Abstract
During the past decade, various types of small non-coding RNAs were found to be expressed in all kingdoms and phyla of life. Intense research efforts have begun to shed light on their biological functions, although much remains to be determined in order to fully characterize their scope of biological action. Typically, small RNAs provide sequence specificity to a protein complex that is driven to silence a long target RNA. MicroRNAs (miRNAs) are small RNAs that are coded in the genome of most eukaryotes, and contribute to the cellular identity by regulating cell-specific gene networks by translational repression or degradation of mRNA. These effects commonly fine-tune gene expression associated with developmental or environmental cues. Different cell types can be characterized by their distinctive cellular miRNA landscape. The human placenta expresses a unique set of miRNAs, a high proportion of which is derived from a large cluster located on chromosome 19, (termed chromosome 19 miRNA cluster, or C19MC). Interestingly, a fraction of these placenta-enriched miRNAs are released to the extracellular environment through exosomes that were recently found to induce an antiviral immunity. In this review, we explore relevant placental viral infections and discuss the antiviral role of exosome-packaged placental C19MC miRNAs in this context.
Keywords: trophoblast, virus, C19MC, trophomiR, exosomes
Introduction
In eutherian organisms, the placenta shields the embryo from hematogenous spread of diverse pathogens. Within the human hemochorial placenta, the physical barrier interfacing the maternal and fetal blood systems includes the trophoblast bilayer, basement membrane, and fetal capillary endothelial cells. Located at the placental villous surface and in direct contact with the maternal blood are the syncytiotrophoblasts, which are multinucleated, terminally differentiated cells that regulate the vital maternal-fetal exchange of gases, nutrients, and waste products. Subjacent to the syncytiotrophoblasts is a discontinuous layer of mononucleated cytotrophoblasts, which are less thoroughly differentiated than their syncytialized counterparts and retain a proliferative capacity. Previously thought to be a passive barrier to pathogens or an inert sieve to nutrients, it is now clear that trophoblasts actively orchestrate an extensive repertoire of signals designed to optimize placental transport functions, produce crucial hormones, and immunologically defend the developing fetus.
Understanding host-pathogen interactions at the maternal-fetal interface is essential for the development of antimicrobial therapies and for the preventive strategies that target fetal infections. Despite intense research efforts and progress over the last decades, the defense mechanisms used by the placenta against microbial infections remain largely unknown. Some level of protection is conferred by the presence of a continuous layer of syncytiotrophoblasts, which harbors relatively few intercellular junctions as well as a reduced level of membrane proteins that may serve as receptors for pathogens (Robbins et al., 2012; Zeldovich et al., 2012). Recent studies also suggest that complex cellular interactions among the compartments of the materno-placental unit create a unique immunological environment for the development and function of placental trophoblasts while avoiding the production of rejection signals.
Pathways involved in the maternal immune adaptation to pregnancy have been described elsewhere (Mor et al., 2010; Erlebacher, 2013; Zenclussen, 2013). Here we focus on viral infection of the feto-placental unit and a recently identified microRNA-based component of an antiviral defense system deployed during pregnancy. The microRNAs (miRNAs) involved in this process are uniquely enriched in trophoblasts (we termed them “trophomiRs”) and are expressed from a large cluster on chromosome 19 referred to as the chromosome 19 miRNA cluster (C19MC). These findings are intriguing, because RNA interference (RNAi) by small RNAs is prevalent in antiviral defense across most phyla, yet it was thought that the majority of antiviral signals in vertebrates are carried out by proteins, not RNAi (review in (Ding, 2010; Yan et al., 2012), a notion that was recently challenged (see below). Thus, the finding that C19MC miRNAs originating from human trophoblasts confer resistance to viral infections in recipient cells (Delorme-Axford et al., 2013) may open a new chapter in the evolving story concerning the functions of miRNAs in human physiology.
Viruses and pregnancy
A long-standing controversy exists regarding the susceptibility of pregnant women to viral infections, with a prevailing speculation that attributes “pregnancy-associated immune deficiency” to suppression of cellular immunity and tolerance of the semiallogeneic fetus and placenta. There are conflicting data regarding specific immune mediators that may explain this tolerance, with some attributed to the high level of steroid hormones during pregnancy, particularly progesterone (Arck et al., 2007). Recent data regarding the role of competent immunity in implantation and intrauterine development, and the intricate immune signals that govern maternal -placental interactions, challenge the gestational immune suppression dogma. Notwithstanding, the level of immunity may vary during pregnancy, with an immunogenic state early in pregnancy, which is attenuated until late in pregnancy, when inflammation is associated with labor (Mor et al., 2010).
Although assessing the likelihood of vertical transmission of viruses to the fetus outside of pregnancy is naturally impossible, the susceptibility of pregnant women to viral pathogens can be compared to that of non-pregnant women. Studies in this field are confounded by the greater medical attention received during pregnancy, and the severity of infections. It is clear that (a) several maternal viral pneumonias, such as influenza, coronavirus (SARS), varicella, rubeola, and other pneumonias, may become more severe during pregnancy compared to their course in non-pregnant individuals (Longman et al., 2007) and (b) transmission of viral pathogens to the intrauterine environment may damage the developing embryo (Kaplan, 1993).
Certain viruses can gain access to the placenta and the fetus and drastically impact the outcome of pregnancy (Duff et al., 2009). Viruses may move into the intrauterine compartment through diverse routes, including (a) infected sperm during fertilization, (b) intrauterine viral infections prior to and during implantation, (c) hematogenous transmission across the placenta by direct infection of trophoblasts or through intercellular routes, (d) placental transfer of infected macrophages from the maternal blood supply, (e) infection during an invasive procedure during pregnancy, (f) ascending infection from the vagina via intact fetal membranes or after membrane rupture, and (g) infection during passage of the fetus at birth through the vaginal canal or through the abdominal incision during cesarean delivery. Mother-to-baby transmission of viruses may also reflect postnatal routes through direct contact, aerosols, and breast milk. Notably, certain viruses may not access the intrauterine compartment or may lack a marked, direct effect on the fetus or the placenta, yet may adversely impact the course of pregnancy by causing significant maternal morbidity, affecting specific organs or causing a general maternal inflammatory response, with secondary consequences to the fetoplacental unit. Viruses may also predispose to bacterial infections, which may entail subsequent complications (Cardenas et al., 2010; Racicot et al., 2013).
Cytomegalovirus (CMV) is one of the most important pathogens during human pregnancy and is a leading cause of congenital viral infection. This DNA virus is a member of the herpesvirus family and can be transmitted to the fetus by either a transplacental or a trans-vaginal route. Fetal infection is most severe in the case of primary maternal CMV infection, particularly during the early stages of pregnancy. Approximately 30–40% of fetuses become infected following a primary maternal infection, and nearly one-half of these fetuses display symptomatic infection at birth (Nigro et al., 2005). The clinical impact of CMV infection during pregnancy ranges from a mild viral syndrome and maternal hepatitis to grave fetal effects, including growth restriction and central nervous system involvement manifested as microcephalus, mental retardation, developmental delay, hearing loss, visual disturbances, or death. The rate of transmission is markedly lower in recurrent maternal CMV infection, and fetal infection, if it occurs, is usually milder (Duff et al., 2009; Syggelou et al., 2010).
Like CMV, infection by rubella, an RNA virus of the togavirus family, can be transmitted to the fetus by a transplacental or trans-vaginal route, with infectivity nearing 50% in the first trimester and declining thereafter. Prior to widespread immunization, infection by rubella caused the congenital rubella syndrome, characterized by deafness, cataracts and chorioretinitis, CNS defects, and cardiac malformations (Best, 2007; Duff et al., 2009). Parvovirus, a DNA virus can also cause significant fetal morbidity. Parvovirus B19 can cross the placenta, bind to red blood cell progenitors and suppresses erythropoiesis, resulting in non-immune hydrops (Ergaz et al., 2006). Like CMV and rubella, the incidence of fetal parvovirus B19 infection declines as pregnancy progresses.
