Abstract
Epigenetic processes, such as changes in DNA methylation, likely mediate the link between environmental exposures in utero and altered gene expression. Differentially methylated regions (DMRs) that regulate imprinted genes may be especially vulnerable to environmental exposures since imprinting is established and maintained largely through DNA methylation, resulting in expression from only one parental chromosome. We used the human embryonic kidney cell line, HEK-293, to investigate the effects of exposure to physiologically relevant doses of lead acetate (Pb) on the methylation status of nine imprinted gene DMRs. We assessed mean methylation after seventy-two hours of Pb exposure (0-25 μg/dL) using bisulfite pyrosequencing. The PEG1/MEST and IGF2 DMRs had maximum methylation decreases of 9.6% (20 μg/dL; p< 0.005) and 3.8% (25 μg/dL; p< 0.005), respectively. Changes at the MEG3 DMRs had a maximum decrease in methylation of 2.9% (MEG3) and 1.8% (MEG3-IG) at 5μg/dL Pb, but were not statistically significant. The H19, NNAT, PEG3, PLAGL1, and SGCE/PEG10 DMRs showed a less than 0.5% change in methylation for (across the dose range used), and were deemed non-responsive to Pb in our model. Pb exposure below reportable/actionable levels increased expression of PEG1/MEST concomitant with decreased methylation. These results suggest that Pb exposure can stably alter the regulatory capacity of multiple imprinted DMRs.
Keywords: DNA methylation, epigenetics, insulin-like growth factor 2 (IGF2), paternally expressed gene 1 / mesoderm specific transcript (PEG1/MEST), lead acetate, bisulfite pyrosequencing
Introduction
The developmental origins of health and disease (DoHaD) hypothesis states that the origin of many diseases manifested later in life is established based on responses to perceived conditions in the in utero environment (Barker et al., 1989). Numerous epidemiological studies support this hypothesis, for example, individuals exposed to severe caloric restriction in utero have a higher incidence of type 2 diabetes (Li et al., 2010; Ravelli et al., 1998) coronary heart disease (Roseboom et al., 2000), neurological disorders such as schizophrenia and ADHD (Kyle and Pichard, 2006), obesity (Ravelli et al., 1999), and breast cancer (Elias et al., 2004; Painter et al., 2006). Because of fetal programming, a stimulus or insult during sensitive periods of early life development can have permanent effects on structure, physiology and metabolism in later life (Godfrey and Barker, 2001; Lucas, 1994). Epigenetics plays a vital role in the ability to respond to nutritional and environmental factors that can lead to persistent changes in regulatory and growth-related gene expression as an important component of fetal programming (Gluckman and Hanson, 2004; Santos and Dean, 2004).
Epigenetic mechanisms, including modifications of histone proteins, DNA methylation, and the action of noncoding RNAs, all contribute to regulating repression or activation of gene expression. Due to its mitotic stability, DNA methylation is of particular interest with regard to determining if patterns of methylation can provide a record of past exposures (Heijmans et al., 2009; Heijmans et al., 2008; Hoyo et al., 2009; Mathers, 2007). The differentially methylated regions (DMRs) that regulate imprinted genes are established during gametogenesis and early embryogenesis and are stably maintained throughout somatic cell division (Hackett and Surani, 2013; Reik and Walter, 2001). These DMRs are reasonable for study as an archival record for several reasons. First, they have a known baseline level of methylation from which deviations are detectable; second, these regions are critical for the appropriate establishment and maintenance of imprinted gene expression; third, imprinted genes are essential for appropriate placental function, early development and growth; fourth, the DMRs exhibit parental origin-specific methylation profiles that are spatially and temporally stable such that methylation changes at imprinted gene DMRs resulting from in utero exposure to famine have been detected six decades post-exposure (Heijmans et al., 2008); and last, these DMRs are known to exhibit shifts in methylation in response to a wide variety of other exposures, including maternal stress (Heijmans et al., 2008; Hochberg et al., 2011; Hoyo et al., 2011; Joubert et al., 2012; Murphy et al., 2012a; Timmermans et al., 2009; Tobi et al., 2009), and are widely deregulated in cancer (Sharma et al., 2010); The DMRs of many imprinted genes in humans have been characterized (Hoyo et al., 2011; Murphy et al., 2012b; Skaar et al., 2012; Woodfine et al., 2011) and provide relevant loci to investigate possible epigenetic responses resulting from environmental exposures (Coolen et al., 2011).
In the US ~24 million households have Pb paint and increased levels of Pb-contaminated dust (CDC, 2012). Children are the most susceptible to Pb exposure and poisoning because they have an extended mouthing and teething phase of development, which can expose them to Pb-containing dust, the most common route of exposure. During childhood there are thought to be windows of vulnerability during which environmental exposure-mediated epigenetic changes can have detrimental consequences for adult health (Martorell et al., 2010; Sullivan, 2010; Victora et al., 2008) In the Cincinnati Lead Study, heightened vulnerability to Pb exposure occurs up to age 7 years, during which imprinted DMRs may become epigenetically deregulated 2. Those at greatest risk for Pb exposure are those children living at or below the poverty level, in older housing, and children who are members of ethnic minority groups (CDC, 2012).
A growing body of research has shown that, even at low levels, Pb exposure has negative biological consequences. Exposure to <5 μg/dL Pb during development has adverse impacts on cognitive and behavioral functions and neurochemical systems, causing permanent deficiencies in learning, memory, attention, and executive function (Cecil et al., 2008; Nigg et al., 2008; Surkan et al., 2007). The link between developmental Pb exposure and cognitive impairment in humans is hypothesized to involve epigenetic modifications and such associations have indeed been demonstrated in whole blood (Hogg et al., 2012; Pilsner et al., 2009). The current reference level for Pb at which the Centers for Disease Control and Prevention recommends public health actions be initiated for children, ages 1-5 years old, is 5 μg/dL, and for adults, anyone 18 years or old, 25 μg/dL (CDC, 2012).
