Abstract
Antibody-based proteomics is an enabling technology that has significant implications for cancer biomarker discovery, diagnostic screening, prognostic and pharmacodynamic evaluation of disease state, and targeted therapeutics. Quantum dot based fluoro-immunoconjugates possess promising features towards realization of this goal such as high photostability, brightness and multispectral tunability. However, current strategies to generate such conjugates are riddled with complications such as improper orientation of antigen binding sites of the antibody, aggregation and stability issues. We report a facile yet effective strategy to conjugate anti epidermal growth factor receptor (EGFR) antibody to quantum dots using copper-free click reaction, and compared them to similar constructs prepared using traditional strategies such as succinimidyl-4-(N-maleimidomethyl) cyclohexane-1-carboxylate (SMCC) and biotin-streptavidin schemes. The Fc and Fab regions of the conjugates retain their binding potential, compared to those generated through the traditional schemes. We further applied the conjugates in testing a novel microsphere array device designed to carry out sensitive detection of cancer biomarkers through fluoroimmunoassays. Using purified EGFR, we determined the limit of detection of the microscopy centric system to be 12.5 ng/ml. The biological assay, in silico, was successfully tested and validated by using tumor cell lysates, as well as human serum from breast cancer patients and the results were compared to normal serum. A pattern consistent with established clinical data was observed, which further validates the effectiveness of the developed conjugates and its successful implementation both in vitro as well as in silico fluoroimmunoassays. The results suggest the potential development of a high throughput in silico paradigm for predicting the class of patient cancer based on EGFR expression levels relative to normal reference levels in blood.
Introduction
Fluoroimmunoassays are sensitive platforms to achieve antibody (Ab)-based detection of tumor biomarkers. The performance of these assays is dependent on the reliable functioning of the molecular recognition and binding probes. Although Ab-fluorophore conjugates are popular and several conjugation strategies available, the low binding efficiency and non-specific labeling is predominant, often leading to erroneous interpretations.1, 2 Therefore, careful optimization of conjugation and binding conditions is critical for the proper evaluation of the biological labeling. Because of their excellent photostability, high quantum yield, and the potential for multiplexing information based on single excitation and multiple emission wavelengths, quantum dots (QDs) are ideal fluorophores for a microscopy centric system design.3 However, the disproportionate dimensions of QD and Ab need careful consideration. Unlike organic fluorophores and Ab conjugates, where multiple dyes can be conjugated to a single Ab without interference with the Ab binding sites, QD-Ab conjugates can possess multiple Abs per nanoparticle.4 This molecular orientation could lead to improper orientation of the biomolecule’s binding sites, consequently attenuating the binding potential of the Ab-QD conjugate.4
Several strategies have been used to conjugate Ab to QD,5, 6 but retention of the biological functions of ligands such as Ab in these QD conjugates remains a challenge. For example, previous reports have shown that succinimidyl-4-(N-maleimidomethyl) cyclohexane-1-carboxylate (SMCC)-based Ab-QD conjugates demonstrated poor stability in aqueous aerated solutions, resulting in low binding and staining efficiency.4, 7 Although biotin-streptavidin based Ab-QD conjugates have demonstrated relatively better performance, they suffer from poor biospecificity because of the low number of functional Ab. Several factors can mediate this inefficiency, including the large dimensions of the functional groups, overall size of the probe, aggregation caused by Ab crosslinking to multiple QDs, and random orientation of the Ab.6
Here, we report the development of Ab-QD conjugates employing copper-free ‘click’ chemistry reaction. Copper (Cu)-free cycloaddition reactions are highly favored over Cu catalyzed reactions because of the fluorescence quenching potential of Cu ions on dyes and QDs.8 The rapid, specific, efficient, stable, facile, modular and aqueous phase conjugation strategy of ‘click’ reaction has proven to be a reliable and powerful technique that is employed widely.9 While this strategy has been used to conjugate transferrin to QDs in the past,10 we have adapted it to conjugating antibodies, both bivalent (whole) and monovalent (half) Abs, with suitable modifications such as the selection of appropriate crosslinkers to ensure a highly modular assembly process. Certain applications and immunochemical techniques require the Ab in its smaller sized analogue, which offers several advantages such as specific binding to thiol (SH) groups for bioconjugation, lower stearic hindrance, higher tissue penetration and lower immunogenicity.11, 12 The versatile nature of the conjugation strategy is applied to generate stable building blocks from both whole and half Ab, which enhanced the efficiency and yield of the Ab-QD constructs. In addition, we evaluated the Ab-QD conjugates further by assessing their binding efficiency and biospecificity both in cellulo as well as in fluoroimmunoassays and found that the Ab on the Ab-QD constructs indeed retains its Fc and Fab binding characteristics. We also compared the Ab-QD construct developed using click reaction with similar constructs prepared through traditional conjugation methods such as SMCC-based amine-thiol and biotin-streptavidin affinity reactions and found superior labeling efficiency of EGFR expressing cancer cells using the constructs developed using click reaction in comparison to the traditional strategies.
Finally, we demonstrate the application of the constructs in silico using a novel microsphere array (3D MSA) designed for highly sensitive detection of cancer biomarkers in serum and other biological fluids. The 3D MSA device has some obvious advantages over traditional arrays such as ordered placement of microspheres for increased sensitivity, simplification of image processing and controlled binding conditions through a microfluidic setup.13,14 We have previously demonstrated controlled trapping of polystyrene microspheres and simultaneously applied advanced signal and image processing techniques to achieve a highly optimized device for performing fluoroimmunoassays.15 Herein, we have implemented the biological protocol of the immunoassay to the device and tested its performance and sensitivity. The versatile and efficient conjugation and analysis used in this study creates a platform for high throughput screening of biological samples.