A number of other pertinent human viral pathogens can affect the newborn, but are less likely to be transmitted transplacentally during pregnancy. Intrauterine spread by these viruses may represent transmission by a different pathway or feto-placental resistance. Although intrauterine infection by herpes simplex virus (HSV), a DNA virus of the herpesvirus family, can cause central nervous system, cutaneous, and ophthalmologic manifestations, congenital infections are rare. Significant infection may occur during delivery, when the mother has active genital HSV infection (Marquez et al., 2011). Similarly, infection of the fetus by HIV, an RNA virus of the retrovirus family, occurs primarily during vaginal delivery or in association with an invasive procedure during pregnancy. Indeed, the rate of perinatal transmission has markedly declined in recent years due to the use of highly active anti-retroviral treatment and because of cesarean delivery in mothers who have a sustained high viral load (Connor et al., 1994).
Other viruses may affect human pregnancy through active maternal infection at the time of delivery or through a systemic maternal infection that secondarily influences the pregnancy and may cause (early or late) pregnancy loss with or without clear evidence of vertical transmission to the feto-placental unit. The RNA paramyxovirus family member mumps increases the risk of spontaneous abortion with first trimester infection, but no clear data that associate it with fetal infection or congenital anomalies have been reported. Similarly, infection by the DNA herpesvirus varicella zoster may increase the risk of early spontaneous abortion, and peripartum maternal infection may be transmitted during delivery, causing neonatal varicella infection.
Peripartum infection by human papilloma virus may be transmitted to the newborn by direct contact during vaginal delivery (Park et al., 2012). Maternal infection by rubeola (measles) may cause maternal pneumonia, and has been associated with first trimester miscarriages, preterm birth later in pregnancy, or neonatal measles associated with peripartum infection. Influenza during pregnancy may also cause serious maternal illness, with peripartum infection causing neonatal influenza (Duff et al., 2009).
Whereas the mechanisms used by viruses to infect the feto-placental unit remain largely unknown, the facts outlined above suggest that virus-specific pathways determine the susceptibility of the fetus and the intrauterine tissues to infections, which may also elicit selective immune responses. Studies designed to decipher viral tropism, invasion into placental cells, and trophoblast response to viral infection yielded variable results, which largely depend on the type of virus studied and the system used for dissecting viral infectivity (Arechavaleta-Velasco et al., 2002; Abrahams et al., 2006). CMV, for example, which readily crosses the placenta, particularly during primary maternal infections, does not infect the placenta in an ex vivo placental perfusion model (Muhlemann et al., 1995). Inconsistent results were also observed with HIV and varicella infection of trophoblasts (Douglas et al., 1992; Qureshi et al., 1996). HIV may infect the placenta and, subsequently, the fetus via placental macrophages, but not through trophoblasts, or through breaks in trophoblast integrity (McGann et al., 1994; Burton et al., 1996). Some viruses (HSV, adeno) may infect only less well-differentiated trophoblasts, with decreased infection once the cells differentiate into syncytiotrophoblasts (Parry et al., 1997a; Parry et al., 1997b).
The process of viral infection may also involve trophoblast gene expression, invasiveness, and/or apoptosis. For example, HSV affects HLA-G transport (Schust et al., 1996), and CMV reduces the invasiveness of extravillous trophoblasts (Fisher et al., 2000). Retroviruses may have an affinity for trophoblasts, and retrovirally integrated genes encode transcripts that are essential for placental formation, such as syncytin (Mi et al., 2000; Dupressoir et al., 2009; Haig, 2012).
The antiviral activity of microRNAs and other small non-coding RNAs
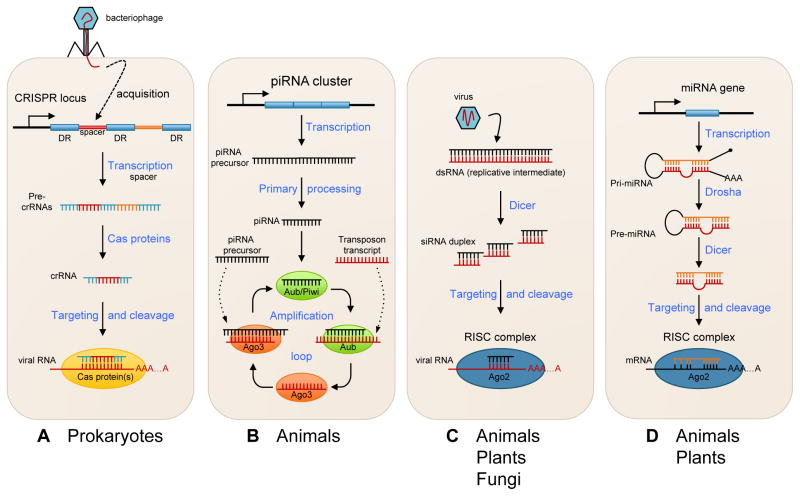
Small non-coding RNAs are ubiquitous in organisms of all kingdoms, from bacteria and archaea to plants and animals. There are various classes, characterized by specific structural features, modes of biogenesis, and biological functions. The biological significance of many small RNA species remains unclear. Recent data suggest that a large fraction of small RNAs are involved in defense mechanisms against genetic parasites such as viruses, plasmids, or transposons (Ding, 2010; Wiedenheft et al., 2012; tenOever, 2013). These pathways are schematically illustrated in Fig. 1. For instance, an important antiviral immune system in prokaryotes is the CRISPR (clustered regularly interspaced short palindromic repeats) mechanism, which is based on the production of CRISPR RNAs (crRNAs), ~60 nt in length, that target and cleave invading nucleic acid from bacteriophages or plasmids. A defining feature of this mechanism includes the incorporation of small invader-derived DNA fragments into the CRISP locus, which will be eventually integrated in the sequence of crRNAs and provide the specificity against invading genetic fragments. The crRNAs then assemble into a ribonucleic complex containing Cas (CRISPR-associated) proteins that guide the target recognition and degradation (reviews in (Sorek et al., 2008; Horvath et al., 2010; Marraffini et al., 2010; Wiedenheft et al., 2012).
Fig. 1. A simplified representation of major small RNA pathways.
(A) In the CRISPR-mediated immunity, a short segment of DNA from a virus or an invading plasmid is integrated into the CRISPR locus containing an array of short direct repeats (DR), separated by short spacer sequences. This locus is transcribed as a long primary transcript (pre-crRNA) containing multiple repeat-spacer elements and processed into short crRNAs by specific Cas (CRISPR-associated) endoribonucleases. CrRNAs are assembled in large protein complexes containing Cas proteins, which can recognize and cleave the foreign DNA. (B) The piRNA pathway in Drosophila melanogaster. A single-strand primary precursor targeting active transposons is transcribed from piRNA loci. After processing, it associates with Aubergine (AUB) or Piwi proteins. This generates secondary piRNAs that are bound by Argonaute 3 (AGO3), which in turn targets and silences transposons and generates new antisense piRNAs from primary transcripts. This amplification process is also referred to as a “ping-pong” cycle. (C) This illustrates siRNA and antiviral defense. Virus-derived siRNAs are processed from viral double-stranded RNA (dsRNA) arising from replication intermediates by members of the Dicer family of endonucleases. These viral siRNAs are then incorporated into the RISC, containing an AGO protein, and guide the targeted cleavage of viral RNAs. (D) The canonical miRNA biogenesis starts with the transcription of a long primary miRNA transcript (pri-miRNA) that can fold back on itself to form a stem loop structure. The stem loop is then released from the primary transcript following cleavage by a protein complex containing the RNase III Drosha. The resulting precursor or pre-miRNA undergoes further processing by Dicer to release a miRNA duplex. Finally, one strand is loaded onto the RISC complex and guides the complex to block translation or degrade the target mRNA.
In animals, certain defense mechanisms against mobile genetic elements or transposons are similar to the CRISPR system, where transposon sequences that have incorporated into specific genomic loci are transcribed and processed into 21- to 30-nucleotide small RNAs by members of the Piwi/Argonaute-class of proteins (review in (Malone et al., 2009). These Piwi-interacting RNAs (piRNAs) are primarily expressed in the germ line, where they control transposable element dissemination through degradation of target RNA (Brennecke et al., 2007; Gunawardane et al., 2007). PiRNAs are also involved in transcriptional silencing via chromatin modifications that can be transmitted over generations (Ashe et al., 2012; Shirayama et al., 2012).