Materials and Methods
Cell culture and lead acetate treatment
The human embryonic kidney cell line, HEK-293, was obtained from American Type Culture Collection (ATCC, Manassas, VA). HEK-293 cells were cultured in Eagle's minimum essential media (EMEM, Gibco, Carlsbad, CA) supplemented with 10% Fetal Bovine Serum (FBS, SAFC BioScience), 5% penicillin streptomycin (Life Technologies, Carlsbad, CA), and 1% non-essential amino acids (Life Technologies, Carlsbad, CA). Cells (1×105) were plated in six-well plates, grown to 40-50% confluence and treated 24 hours post plating with lead (IV) acetate (Sigma; St. Louis, MO) at concentrations from 0-25 μg/dL. Seventy-two hours post treatment cells were 90-95% confluent and harvested. Genomic DNA was isolated using the Qiagen DNeasy Blood and Tissue Kit (Qiagen; Valencia, CA). Experiments were performed in four experimental replicates plated in triplicate for independent biological and technical replicates.
Methylation Analysis
Genomic DNA (800 ng) from the cells was treated with sodium bisulfite using the Zymo EZ DNA methylation kit (Zymo Research; Irvine, CA). Bisulfite treatment modifies the DNA by converting unmethylated cytosines to uracils, leaving methylated cytosines unchanged. Pyrosequencing was performed using a Qiagen Pyromark Q96 MD Pyrosequencer.
The assay conditions, primers and genomic coordinates for the DMRs analyzed herein, including IGF2, PEG1/MEST, MEG3-IG, MEG3, SGCE/PEG10, NNAT, H19 and PEG3 have been reported previously (Murphy et al., 2012b; Nye et al., 2013). Mixtures of fully methylated and fully unmethylated DNA run in duplicate/triplicate that included 0%, 25%, 50%, 75% and 100% methylated DNA showed a linear increase in methylation detected across the full range of possible values (R > 0.96). Pyrosequencing assay design was performed using PSQ Assay design software (Qiagen; Valencia, CA). The percentage of methylation for each CpG in the target sequence was calculated using PyroQ CpG software (Qiagen). All pyrosequencing assays were performed in duplicate, and values shown represent the mean methylation of the CpG sites contained within the region analyzed.
qRT-PCR assay
We used TaqMan Assays for PEG1/MEST (Hs00853380_g1), IGF2 (HS00171254) and beta-2-microglobulin (HS00187842) (Invitrogen, Carlsbad, CA). Using reverse transcriptase qPCR, cDNA was synthesized from 2 μg total RNA generated by oligo-dT priming using the SuperScript II Reverse Transcriptase first-strand synthesis kit (Invitrogen, Carlsbad, CA). The Applied Biosystems 7900H Fast Real Time PCR system (Foster City, CA) was used for real-time qRT-PCR. Relative PEG1/MEST and IGF2 expression levels were calculated using the delta delta Ct method and normalized to beta-2-microglobulin. For relative gene expression values each dose of Pb was normalized to the control and multiplied by 100 for graphical representation of the data.
Data analysis
For each DMR, we measured methylation at multiple CpG sites: four for H19, three for IGF2, eight for MEG3, four for MEG3-IG, four for PEG1/MEST, three for NNAT, ten for PEG3, six for PLAGL1, and six for SGCE/PEG10. We then calculated the mean methylation for these CpG sites at each DMR. The mean DNA methylation fractions at the individual CGs were analyzed and compared among the different treatment groups. We have previously reported that Cronbach's alphas for all DMRs were >0.89 allowing the use of mean methylation as the variable (Liu et al., 2012). Analysis of variance (ANOVA) was performed to determine the difference between groups for mean methylation followed by post-hoc Dunnett's test for multiple comparisons. GraphPad Prism 6 statistical software was used to perform all analyses (GraphPad Prism, CA).
Results
Exposure of the human embryonic kidney cell line HEK-293 to physiologically relevant doses of lead acetate (Pb) were investigated to determine if these doses elicit changes in DNA methylation profiles for nine imprinted gene DMRs. We used bisulfite pyrosequencing to quantitatively measure the level of methylation at CpG sites within these regions, including DMRs that regulate the genes H19 Imprinted maternally expressed transcript (H19), Insulin-like Growth Factor 2 (IGF-2), Paternally Expressed Gene 1/Mesoderm-Specific Transcript (PEG1/MEST), Neuronatin (NNAT), Paternally Expressed Gene 3 (PEG3), Pleiomorphic Adenoma Gene-Like 1 (PLAGL1), Epsilon Sarcoglycan and Paternally Expressed Gene 10 (SGCE/PEG10), and two DMRs within the chromosome 14q32.2 imprinted domain: the Maternally Expressed Gene 3 (MEG3) promoter region (MEG3 DMR); and the DMR in the intergenic region between Delta-like 1 homolog (DLK1) and MEG3 (MEG3-IG DMR).