Results and Discussion
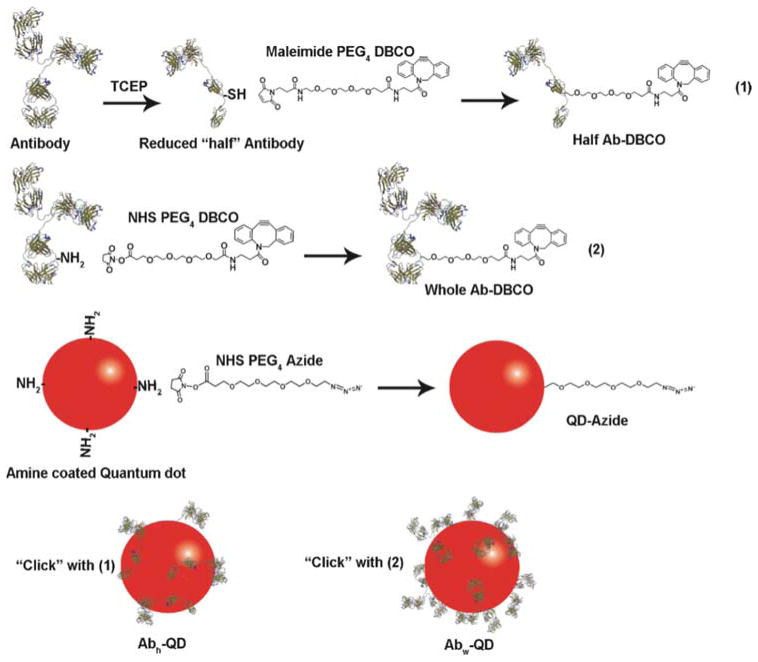
The conceptual design of the bioconjugation of Ab to QDs using copper-free cycloaddition is illustrated in Figure 1. Two schemes to ‘click’ Abs on QD in aqueous buffer conditions were implemented. Firstly, selective cleavage of disulfide linkage of whole Ab to obtain half Ab was performed using tris (2-carboxyethyl) phosphine (TCEP). Cleaved, ‘half’-Ab presenting thiol groups was functionalized with dibenzocyclooctyne (DBCO) via a maleimide-PEG-DBCO crosslinker. Secondly, ‘whole’ Ab was functionalized with DBCO using an N-hydroxysuccinimide (NHS)-based crosslinker, NHS-PEG-DBCO. Mouse anti-human EGFR monoclonal Ab was used throughout the study. Concurrently, QD-Azide adducts were prepared by reacting amine coated QD with NHS-PEG-Azide crosslinker. The whole Ab or half Ab, and QD adducts were subsequently ‘clicked’ to obtain Abw-QD or Abh-QD conjugates, respectively, with high yield (88%). We also prepared Ab-QD conjugates by using traditional strategies for comparative analysis with the click reaction. Thus, Abs-QD was prepared by reacting SMCC with the PEG linker on half Ab, and Abb-QD was obtained by mixing biotinylated whole Ab with streptavidin coated QDs, which forms strong non-covalent intermolecular binding.6
Figure 1.
Schematic of the Ab-QD conjugation strategy using Cu-free click reaction.
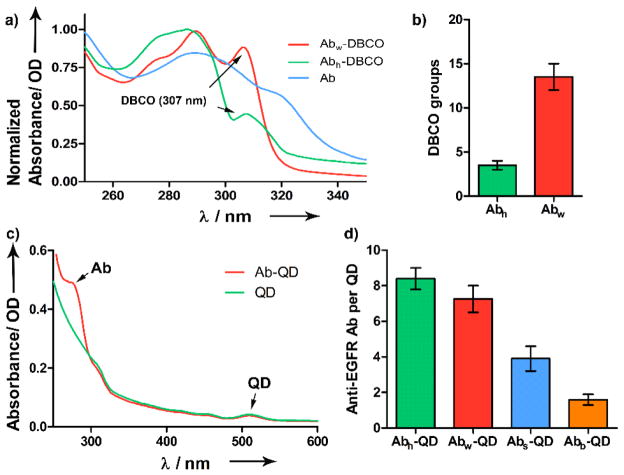
The degree of DBCO incorporation on the half and whole Ab was determined using UV-vis absorption scans. Based on the relative absorbance values of DBCO (λmax = 307 nm) and Ab (λmax = 280 nm) (Figure 2a), the number of DBCO per half Ab and whole Ab was estimated to be 3.4 and 13.5, respectively (Figure 2b). Incorporation of the azide groups on QDs were determined by zeta potential and the hydrodynamic size of the nanoparticles was determined by dynamic light scattering (DLS) analysis (Table 1). The successful conjugation of QD525 (λmax = 515 nm) to Ab, after purification to remove unbound Ab, was confirmed using UV-vis absorption data (Figure 2c). In addition, the hydrodynamic diameter and zeta potential values increased from 23.9 ± 1.8 nm to 32.4 ± 2.1 nm and from −16.7 mV to −5.7 mV, respectively. The yield of Ab-QD conjugates was high, which can be attributed to the stability of the DBCO and azide groups in aqueous solution and the efficiency of the reaction. Using Bradford protein assay, the number of Ab per QD were estimated to be approximately 8.4 for Abh-QD conjugates and 7.2 for Abw-QD (Figure 2d). In comparison, we observed the overall yield of Abs-QD was modest after purification (~60%), with 3.9 Abs per QD; and that of Abb-QD was appreciably higher (~82%), with 1.6 Abs per QD. This suggests that during the two-step conjugation, some of the maleimide groups on SMCC could have undergone hydrolysis, thus rendering the functional group inactive for reaction with the SH groups on Ab.16 However, using crosslinkers such as DBCO and azide allowed highly selective reaction, resulting in high yields with minimal purification steps. Due to the unique nature of Ab and QD interaction, the various Ab-QD constructs would still have a single dedicated QD, irrespective of the loading number. However, Ab-QD conjugates with high loading number such as Abh-QD and Abw-QD, could possess higher number of Abs with the optimal orientation for functional binding than those with low loading numbers. Essentially, this means that successful binding of any Ab-QD conjugate to its target, irrespective of the loading number, will yield similar brightness. This suggests that the performance of a particular Ab-QD construct will depend on the number of optimally oriented and functional Ab decorated on QD surface, which to a certain degree is dependent on the loading number and efficiency of the conjugation strategy. Additionally, variables such as cleaved Ab with half the number of antigen binding sites as whole Ab, also could significantly impact the functionality of the Ab-QD conjugates.