Viral infection often results in the formation of double-stranded RNA (dsRNA) intermediates produced during replication, which may become potent triggers of antiviral immunity. In plants, fungi, and invertebrates, one of the main systems activated to ward off viral infections invokes another class of small RNAs called small interfering RNAs (siRNAs). These viral dsRNAs are typically processed into 21- to 25-nucleotide-long siRNAs by the RNA endonuclease Dicer and are loaded into the RNA-induced silencing complex (RISC) to guide the degradation of the complementary RNA. As with piRNAs, there is amplification of secondary siRNAs that expand the pool of primary siRNAs and ensure efficient silencing of the invading nucleic acids (Pak et al., 2007; Sijen et al., 2007). Consequently, virus-induced siRNAs can accumulate in infected host cells and efficiently suppress viral propagation. Notably, transposon-derived plant dsRNAs also spawn siRNAs, which can induce target degradation as well as epigenetic changes resembling animal piRNAs. Recent data establish the presence of functional antiviral RNAi pathways in vertebrates, which counteract virus-induced suppression of cellular RNAi (Li et al., 2013a; Maillard et al., 2013; tenOever, 2013).
MiRNAs represent the best-characterized class of small non-coding RNAs. These approximately 22-nucleotide-long RNAs are found in all eukaryotes with the notable exception of fungi, and are known to regulate the expression of numerous genes through inhibiting the translation of targeted mRNAs and enhancing their degradation (Bartel, 2009). Most miRNAs are derived from a limited number of genes located in independent transcription units or are embedded in introns of protein-coding genes. Transcription by RNA polymerase II gives rise to long RNA transcripts that are successively cleaved by two RNase III enzymes, Drosha and Dicer, yielding miRNA duplexes. One of the two strands is then loaded onto RISC, which contains Ago proteins as a silencing effector.
The physiological influence of miRNA action at the molecular level is usually subtle, with most miRNAs modulating low-magnitude gene expression changes, possibly to diminish stochastic transcriptional bursts (Baek et al., 2008; Selbach et al., 2008). Nonetheless, miRNAs exhibit a wide range of biological actions, affecting many cellular pathways, including the regulation of the immune response, by controlling the development of immune cells and immune signaling networks (Xiao et al., 2009; Baumjohann et al., 2013). Although miRNAs have the potential to modulate the immune response, they do not appear to significantly silence viral RNAs, possibly reflecting the relatively low level of expression when compared to viral RNAs, the low silencing capacity of miRNAs, miRNA kinetics relative to the half-life of relevant target protein, and the fact that only a limited number of miRNAs are encoded in the genome (tenOever, 2013). Intriguingly, there are examples of viruses able to subvert the RNAi machinery for their own benefit (Cullen, 2013). For example, many viruses of the herpes and picornavirus families encode viral miRNAs that target host or viral transcripts to promote their replication (Grundhoff et al., 2011).
Certain virus-encoded miRNAs are mimics of specific host miRNAs, such as miR-155, an important regulator of myeloid and lymphoid cells in the immune system (Gottwein et al., 2007; Skalsky et al., 2007; Boss et al., 2011). In addition, the host liver-specific miR-122 promotes the replication of the hepatitis C virus by protecting the viral genome from degradation by cellular nucleases (Jopling et al., 2005; Li et al., 2013b), and members of the poxvirus family have the ability to degrade host cellular miRNAs (Backes et al., 2012). Some viruses can also exploit specialized enzymatic activities of the RNAi machinery in order to process viral mRNAs critical to their replication but in an RNAi-independent manner (Bouttier et al., 2012). Together, these observations point to the vast complexity and plasticity of host RNAi pathways that can interact with invading viruses to modulate their pathogenesis.
Chromosome 19 miRNA cluster (C19MC)
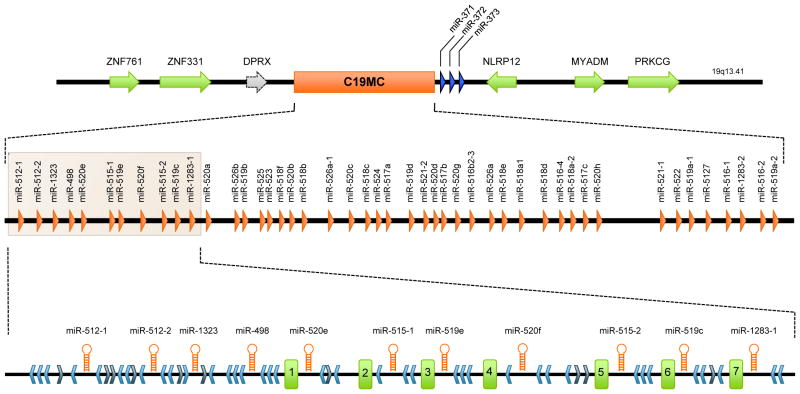
Human placental trophoblasts produce miRNAs throughout pregnancy. A large fraction of these trophoblastic miRNAs are derived from a single cluster located on chromosome 19 (Bentwich et al., 2005; Donker et al., 2012). This chromosome 19 miRNA cluster (C19MC, Fig. 2) spans approximately 100 kb of genomic DNA, and harbors 46 intronic miRNA genes flanked by numerous Alu repeats and 37 highly repetitive short DNA segments (~120 nt) that could be spliced exons of non-coding, short-lived RNA transcripts of unknown function (Bortolin-Cavaille et al., 2009).
Fig. 2. The genomic organization of the C19MC miRNA cluster.
The figure depicts the genomic region of the C19MC cluster, with the middle section depicting the position of the miRNA genes. At the bottom, an enlargement of the 5′ end of the region details the structural organization of the elements constituting the cluster. The light blue chevrons represent Alu elements in an antisense orientation relative to the miRNA genes while the dark blue chevrons correspond to sense Alu sequences. The seven first-expressed exons are represented by green rectangles numbered 1-7. The figure was modified from (Bortolin-Cavaille et al., 2009).
Recombination events among Alu repeats have been implicated in duplication events of miRNA, leading to the evolution and expansion of these clustered miRNA genes (Zhang et al., 2008; Lehnert et al., 2009). While the contribution of Alu repeats to the structural organization of this region is well delineated, the functional interaction between the C19MC miRNAs and resident Alu repeats is unclear. Interestingly, several miRNA species from C19MC share high sequence complementarity with Alu sense repeats, but not antisense sequences, suggesting that these miRNAs could target transcribed Alu sequences and participate in a defense mechanism against deleterious Alu insertions into the genome (Lehnert et al., 2009). Sequence similarity among other miRNAs and transposable elements suggested the interesting yet unproven role of miRNAs in protection of the genome against foreign genomic elements (Shalgi et al., 2010; Ahn et al., 2013).
The C19MC miRNAs are primate-specific, with an expression pattern that is highly tissue-selective and mostly restricted to the placenta. C19MC miRNAs can also be detected in the testes and the fetal brain. In cell lines, C19MC miRNAs seem to be exclusively expressed in cells derived from trophoblasts, including choriocarcinoma cell lines JEG3, JAr, and BeWo, with the notable exception of extravillous trophoblast line HTR8/SVneo (Donker et al., 2012; Morales-Prieto et al., 2012). Expression of these miRNAs can be reactivated in cell lines by treatment with 5-aza-2′-deoxycytidine (5-Aza-CdR), a DNA methylation inhibitor (Saito et al., 2009; Tsai et al., 2009), suggesting that expression of the C19MC miRNAs is epigenetically controlled, at least in part, by mechanisms involving methylation. Interestingly, a CpG-rich region located ~17 kb up-stream of the first miRNA gene was found to be hypermethylated in cells that do not express these miRNAs (Tsai et al., 2009). The fact that the C19MC cluster is imprinted and exclusively expressed in the placenta from the paternally inherited chromosome provides further support to the epigenetic regulation of C19MC miRNAs (Noguer-Dance et al., 2010).
