Abstract
Failure to undergo activation-induced cell death due to global dysregulation of apoptosis is the pathogenic hallmark of large granular lymphocyte (LGL) leukemia. Consequently, immunosuppressive agents are rational choices for treatment. This first prospective trial in LGL leukemia was a multicenter, phase 2 clinical trial evaluating methotrexate at 10 mg/m2 orally weekly as initial therapy (Step 1). Patients failing methotrexate were eligible for treatment with cyclophosphamide at 100 mg orally daily (Step 2). The overall response in Step 1 was 38% with 95% confidence interval (CI): 26%, 53%. The overall response in Step 2 was 64% with 95% CI: 35%, 87%. The median overall survival for patients with anemia was 69 months with a 95% CI lower bound of 46 months and an upper bound not yet reached. The median overall survival for patients with neutropenia has not been reached 13 years from study activation. Serum biomarker studies confirmed the inflammatory milieu of LGL but were not a priori predictive of response. We identify a gene expression signature that correlates with response and may be STAT3 mutation driven. Immunosuppressive therapies have efficacy in LGL leukemia. Gene signature and mutational profiling may be an effective tool in determining whether methotrexate is appropriate therapy.
Keywords: Leukemia, T-Cell Large Granular Lymphocytic; Methotrexate; Cyclophosphamide; STAT3 Transcription Factor
INTRODUCTION
Large granular lymphocyte (LGL) leukemia is characterized by clonal expansion of cytotoxic T cells (CTL).(1, 2) Prominent clinical features include neutropenia, anemia, and rheumatoid arthritis. The terminal effector memory phenotype (CD3+/CD8+/CD57+/CD45RA+/CD62L−) of leukemic LGL suggests a pivotal chronic antigen driven immune response.(3) Leukemic LGL survival is promoted by PDGF and IL-15, resulting in global dysregulation of apoptosis and resistance to normal pathways of activation-induced cell death.(4–8) These pathogenic features explain in part why treatment of LGL leukemia is based on immunosuppressive therapy.
No standard therapy for LGL leukemia has been established due to the absence of large prospective trials. There have been six large retrospective studies (>40 patients) of immunosuppressive treatment in LGL leukemia.(1, 9–13) The three most commonly used have been methotrexate (MTX), cyclophosphamide (Cy), and cyclosporine (CyA). Overall response rates of 56% for MTX (n = 96), 61% for Cy (n = 85), and 56% for CyA (n = 123) have been reported.(14) We present herein results of the only large prospective trial of immunosuppressive therapy in LGL leukemia. Correlative laboratory studies were conducted to determine if biomarkers or genetic analysis could predict therapeutic response. We were particularly interested in the STAT3 pathway as we had found STAT3 to be constitutively activated in leukemic LGL.(6) Moreover, STAT3 was predicted to be a key node in a network model of leukemic LGL survival.(4) Most recently, we demonstrated somatic mutations that activate STAT3 in 40% of LGL leukemia patients(15) and subsequently analyzed their potential association with response to immunosuppressives in this study.
METHODS
Eligibility Criteria
Eligibility for the Eastern Cooperative Oncology Group E5998 study (Clinicaltrials.gov NCT00003910; Methotrexate With or Without Cyclophosphamide in Treating Patients With Lymphocytic Leukemia) included a diagnosis of the T cell form of LGL leukemia (performed locally) as determined by: 1) phenotypic studies from peripheral blood showing CD3+CD57+ cells greater than 400/mm3 or CD8+ cells greater than 650/mm3 in the eight weeks prior to registration and 2) evidence for clonal T cell receptor gene rearrangement within one year prior to registration. Patients needed to meet one of the following indications for treatment: a) severe neutropenia less than 500/mm3; b) neutropenia associated with recurrent infection; c) symptomatic anemia; and/or d) transfusion-dependent anemia. Decisions to start treatment were not based on any other factor, such as autoimmune disease or thrombocytopenia. Patients had no prior therapy with oral methotrexate or oral cyclophosphamide, were 18 years or older, and signed institutional review board informed consent in accordance with the Declaration of Helsinki. Other eligibility criteria included bilirubin <2.0 mg/dl, SGOT (AST) < 1.5 times normal, creatine < 2.0 mg/dl, ECOG performance status of 0–2, no previous or concurrent malignancies (except inactive non-melanoma skin cancer, in situ carcinoma of the cervix, or other cancer if the patient had been disease free for over five years), no other serious medical illness, and for female patients, not pregnant or breastfeeding.