Figure 2.
a) Absorption spectra of Ab functionalized with DBCO groups, compared to non-functionalized Ab. b) Comparison of degree of DBCO incorporation to Abh and Abw estimated using Bradford assay. c) Absorption spectra of Ab-QD construct compared to QD alone. d) Comparison of the number of anti-EGFR Ab conjugated per QD among the four Ab-QD conjugates.
Table 1.
Physical characterization of the Ab-QD constructs
| Sample | Size (nm) | Zeta Potential (mV) |
|---|---|---|
| QD-Amine | 23.9 ± 1.8 | − 16.7 |
| QD-Azide | 27.8 ± 1.4 | − 23.7 |
| Abw-QD | 32.4 ± 2.1 | − 5.7 |
| ABh-QD | 31.1 ± 1.8 | − 6.1 |
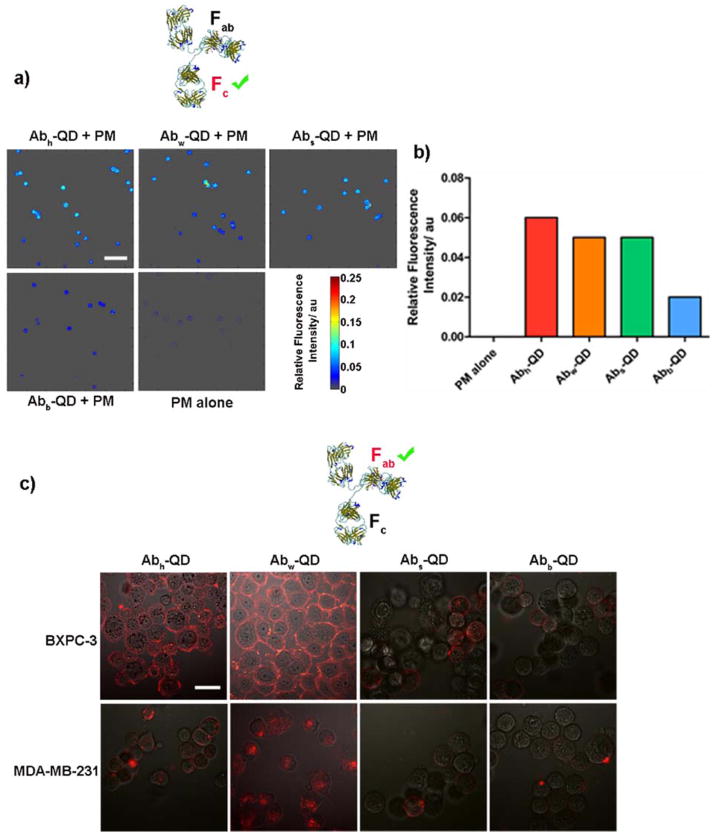
To determine if the binding potential of Fc region of the Ab-QD conjugates was retained, we employed Protein G coated PM beads (PGPM), which has an average diameter of 15 μm. Protein G is an immunoglobulin binding protein of bacterial origin with very high specificity and affinity to the Fc region of Ab.17 Successful binding of Ab-QD to protein G was determined by evaluating the fluorescence of the PM beads (see Supporting Information) by using confocal laser scanning microscope (CLSM). For quantification of fluorescence intensity and to prevent subjective data analysis, we have developed MATLAB-based algorithms for signal and image processing of the five constructs, including the control product, PGPM alone (Figure 3a). Our results show that Abh-QD, Abw-QD and Abs-QD constructs exhibit appreciable binding to the PGPM surface relative to Abb-QD and the control product (Figure 3b). This suggests that the biotin-streptavidin interaction might have blocked the binding domains in the Fc region, thus decreasing the binding efficiency. Given the large size of streptavidin, it is likely that proximity and stearic effects interfered with the availability of the binding domains for ligand-receptor molecular interaction. To ensure that the binding of Ab to Protein G is through the Fc region and not by non-specific binding, we also employed nonfunctionalized PM beads and Protein A coated PM beads. Protein A is a subtype of Protein G, but with lower binding specificity and affinity for Ab, than Protein G.18 None of the constructs demonstrated any appreciable binding to the plain PM beads as well as Protein A coated PM beads, confirming the specific binding of the Ab to the PGPM beads through the Fc region.
Figure 3.
a) MATLAB generated fluorescence intensity images showing the binding of the Fc region of the four Ab-QD constructs to Protein G coated PM. Scale bar represents 100 μm and is unchanged across all images. b) Quantitation of the relative fluorescence intensity of Ab-QD-PM assembly. c) CLSM images showing binding characteristics of the Fab region of Ab-QD constructs to BxPC-3 and MDA-MB-231 cells after 4h. The reflectance and fluorescence channels were merged. Scale bar represents 30 μm and is unchanged across all images.
Next, we sought to determine retention of antigen recognition and binding of the Fab region of the Ab-QD constructs. The anti-human EGFR Ab based constructs were evaluated for EGFR binding in cellulo, using MDA-MB-231 (EGFR positive) and BxPc-3 (EGFR positive) cancer cell lines. MDA-MB-231 cells are known to internalize the Ab-receptor complex after binding of the ligands to the receptors, whereas BxPc-3 resist such internalization.19, 20 We investigated if the constructs retain the same pattern of internalization for the respective cell lines. This strategy was chosen over using EGFR negative cell lines because most cell lines express a basal level of EGFR, which could confound data analysis. Moreover, our approach will also facilitate the delineation of specific binding of the constructs to EGFR from cross reactions to analogous surface receptors. Click conjugated Ab-QD constructs, particularly Abw-QD, showed high degree of binding to the cells and conformed to the internalization characteristics of the respective cell lines (Figure 3c). In contrast, Abs-QD and Abb-QD displayed low binding affinity and poor correlation with the expected internalization pattern, suggesting improper orientation of Ab on the QD surface. Interestingly, Abh-QD demonstrated good binding affinity to BxPc-3 cells and poor internalization in MDA-MB-231 cells. Typically, internalization of EGF receptor-ligand complex is preceded by dimerization of the surface receptors, essentially by bivalent antibodies (whole Ab with both Fab regions intact).21 Monovalent Ab (half Ab with single Fab region) can prevent receptor dimerization and activation of EGFR.22 Although Abh-QD is capable of successfully binding to EGFR on the cell surface, it is likely that this conjugate is not able to initiate receptor dimerization, thus preventing internalization of the construct. It is well known that nanoparticles can show high degree of nonspecific uptake by cells. To assess this potential effect, we performed blocking studies with the anti-EGFR Ab. The results demonstrate minimal binding and internalization of Abh-QD and Abw-QD in the respective cell lines (see Supporting Information).