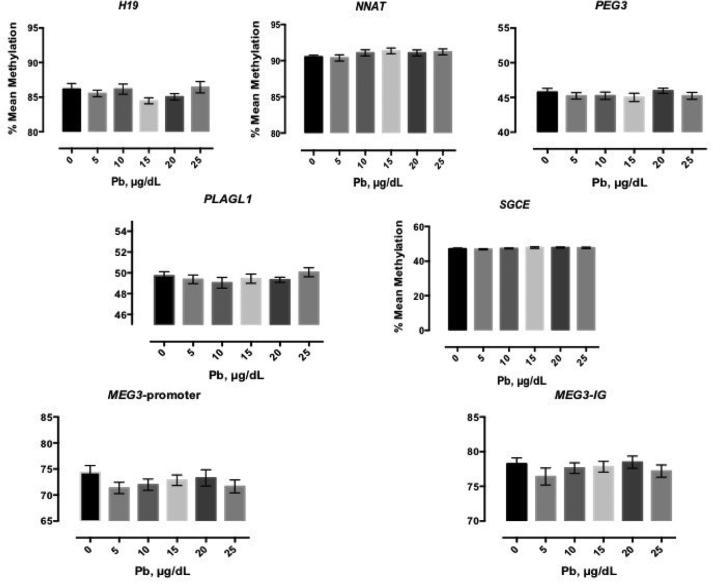
There were five DMRs (H19, NNAT, PEG3, PLAGL1, and SGCE) that showed a 0.5% fluctuation in methylation levels across the full range of Pb exposures, relative to the mock treated control; we classified them as non-Pb-responsive in our model (Figure 1). Furthermore, Pb doses from 35-55 μg/dL also showed no significant change in methylation at these regions (data not shown), indicating these particular DMRs are only minimally malleable to Pb exposure in this cell type.
Figure 1. Mean methylation of the non-responsive to Pb DMRs: H19, NNAT, PEG3, PLAGL1, SGCE, MEG3-p-promoter, MEG3-IG.
H19, NNAT, PEG3, PLAGL1, SGCE show little change in response to Pb treatment in HEK-293 cells. MEG3-promoter andMEG3-IG show slight change in response to Pb treatment. HEK-293 cells were treated for 72 hours. Shown is the mean +/− standard error for four independent experiments plated in triplicate (n=12) for each dose of Pb.
Two DMRs, MEG3- promoter and MEG3-IG, had methylation profiles that showed more variability in response to Pb exposure, but did not reach statistical significance (Figure 1, bottom graphs). For the MEG3 promoter, the mean methylation decreased up to 2.1% with Pb exposure (Figure 1, bottom left). MEG3-IG DMR had a maximum reduction of 1.8% (Figure 1, bottom right).
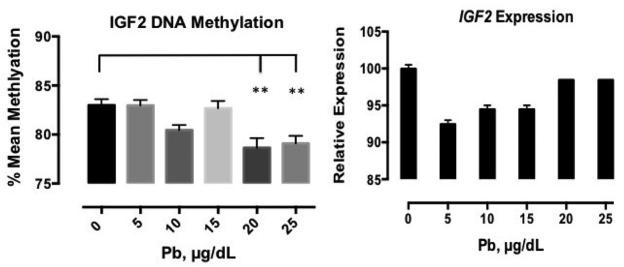
The IGF2 DMR showed a significant reduction in mean methylation in response to Pb treatment; the data after 72 hours of Pb treatment are presented in Figure 2. This decrease was most pronounced at 20 and 25 μg/dL relative to the control, a decrease of 4.3% (p< 0.001) was observed at 20 μg/dL, and at the highest dose, there was a 3.9% decrease in mean methylation of the IGF2 DMR (p< 0.001). We investigated the effects of Pb treatment on IGF2 gene expression using qPCR, and observed an overall decrease in expression over the full range of exposure. Between 0 μg/dL and 5 μg/dL, relative gene expression decreased by 7.5% but then increased 2% comparing the 5 μg/dL to the 10μg/dL and 15 μg/dL doses. At 20 and 25 μg/dL, relative expression was similar to that of the control. This data suggest that lower levels of Pb alter the expression of IGF2 more so than higher Pb doses.
Figure 2. IGF2 mean methylation and gene expression significantly altered.
Mean methylation of the IGF2 DMR and relative IGF2 gene expression in response to 0-25μg/dl Pb for 72 hours. Shown is the mean methylation +/− standard error for four independent experiments plated in triplicate, and for duplicate measures of IGF2 gene expression, which was normalized to B2M. Relative gene expression values were normalized to the control and multiplied by 100 for graphical representation of the data. ** p <0.001
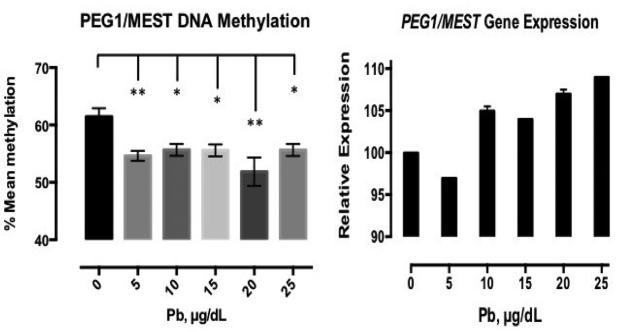
After Pb treatment, the methylation profile of PEG1/MEST showed the most dramatic shifts of all the DMRs in mean methylation (Figure 3). There was a reduction in mean methylation at all doses, from 61.5% in the control to 54.6% and 55.7% methylation at the two lowest doses. However, the most dramatic change was at the 20μg/dL dose, where there was a 9.6% reduction in mean methylation (p< 0.0001). At the 25μg/dL dose methylation was 55.7%. We next investigated the effects of Pb treatment on PEG1/MEST expression and found that expression increased at doses 10 μg/dL at the same time that methylation decreased in response to Pb treatment (Figure 3). We observed the most prominent change in PEG1/MEST expression between 5 and 10 μg/dL Pb.