Objectives
The primary objectives of this study were 1) to estimate the complete response (CR) rate, partial response (PR) rate, and overall response rate of MTX therapy in LGL leukemia patients treated for neutropenia or anemia and 2) to estimate CR rate, PR rate, and overall response rate of Cy treatment in patients failing MTX, for treatment indications of neutropenia or anemia. Secondary objectives were to conduct correlative s tudies to better define LGL leukemia pathogenesis as well as to correlate with therapeutic response.
Study Design
Step 1 consisted of MTX at 10 mg/m2 orally in divided doses once weekly. One cycle of therapy consisted of four weeks of treatment. Prednisone was given at 1 mg/kg orally daily × 30 days and then tapered off in the subsequent 24 days. Patients not responding to MTX received Cy at 100 mg orally daily with the same prednisone schedule (Step 2). Patients achieving PR in either step received MTX or Cy, respectively, for a maximum of one year. Patients achieving CR in either step received MTX or Cy, respectively, for one additional month after documentation of CR. Protocol treatment was discontinued in patients failing step 2 therapy. Since the primary treatment indication was neutropenia or anemia and there was a potential for differential response rates, we conducted studies of identical design in each stratum defined by the primary symptom. Simon’s optimal two stage design was employed to allow for early termination of the study if this treatment demonstrated no beneficial effects with respect to response. The study was designed to terminate if fewer than 4 patients of 17 in each stratum achieved a complete or partial response in the first stage. Since 4 or more achieved a response, the study continued to the second stage. The study was designed to test the null hypothesis of a 20% response rate versus an alternative of 40% with 90% power and an overall type I error rate of 0.08. Materials and methods for correlative laboratory studies, additional figures, and the complete clinical trial protocol are included as supplemental information.
Response Criteria
Treatment response was assessed after four cycles of therapy. CR was defined as attainment of normal CBC (ANC > 1500/mm3; lymphocyte count < 4,000/mm3; hemoglobin > 11g/dl; platelet count > 100,000/mm3). In addition, LGL counts as determined by repeat flow cytometry needed to be in the normal range. PR was defined as improvement in hematologic parameters in the absence of CR: 1) ANC > 500, as long as this represented 50% increase (treatment category a); 2) improvement in ANC > 50% over baseline (treatment category b); 3) increase in hemoglobin by > 1g/dl for at least four months duration (treatment category c); and 4) decrease in monthly transfusion requirements of > 50% for at least four months duration (treatment category d). Progressive disease was defined as worsening of hematologic parameters in patients previously achieving PR/CR. No response was defined as lack of CR/PR. Complete molecular remission was determined by showing absence of T cell clone using repeat T cell receptor gene rearrangement studies.
Statistical Methods
Univariate associations between dichotomous variables were evaluated by Fisher’s Exact test (1990). Associations involving ordered categorical variables were evaluated by the Wilcoxon Rank Sum test. Overall survival (OS) was defined as the time from study registration to death from any cause or date last known alive. Progression free survival (PFS) was defined as the time from registration to progression or to death without documentation of progression (censored). Patients who were alive without a progression were censored at the date of last contact. The methods of Kaplan and Meier (1958) were used to estimate survival curves and the significance was tested by logrank tests. P-values were reported for two-sided tests. No adjustments were made for multiple comparisons. The median follow-up was 6.3 years (range, 0.5–12 years) for the 34 surviving patients.
Laboratory Correlates
Array Data
Array results are deposited under Gene Expression Omnibus accession number GSE42664. Patient samples were prepared from peripheral blood collected immediately prior to the start of treatment. CD8+ T cells were prepared from normal donor lymphocyte filters from blood donations utilizing a Rosette-Sep negative isolation protocol (Stemcell Technologies, Vancouver, Canada) and confirmed to be >85% CD3+/CD8+ by flow cytometry. Terminal Effector Memory CD45RA+ (TEMRA) samples were additionally depleted of CD45RO+ cells by the addition of anti-CD45RO tetrameric antibody complexes and magnetic particles (Stemcell Technologies) to yield CD8+, CD3+, CD45RA+ and CCR7-cells. The use of age-matched controls limited the amount of contaminating naïve cells to less than 15 percent by flow cytometry. The comparison of LGL to normal CD8 has been previously shown(5), but the comparison to TEMRA is novel to this study. In concordance with that earlier study, LGL samples were not enriched prior to RNA isolation. Less than 25% of samples on this new array were represented in that study. Additional array information and validationare available in the supplement.
