, Yokoyama N, Matsumoto S. Physiological status and change of cytoplasmic lipid droplets in the pheromone-producing cells of the silkmoth, Bombyx mori (Lepidoptera, Bombycidae) Arthropod structure & development. 2001;30:113–123. doi: 10.1016/s1467-8039(01)00027-5. [DOI] [PubMed] [Google Scholar]
- Fredrickson DS, Gordon RS., Jr The metabolism of albumin-bound C14-labeled unesterified fatty acids in normal human subjects. J Clin Invest. 1958;37:1504–1515. doi: 10.1172/JCI103742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris CA, Haas JT, Streeper RS, Stone SJ, Kumari M, Yang K, Han X, Brownell N, Gross RW, Zechner R, Farese RV., Jr DGAT enzymes are required for triacylglycerol synthesis and lipid droplets in adipocytes. Journal of lipid research. 2011;52:657–667. doi: 10.1194/jlr.M013003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman AGD, Downer RGH. Synthesis of diacylglycerols by monoacylglycerol acyltransferase from crop, midgut and fat body tissues of the american cockroach, Periplaneta americana L. Insect Biochemistry. 1979;9:129–134. [Google Scholar]
- Hofmann K. A superfamily of membrane-bound O-acyltransferases with implications for wnt signaling. Trends Biochem Sci. 2000;25:111–112. doi: 10.1016/s0968-0004(99)01539-x. [DOI] [PubMed] [Google Scholar]
- Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Computer applications in the biosciences : CABIOS. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- Kawooya JK, Law JH. Role of lipophorin in lipid transport to the insect egg. The Journal of biological chemistry. 1988;263:8748–8753. [PubMed] [Google Scholar]
- Kawooya JK, Osir EO, Law JH. Uptake of the major hemolymph lipoprotein and its transformation in the insect egg. The Journal of biological chemistry. 1988;263:8740–8747. [PubMed] [Google Scholar]
- Lardizabal KD, Mai JT, Wagner NW, Wyrick A, Voelker T, Hawkins DJ. DGAT2 is a new diacylglycerol acyltransferase gene family: purification, cloning, and expression in insect cells of two polypeptides from Mortierella ramanniana with diacylglycerol acyltransferase activity. The Journal of biological chemistry. 2001;276:38862–38869. doi: 10.1074/jbc.M106168200. [DOI] [PubMed] [Google Scholar]
- Liu Q, Siloto RM, Lehner R, Stone SJ, Weselake RJ. Acyl-CoA:diacylglycerol acyltransferase: molecular biology, biochemistry and biotechnology. Prog Lipid Res. 2012;51:350–377. doi: 10.1016/j.plipres.2012.06.001. [DOI] [PubMed] [Google Scholar]
- Lockey KH. Lipids of the insect cuticle: origin, composition and function. Comparative Biochemistry and Physiology Part B: Comparative Biochemistry. 1988;89:595–645. [Google Scholar]
- Lok CM, van der Horst DJ. Chiral 1,2-diacylglycerols in the haemolymph of the locust, Locusta migratoria. Biochimica et biophysica acta. 1980;618:80–87. doi: 10.1016/0005-2760(80)90055-7. [DOI] [PubMed] [Google Scholar]
- McFie PJ, Stone SL, Banman SL, Stone SJ. Topological orientation of acyl-CoA:diacylglycerol acyltransferase-1 (DGAT1) and identification of a putative active site histidine and the role of the n terminus in dimer/tetramer formation. The Journal of biological chemistry. 2010;285:37377–37387. doi: 10.1074/jbc.M110.163691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peled Y, Tietz A. Acylation of monoglycerides by locust fat-body microsomes. FEBS letters. 1974;41:65–68. doi: 10.1016/0014-5793(74)80955-5. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Molecular biology and evolution. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Smith SJ, Cases S, Jensen DR, Chen HC, Sande E, Tow B, Sanan DA, Raber J, Eckel RH, Farese RV., Jr Obesity resistance and multiple mechanisms of triglyceride synthesis in mice lacking Dgat. Nature genetics. 2000;25:87–90. doi: 10.1038/75651. [DOI] [PubMed] [Google Scholar]
- Soulages JL, Rimoldi OJ, Peluffo OR, Brenner RR. Transport and utilization of free fatty acids in Triatoma infestans. Biochem Biophys Res Commun. 1988;157:465–471. doi: 10.1016/s0006-291x(88)80272-9. [DOI] [PubMed] [Google Scholar]
- Soulages JL, Wells MA. Metabolic-fate and turnover rate of hemolymph free fatty-acids in adult Manduca sexta. Insect Biochemistry and Molecular Biology. 1994;24:79–86. [Google Scholar]
- Stone SJ, Myers HM, Watkins SM, Brown BE, Feingold KR, Elias PM, Farese RV., Jr Lipopenia and skin barrier abnormalities in DGAT2-deficient mice. The Journal of biological chemistry. 2004;279:11767–11776. doi: 10.1074/jbc.M311000200. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular biology and evolution. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tietz A, Weintraub H, Peled Y. Utilization of 2-acyl-sn-glycerol by locust fat body microsomes. Specificity of the acyltransferase system. Biochimica et biophysica acta. 1975;388:165–170. doi: 10.1016/0005-2760(75)90120-4. [DOI] [PubMed] [Google Scholar]
- Turchetto-Zolet AC, Maraschin FS, de Morais GL, Cagliari A, Andrade CM, Margis-Pinheiro M, Margis R. Evolutionary view of acyl-CoA diacylglycerol acyltransferase (DGAT), a key enzyme in neutral lipid biosynthesis. BMC evolutionary biology. 2011;11:263. doi: 10.1186/1471-2148-11-263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Horst DJ, van Hoof D, van Marrewijk WJ, Rodenburg KW. Alternative lipid mobilization: the insect shuttle system. Molecular and cellular biochemistry. 2002;239:113–119. [PubMed] [Google Scholar]
- Xu J, Francis T, Mietkiewska E, Giblin EM, Barton DL, Zhang Y, Zhang M, Taylor DC. Cloning and characterization of an acyl-CoA-dependent diacylglycerol acyltransferase 1 (DGAT1) gene from Tropaeolum majus, and a study of the functional motifs of the DGAT protein using site-directed mutagenesis to modify enzyme activity and oil content. Plant biotechnology journal. 2008;6:799–818. doi: 10.1111/j.1467-7652.2008.00358.x. [DOI] [PubMed] [Google Scholar]
- Yen CL, Monetti M, Burri BJ, Farese RV., Jr The triacylglycerol synthesis enzyme DGAT1 also catalyzes the synthesis of diacylglycerols, waxes, and retinyl esters. Journal of lipid research. 2005;46:1502–1511. doi: 10.1194/jlr.M500036-JLR200. [DOI] [PubMed] [Google Scholar]
- Yen CL, Stone SJ, Koliwad S, Harris C, Farese RV., Jr Thematic review series: glycerolipids. DGAT enzymes and triacylglycerol biosynthesis. Journal of lipid research. 2008;49:2283–2301. doi: 10.1194/jlr.R800018-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler R, Van Antwerpen R. Lipid uptake by insect oocytes. Insect biochemistry and molecular biology. 2006;36:264–272. doi: 10.1016/j.ibmb.2006.01.014. [DOI] [PubMed] [Google Scholar]