Figure 3. Mean methylation of PEG1/MEST is decreased with Pb treatment while PEG1/MEST gene expression is increased.
Pb treatments, 0-25μg/dL, for 72 hours results in a significant decrease in mean methylation while gene expression is increasing. Shown is the mean methylation +/− standard error for four independent experiments plated in triplicate, and duplicate measures of PEG1/MEST gene expression, which, were normalized to B2M. Relative gene expression values were normalized to the control and multiplied by 100 for graphical representation of the data. * p<0.005, ** p<0.001.
Discussion
Our key findings are that doses of Pb below the actionable levels recommended by the CDC alter regulatory methylation with consequent changes in expression for two imprinted genes using a human cell culture model of Pb exposure. We found that the PEG1/MEST DMR was the most vulnerable to Pb exposure with a 5.8%-9.6% reduction in mean methylation across the 0-25 μg/dL dose range. For the IGF2 DMR, Pb exposure reduced methylation up to 4.3% across the same dose range. We observed a trend towards decreased methylation for the MEG3 and MEG3-IG DMRs, although changes at these DMRs did not reach statistical significance. There was no change in mean methylation at the H19, NNAT, PEG3, PLAGL1, and SGCE/PEG10 DMRs and therefore we consider them non-responsive to Pb in this model. Overall these findings suggest that, at the actionable Pb levels for children and adults, there are certain DMRs that are vulnerable to Pb treatment and others for which methylation levels are stably maintained within this cell model. The public health significance for these findings s that people who were exposed to Pb in early life and chronically exposed to low levels throughout adulthood are more likely to have altered or abnormal methylation at regions regulating imprinted genes if the results obtained here are also relevant in other cell types and what occurs in vivo.
Many imprinted genes are expressed in the brain and play a role in behavior and cognition in humans and animals (Davies et al., 2005; Isles and Wilkinson, 2000; Lefebvre et al., 1998). Epidemiological studies suggest that environmental exposures during development have a role in susceptibility to neurodevelopmental diseases and disorders in later life (Hou et al., 2012). Evidence from animal studies further supports the link between early environmental exposure and epigenetic shifts in conferring disease susceptibility (Hou et al., 2012). We investigated Pb as an environmental toxicant and showed that human cells exposed to this heavy metal exhibit functional methylation shifts at imprinted gene DMRs, including significant changes in mean methylation at the PEG1/MEST and IGF2 DMRs with consequent alterations in PEG1/MEST and IGF2 transcriptional activity.
We observed non-monotonic dose responses for IGF2, MEG3, MEG3-IG, and SGCE/PEG10, where lower doses of Pb had a greater effect on DNA methylation (Figures 1 & 2) and on IGF2 and PEG1/MEST gene expression (Figures 2 & 3). A common theme in toxicology is ‘dose makes the poison’, where it has been widely accepted that higher doses cause greater effects. As we have found here this may not always be the case. There are some biological responses that are more prominent at low doses of exposure, suggesting the need to further study low-level exposure to toxicants. In hormone receptor studies, endocrine-disrupting chemicals (EDCs) have been shown to produce non-monotonic dose response curves, and in the past 20 years several studies have identified EDCs that mimic or disrupt hormone function at low doses in ways not predicted by high-dose studies (Myers et al., 2009). Heavy metals have been classified as EDCs, so it is likely that the observed low dose effects on DNA methylation and gene expression in our study can be explained by this lack of a clear dose-response relationship (Schug et al., 2011). We hypothesize that, in addition to DNA methylation, histone modification and/or miRNA regulation may also contribute to the non-monotonic dose responses observed.
PEG1/MEST is an imprinted gene located at chromosome 7q32.2 that encodes a member of the α/β-hydrolase fold family. In mice, disruption of Peg1/Mest expression causes growth retardation, defective maternal nurturing and increased pup lethality (Lefebvre et al., 1998). It is not currently known if PEG1/MEST has similar functions in humans. However, deregulated methylation at this DMR in humans has been associated with invasive breast cancer (Pedersen et al., 1999) and cervical cancer (Vidal, 2012). Paternally expressed Insulin-like Growth Factor 2 (IGF2) is the most studied genomically imprinted gene and is located at chromosome 11p15.5. It encodes a mitogenic growth factor critically important to early growth and development and is frequently overexpressed in many types of cancer. Loss of imprinting (activation of the normally silent maternal allele) within the 11p15 chromosomal region has been shown to occur in many cancers, including colorectal (Cui et al., 2002), pancreatic, ovarian, and Wilms’ tumors (reviewed in (Feinberg, 2004)). In addition, IGF2 DMR methylation is altered in response to in utero exposures such as famine (Heijmans et al., 2008), folic acid intake (Hoyo et al., 2011), and exposure to tobacco smoke (Murphy et al., 2012a). Murphy et. al measured IGF2 DMR methylation and expression in umbilical cord blood and found a 1% change in methylation correlates with a two-fold change in transcription (Murphy et al., 2012a); this suggests that as little as a 1% decrease in methylation at this DMR can produce an expression change theoretically equivalent to that observed with complete loss of IGF2 imprinting.