Cytokine Studies
Sera from healthy anonymous donors were collected by Florida Blood Services (St Petersburg, FL) and by the Hershey Medical Center Blood Bank (Hershey, PA) and consisted of 39 normal donors, 16 females (average age: 57, range: 23–83) and 23 males (average age: 68, range: 49–100). All trial participant sera analyzed in these studies were from baseline draws prior to the initiation of the treatment regimen.
A total of 27 analytes were grouped into 4 panels which are noted in the supplemental methods. All manufacturer’s recommendations were followed in regard to serum dilution with appropriate buffers, setting of gates, and reporter PMT settings for the Bio-Plex 200. Standards of known concentration for each analyte were used to construct a five-parameter linear regression model. Mean fluorescent intensity values for each bead region that corresponded to the analytes studied were fit to this model to determine concentrations in picograms per milliliter for each analyte measured.
STAT3 Testing
STAT3 mutation sequencing, cloning and mutagenesis, and reporter assays were carried out in a similar manner to earlier publications (15) with complete methods noted in the supplemental material.
Western Blot
Samples were lysed with RIPA buffer (Sigma, St. Louis, Missouri) and run on 10% Bis-Tris gels and then transferred onto PVDF membranes using a Trans Blot SD transfer cell (Bio-Rad). Blots were washed in 0.1% TBS-Tween and incubated with antibodies in either 5% milk or BSA TBS-T. Antibodies to E2F-1 (3742), C-MYC (9402) and GAPDH (2118) were purchased from Cell Signaling (Danvers, Massachusetts). Blots were developed with the Clarity ECL reagent (Bio-Rad) and imaged using the Chemidoc XRS+ system (Bio-Rad). Bands were quantified with the Quantity One software (Bio-Rad).
RESULTS
Demographics
The study accrued 59 patients between July 16, 1999 and March 24, 2009. Per two stage design, a response analysis was conducted after the first 17 eligible patients were accrued to each stratum. Since there were more than four responses, accrual continued to a total of 59 patients. The study terminated with 59 patients on March 24, 2009 due to slower than expected accrual. Of the 59 patients enrolled, four patients were ineligible for Step 1: one patient did not satisfy indications for treatment; two patients did not meet eligibility criteria for diagnosis, having too few LGL; and in one patient, eligibility labs were performed more than four weeks after registration. There were 16 patients enrolled in Step 2 with two patients being ineligible: one patient did not satisfy indications for treatment and the other patient received less than four cycles of MTX therapy in Step 1. Therefore, response and survival analyses were based on 55 eligible p atients with data available as of August 2012. Median time from diagnosis to registration for eligible patients was 2.7 months (range 0 to 95 months).
Baseline demographic characteristics of the study patients are shown in Table 1. Treatment indication was anemia in 29 (53%) patients and neutropenia in 26 (47%). There was some degree of overlap of cytopenias in these patients (Table 1). Importantly, however, none of the patients with anemia as treatment indication had severe neutropenia (ANC < 500 mm3) and similarly none of the patients with neutropenia as treatment indication had severe anemia (transfusion dependent). There were 7 patients with rheumatoid arthritis, 5 presented with neutropenia and 2 presented with anemia.
Table 1.