C19MC miRNAs, exosomes and viral resistance
We recently found that primary human trophoblast (PHT) cells are resistant to infection by a diverse and unrelated panel of DNA and RNA viruses, including coxsackievirus B, poliovirus, vesicular stomatitis virus (VSV), vaccinia virus, herpes simplex virus-1, and human CMV (details about this and other findings summarized in this section are discussed in (Delorme-Axford et al., 2013) and schematically illustrated in Fig. 3). Importantly, we found that, in addition to their own resistance, PHT cells produced an antiviral component that was released into the culture medium and conferred viral resistance to non-placental cells, including Vero (African green monkey kidney), U2OS (human osteosarcoma), Caco-2 (human colorectal adenocarcinoma), and RL-95 (human endometrial adenocarcinoma), as well as to several physiologically relevant maternal and fetal primary cell types, such as human umbilical vein endothelial cells (HUVEC), human uterine microvascular endothelial cells (HUtMVEC), and human placental and human foreskin fibroblasts. Surprisingly, sonication of PHT conditioned media ablated its antiviral properties, which implied that PHT-derived lipid vesicles, which are known to be sensitive to sonication, might mediate the viral resistance.
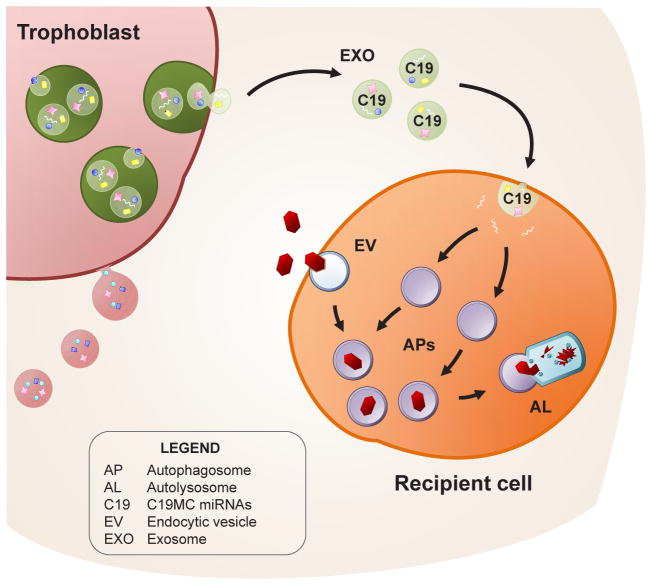
Fig. 3. Representation of exosome-mediated transfer of C19MC miRNAs (C19) to recipient cells, and the induction of viral resistance.
Primary placental trophoblasts release exosomes (EXO) containing C19MC miRNAs, which are taken up by recipient cells. These C19MC miRNAs stimulate autophagy in the recipient cells. Incoming viral particles (in red), are trafficked in endocytic vesicles (EV) from the endosomal pathway, and guided into C19MC-induced autophagosomes (APs). These autophagosomes fuse with lysosomes to form autolysosomes (AL), as a mechanism to degrade virus-containing vesicles. Used with permission, (Delorme-Axford et al., 2013).
Consistent with a role for PHT-derived exosomes in mediating the antiviral effects of PHT conditioned medium, we observed decreased viral replication in cells exposed to exosomes that were isolated and purified from PHT cell conditioned medium, but not from exosome-depleted conditioned media. Because exosomes have previously been shown to play an important role in intercellular communication through the transfer of mRNAs and miRNAs, and because PHT exosomes contain placental miRNAs that are enriched for C19MC species (Bentwich et al., 2005; Donker et al., 2012), we sought to assess the role of C19MC miRNAs in the antiviral property of PHT exosomes. We stably transfected non-placental cells that do not express members of the C19MC endogenously with a BAC that harbors the entire C19MC, and found that these cells became resistant to VSV infection compared to cells expressing a control BAC. This effect was recapitulated when miRNA mimics of select groups and/or individual members of the C19MC family (miR-517-3p, -516b-5p, and -512-3p, but not non-C19MC miRNAs) were transfected into non-placental cells.
Our findings with respect to the function of extracellular RNAs are consistent with the notion that extracellular miRNAs (ex-miRNAs) are present in a variety of human body fluids such as blood, breast milk, and urine (Chen et al., 2012). Importantly, ex-miRNAs are functionally active in reshaping the transcriptome landscape of recipient cells (Valadi et al., 2007; Vickers et al., 2011), implying that ex-miRNAs, which may be vesicle-encapsulated or bound to protein carriers in extracellular fluids, are key players in a non-hormonal modes of intercellular communication (Arroyo et al., 2011; Mittelbrunn et al., 2012; Raposo et al., 2013).
Exosomes, lipid bilayer vesicles with a median size of 100 nm, are known to mediate local or distant cross talk among diverse cells and tissues. They are involved in diverse physiological and pathological processes, including immune surveillance, virus and host interaction, and tumor niche establishment (Mittelbrunn et al., 2011; Peinado et al., 2012; Feng et al., 2013; Ramakrishnaiah et al., 2013). Prior to their release into the extracellular space, exosomes form intraluminal vesicles within multivesicular bodies (Scita et al., 2010; Hanson et al., 2012). Several components of the endosomal sorting complexes required for transport (ESCRT) and members of the Rab family, such as ALIX and Rab27, are required for exosome production, highlighting the functional connection of endocytosis with exosome biogenesis (Ostrowski et al., 2010; Baietti et al., 2012). ESCRT-independent, ceramide-dependent cascades for exosome production have been implicated in certain tissues, such as oligodendrocytes (Trajkovic et al., 2008).
Microvesicles (100–1,000 nm in diameter) are typically larger than exosomes, and originate from the plasma membrane via direct shedding. In this, they are strikingly distinct from exosomes of endocytic origin. It is therefore plausible that these two types of lipid vesicles harbor distinct cargo content and surface proteins, which might be functionally correlated with the transmission of viral resistance. Our recent work (unpublished) suggests that circulating microvesicles may also carry C19MC miRNAs, implying that additional characteristics of trophoblast vesicles determine transmission of antiviral miRNAs. Lastly, circulating ex-miRNAs are also tightly associated with certain proteins, such as high-density lipoprotein and Ago2 (Arroyo et al., 2011; Vickers et al., 2011). These miRNAs are taken up by recipient cells, where they regulate their respective mRNA targets (Vickers et al., 2011). The functional significance of this type of protein-bound ex-miRNAs in the context of antiviral activity remains to be established.
Mechanistically, we found that exposure of non-placental cells to PHT conditioned media, as well as to purified PHT exosomes, markedly stimulated autophagy (Delorme-Axford et al., 2013). An evolutionarily conserved process, autophagy utilizes the lysosomal degradation pathway to degrade foreign microbes (Choi et al., 2013). We showed that, after incubation with conditioned media or PHT-derived exosomes, there was a significant induction of autophagosomes, demonstrated by electron microscopy and LC3b punctae formation by fluorescence microscopy. In addition, we found that miRNA mimics from members of the C19MC capable of inducing viral resistance also induced autophagy. The induction of autophagy by members of the C19MC appears to be the result of increased flux through the autophagic pathway, as we observed significant degradation of p62 in cells expressing the entire C19MC. Importantly, pharmacological or siRNA-medicated inhibition of autophagy abrogated the antiviral effect of C19MC miRNAs, and viral infection was restored. Together, these observations strongly suggest a unique role for trophoblast-derived C19MC miRNAs in the conferral of resistance to viral infection to non-placental cells and implicate autophagy in this form of antiviral signaling.
Other functions of C19MC miRNAs
Although members of the C19MC represent the most abundant family of placental miRNAs, much remains to be clarified about their function beyond an antiviral immunity. Several lines of evidence suggest a role in cell differentiation and proliferation. For example, high levels of C19MC miRNAs have been observed in embryonic and induced stem cells (Bar et al., 2008; Ren et al., 2009; Stadler et al., 2010). Interestingly, their expression drops considerably when these cells differentiate (Ren et al., 2009; Stadler et al., 2010), which may indicate a role in the maintenance of an undifferentiated state. Furthermore, several reports indicate that certain C19MC miRNAs are aberrantly expressed in specific tumors, possibly reflecting reactivation of the C19MC cluster as the consequence of chromosomal rearrangement or epigenetic modifications (Saito et al., 2009; Tsai et al., 2009; Rippe et al., 2010; Liu et al., 2013). Li and colleagues showed that the C19MC locus was frequently amplified in certain pediatric brain tumors and suggested that members of this family of miRNAs had oncogenic properties in vitro and in vivo (Li et al., 2009). In contrast, miR-519, a C19MC miRNA, was shown to exhibit strong tumor-suppressive activities (Marasa et al., 2010; Abdelmohsen et al., 2012). Two other C19MC members, miR517a and miR-517c, were recently implicated in inhibition of cell proliferation and tumor-suppressing activity (Liu et al., 2013).