Epigenetic mechanisms are proposed to link early exposure to adult-onset disease. The lack of suitable epigenetic targets to ascertain periconceptional, prenatal, and early postnatal environmental exposures has limited the ability to infer causality in retrospective epidemiologic studies. We have previously shown stability of DNA methylation at imprinted gene DMRs across fetal tissues and between birth and one year of age (Murphy et al., 2012b). To our knowledge this is the first study to demonstrate a connection between alterations of an imprinted gene's methylation profile and Pb exposure in an in vitro model. Although Pb levels as high as 25 μg/dL are no longer commonly detected, such levels were more common several decades ago. This may have led to permanent alterations in the epigenetic profile of imprinted genes in humans that make relevant the study of the effects of high versus low Pb exposure and more importantly, may have serious implications for the health of those historically exposed to such doses.
In 1991, the Secretary of the Department of Health and Human Services called Pb the “number one environmental threat to the health of children in the United States” (EPA). Pb exposure is ubiquitous in numerous ways: through air, drinking water, food, contaminated soil, deteriorating paint, and household dust. High concentrations of airborne Pb particles in the home can come from Pb-contaminated dust from outdoor sources, including contaminated soil tracked inside and use of Pb in soldering and stained-glass making. The effects of high Pb exposure on fetuses and young children can be severe, and include delays in physical and mental development, lower IQ levels, shortened attention spans, and increased behavioral problems (Canfield et al., 2003a; Canfield et al., 2003b; Needleman, 1989). Our findings that threshold actionable levels of Pb exposure resulted in DNA methylation and gene expression changes at the IGF2 and PEG1/MEST DMRs support the idea that environmental exposures can be reflected in aberrantly methylated imprinted genes. The stability of imprinted gene DMR methylation, along with their critical role in growth and development, make these DMRs suitable for further study as biomarkers of, or intermediate endpoints for, early life environmental exposures.
We used human embryonic kidney cells for all assays, cells that are derived from the mesodermal germ layer. The mesoderm also gives rise to blood cells, the circulatory and reproductive systems, muscles, connective tissues, adipose tissue, the peritoneum, cartilage, bone and blood vessels. Since ~10% of Pb is excreted in urine through the action of the kidneys, employing a kidney cell line for these studies supports that our findings have direct biologic relevance. However, the molecular mechanisms or biological importance of Pb exposure changing the methylation status of these particular genes in kidney cells is not fully understood. The use of only one cell line is a limitation of this study, and future studies should be performed using other cell types, for example human neural stem cells may allow for understanding the biological mechanisms of Pb exposure and effects on epigenetic reprogramming and differentiation.
Other groups have reported that Pb exposure increases the risk of developing renal cell carcinoma (Boffetta et al., 2011) and IGF2 dysregulation has previously been implicated in this disease (Nonomura et al., 1997; Oda et al., 1998) as well as in Wilm's tumor (Cui et al., 2001), another cancer of the kidney. In addition, PEG1/MEST is involved in maintenance of stem cell self-renewal in the kidney (Dekel et al., 2006). These reports highlight the importance of these particular genes to kidney function and relevance to disease. However, our limited focus on DNA methylation restricts our ability to fully understand how in vitro Pb exposure relates to altered gene expression via other epigenetic mechanisms. DNA methylation is often highly coordinated with chromatin remodeling and methyl-DNA binding factors, and thus it is likely that Pb might also alter histone modifications and/or the function on noncoding RNA molecules.
Conclusions
In conclusion we modeled Pb exposure in a human cell culture system and found significant alterations of DNA methylation at the DMRs regulating PEG1/MEST and IGF2 and changes in gene expression. These findings suggest that Pb is able to alter the regulatory capacity of these imprinted DMRs. A better understanding of the molecular mechanisms that are involved in epigenetic profile shifts at imprinted gene DMRs following Pb exposure are needed for causal connections between environmental Pb exposure in utero and phenotypic consequences to be established.
Supplementary Material
Highlights.
In vitro model of lead (Pb) exposure and the effects on DNA methylation of nine imprinted genes.
Significant alterations at PEG1/MEST and IGF2 DMRs after Pb treatment.
In vitro Pb exposure decreases methylation of PEG1/MEST by 10.7% and IGF2 by 5%.
Pb exposure increased gene expression of PEG1/MEST concomitant with decreased methylation.
Acknowledgements
We thank Zhiqing Huang for excellent technical contributions.
This work was supported by NIH grants R25CA057726, R01DK085173, R01ES016772, P01ES022831 and by US EPA grant RD-83543701. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH or the US EPA.
Abbreviations
- DMRs
Differentially Methylated Regions
- HEK-293
Human Embryonic Kidney Cells
- Pb
Lead (IV) acetate
- H19
H19 Imprinted maternally expressed transcript
- IGF2
Insulin-like Growth Factor 2
- PEG1/MEST
Paternally Expressed Gene 1 / Mesoderm-Specific Transcript
- NNAT
Neuronatin
- PEG3
Paternally Expressed Gene 3
- PLAGL1
Pleiomorphic Adenoma Gene-Like 1
- SGCE/PEG10
Epsilon Sarcoglycan / Paternally Expressed Gene 10 (SGCE/PEG10)
- EDCs
endocrine-disrupting chemicals
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Authors’ contributions
MDN participated in the design of the study, performed the experiments, and drafted the manuscript. CH conceived the idea, participated in the design of the study, and revised it critically for intellectual content. SKM conceived the idea, participated in the design and coordination of the study, and revised it critically for intellectual content. All authors read and approved the final manuscript.
Li Y, Xie C, Murphy SK, Skaar D, Nye MD, Vidal AC, Alvaro K, Jirtle RL, Hoyo C. Early Lead Exposure and DNA Methylation Alterations in Imprinted Domains.