Step 1 On-Study Characteristics
| Variable | Category | Neutropenia | Anemia | Total |
|---|---|---|---|---|
| Number of Patients | 26 | 29 | 55 | |
| Age | Mean (SD) | 59.0 (15.8) | 71.2 (17.3) | 65.4 (17.6) |
| Median (Q1,Q3) | 60 (47, 73) | 74 (67, 84) | 70 (54, 80) | |
| [Min, Max] | [24, 81] | [20, 89] | [20, 89] | |
| Gender | Male | 13 (50.0%) | 17 (58.6%) | 30 (54.5%) |
| Female | 13 (50.0%) | 12 (41.4%) | 25 (45.5%) | |
| Race | White | 24 (92.3%) | 28 (96.6%) | 52 (94.5%) |
| Hispanic | 1 (3.8%) | 0 (0.0%) | 1 (1.8%) | |
| Black | 1 (3.8%) | 0 (0.0%) | 1 (1.8%) | |
| Asian | 0 (0.0%) | 1 (3.4%) | 1 (1.8%) | |
| ECOG Performance Status | 0 | 11 (42%) | 5 (17%) | 16 (29%) |
| 1 | 10 (38%) | 23 (79%) | 33 (60%) | |
| 2 | 5 (19%) | 1 (3%) | 6 (11%) | |
| Rheumatoid Arthritis | No | 21 (81%) | 27 (93%) | 48 (87%) |
| Yes | 5 (19%) | 2 (7%) | 7 (13%) | |
| Months from Diagnosis to Study Entry | Mean (SD) | 10.37 (15.13) | 15.23 (25.10) | 12.93 (20.94) |
| Median (Q1,Q3) | 2.7 (1.3, 9.0) | 1.9 (1.6, 16.0) | 2.7 (1.4, 14.1) | |
| [Min, Max] | [0.4, 53.5] | [0.0, 95.0] | [0.0, 95.0] | |
| Hemoglobin (g/dl) | Mean (SD) | 12.27 (2.01) | 9.24 (1.16) | 10.67 (2.21) |
| Median (Q1,Q3) | 12.4 (11.0, 13.5) | 9.3 (8.6, 9.9) | 10.3 (9.0, 12.3) | |
| [Min, Max] | [8.4, 16.3] | [7.0, 11.7] | [7.0, 16.3] | |
| ANC (cells/mm3) | Mean (SD) | 248.4 (192.2) | 1554.7 (1000.4) | 937.2 (984.5) |
| Median (Q1,Q3) | 212 (98, 400) | 1240 (986, 1862) | 700 (262, 1322) | |
| [Min, Max] | [0, 620] | [693, 5350] | [0, 5350] | |
| Platelets (K/mm3) | Mean (SD) | 170.4 (78.8) | 248.9 (117.4) | 211.8 (107.6) |
| Median (Q1,Q3) | 174 (102, 211) | 228 (181, 310) | 205 (154, 254) | |
| [Min, Max] | [49, 370] | [59, 556] | [49, 556] | |
| WBC (K/mm3) | Mean (SD) | 5.6 (4.1) | 8.3 (5.2) | 7.0 (4.8) |
| Median (Q1,Q3) | 4 (3, 8) | 6 (5, 9) | 6 (4, 9) | |
| [Min, Max] | [1, 18] | [3, 25] | [1, 25] | |
| Total # of LGL cells/mm3 | Mean (SD) | 2515.7 (2603.9) | 2564.5 (2402.9) | 2542.0 (2471.5) |
| Median (Q1,Q3) | 1339 (730, 2797) | 1800 (1024, 2770) | 1724 (758, 2824) | |
| [Min, Max] | [231, 8500] | [266, 9856] | [231, 9856] | |
| Freq. of Missing | 3 | 2 | 5 | |
| Percentage of cells CD3+/CD57+ | Mean (SD) | 52.3 (25.8) | 41.3 (24.2) | 46.5 (25.3) |
| Median (Q1,Q3) | 48 (35, 67) | 43 (21, 56) | 46 (28, 61) | |
| [Min, Max] | [3, 100] | [7, 90] | [3, 100] | |
| Freq. of Missing | 3 | 3 | 6 |
Treatment Compliance/Toxicity
Fifty-four patients began MTX therapy while one patient was excluded because of insufficient data. Of patients on therapy, 81% received at least 4 cycles of MTX treatment and 57% received at least 4 cycles without dose adjustment or omission. The median number of cycles for Step 1 was 5 (range: 1–14). Fourteen (26%) patients began Step 2 therapy and received a median of 3.5 (range: 1–12) cycles of Cy treatment. Seven (50%) of the patients received at least 4 cycles of Cy and 5 (36%) received at least 4 cycles without dose adjustment or omission.
During Step1, excessive complication or toxicity was the predominant off-treatment reason, followed by progressive disease and patient withdrawal; during Step 2, excessive complication or toxicity was the most common off-treatment reason.
Toxicity was assessed separately for patients wi th neutropenia versus anemia. Hematologic toxicities were excluded in the calculation of the worst degree for all toxicity as it is difficult to distinguish treatment related nadirs from cytopenia due to disease. In Step 1 therapy for 25 neutropenic patients, there was one grade 5 toxicity in a patient with infection associated with neutropenia. Grade 4 toxicities included infection without neutropenia (one patient) and dyspnea (one patient). In Step 1 therapy for 29 anemia patients, there were three grade 5 toxicities (pneumonitis/pulmonary infiltrates). Grade 4 non-hematologic toxicities included fatigue (two), hyperglycemia (one), hyponatremia (one), dyspnea (one), hypoxia (one), and pneumonitis/pulmonary infiltrates (one).