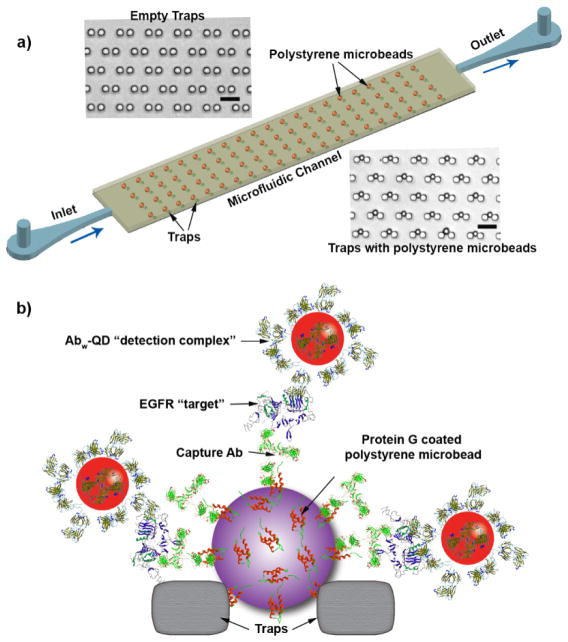
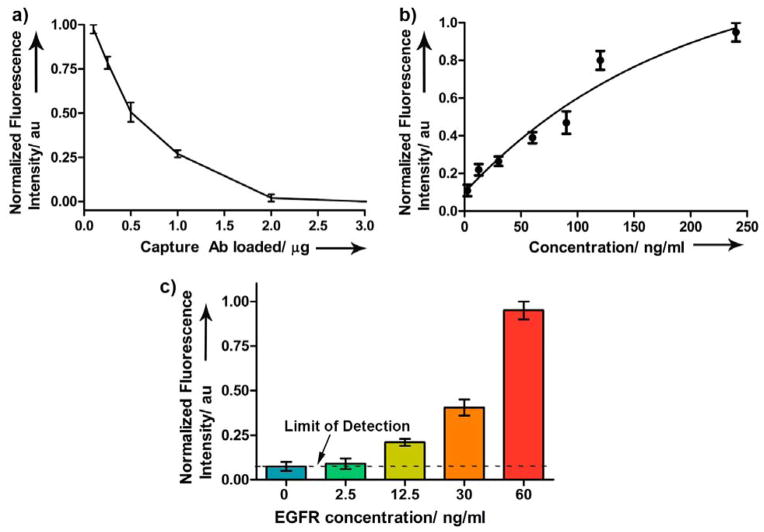
Motivated by the performance of Abw-QD, we carried out fluoroimmunoassays in silico, using the 3D MSA system we recently developed to detect EGFR from biologically relevant samples (Figure 4a). Optimal trapping of 15 μm plain PM beads has previously been demonstrated.23 In this study, we implemented the same strategy to trap PGPM beads preincubated with an anti-EGFR polyclonal Ab, as the capture Ab. A similar trapping efficiency to plain PM beads in the MSA device was achieved (Figure 4b). To prevent nonspecific binding of Ab-QD conjugates in subsequent steps, we saturated all the Protein G binding sites on the PGPM with capture Ab. We then determined the concentration of mouse anti-EGFR polyclonal Ab required to saturate the 15 μm PGPM beads by introducing Abw-QD. and measuring fluorescence intensity from Abw-QD to detect potential false positives (Figure 5a). We did not observe any fluorescence signal after adding 2 μg of capture Ab (~ 1.5 × 105 IgG/bead), suggesting successful saturation and blocking of Fc binding sites on PGPM. We then tested the capture and proper functioning of the fluoroimmunoassay in solution using purified sample of EGFR at various titrations. The capture and detection Abs used in this assay bind to different epitopes of the extracellular domain of EGFR. As shown in Figure 5b, we observed a strong positive non-linear correlation between fluorescence intensity and EGFR concentration, in solution phase. Saturation limit of the capture Ab-PGPM complex can simply be extended by addition of more beads, thus improving the detection range of the target protein.
Figure 4.
a) Illustration of 3D microsphere array device with image of actual traps (top) and traps filled with PM (bottom). Scale bars represent 30 μm. b) Schematic of the fluoroimmunoassay involving Protein G coated PM, monoclonal anti-EGFR capture antibody, EGFR as target protein and Abw-QD constructs as detection complex.
Figure 5.
a) Estimation of the binding capacity of anti-EGFR Ab on PGPM to ensure saturation of all Fc binding sites on Protein G by capture Ab. b) Binding studies with Abw-QD showing the response of the assay to increasing concentrations of EGFR on a Protein G based PM platform in solution phase. b) Sensitivity of the fluoroimmunoassay using purified EGFR in solution phase, defining the limit of detection.
Because the in silico fluoroimmunoassay is designed for CLSM based readout, we carried out sensitivity studies first in solution phase to determine the limit of detection (LOD) of the assay using CLSM. The intensity of fluorescent halo surrounding the bead was used as a measure of varying EGFR concentration. There was no detectable fluorescence at < 12.5 ng/ml EGFR concentration (Figure 5c). For reference, normal serum EGFR levels in healthy individuals is 75.3 ng/ml and those with cancer can either have an elevated or lower basal serum EGFR values depending on the cancer type.24 Therefore, it is envisaged that the implementation of such assays will be towards determining the treatment response and prognosis rather than diagnosis of primary cancer.