REFERENCES
- Barker DJ, Osmond C, Law CM. The intrauterine and early postnatal origins of cardiovascular disease and chronic bronchitis. J Epidemiol Community Health. 1989;43:237–240. doi: 10.1136/jech.43.3.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boffetta P, Fontana L, Stewart P, Zaridze D, Szeszenia-Dabrowska N, Janout V, Bencko V, Foretova L, Jinga V, Matveev V, Kollarova H, Ferro G, Chow WH, Rothman N, van Bemmel D, Karami S, Brennan P, Moore LE. Occupational exposure to arsenic, cadmium, chromium, lead and nickel, and renal cell carcinoma: a case-control study from Central and Eastern Europe. Occup Environ Med. 2011;68:723–728. doi: 10.1136/oem.2010.056341. [DOI] [PubMed] [Google Scholar]
- Brewster UC, Perazella MA. A review of chronic lead intoxication: an unrecognized cause of chronic kidney disease. Am J Med Sci. 2004;327:341–347. doi: 10.1097/00000441-200406000-00008. [DOI] [PubMed] [Google Scholar]
- Canfield RL, Henderson CR, Jr., Cory-Slechta DA, Cox C, Jusko TA, Lanphear BP. Intellectual impairment in children with blood lead concentrations below 10 microg per deciliter. N Engl J Med. 2003a;348:1517–1526. doi: 10.1056/NEJMoa022848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canfield RL, Kreher DA, Cornwell C, Henderson CR., Jr. Low-level lead exposure, executive functioning, and learning in early childhood. Child Neuropsychol. 2003b;9:35–53. doi: 10.1076/chin.9.1.35.14496. [DOI] [PubMed] [Google Scholar]
- CDC 2012 http://www.cdc.gov/nceh/lead/default.htm.
- Cecil KM, Brubaker CJ, Adler CM, Dietrich KN, Altaye M, Egelhoff JC, Wessel S, Elangovan I, Hornung R, Jarvis K, Lanphear BP. Decreased brain volume in adults with childhood lead exposure. PLoS Med. 2008;5:e112. doi: 10.1371/journal.pmed.0050112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coolen MW, Statham AL, Qu W, Campbell MJ, Henders AK, Montgomery GW, Martin NG, Clark SJ. Impact of the genome on the epigenome is manifested in DNA methylation patterns of imprinted regions in monozygotic and dizygotic twins. PLoS One. 2011;6:e25590. doi: 10.1371/journal.pone.0025590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui H, Niemitz EL, Ravenel JD, Onyango P, Brandenburg SA, Lobanenkov VV, Feinberg AP. Loss of imprinting of insulin-like growth factor-II in Wilms' tumor commonly involves altered methylation but not mutations of CTCF or its binding site. Cancer Res. 2001;61:4947–4950. [PubMed] [Google Scholar]
- Cui H, Onyango P, Brandenburg S, Wu Y, Hsieh CL, Feinberg AP. Loss of imprinting in colorectal cancer linked to hypomethylation of H19 and IGF2. Cancer Res. 2002;62:6442–6446. [PubMed] [Google Scholar]
- Davies W, Isles A, Smith R, Karunadasa D, Burrmann D, Humby T, Ojarikre O, Biggin C, Skuse D, Burgoyne P, Wilkinson L. Xlr3b is a new imprinted candidate for X- linked parent-of-origin effects on cognitive function in mice. Nat Genet. 2005;37:625–629. doi: 10.1038/ng1577. [DOI] [PubMed] [Google Scholar]
- Dekel B, Metsuyanim S, Schmidt-Ott KM, Fridman E, Jacob-Hirsch J, Simon A, Pinthus J, Mor Y, Barasch J, Amariglio N, Reisner Y, Kaminski N, Rechavi G. Multiple imprinted and stemness genes provide a link between normal and tumor progenitor cells of the developing human kidney. Cancer Res. 2006;66:6040–6049. doi: 10.1158/0008-5472.CAN-05-4528. [DOI] [PubMed] [Google Scholar]
- Elias SG, Peeters PH, Grobbee DE, van Noord PA. Breast cancer risk after caloric restriction during the 1944-1945 Dutch famine. J Natl Cancer Inst. 2004;96:539–546. doi: 10.1093/jnci/djh087. [DOI] [PubMed] [Google Scholar]
- EPA http://www.epa.gov/iaq/homes/hip-lead.html.