Supporting a role for C19MC in trophoblast differentiation, recent data in our lab (Xie L, Mouillet JF, unpublished) suggest that members of the C19MC cluster are expressed at much higher levels in villous trophoblasts compared to extravillous trophoblasts (EVT) or EVT-derived cell lines. Further research is needed to confirm a role for C19MC miRNAs in the control of trophoblast proliferation, migration, and invasion.
Conclusions and perspectives
A number of experimental observations indicate that trophoblastic C19MC miRNAs, particularly when packaged in exosomes derived from human trophoblasts, exhibit cell autonomous and transferrable antiviral activity. The precise mechanisms underlying these observations remain unknown. Moreover, it is not clear whether the antiviral activity of vesicle-packaged miRNAs intersects with other known antiviral pathways that attenuate viral infections, such as type-I interferon. Although it was recently demonstrated that miRNAs could activate RNA-sensing pathways and trigger a range of immune responses (Fabbri et al., 2012; Lehmann et al., 2012), the relevance of these mechanisms to C19MC miRNAs remains to be shown. Intriguingly, RNAi-based mechanisms represent a major antiviral defense in prokaryotes as well as in plants and invertebrates (Ding, 2010; Wiedenheft et al., 2012; Yan et al., 2012). In these organisms there is a direct interaction between small RNAs and the invading nucleic acids, which usually results in elimination of the foreign genetic elements.
Although the molecular mechanisms underlying the action of the C19MC miRNAs are currently unknown, it seems unlikely that they involve the recognition and silencing of specific viral RNAs. Instead, C19MC miRNAs may enhance cellular defense mechanisms, including autophagy (Delorme-Axford et al., 2013), via a signaling pathway that remains to be elucidated. The mechanism underlying the transport of C19MC miRNAs to other cells, the induction of an antiviral effect, and the possible selectivity of this process are also unknown.
Acknowledgments
The authors thank Lori Rideout for assistance during preparation of the manuscript and Bruce Campbell for editing. The project was supported by Pennsylvania Department of Health Research Formula Funds (to JFM), NIH R01HD065893 and NIH R21HD071707 (to YS), NIH R01AI081759 (to CBC), NIH R01HD075665 (to CBC and YS), and the Burroughs Wellcome Investigators in the Pathogenesis of Infectious Disease Award (to CBC). The project was also supported through University of Pittsburgh core resources through NIH UL1RR024153 and NIH UL1TR000005.
Abbreviations used in this paper
- C19MC
chromosome 19 miRNA cluster
- miRNA
microRNA
- PHT
primary human trophoblast
References
- ABDELMOHSEN K, SRIKANTAN S, TOMINAGA K, KANG MJ, YANIV Y, MARTINDALE JL, YANG X, PARK SS, BECKER KG, SUBRAMANIAN M, MAUDSLEY S, LAL A, GOROSPE M. Growth inhibition by miR-519 via multiple p21-inducing pathways. Mol Cell Biol. 2012;32:2530–2548. doi: 10.1128/MCB.00510-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ABRAHAMS VM, SCHAEFER TM, FAHEY JV, VISINTIN I, WRIGHT JA, ALDO PB, ROMERO R, WIRA CR, MOR G. Expression and secretion of antiviral factors by trophoblast cells following stimulation by the TLR-3 agonist, Poly(I: C) Hum Reprod. 2006;21:2432–2439. doi: 10.1093/humrep/del178. [DOI] [PubMed] [Google Scholar]
- AHN K, GIM JA, HA HS, HAN K, KIM HS. The novel MER transposon-derived miRNAs in human genome. Gene. 2013;512:422–428. doi: 10.1016/j.gene.2012.08.028. [DOI] [PubMed] [Google Scholar]
- ARCK P, HANSEN PJ, MULAC JERICEVIC B, PICCINNI MP, SZEKERES-BARTHO J. Progesterone during pregnancy: endocrine-immune cross talk in mammalian species and the role of stress. Am J Reprod Immunol. 2007;58:268–279. doi: 10.1111/j.1600-0897.2007.00512.x. [DOI] [PubMed] [Google Scholar]
- ARECHAVALETA-VELASCO F, KOI H, STRAUSS JF, 3RD, PARRY S. Viral infection of the trophoblast: time to take a serious look at its role in abnormal implantation and placentation? J Reprod Immunol. 2002;55:113–121. doi: 10.1016/s0165-0378(01)00143-7. [DOI] [PubMed] [Google Scholar]
- ARROYO JD, CHEVILLET JR, KROH EM, RUF IK, PRITCHARD CC, GIBSON DF, MITCHELL PS, BENNETT CF, POGOSOVA-AGADJANYAN EL, STIREWALT DL, TAIT JF, TEWARI M. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci USA. 2011;108:5003–5008. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ASHE A, SAPETSCHNIG A, WEICK EM, MITCHELL J, BAGIJN MP, CORDING AC, DOEBLEY AL, GOLDSTEIN LD, LEHRBACH NJ, LE PEN J, PINTACUDA G, SAKAGUCHI A, SARKIES P, AHMED S, MISKA EA. piRNAs can trigger a multigenerational epigenetic memory in the germline of C. elegans. Cell. 2012;150:88–99. doi: 10.1016/j.cell.2012.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BACKES S, SHAPIRO JS, SABIN LR, PHAM AM, REYES I, MOSS B, CHERRY S, TENOEVER BR. Degradation of host microRNAs by poxvirus poly(A) polymerase reveals terminal RNA methylation as a protective antiviral mechanism. Cell Host Microbe. 2012;12:200–210. doi: 10.1016/j.chom.2012.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BAEK D, VILLEN J, SHIN C, CAMARGO FD, GYGI SP, BARTEL DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BAIETTI MF, ZHANG Z, MORTIER E, MELCHIOR A, DEGEEST G, GEERAERTS A, IVARSSON Y, DEPOORTERE F, COOMANS C, VERMEIREN E, ZIMMERMANN P, DAVID G. Syndecan-syntenin-ALIX regulates the biogenesis of exosomes. Nat Cell Biol. 2012;14:677–685. doi: 10.1038/ncb2502. [DOI] [PubMed] [Google Scholar]
- BAR M, WYMAN SK, FRITZ BR, QI J, GARG KS, PARKIN RK, KROH EM, BENDORAITE A, MITCHELL PS, NELSON AM, RUZZO WL, WARE C, RADICH JP, GENTLEMAN R, RUOHOLA-BAKER H, TEWARI M. MicroRNA discovery and profiling in human embryonic stem cells by deep sequencing of small RNA libraries. Stem Cells. 2008;26:2496–2505. doi: 10.1634/stemcells.2008-0356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BARTEL DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BAUMJOHANN D, ANSEL KM. MicroRNA-mediated regulation of T helper cell differentiation and plasticity. Nat Rev Immunol. 2013;13:666–678. doi: 10.1038/nri3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BENTWICH I, AVNIEL A, KAROV Y, AHARONOV R, GILAD S, BARAD O, BARZILAI A, EINAT P, EINAV U, MEIRI E, SHARON E, SPECTOR Y, BENTWICH Z. Identification of hundreds of conserved and nonconserved human microRNAs. Nat Genet. 2005;37:766–770. doi: 10.1038/ng1590. [DOI] [PubMed] [Google Scholar]
- BEST JM. Rubella. Semin Fetal Neonatal Med. 2007;12:182–192. doi: 10.1016/j.siny.2007.01.017. [DOI] [PubMed] [Google Scholar]