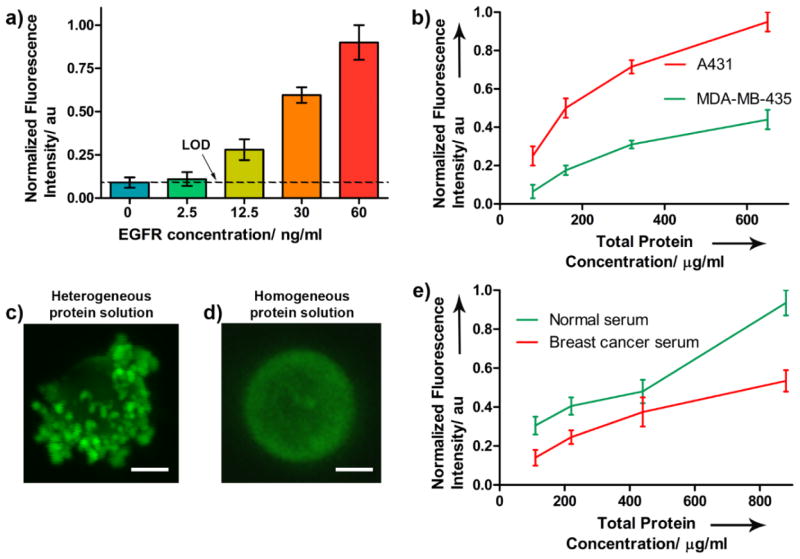
Implementation of the fluoroimmunoassay in silico was carried out by sequential injection of the PGPM-capture Ab complex, followed by injection of EGFR and finally Abw-QD, allowing an incubation time of 5 min each. The sequential loading and flow-through design facilitated effective washing of unreacted components (Figure 4a). The system also enabled rapid binding of reactants within the confined volume. We did not observe significant changes in fluorescence readout upon longer incubation times, suggesting optimal binding of EGFR and detection complex at shorter incubation times (see Supporting Information). A MATLAB algorithm (see Supporting Information) was generated to process the CLSM images. This will provide accurate quantitation of fluorescence intensity from the PM surface. The algorithm takes into account imaging artifacts such as non-specific fluorescence from sources other than the PM beads, provides accurate and unbiased information on a huge number of beads and images, and enables automated signal processing, thus simplifying data interpretation. We observed similar sensitivity (12.5 ng/ml) in silico (Figure 6a) relative to the solution phase method. Correlation of the sensitivities of these two methods suggests successful translation and implementation of the fluoroimmunoassay in the MSA device. While efforts to improve the LOD and sensitivity of the in silico method is an ongoing study, the current sensitivity is sufficient to investigate patterns in differential expression of serum EGFR in various tumor models.
Figure 6.
a) Sensitivity of the fluoroimmunoassay using purified EGFR in silico, defining the limit of detection. b) Plot showing the assay response to EGFR receptor density from cell culture lysates of A431 and MDA-MB-435 cell lines. c) CLSM image of single PM after assay performed using heterogeneous protein samples such as culture lysates and human serum. Scale bar represents 7.5 μm. d) CLSM image of single PM after assay performed using purified EGFR. Scale bar represents 7.5 μm. e) Plot showing the assay response to serum EGFR levels from breast cancer patient and compared to normal serum.
For final validation of the microfluidic protocol, the challenge was to determine the selectivity of the assay for detecting EGFR in a heterogeneous mixture of proteins such as in vitro tumor cell lysates, and in a more clinically relevant samples such as human serum derived from breast cancer patients. As in vitro models of biomarker expression, EGFR overexpressing tumor cell line A431 and lower EGFR expressing tumor cell line MDA-MB-435 were compared. MDA-MB-435 expresses 2 × 104 EGFR receptors per cell compared to 2 × 106 receptors per cell expressed by A431.25, 26 Therefore, MDA-MB-435 cell line can be classified as a representative model of “normal” cells. Cell lysates for each cell type were prepared using standard protocols and their total protein concentration of 78±3 mg/ml was determined. Different dilutions of the lysates were then injected into the MSA and assayed to determine the relative contribution of EGFR from the protein mixture to the fluorescence intensity. We observed a positive non-linear correlation between fluorescence intensity and EGFR density as well as EGFR concentration across the two cell types (Figure 6b). This suggests the involvement of EGFR selective recognition and binding to Ab-based PM and QD constructs, and also demonstrates successful signal collection and processing. CLSM images revealed dense aggregations on the PM surface (Figure 6c). Similar images of PM taken with purified EGFR do not exhibit these dense surface deposits (Figure 6d). This is not surprising because the complex and heterogeneous solutions are more likely to contain proteins that have a high tendency for nonspecific interactions on the bead surface. Thus, attempts to quantitate the proteins of interest from such mixtures are complicated and remain a challenge. However, the dense aggregates did not influence the fluorescence signal output and the LOD remained unchanged.
Finally, we evaluated human serum samples to confirm the selectivity of the assay in a more complex and clinically relevant heterogeneous protein mixture than the preceding study. Clinical serum samples from breast cancer patients and normal individuals were used for comparison. A total protein concentration of 107±4 mg/ml was estimated for both samples. Diluted mixtures were injected into the MSA and assayed. We observed a positive non-linear correlation between fluorescence intensity and EGFR concentration. However, serum samples from breast cancer patients exhibited two-fold lower EGFR levels compared to normal serum (Figure 6e). This result is consistent with previous studies that have reported significantly lower serum levels of EGFR among patients with breast and ovarian cancer.27,28 The median values are 56.3 ng/ml and 30.9 ng/ml at time of primary diagnosis and metastasis, respectively, when compared to normal EGFR levels of 75.3 ng/ml.24 In contrast, cancers of brain and lung are known to have elevated serum EGFR.29 These data provide further validation of the accuracy and selectivity of the assay in determining the relative levels of EGFR in representative tumor models and clinical samples.