Sixteen patients were analyzed for toxicity in Step 2 therapy. There were no grade 5 toxicities. Grade 4 non-hematologic toxicity included melena/GI bleeding (one) and increased SGOT (one).
Response
Tables 2A and 2B summarize the best confirmed response for patients by treatment indication for Step 1 and 2, respectively. Among the 55 eligible patients in Step 1, three (5%) achieved a complete response, eighteen (33%) had a partial response, twenty three (42%) had stable disease, one (2%) had progressive disease, nine (16%) were unevaluable, and one (2%) was unknown. Unevaluable patients did not complete four cycles of therapy for noted reasons: early death, ie death before response assessment at end of cycle 4 (three, including two of the grade 5 toxicities in step 1), stroke (one), patient withdrawal (one), loss to follow-up (one), progressive disease after one cycle (one), and toxicity (two). The estimated overall response rate to MTX was 38% with 95% CI: 25%, 52%. For patients with neutropenia, the overall response rate was 42%. Patients with anemia had an overall response rate of 34%.
Table 2.
| A: Best Confirmed Response in Step 1 (Eligible Patients) | ||||||
|---|---|---|---|---|---|---|
| Best Confirmed Response | Neutropenia (n=26) | Anemia (n=29) | All Patients (n=55) | |||
| N | Percent | N | Percent | N | Percent | |
| Complete Response | 2 | 8% | 1 | 3% | 3 | 5% |
| Partial Response | 9 | 35% | 9 | 31% | 18 | 33% |
| No Change/Stable | 11 | 42% | 12 | 41% | 23 | 42% |
| Progressive Disease | 0 | 0% | 1 | 3% | 1 | 2% |
| Unevaluable | 3 | 12% | 6 | 21% | 9 | 16% |
| Unknown | 1 | 4% | 0 | 0% | 1 | 2% |
| B: Best Confirmed Response in Step 2 (Eligible Patients) | ||||||
|---|---|---|---|---|---|---|
| Best Confirmed Response | Neutropenia (n=8) | Anemia (n=6) | All Patients (n=14) | |||
| N | Percent | N | Percent | N | Percent | |
| Complete Response | 1 | 13% | 2 | 33% | 3 | 21% |
| Partial Response | 3 | 38% | 3 | 50% | 6 | 43% |
| No Change/Stable | 2 | 25% | 0 | 0 | 2 | 14% |
| Progressive Disease | 0 | 0% | 0 | 0 | 0 | 0% |
| Unevaluable | 2 | 25% | 1 | 17% | 3 | 21% |
| Unknown | 0 | 0% | 0 | 0 | 0 | 0% |
Among the 14 eligible patients in Step 2, three (21%) achieved a complete response, six (43%) achieved a partial response, two (14%) had stable disease, and three (21%) were unevaluable. The overall response rate for Cy was 64% with 95% CI: 35%, 87%. For patients with neutropenia, the overall response rate was 50%. Patients with anemia had an overall response rate of 83%. Of the six eligible patients who achieved a CR in Step 1 or Step 2, one patient had a molecular remission.
The response rate to MTX in patients with rheumatoid arthritis was not different statistically than the overall group.
Two of the 7 patients (28.6%) with rheumatoid arthritis had a response (one complete response and one partial response) to MTX, with 95% CI: 3.7%, 71.0%.