- Feinberg AP. The epigenetics of cancer etiology. Semin Cancer Biol. 2004;14:427–432. doi: 10.1016/j.semcancer.2004.06.005. [DOI] [PubMed] [Google Scholar]
- Gluckman PD, Hanson MA. Living with the past: evolution, development, and patterns of disease. Science. 2004;305:1733–1736. doi: 10.1126/science.1095292. [DOI] [PubMed] [Google Scholar]
- Godfrey KM, Barker DJ. Fetal programming and adult health. Public Health Nutr. 2001;4:611–624. doi: 10.1079/phn2001145. [DOI] [PubMed] [Google Scholar]
- Hackett JA, Surani MA. DNA methylation dynamics during the mammalian life cycle. Philos Trans R Soc Lond B Biol Sci. 2013;368:20110328. doi: 10.1098/rstb.2011.0328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heijmans BT, Tobi EW, Lumey LH, Slagboom PE. The epigenome: archive of the prenatal environment. Epigenetics. 2009;4:526–531. doi: 10.4161/epi.4.8.10265. [DOI] [PubMed] [Google Scholar]
- Heijmans BT, Tobi EW, Stein AD, Putter H, Blauw GJ, Susser ES, Slagboom PE, Lumey LH. Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proc Natl Acad Sci U S A. 2008;105:17046–17049. doi: 10.1073/pnas.0806560105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochberg Z, Feil R, Constancia M, Fraga M, Junien C, Carel JC, Boileau P, Le Bouc Y, Deal CL, Lillycrop K, Scharfmann R, Sheppard A, Skinner M, Szyf M, Waterland RA, Waxman DJ, Whitelaw E, Ong K, Albertsson-Wikland K. Child health, developmental plasticity, and epigenetic programming. Endocr Rev. 2011;32:159–224. doi: 10.1210/er.2009-0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogg K, Price EM, Hanna CW, Robinson WP. Prenatal and perinatal environmental influences on the human fetal and placental epigenome. Clin Pharmacol Ther. 2012;92:716–726. doi: 10.1038/clpt.2012.141. [DOI] [PubMed] [Google Scholar]
- Hou L, Zhang X, Wang D, Baccarelli A. Environmental chemical exposures and human epigenetics. Int J Epidemiol. 2012;41:79–105. doi: 10.1093/ije/dyr154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyo C, Murphy SK, Jirtle RL. Imprint regulatory elements as epigenetic biosensors of exposure in epidemiological studies. J Epidemiol Community Health. 2009;63:683–684. doi: 10.1136/jech.2009.090803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyo C, Murtha AP, Schildkraut JM, Jirtle RL, Demark-Wahnefried W, Forman MR, Iversen ES, Kurtzberg J, Overcash F, Huang Z, Murphy SK. Methylation variation at IGF2 differentially methylated regions and maternal folic acid use before and during pregnancy. Epigenetics. 2011;6:928–936. doi: 10.4161/epi.6.7.16263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isles AR, Wilkinson LS. Imprinted genes, cognition and behaviour. Trends Cogn Sci. 2000;4:309–318. doi: 10.1016/s1364-6613(00)01504-7. [DOI] [PubMed] [Google Scholar]
- Joubert BR, Haberg SE, Nilsen RM, Wang X, Vollset SE, Murphy SK, Huang Z, Hoyo C, Midttun O, Cupul-Uicab LA, Ueland PM, Wu MC, Nystad W, Bell DA, Peddada SD, London SJ. 450K epigenome-wide scan identifies differential DNA methylation in newborns related to maternal smoking during pregnancy. Environ Health Perspect. 2012;120:1425–1431. doi: 10.1289/ehp.1205412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyle UG, Pichard C. The Dutch Famine of 1944-1945: a pathophysiological model of long-term consequences of wasting disease. Curr Opin Clin Nutr Metab Care. 2006;9:388–394. doi: 10.1097/01.mco.0000232898.74415.42. [DOI] [PubMed] [Google Scholar]
- Lefebvre L, Viville S, Barton SC, Ishino F, Keverne EB, Surani MA. Abnormal maternal behaviour and growth retardation associated with loss of the imprinted gene Mest. Nat Genet. 1998;20:163–169. doi: 10.1038/2464. [DOI] [PubMed] [Google Scholar]
- Li Y, He Y, Qi L, Jaddoe VW, Feskens EJ, Yang X, Ma G, Hu FB. Exposure to the Chinese famine in early life and the risk of hyperglycemia and type 2 diabetes in adulthood. Diabetes. 2010;59:2400–2406. doi: 10.2337/db10-0385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Murphy SK, Murtha AP, Fuemmeler BF, Schildkraut J, Huang Z, Overcash F, Kurtzberg J, Jirtle R, Iversen ES, Forman MR, Hoyo C. Depression in pregnancy, infant birth weight and DNA methylation of imprint regulatory elements. Epigenetics. 2012;7:735–746. doi: 10.4161/epi.20734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas A. Role of nutritional programming in determining adult morbidity. Arch Dis Child. 1994;71:288–290. doi: 10.1136/adc.71.4.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martorell R, Melgar P, Maluccio JA, Stein AD, Rivera JA. The nutrition intervention improved adult human capital and economic productivity. J Nutr. 2010;140:411–414. doi: 10.3945/jn.109.114504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathers JC. Early nutrition: impact on epigenetics. Forum Nutr. 2007;60:42–48. doi: 10.1159/000107066. [DOI] [PubMed] [Google Scholar]