Conclusion
We have demonstrated a facile and effective strategy to conjugate whole and half Ab to QDs using Cu-free click reaction. In doing so, we have demonstrated the functional and binding sites, Fc and Fab regions, of the Ab remains unaffected. We have compared the Ab-QD constructs prepared herein with similar constructs using traditional conjugation strategies through EGFR-specific cell binding studies. The efficient binding of the constructs, particularly Abw-QD, prompted their use to evaluate of a novel microsphere array microfluidic device. Fluoroimmunoassays were carried out on the device with CLSM based readouts and MATLAB-based algorithms for signal and image processing. Using EGFR as model biomarker, we estimated the sensitivity and limit of detection of the system to be 12.5 ng/ml. The robustness of the device and the assay was further tested on heterogeneous protein mixtures derived from cell culture lysates of EGFR overexpressing and under expressing cell lines, which showed a positive correlation between EGFR receptor density and fluorescence intensity. Similarly, clinically relevant human serum samples obtained from breast cancer patients was tested and compared to normal serum for selective and sensitive detection of EGFR. Interestingly, lower EGFR levels were found in serum derived from breast cancer patients, which is consistent with recent clinical studies. Therefore, this highly versatile and efficient conjugation strategy can enable molecular imaging of disease biomarkers both in cellulo and in silico as demonstrated here, with the potential for translation to in vivo systems for antibody-based imaging and biomarker characterization.
Experimental Methods
I. Conjugation of Antibodies to Quantum dots
Anti-human EGFR monoclonal Antibodies (Ab) were purchased from Sigma Aldrich Inc. (St. Louis, MO). The Ab were cleaved using a reducing agent, TCEP from Thermo Scientific Inc. (Rockford, IL), to generate reduced ‘half’ Ab. Briefly, to 20 μM of Ab in 100 μM of phosphate buffered saline (PBS) with a pH of 7.2, 50 μM of TCEP was added and incubated at room temperature (RT) for 1 h. Unlike other disulfide bond reducing agents such as dithiothreitol (DTT), TCEP does not need removal before performing the cross-linking reactions. Immediately after, 100 μM of DBCO-PEG4-maleimide purchased from Click Chemistry Tools (Scottsdale, AZ) was added and incubated at RT for 2 h. The Abh-DBCO conjugates thus formed are purified to remove unreacted reagents and excess TCEP, using Slide-A-Lyzer MINI Dialysis Device, 10k MWCO from Thermo Scientific Inc. (Rockford, IL) overnight. Removal of TCEP is necessary before performing click reaction since azide groups are reduced by TCEP. Similarly, 20 μM of whole Ab were reacted with 100 μM of DBCO-PEG4-NHS crosslinker from Click Chemistry Tools, in 50 mM borate buffer pH 8.5 at RT for 2 h to generate Abw-DBCO. The conjugates were purified using dialysis as mentioned above. To prepare QD-Azide conjugates, 300 nM of amine functionalized QD525, with an excitation and emission wavelength of 515 nm and 525 nm, respectively, from Life Technologies Inc. (Carlsbad, CA) in borate buffer was incubated with 3 μM of Azide-PEG4-NHS from Click Chemistry Tools, at RT for 2 h. The QD-Azide conjugates were purified overnight using dialysis. Click reaction was performed using the conjugates by incubating Abh-DBCO and QD-Azide at 37°C for 6 h in a water bath to generate Abh-QD. Similarly, Abw-DBCO and QD-Azide were clicked to generate Abw-QD constructs. Conjugation of Ab to QD using SMCC and biotin-streptavidin strategies was adopted from previously well characterized protocols.6
II. Physicochemical characterization
The absorption spectra to determine DBCO incorporation and successful conjugation of Ab to QDs was recorded on a Beckman Coulter DU 640 UV-visible spectrophotometer (Brea, CA) and analyzed using Graphpad Prism statistical software. At λ = 307 nm, the characteristic DBCO peak can be distinguished from the protein peak at λ = 280 nm. The respective absorbance readings for Abh-DBCO and Abw-DBCO were recorded. The number of DBCO per IgG was calculated using the formula MolarityDBCO/MolarityIgG, where MolarityDBCO = AbsorbanceDBCO/extinction coefficientDBCO and MolarityIgG = AbsorbanceIgG/extinction coefficientIgG. The extinction coefficient of DBCO and IgG are 12,000 and 204,000 M−1L cm−1, respectively. The number of IgG per QD was calculated using Bio-Rad protein assay dye (Hercules, CA) according to the manufacturer’s protocol, where the extinction coefficient of QD525 was taken as 130,000 M−1Lcm−1. The absorbance values were corrected for background using unconjugated QDs, including QD525-strepatavidin. Dynamic light scattering measurements were taken using a Malvern Zetasizer Nano ZS (Westborough, MA) instrument at 25°C. Two measurements were conducted for each sample with at least 10 runs and each run lasting 10 s. All sizes reported were based on intensity average.
III. Fc and Fab binding assays
For Fc binding studies, Protein G coated polystyrene beads from Spherotech Inc. (Lake Forest, IL) with an average diameter of 15 μm was used. As control, Protein A coated PM beads purchased from Polysciences Inc. (Warrington, PA), with an average diameter of 10 μm was used. Equimolar solutions of the Ab-QD constructs and PGPM were incubated at 25°C for 15 m in a shaker and fluorescence images were acquired using an Olympus FV1000 confocal laser scanning microscope (Center Valley, PA). Fluorescence/reflectance images were taken with a 20X objective using He:Ne 488 nm excitation laser and emission range of dichroic mirrors set to 455–675 nm. MATLAB® from MathWorks (Natick, MA) was used to process the images using standard algorithms to quantitate the data. An average of 100 particles were counted for each group.
MDA-MB-231 and BxPc-3 cell lines from American Type Culture Collection – ATCC (Manassas, VA) were cultured under recommended standard conditions. MDA-MB-231 were cultured in Dulbecco’s Modified Eagle’s Medium containing 10% fetal bovine serum (FBS), L-glutamine (2 mM), penicillin (100 units/ml), and streptomycin (100 μg/ml), incubated at 37 °C in a humidified atmosphere of 5% CO2 and 95% air. BxPc-3 cells were cultured in RPMI-1640 under similar conditions. 5 × 105 cells per well were grown in in an 8 well chamber culture slide from BD Biosciences (San Jose, CA) to which 100 nM of Ab-QD was added and incubated at 37°C in a humidified, 5% CO2 atmosphere for 4 h. The slide substrates were gently washed 2X with PBS before imaging with CLSM. Fluorescence/reflectance images were taken with a 60X objective using He:Ne 488 nm excitation laser and emission range of dichroic mirrors set to 455–575 nm. Fluorescence and reflectance image overlay with false color was performed using Fluoview FV10-ASW software from Olympus (Center Valley, PA).