Survival and Progression-Free Survival
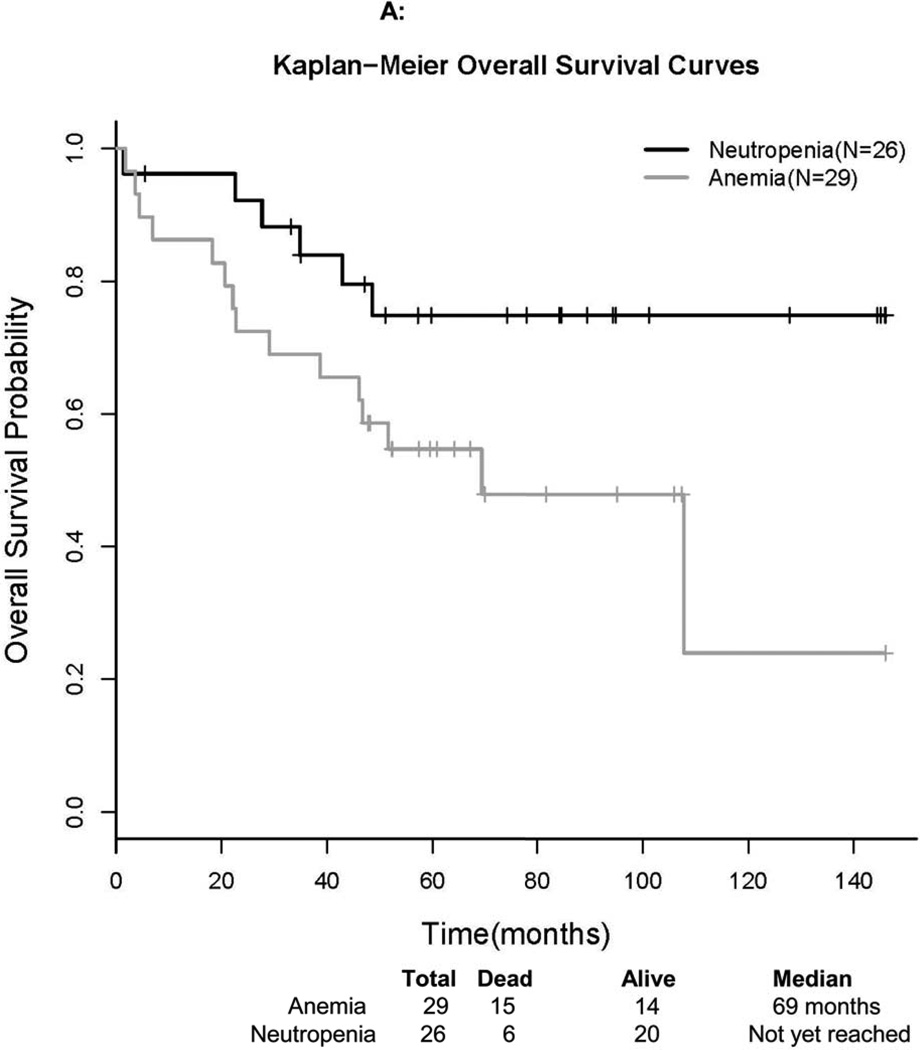
Survival time was defined as the time from registration to the date of death or the date last known alive (censored). The median overall survival for patients presenting with anemia was 69 months. The lower bound of the 95% CI was 46 months, but the upper bound has not been reached to date. The median OS for patients presenting with neutropenia has not been reached as of August 2012. The OS Kaplan-Meier plot by treatment indication is presented in Figure 1A. The median follow-up time of 34 surviving patients was 76 months. For the 15 deaths among patients presenting with anemia, there were 7 due to disease, 3 due to treatment, 4 due to other causes, and 1 due to unknown cause. For the 6 deaths among patients presenting with neutropenia, there was 1 due to disease, 1 due to treatment, 2 due to other causes (neither disease nor treatment), and 2 due to unknown causes.
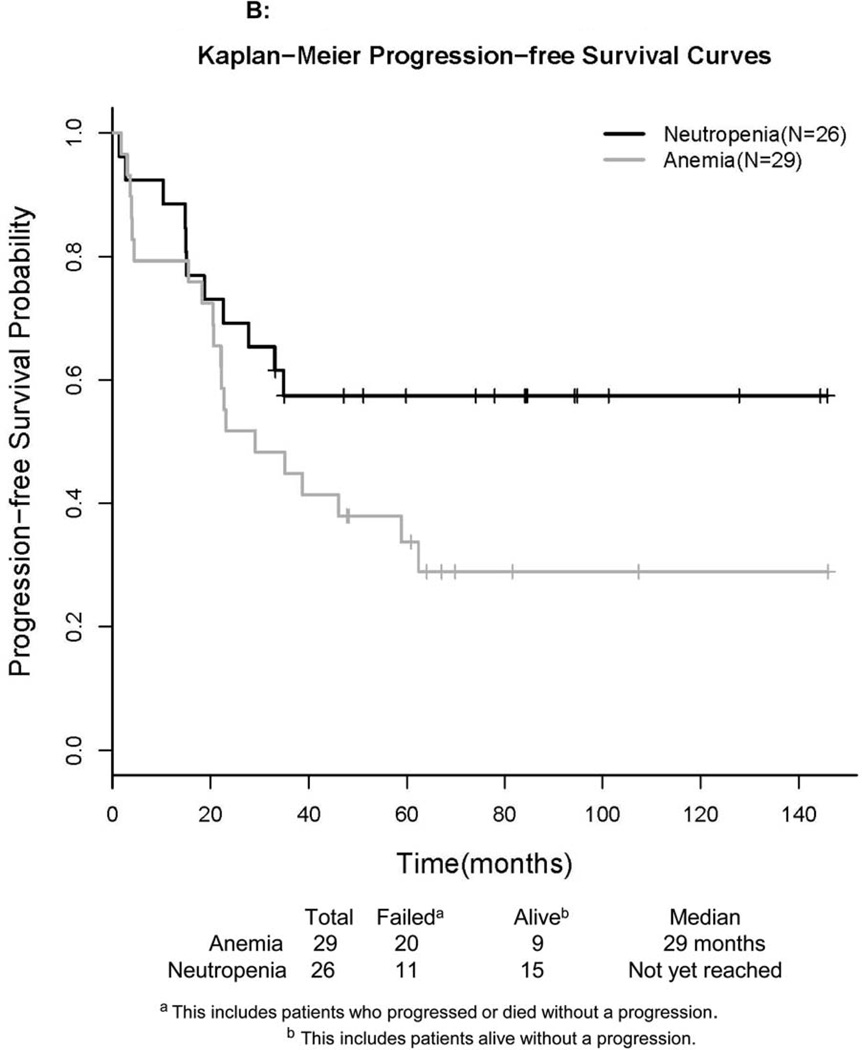
Figure 1. Survival Curves of Study Participants.
Kaplan-Meier curves are presented for overall (A) and progression-free (B) survival, stratified by reason for treatment.
PFS was defined as the time from registration to progression or death without documentation of progression (censored). The Kaplan-Meier plot for PFS is presented in Figure 1B for 55 eligible patients in Step 1. The median PFS for patients with anemia was 29 months with a 95% CI: 21, 62 months. For patients presenting with neutropenia, the median PFS has not been reached as of August 2012, but the lower bound of the 95% CI was 23 months. Patients who were alive without a progression were censored at the date of last contact.
Laboratory Correlates
Serum Biomarkers