- Murphy SK, Adigun A, Huang Z, Overcash F, Wang F, Jirtle RL, Schildkraut JM, Murtha AP, Iversen ES, Hoyo C. Gender-specific methylation differences in relation to prenatal exposure to cigarette smoke. Gene. 2012a;494:36–43. doi: 10.1016/j.gene.2011.11.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy SK, Huang Z, Hoyo C. Differentially methylated regions of imprinted genes in prenatal, perinatal and postnatal human tissues. PLoS One. 2012b;7:e40924. doi: 10.1371/journal.pone.0040924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers JP, Zoeller RT, vom Saal FS. A clash of old and new scientific concepts in toxicity, with important implications for public health. Environ Health Perspect. 2009;117:1652–1655. doi: 10.1289/ehp.0900887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Needleman HL. The persistent threat of lead: a singular opportunity. Am J Public Health. 1989;79:643–645. doi: 10.2105/ajph.79.5.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigg JT, Knottnerus GM, Martel MM, Nikolas M, Cavanagh K, Karmaus W, Rappley MD. Low blood lead levels associated with clinically diagnosed attention- deficit/hyperactivity disorder and mediated by weak cognitive control. Biol Psychiatry. 2008;63:325–331. doi: 10.1016/j.biopsych.2007.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonomura N, Nishimura K, Miki T, Kanno N, Kojima Y, Yokoyama M, Okuyama A. Loss of imprinting of the insulin-like growth factor II gene in renal cell carcinoma. Cancer Res. 1997;57:2575–2577. [PubMed] [Google Scholar]
- Nye MD, Hoyo C, Huang Z, Vidal AC, Wang F, Overcash F, Smith JS, Vasquez B, Hernandez B, Swai B, Oneko O, Mlay P, Obure J, Gammon MD, Bartlett JA, Murphy SK. Associations between methylation of paternally expressed gene 3 (PEG3), cervical intraepithelial neoplasia and invasive cervical cancer. PLoS One. 2013;8:e56325. doi: 10.1371/journal.pone.0056325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda H, Kume H, Shimizu Y, Inoue T, Ishikawa T. Loss of imprinting of igf2 in renal-cell carcinomas. Int J Cancer. 1998;75:343–346. doi: 10.1002/(sici)1097-0215(19980130)75:3<343::aid-ijc3>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- Painter RC, De Rooij SR, Bossuyt PM, Osmond C, Barker DJ, Bleker OP, Roseboom TJ. A possible link between prenatal exposure to famine and breast cancer: a preliminary study. Am J Hum Biol. 2006;18:853–856. doi: 10.1002/ajhb.20564. [DOI] [PubMed] [Google Scholar]
- Pedersen IS, Dervan PA, Broderick D, Harrison M, Miller N, Delany E, O'Shea D, Costello P, McGoldrick A, Keating G, Tobin B, Gorey T, McCann A. Frequent loss of imprinting of PEG1/MEST in invasive breast cancer. Cancer Res. 1999;59:5449–5451. [PubMed] [Google Scholar]
- Pilsner JR, Hu H, Ettinger A, Sanchez BN, Wright RO, Cantonwine D, Lazarus A, Lamadrid-Figueroa H, Mercado-Garcia A, Tellez-Rojo MM, Hernandez-Avila M. Influence of prenatal lead exposure on genomic methylation of cord blood DNA. Environ Health Perspect. 2009;117:1466–1471. doi: 10.1289/ehp.0800497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravelli AC, van der Meulen JH, Michels RP, Osmond C, Barker DJ, Hales CN, Bleker OP. Glucose tolerance in adults after prenatal exposure to famine. Lancet. 1998;351:173–177. doi: 10.1016/s0140-6736(97)07244-9. [DOI] [PubMed] [Google Scholar]
- Ravelli AC, van Der Meulen JH, Osmond C, Barker DJ, Bleker OP. Obesity at the age of 50 y in men and women exposed to famine prenatally. Am J Clin Nutr. 1999;70:811–816. doi: 10.1093/ajcn/70.5.811. [DOI] [PubMed] [Google Scholar]
- Reik W, Walter J. Genomic imprinting: parental influence on the genome. Nat Rev Genet. 2001;2:21–32. doi: 10.1038/35047554. [DOI] [PubMed] [Google Scholar]
- Roseboom TJ, van der Meulen JH, Osmond C, Barker DJ, Ravelli AC, Schroeder-Tanka JM, van Montfrans GA, Michels RP, Bleker OP. Coronary heart disease after prenatal exposure to the Dutch famine, 1944-45. Heart. 2000;84:595–598. doi: 10.1136/heart.84.6.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos F, Dean W. Epigenetic reprogramming during early development in mammals. Reproduction. 2004;127:643–651. doi: 10.1530/rep.1.00221. [DOI] [PubMed] [Google Scholar]
- Schug TT, Janesick A, Blumberg B, Heindel JJ. Endocrine disrupting chemicals and disease susceptibility. J Steroid Biochem Mol Biol. 2011;127:204–215. doi: 10.1016/j.jsbmb.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Kelly TK, Jones PA. Epigenetics in cancer. Carcinogenesis. 2010;31:27–36. doi: 10.1093/carcin/bgp220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skaar DA, Li Y, Bernal AJ, Hoyo C, Murphy SK, Jirtle RL. The human imprintome: regulatory mechanisms, methods of ascertainment, and roles in disease susceptibility. ILAR J. 2012;53:341–358. doi: 10.1093/ilar.53.3-4.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan LM. 2010 http://www.thousanddays.org/
- Surkan PJ, Zhang A, Trachtenberg F, Daniel DB, McKinlay S, Bellinger DC. Neuropsychological function in children with blood lead levels <10 microg/dL. Neurotoxicology. 2007;28:1170–1177. doi: 10.1016/j.neuro.2007.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmermans S, Jaddoe VW, Hofman A, Steegers-Theunissen RP, Steegers EA. Periconception folic acid supplementation, fetal growth and the risks of low birth weight and preterm birth: the Generation R Study. Br J Nutr. 2009;102:777–785. doi: 10.1017/S0007114509288994. [DOI] [PubMed] [Google Scholar]
- Tobi EW, Lumey LH, Talens RP, Kremer D, Putter H, Stein AD, Slagboom PE, Heijmans BT. DNA methylation differences after exposure to prenatal famine are common and timing- and sex-specific. Hum Mol Genet. 2009;18:4046–4053. doi: 10.1093/hmg/ddp353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Victora CG, Adair L, Fall C, Hallal PC, Martorell R, Richter L, Sachdev HS, Maternal, Child Undernutrition Study, G. Maternal and child undernutrition: consequences for adult health and human capital. Lancet. 2008;371:340–357. doi: 10.1016/S0140-6736(07)61692-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodfine K, Huddleston JE, Murrell A. Quantitative analysis of DNA methylation at all human imprinted regions reveals preservation of epigenetic stability in adult somatic tissue. Epigenetics Chromatin. 2011;4:1. doi: 10.1186/1756-8935-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.