IV. 3D Microsphere Array studies
The binding capacity and concentration of mouse anti-human EGFR polyclonal Ab from Sigma Aldrich (St. Louis, MO) as capture Ab, on PGPM was first determined using different titrations of the Ab (0.1, 0.25, 0.5, 1, 2 & 3 μg) on 0.25 mg/ml of PGPM. After incubation for 15 m at RT, the beads were centrifuged at 10,000 rpm for 5 m to remove unbound Ab. Abw-QD (0.25 mM) was added and incubated for 15 m at RT. After centrifugation to remove unbound Abw-QD, the final mixture was added to 96-well plates and endpoint fluorescence readings were recorded using Synergy HT multimode plate reader from BioTek Instruments Inc. (Winooski, VT). The values were normalized relative to the highest fluorescence reading. Based on the binding capacity, number of PGPM beads per unit mass (~5.2 × 107/mg) and the number of Protein G per bead (~5 × 106), the number of Ab/bead was calculated. The microfluidic trap-based array device was fabricated in polydimethylsiloxane (PDMS) using soft lithography techniques as shown previously.23 The device was mounted on the CLSM with the inlet connected to a syringe pump from Harvard Apparatus (Holliston, MA) set to an infusion rate of 100 μl/min. PGPM beads (0.5% w/v) were diluted 50x and 2 μg of anti-human EGFR polyclonal Ab from Sigma Aldrich (St. Louis, MO) was added to a final volume of 100 μl and incubated for 15 m at 25°C. The PGPM-Ab complex was injected into the device and trapping process was monitored in real time using CLSM set to a fast scan rate in reflectance mode. A wash cycle with PBS was initiated to remove unbound Ab using the microfluidic-syringe pump setup. Subsequent cycles consisted of injecting purified EGFR at various dilutions from 0–250 ng/ml into the system and performing wash steps to remove unbound protein, after allowing an incubation time of 5 m. Finally, Abw-QD was injected and incubated for an additional 5 m and followed by a wash step to remove unreacted components. Fluorescence/reflectance images were recorded using CLSM using the 488 nm laser with emission set to 455–575 nm. A unique MATLAB algorithm (in supporting information) was used to extract data such as number of beads, number of fluorescent beads, intensity of fluorescence, background fluorescence and graphical representation of the accumulated data. Similarly, fluoroimmunoassays using lysed tumor cells and human serum were performed after dilutions to the order of 500–1000x, to prevent clogging of the microfluidic channels. Lysis of A431 and MDA-MB-435 cells, grown in DMEM, were performed using NP40 cell lysis buffer from Life Technologies Inc. (Carlsbad, CA) following the manufacturer’s instructions. Human serum from breast cancer patients and normal serum were purchased from Bioreclamation Inc. (Liverpool, NY). Dilutions were performed in PBS before injection.
Solution phase fluoroimmunoassays were performed by first mixing PGPM-Ab and purified EGFR and incubating for 5 m at 25°C followed by centrifugation at 10,000 rpm for 2 m. The supernatant was carefully decanted and the process repeated 2x. Subsequently, Abw-QD was added to the mixture and incubated for 5 m and similarly washed using PBS to remove excess reactants. The final mixture was added to 96-well plates and endpoint fluorescence readings were recorded using a plate reader and analyzed using Graphpad Prism statistical software.
Supplementary Material
Acknowledgments
This study was supported by grant from National Science Foundation (CCF0963742). We thank Dr. Pinaki Sarder for helpful discussions and assistance with data analysis.
Footnotes
Supporting Information Available: This information is available free of charge via the Internet at http://pubs.acs.org/.
References
- 1.McCormack T, O’Keeffe G, Mac Craith B, O’Kennedy R. Assessment of the Effect of Increased Fluorophore Labelling on the Binding Ability of an Antibody. Analytical Letters. 1996;29:953–968. [Google Scholar]
- 2.Vira S, Mekhedov E, Humphrey G, Blank PS. Fluorescent-labeled antibodies: Balancing functionality and degree of labeling. Anal Biochem. 2010;402:146–150. doi: 10.1016/j.ab.2010.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Michalet X, Pinaud FF, Bentolila LA, Tsay JM, Doose S, Li JJ, Sundaresan G, Wu AM, Gambhir SS, Weiss S. Quantum dots for live cells, in vivo imaging, and diagnostics. Science. 2005;307:538–544. doi: 10.1126/science.1104274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pathak S, Davidson MC, Silva GA. Characterization of the functional binding properties of antibody conjugated quantum dots. Nano Lett. 2007;7:1839–1845. doi: 10.1021/nl062706i. [DOI] [PubMed] [Google Scholar]
- 5.Medintz IL, Uyeda HT, Goldman ER, Mattoussi H. Quantum dot bioconjugates for imaging, labelling and sensing. Nat Mater. 2005;4:435–446. doi: 10.1038/nmat1390. [DOI] [PubMed] [Google Scholar]
- 6.Xing Y, Chaudry Q, Shen C, Kong KY, Zhau HE, Chung LW, Petros JA, O’Regan RM, Yezhelyev MV, Simons JW, Wang MD, Nie S. Bioconjugated quantum dots for multiplexed and quantitative immunohistochemistry. Nat Protoc. 2007;2:1152–1165. doi: 10.1038/nprot.2007.107. [DOI] [PubMed] [Google Scholar]
- 7.Lee J, Choi Y, Kim K, Hong S, Park HY, Lee T, Cheon GJ, Song R. Characterization and cancer cell specific binding properties of anti-EGFR antibody conjugated quantum dots. Bioconjug Chem. 2010;21:940–946. doi: 10.1021/bc9004975. [DOI] [PubMed] [Google Scholar]