Of the 27 serum cytokines measured in 41 patients and 37 age-matched normals, 9 of them differed from the normal samples tested (Wilcoxon p-value less than .0019) (Table S1, online only). We confirmed the involvement of proteins known to be dysregulated in LGL Leukemia such as Fas Ligand (FASL) and Interleukin 18.(16, 17) We also found elevated serum levels of soluble ICAM and VCAM which have not been previously reported in LGL leukemia. None of these biomarkers were predictive of therapeutic response. We were also interested in knowing whether there were differences in biomarker expression when comparing LGL leukemia patients with neutropenia to those with anemia. Of the 9 serum cytokines shown to be different between LGL and normal we observed higher levels of FASL in anemic patients (unadjusted Wilcoxon p=.051).
STAT3 Mutational Analyses
Sanger sequencing of DNA or RNA samples from 50 of 55 eligible samples was performed to detect the presence of recently discovered mutations in exon 21 of the transcription factor STAT3.(15) In this cohort, 24 of 50 patients had STAT3 mutations (48%). Mutations resulting in variant protein were predominantly amino acid changes Y640F (22%) or D661Y (16%). Other mutations included D661V (4%), N647I (4%) and D661H (2%). Overall, patients with STAT3 mutations were more likely to respond to treatment (p = 0.044). Of interest was whether the dominant mutations in this study, D661Y or Y640F, correlated with response. We found 8 of 11 (73%) patients with Y640F mutations responded to MTX (Fisher exact p-value 0.036), whereas 3 of 8 with D661Y mutations had a response (p-value 0.67). All non-responsive patients with the Y640F mutation were of the unevaluable class; the inability to complete at least four courses of MTX. Therefore, the mutation was 100% predictive of response in those instances where a full course of MTX was administered.