- BORTOLIN-CAVAILLE ML, DANCE M, WEBER M, CAVAILLE J. C19MC microRNAs are processed from introns of large Pol-II, non-protein-coding transcripts. Nucleic Acids Res. 2009;37:3464–3473. doi: 10.1093/nar/gkp205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BOSS IW, NADEAU PE, ABBOTT JR, YANG Y, MERGIA A, RENNE R. A Kaposi’s sarcoma-associated herpesvirus-encoded ortholog of microRNA miR-155 induces human splenic B-cell expansion in NOD/LtSz-scid IL2Rgammanull mice. J Virol. 2011;85:9877–9886. doi: 10.1128/JVI.05558-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BOUTTIER M, SAUMET A, PETER M, COURGNAUD V, SCHMIDT U, CAZEVIEILLE C, BERTRAND E, LECELLIER CH. Retroviral GAG proteins recruit AGO2 on viral RNAs without affecting RNA accumulation and translation. Nucleic Acids Res. 2012;40:775–786. doi: 10.1093/nar/gkr762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BRENNECKE J, ARAVIN AA, STARK A, DUS M, KELLIS M, SACHIDANANDAM R, HANNON GJ. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- BURTON GJ, O’SHEA S, ROSTRON T, MULLEN JE, AIYER S, SKEPPER JN, SMITH R, BANATVALA JE. Significance of placental damage in vertical transmission of human immunodeficiency virus. J Med Virol. 1996;50:237–243. doi: 10.1002/(SICI)1096-9071(199611)50:3<237::AID-JMV5>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- CARDENAS I, MEANS RE, ALDO P, KOGA K, LANG SM, BOOTH CJ, MANZUR A, OYARZUN E, ROMERO R, MOR G. Viral infection of the placenta leads to fetal inflammation and sensitization to bacterial products predisposing to preterm labor. J Immunol. 2010;185:1248–1257. doi: 10.4049/jimmunol.1000289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHEN X, LIANG H, ZHANG J, ZEN K, ZHANG CY. Secreted microRNAs: a new form of intercellular communication. Trends Cell Biol. 2012;22:125–132. doi: 10.1016/j.tcb.2011.12.001. [DOI] [PubMed] [Google Scholar]
- CHOI AM, RYTER SW, LEVINE B. Autophagy in human health and disease. N Engl J Med. 2013;368:651–662. doi: 10.1056/NEJMra1205406. [DOI] [PubMed] [Google Scholar]
- CONNOR EM, SPERLING RS, GELBER R, KISELEV P, SCOTT G, O’SULLIVAN MJ, VANDYKE R, BEY M, SHEARER W, JACOBSON RL, et al. Reduction of maternal-infant transmission of human immunodeficiency virus type 1 with zidovudine treatment. Pediatric AIDS Clinical Trials Group Protocol 076 Study Group. N Engl J Med. 1994;331:1173–1180. doi: 10.1056/NEJM199411033311801. [DOI] [PubMed] [Google Scholar]
- CULLEN BR. MicroRNAs as mediators of viral evasion of the immune system. Nat Immunol. 2013;14:205–210. doi: 10.1038/ni.2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DELORME-AXFORD E, DONKER RB, MOUILLET JF, CHU T, BAYER A, OUYANG Y, WANG T, STOLZ DB, SARKAR SN, MORELLI AE, SADOVSKY Y, COYNE CB. Human placental trophoblasts confer viral resistance to recipient cells. Proc Natl Acad Sci USA. 2013;110:12048–12053. doi: 10.1073/pnas.1304718110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DING SW. RNA-based antiviral immunity. Nat Rev Immunol. 2010;10:632–644. doi: 10.1038/nri2824. [DOI] [PubMed] [Google Scholar]
- DONKER RB, MOUILLET JF, CHU T, HUBEL CA, STOLZ DB, MORELLI AE, SADOVSKY Y. The expression profile of C19MC microRNAs in primary human trophoblast cells and exosomes. Mol Hum Reprod. 2012;18:417–424. doi: 10.1093/molehr/gas013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DOUGLAS GC, KING BF. Maternal-fetal transmission of human immunodeficiency virus: a review of possible routes and cellular mechanisms of infection. Clin Infect Dis. 1992;15:678–691. doi: 10.1093/clind/15.4.678. [DOI] [PubMed] [Google Scholar]
- DUFF P, SWEET RL, EDWARDS RK. In: Maternal and fetal infections. Creasy and Resnik’s Maternal Fetal Medicine. CREASY RK, RESNIK R, IAMS JD, LOCKWOOD CJ, MOORE TR, editors. Philadelphia: Saunders-Elsevier; 2009. pp. 739–798. [Google Scholar]
- DUPRESSOIR A, VERNOCHET C, BAWA O, HARPER F, PIERRON G, OPOLON P, HEIDMANN T. Syncytin-A knockout mice demonstrate the critical role in placentation of a fusogenic, endogenous retrovirus-derived, envelope gene. Proc Natl Acad Sci USA. 2009;106:12127–12132. doi: 10.1073/pnas.0902925106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ERGAZ Z, ORNOY A. Parvovirus B19 in pregnancy. Reprod Toxicol. 2006;21:421–435. doi: 10.1016/j.reprotox.2005.01.006. [DOI] [PubMed] [Google Scholar]
- ERLEBACHER A. Mechanisms of T cell tolerance towards the allogeneic fetus. Nat Rev Immunol. 2013;13:23–33. doi: 10.1038/nri3361. [DOI] [PubMed] [Google Scholar]
- FABBRI M, PAONE A, CALORE F, GALLI R, GAUDIO E, SANTHANAM R, LOVAT F, FADDA P, MAO C, NUOVO GJ, ZANESI N, CRAWFORD M, OZER GH, WERNICKE D, ALDER H, CALIGIURI MA, NANA-SINKAM P, PERROTTI D, CROCE CM. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc Natl Acad Sci USA. 2012;109:E2110–2116. doi: 10.1073/pnas.1209414109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FENG Z, HENSLEY L, MCKNIGHT KL, HU F, MADDEN V, PING L, JEONG SH, WALKER C, LANFORD RE, LEMON SM. A pathogenic picornavirus acquires an envelope by hijacking cellular membranes. Nature. 2013;496:367–371. doi: 10.1038/nature12029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FISHER S, GENBACEV O, MAIDJI E, PEREIRA L. Human cytomegalovirus infection of placental cytotrophoblasts in vitro and in utero: implications for transmission and pathogenesis. J Virol. 2000;74:6808–6820. doi: 10.1128/jvi.74.15.6808-6820.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GOTTWEIN E, MUKHERJEE N, SACHSE C, FRENZEL C, MAJOROS WH, CHI JT, BRAICH R, MANOHARAN M, SOUTSCHEK J, OHLER U, CULLEN BR. A viral microRNA functions as an orthologue of cellular miR-155. Nature. 2007;450:1096–1099. doi: 10.1038/nature05992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GRUNDHOFF A, SULLIVAN CS. Virus-encoded microRNAs. Virology. 2011;411:325–343. doi: 10.1016/j.virol.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GUNAWARDANE LS, SAITO K, NISHIDA KM, MIYOSHI K, KAWAMURA Y, NAGAMI T, SIOMI H, SIOMI MC. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science. 2007;315:1587–1590. doi: 10.1126/science.1140494. [DOI] [PubMed] [Google Scholar]
- HAIG D. Retroviruses and the placenta. Curr Biol. 2012;22:R609–613. doi: 10.1016/j.cub.2012.06.002. [DOI] [PubMed] [Google Scholar]
- HANSON PI, CASHIKAR A. Multivesicular body morphogenesis. Annu Rev Cell Dev Biol. 2012;28:337–362. doi: 10.1146/annurev-cellbio-092910-154152. [DOI] [PubMed] [Google Scholar]
- HORVATH P, BARRANGOU R. CRISPR/Cas, the immune system of bacteria and archaea. Science. 2010;327:167–170. doi: 10.1126/science.1179555. [DOI] [PubMed] [Google Scholar]
- JOPLING CL, YI M, LANCASTER AM, LEMON SM, SARNOW P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309:1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- KAPLAN C. The placenta and viral infections. Semin Diagn Pathol. 1993;10:232–250. [PubMed] [Google Scholar]