- 8.Kotagiri N, Niedzwiedzki DM, Ohara K, Achilefu S. Activatable probes based on distance-dependent luminescence associated with Cerenkov radiation. Angew Chem Int Ed Engl. 2013;52:7756–7760. doi: 10.1002/anie.201302564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Best MD. Click chemistry and bioorthogonal reactions: unprecedented selectivity in the labeling of biological molecules. Biochemistry. 2009;48:6571–6584. doi: 10.1021/bi9007726. [DOI] [PubMed] [Google Scholar]
- 10.Schieber C, Bestetti A, Lim JP, Ryan AD, Nguyen TL, Eldridge R, White AR, Gleeson PA, Donnelly PS, Williams SJ, Mulvaney P. Conjugation of transferrin to azide-modified CdSe/ZnS core-shell quantum dots using cyclooctyne click chemistry. Angew Chem Int Ed Engl. 2012;51:10523–10527. doi: 10.1002/anie.201202876. [DOI] [PubMed] [Google Scholar]
- 11.Jain RK. Physiological barriers to delivery of monoclonal antibodies and other macromolecules in tumors. Cancer Res. 1990;50:814s–819s. [PubMed] [Google Scholar]
- 12.Yokota T, Milenic DE, Whitlow M, Schlom J. Rapid tumor penetration of a single-chain Fv and comparison with other immunoglobulin forms. Cancer Res. 1992;52:3402–3408. [PubMed] [Google Scholar]
- 13.Walt DR. Techview: molecular biology. Bead-based fiber-optic arrays. Science. 2000;287:451–452. doi: 10.1126/science.287.5452.451. [DOI] [PubMed] [Google Scholar]
- 14.Meza MB. Bead-based HTS applications in drug discovery. Drug Discovery Today. 2000;5(Supplement 1):38–41. [Google Scholar]
- 15.Xu X, Sarder P, Kotagiri N, Achilefu S, Nehorai A. Performance analysis and design of position-encoded microsphere arrays using the Ziv-Zakai bound. IEEE Trans Nanobioscience. 2013;12:29–40. doi: 10.1109/TNB.2012.2230116. [DOI] [PubMed] [Google Scholar]
- 16.Sharpless NE, Flavin M. The reactions of amines and amino acids with maleimides. Structure of the reaction products deduced from infrared and nuclear magnetic resonance spectroscopy. Biochemistry. 1966;5:2963–2971. doi: 10.1021/bi00873a028. [DOI] [PubMed] [Google Scholar]
- 17.Kato K, Lian LY, Barsukov IL, Derrick JP, Kim H, Tanaka R, Yoshino A, Shiraishi M, Shimada I, Arata Y, et al. Model for the complex between protein G and an antibody Fc fragment in solution. Structure. 1995;3:79–85. doi: 10.1016/s0969-2126(01)00136-8. [DOI] [PubMed] [Google Scholar]
- 18.Akerstrom B, Brodin T, Reis K, Bjorck L. Protein G: a powerful tool for binding and detection of monoclonal and polyclonal antibodies. J Immunol. 1985;135:2589–2592. [PubMed] [Google Scholar]
- 19.Patel D, Lahiji A, Patel S, Franklin M, Jimenez X, Hicklin DJ, Kang X. Monoclonal antibody cetuximab binds to and down-regulates constitutively activated epidermal growth factor receptor vIII on the cell surface. Anticancer Res. 2007;27:3355–3366. [PubMed] [Google Scholar]
- 20.Arnoletti JP, Buchsbaum DJ, Huang ZQ, Hawkins AE, Khazaeli MB, Kraus MH, Vickers SM. Mechanisms of resistance to Erbitux (anti-epidermal growth factor receptor) combination therapy in pancreatic adenocarcinoma cells. J Gastrointest Surg. 2004;8:960–969. doi: 10.1016/j.gassur.2004.09.021. [DOI] [PubMed] [Google Scholar]
- 21.Schlessinger J. Ligand-induced, receptor-mediated dimerization and activation of EGF receptor. Cell. 2002;110:669–672. doi: 10.1016/s0092-8674(02)00966-2. [DOI] [PubMed] [Google Scholar]
- 22.Spaargaren M, Defize LH, Boonstra J, de Laat SW. Antibody-induced dimerization activates the epidermal growth factor receptor tyrosine kinase. J Biol Chem. 1991;266:1733–1739. [PubMed] [Google Scholar]
- 23.Xu X, Sarder P, Li Z, Nehorai A. Optimization of microfluidic microsphere-trap arrays. Biomicrofluidics. 2013;7 doi: 10.1063/1.4793713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Asgeirsson KS, Agrawal A, Allen C, Hitch A, Ellis IO, Chapman C, Cheung KL, Robertson JF. Serum epidermal growth factor receptor and HER2 expression in primary and metastatic breast cancer patients. Breast Cancer Res. 2007;9 doi: 10.1186/bcr1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Buss JE, Kudlow JE, Lazar CS, Gill GN. Altered epidermal growth factor (EGF)-stimulated protein kinase activity in variant A431 cells with altered growth responses to EGF. Proc Natl Acad Sci U S A. 1982;79:2574–2578. doi: 10.1073/pnas.79.8.2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weigum SE, Floriano PN, Christodoulides N, McDevitt JT. Cell-based sensor for analysis of EGFR biomarker expression in oral cancer. Lab Chip. 2007;7:995–1003. doi: 10.1039/b703918b. [DOI] [PubMed] [Google Scholar]
- 27.Baron AT, Cora EM, Lafky JM, Boardman CH, Buenafe MC, Rademaker A, Liu D, Fishman DA, Podratz KC, Maihle NJ. Soluble epidermal growth factor receptor (sEGFR/sErbB1) as a potential risk, screening, and diagnostic serum biomarker of epithelial ovarian cancer. Cancer Epidemiol Biomarkers Prev. 2003;12:103–113. [PubMed] [Google Scholar]
- 28.Sandri MT, Johansson HA, Zorzino L, Salvatici M, Passerini R, Maisonneuve P, Rocca A, Peruzzotti G, Colleoni M. Serum EGFR and serum HER-2/neu are useful predictive and prognostic markers in metastatic breast cancer patients treated with metronomic chemotherapy. Cancer. 2007;110:509–517. doi: 10.1002/cncr.22825. [DOI] [PubMed] [Google Scholar]
- 29.Quaranta M, Divella R, Daniele A, Di Tardo S, Venneri MT, Lolli I, Troccoli G. Epidermal growth factor receptor serum levels and prognostic value in malignant gliomas. Tumori. 2007;93:275–280. doi: 10.1177/030089160709300308. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.