- LEHMANN SM, KRUGER C, PARK B, DERKOW K, ROSENBERGER K, BAUMGART J, TRIMBUCH T, EOM G, HINZ M, KAUL D, HABBEL P, KALIN R, FRANZONI E, RYBAK A, NGUYEN D, VEH R, NINNEMANN O, PETERS O, NITSCH R, HEPPNER FL, GOLENBOCK D, SCHOTT E, PLOEGH HL, WULCZYN FG, LEHNARDT S. An unconventional role for miRNA: let-7 activates Toll-like receptor 7 and causes neurodegeneration. Nat Neurosci. 2012;15:827–835. doi: 10.1038/nn.3113. [DOI] [PubMed] [Google Scholar]
- LEHNERT S, VAN LOO P, THILAKARATHNE PJ, MARYNEN P, VERBEKE G, SCHUIT FC. Evidence for co-evolution between human microRNAs and Alu-repeats. PLoS One. 2009;4:e4456. doi: 10.1371/journal.pone.0004456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LI M, LEE KF, LU Y, CLARKE I, SHIH D, EBERHART C, COLLINS VP, VAN METER T, PICARD D, ZHOU L, BOUTROS PC, MODENA P, LIANG ML, SCHERER SW, BOUFFET E, RUTKA JT, POMEROY SL, LAU CC, TAYLOR MD, GAJJAR A, DIRKS PB, HAWKINS CE, HUANG A. Frequent amplification of a chr19q13.41 microRNA polycistron in aggressive primitive neuroectodermal brain tumors. Cancer Cell. 2009;16:533–546. doi: 10.1016/j.ccr.2009.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LI Y, LU J, HAN Y, FAN X, DING SW. RNA interference functions as an antiviral immunity mechanism in mammals. Science. 2013a;342:231–234. doi: 10.1126/science.1241911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LI Y, MASAKI T, YAMANE D, MCGIVERN DR, LEMON SM. Competing and noncompeting activities of miR-122 and the 5′ exonuclease Xrn1 in regulation of hepatitis C virus replication. Proc Natl Acad Sci USA. 2013b;110:1881–1886. doi: 10.1073/pnas.1213515110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LIU RF, XU X, HUANG J, FEI QL, CHEN F, LI YD, HAN ZG. Down-regulation of miR-517a and miR-517c promotes proliferation of hepatocellular carcinoma cells via targeting Pyk2. Cancer Lett. 2013;329:164–173. doi: 10.1016/j.canlet.2012.10.027. [DOI] [PubMed] [Google Scholar]
- LONGMAN RE, JOHNSON TR. Viral respiratory disease in pregnancy. Curr Opin Obstet Gynecol. 2007;19:120–125. doi: 10.1097/GCO.0b013e328028fdc7. [DOI] [PubMed] [Google Scholar]
- MAILLARD PV, CIAUDO C, MARCHAIS A, LI Y, JAY F, DING SW, VOINNET O. Antiviral RNA interference in mammalian cells. Science. 2013;342:235–238. doi: 10.1126/science.1241930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MALONE CD, HANNON GJ. Small RNAs as guardians of the genome. Cell. 2009;136:656–668. doi: 10.1016/j.cell.2009.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MARASA BS, SRIKANTAN S, MARTINDALE JL, KIM MM, LEE EK, GOROSPE M, ABDELMOHSEN K. MicroRNA profiling in human diploid fibroblasts uncovers miR-519 role in replicative senescence. Aging (Albany NY) 2010;2:333–343. doi: 10.18632/aging.100159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MARQUEZ L, LEVY ML, MUNOZ FM, PALAZZI DL. A report of three cases and review of intrauterine herpes simplex virus infection. Pediatr Infect Dis J. 2011;30:153–157. doi: 10.1097/INF.0b013e3181f55a5c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MARRAFFINI LA, SONTHEIMER EJ. CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea. Nat Rev Genet. 2010;11:181–190. doi: 10.1038/nrg2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MCGANN KA, COLLMAN R, KOLSON DL, GONZALEZ-SCARANO F, COUKOS G, COUTIFARIS C, STRAUSS JF, NATHANSON N. Human immunodeficiency virus type 1 causes productive infection of macrophages in primary placental cell cultures. J Infect Dis. 1994;169:746–753. doi: 10.1093/infdis/169.4.746. [DOI] [PubMed] [Google Scholar]
- MI S, LEE X, LI X, VELDMAN GM, FINNERTY H, RACIE L, LAVALLIE E, TANG XY, EDOUARD P, HOWES S, KEITH JC, JR, MCCOY JM. Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature. 2000;403:785–789. doi: 10.1038/35001608. [DOI] [PubMed] [Google Scholar]
- MITTELBRUNN M, GUTIERREZ-VAZQUEZ C, VILLARROYA-BELTRI C, GONZALEZ S, SANCHEZ-CABO F, GONZALEZ MA, BERNAD A, SANCHEZ-MADRID F. Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nat Commun. 2011;2:282. doi: 10.1038/ncomms1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MITTELBRUNN M, SANCHEZ-MADRID F. Intercellular communication: diverse structures for exchange of genetic information. Nat Rev Mol Cell Biol. 2012;13:328–335. doi: 10.1038/nrm3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MOR G, CARDENAS I. The immune system in pregnancy: a unique complexity. Am J Reprod Immunol. 2010;63:425–433. doi: 10.1111/j.1600-0897.2010.00836.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MORALES-PRIETO DM, CHAIWANGYEN W, OSPINA-PRIETO S, SCHNEIDER U, HERRMANN J, GRUHN B, MARKERT UR. MicroRNA expression profiles of trophoblastic cells. Placenta. 2012;33:725–734. doi: 10.1016/j.placenta.2012.05.009. [DOI] [PubMed] [Google Scholar]
- MUHLEMANN K, MENEGUS MA, MILLER RK. Cytomegalovirus in the perfused human term placenta in vitro. Placenta. 1995;16:367–373. doi: 10.1016/0143-4004(95)90094-2. [DOI] [PubMed] [Google Scholar]
- NIGRO G, ADLER SP, LA TORRE R, BEST AM. Passive immunization during pregnancy for congenital cytomegalovirus infection. N Engl J Med. 2005;353:1350–1362. doi: 10.1056/NEJMoa043337. [DOI] [PubMed] [Google Scholar]
- NOGUER-DANCE M, ABU-AMERO S, AL-KHTIB M, LEFEVRE A, COULLIN P, MOORE GE, CAVAILLE J. The primate-specific microRNA gene cluster (C19MC) is imprinted in the placenta. Hum Mol Genet. 2010;19:3566–3582. doi: 10.1093/hmg/ddq272. [DOI] [PubMed] [Google Scholar]
- OSTROWSKI M, CARMO NB, KRUMEICH S, FANGET I, RAPOSO G, SAVINA A, MOITA CF, SCHAUER K, HUME AN, FREITAS RP, GOUD B, BENAROCH P, HACOHEN N, FUKUDA M, DESNOS C, SEABRA MC, DARCHEN F, AMIGORENA S, MOITA LF, THERY C. Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat Cell Biol. 2010;12:19–30. doi: 10.1038/ncb2000. sup pp 11–13. [DOI] [PubMed] [Google Scholar]
- PAK J, FIRE A. Distinct populations of primary and secondary effectors during RNAi in C. elegans. Science. 2007;315:241–244. doi: 10.1126/science.1132839. [DOI] [PubMed] [Google Scholar]
- PARK H, LEE SW, LEE IH, RYU HM, CHO AR, KANG YS, HONG SR, KIM SS, SEONG SJ, SHIN SM, KIM TJ. Rate of vertical transmission of human papillomavirus from mothers to infants: relationship between infection rate and mode of delivery. Virol J. 2012;9:80. doi: 10.1186/1743-422X-9-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PARRY S, HOLDER J, STRAUSS JF., 3RD Mechanisms of trophoblast-virus interaction. J Reprod Immunol. 1997a;37:25–34. doi: 10.1016/s0165-0378(97)00071-5. [DOI] [PubMed] [Google Scholar]
- PARRY S, MACCALMAN CD, STRAUSS JF., 3RD Recombinant virus-mediated gene transfer in trophoblast cells. Ann N Y Acad Sci. 1997b;828:123–130. doi: 10.1111/j.1749-6632.1997.tb48530.x. [DOI] [PubMed] [Google Scholar]
- PEINADO H, ALECKOVIC M, LAVOTSHKIN S, MATEI I, COSTA-SILVA B, MORENO-BUENO G, HERGUETA-REDONDO M, WILLIAMS C, GARCIA-SANTOS G, GHAJAR C, NITADORI-HOSHINO A, HOFFMAN C, BADAL K, GARCIA BA, CALLAHAN MK, YUAN J, MARTINS VR, SKOG J, KAPLAN RN, BRADY MS, WOLCHOK JD, CHAPMAN PB, KANG Y, BROMBERG J, LYDEN D. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat Med. 2012;18:883–891. doi: 10.1038/nm.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- QURESHI F, JACQUES SM. Maternal varicella during pregnancy: correlation of maternal history and fetal outcome with placental histopathology. Hum Pathol. 1996;27:191–195. doi: 10.1016/s0046-8177(96)90374-3. [DOI] [PubMed] [Google Scholar]
- RACICOT K, CARDENAS I, WUNSCHE V, ALDO P, GULLER S, MEANS RE, ROMERO R, MOR G. Viral infection of the pregnant cervix predisposes to ascending bacterial infection. J Immunol. 2013;191:934–941. doi: 10.4049/jimmunol.1300661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RAMAKRISHNAIAH V, THUMANN C, FOFANA I, HABERSETZER F, PAN Q, DE RUITER PE, WILLEMSEN R, DEMMERS JA, STALIN RAJV, JENSTER G, KWEKKEBOOM J, TILANUS HW, HAAGMANS BL, BAUMERT TF, VAN DER LAAN LJ. Exosome-mediated transmission of hepatitis C virus between human hepatoma Huh7.5 cells. Proc Natl Acad Sci USA. 2013;110:13109–13113. doi: 10.1073/pnas.1221899110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RAPOSO G, STOORVOGEL W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol. 2013;200:373–383. doi: 10.1083/jcb.201211138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- REN J, JIN P, WANG E, MARINCOLA FM, STRONCEK DF. MicroRNA and gene expression patterns in the differentiation of human embryonic stem cells. J Transl Med. 2009;7:20. doi: 10.1186/1479-5876-7-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RIPPE V, DITTBERNER L, LORENZ VN, DRIESCHNER N, NIMZYK R, SENDT W, JUNKER K, BELGE G, BULLERDIEK J. The two stem cell microRNA gene clusters C19MC and miR-371–3 are activated by specific chromosomal rearrangements in a subgroup of thyroid adenomas. PLoS One. 2010;5:e9485. doi: 10.1371/journal.pone.0009485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ROBBINS JR, BAKARDJIEV AI. Pathogens and the placental fortress. Curr Opin Microbiol. 2012;15:36–43. doi: 10.1016/j.mib.2011.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SAITO Y, SUZUKI H, TSUGAWA H, NAKAGAWA I, MATSUZAKI J, KANAI Y, HIBI T. Chromatin remodeling at Alu repeats by epigenetic treatment activates silenced microRNA-512-5p with downregulation of Mcl-1 in human gastric cancer cells. Oncogene. 2009;28:2738–2744. doi: 10.1038/onc.2009.140. [DOI] [PubMed] [Google Scholar]
- SCHUST DJ, HILL AB, PLOEGH HL. Herpes simplex virus blocks intracellular transport of HLA-G in placentally derived human cells. J Immunol. 1996;157:3375–3380. [PubMed] [Google Scholar]
- SCITA G, DI FIORE PP. The endocytic matrix. Nature. 2010;463:464–473. doi: 10.1038/nature08910. [DOI] [PubMed] [Google Scholar]
- SELBACH M, SCHWANHAUSSER B, THIERFELDER N, FANG Z, KHANIN R, RAJEWSKY N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- SHALGI R, PILPEL Y, OREN M. Repression of transposable-elements - a microRNA anti-cancer defense mechanism? Trends Genet. 2010;26:253–259. doi: 10.1016/j.tig.2010.03.006. [DOI] [PubMed] [Google Scholar]
- SHIRAYAMA M, SETH M, LEE HC, GU W, ISHIDATE T, CONTE D, JR, MELLO CC. piRNAs initiate an epigenetic memory of nonself RNA in the C. elegans germline. Cell. 2012;150:65–77. doi: 10.1016/j.cell.2012.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SIJEN T, STEINER FA, THIJSSEN KL, PLASTERK RH. Secondary siRNAs result from unprimed RNA synthesis and form a distinct class. Science. 2007;315:244–247. doi: 10.1126/science.1136699. [DOI] [PubMed] [Google Scholar]
- SKALSKY RL, SAMOLS MA, PLAISANCE KB, BOSS IW, RIVA A, LOPEZ MC, BAKER HV, RENNE R. Kaposi’s sarcoma-associated herpesvirus encodes an ortholog of miR-155. J Virol. 2007;81:12836–12845. doi: 10.1128/JVI.01804-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SOREK R, KUNIN V, HUGENHOLTZ P. CRISPR--a widespread system that provides acquired resistance against phages in bacteria and archaea. Nat Rev Microbiol. 2008;6:181–186. doi: 10.1038/nrmicro1793. [DOI] [PubMed] [Google Scholar]
- STADLER B, IVANOVSKA I, MEHTA K, SONG S, NELSON A, TAN Y, MATHIEU J, DARBY C, BLAU CA, WARE C, PETERS G, MILLER DG, SHEN L, CLEARY MA, RUOHOLA-BAKER H. Characterization of microRNAs involved in embryonic stem cell states. Stem Cells Dev. 2010;19:935–950. doi: 10.1089/scd.2009.0426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SYGGELOU A, IACOVIDOU N, KLOUDAS S, CHRISTONI Z, PAPAEVANGELOU V. Congenital cytomegalovirus infection. Ann N Y Acad Sci. 2010;1205:144–147. doi: 10.1111/j.1749-6632.2010.05649.x. [DOI] [PubMed] [Google Scholar]
- TENOEVER BR. RNA viruses and the host microRNA machinery. Nat Rev Microbiol. 2013;11:169–180. doi: 10.1038/nrmicro2971. [DOI] [PubMed] [Google Scholar]
- TRAJKOVIC K, HSU C, CHIANTIA S, RAJENDRAN L, WENZEL D, WIELAND F, SCHWILLE P, BRUGGER B, SIMONS M. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science. 2008;319:1244–1247. doi: 10.1126/science.1153124. [DOI] [PubMed] [Google Scholar]
- TSAI KW, KAO HW, CHEN HC, CHEN SJ, LIN WC. Epigenetic control of the expression of a primate-specific microRNA cluster in human cancer cells. Epigenetics. 2009;4:587–592. doi: 10.4161/epi.4.8.10230. [DOI] [PubMed] [Google Scholar]
- VALADI H, EKSTROM K, BOSSIOS A, SJOSTRAND M, LEE JJ, LOTVALL JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- VICKERS KC, PALMISANO BT, SHOUCRI BM, SHAMBUREK RD, REMALEY AT. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol. 2011;13:423–433. doi: 10.1038/ncb2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WIEDENHEFT B, STERNBERG SH, DOUDNA JA. RNA-guided genetic silencing systems in bacteria and archaea. Nature. 2012;482:331–338. doi: 10.1038/nature10886. [DOI] [PubMed] [Google Scholar]
- XIAO C, RAJEWSKY K. MicroRNA control in the immune system: basic principles. Cell. 2009;136:26–36. doi: 10.1016/j.cell.2008.12.027. [DOI] [PubMed] [Google Scholar]
- YAN N, CHEN ZJ. Intrinsic antiviral immunity. Nat Immunol. 2012;13:214–222. doi: 10.1038/ni.2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZELDOVICH VB, BAKARDJIEV AI. Host defense and tolerance: unique challenges in the placenta. PLoS Pathog. 2012;8:e1002804. doi: 10.1371/journal.ppat.1002804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZENCLUSSEN AC. Adaptive immune responses during pregnancy. Am J Reprod Immunol. 2013;69:291–303. doi: 10.1111/aji.12097. [DOI] [PubMed] [Google Scholar]
- ZHANG R, WANG YQ, SU B. Molecular evolution of a primate-specific microRNA family. Mol Biol Evol. 2008;25:1493–1502. doi: 10.1093/molbev/msn094. [DOI] [PubMed] [Google Scholar]