Gene expression correlation with response and mutation status
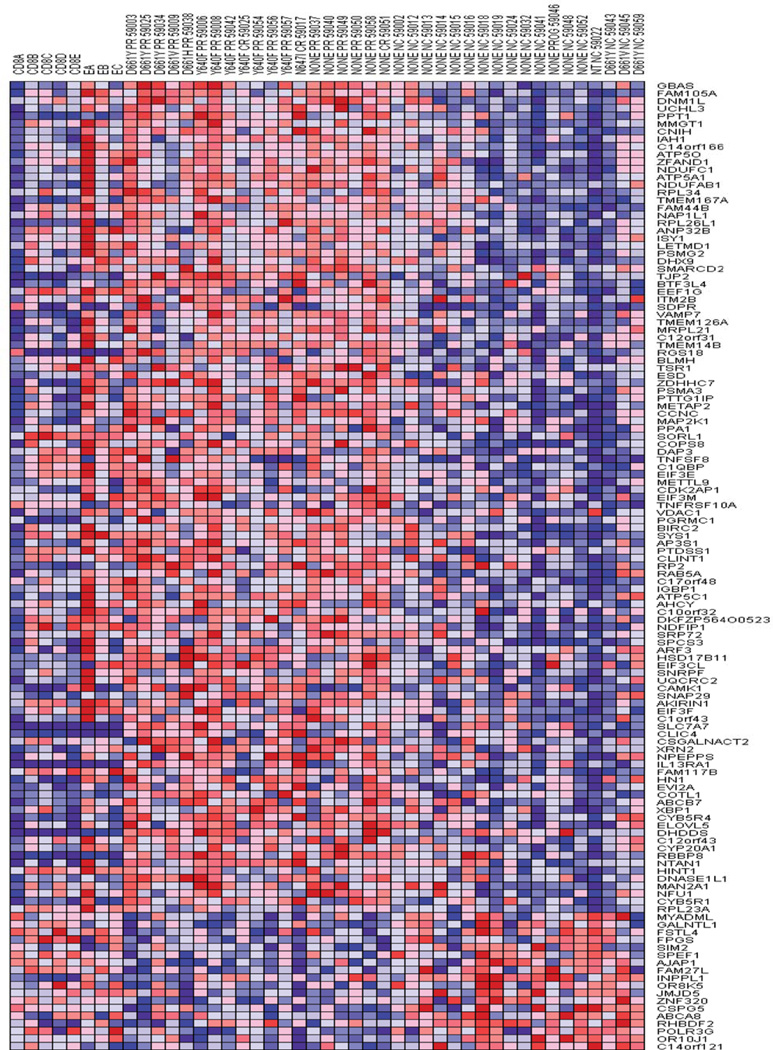
Gene Set Enrichment Analysis (GSEA) of microarray data has been informative in elucidating survival signaling pathways in LGL leukemia.(5) Therefore, microarray analysis was performed on 37 patient samples including 19 MTX responders and 18 non-responders. Ranking gene changes in Gene Pattern, we identified 127 genes with a Z-score greater than 3. The overwhelming majority of the genes ranked as being significantly changed represented upregulation in the responder phenotype (Figure 2). GSEA of KEGG, BioCarta and GenMAPP pathways identified a number of pathways as being enriched in patients that responded to MTX. Gene sets with a normalized enrichment score greater than 1.6 are listed in Table 3A. MYC is a common constituent of several highly ranked gene sets. MYC, FOS and PIM1 have been shown to be transcriptional targets of Stat3 that mediate cell growth and proliferation.(18, 19) Our array results indicated these genes tended to be up-regulated in the responders by at least 50 percent (FOS 1.58 fold, MYC 1.76 and PIM1 1.57). Additional array comparisons and validations are present in the Supplement.
Figure 2. Heatmap of baseline sample gene expression in Methotrexate responders versus nonresponders.
The top 126 genes by Z-score are displayed. Red shading indicates upregulation of mRNAs which are listed by official gene symbol at right. Samples are divided into 4 main groups from left to right, normal CD8+, normal Temra, patients with response to Methotrexate and lastly those not responding. Individual patient lanes are labeled by mutation type, type of response and trial accession number.
PR = partial response, CR = complete response, NC = no change, and PROG = progression
Table 3.
| A. Pathways Upregulated in Responders to Methotrexate in Baseline Samples. | ||||||
|---|---|---|---|---|---|---|
| PATHWAY NAME | SIZE | ES | NES | NOM p-val | FDR q-val | Myc |
| HSA00190_OXIDATIVE_PHOSPHORYLATION | 94 | 0.612149 | 2.343721 | 0 | 0 | |
| OXIDATIVE_PHOSPHORYLATION | 46 | 0.615098 | 2.107513 | 0 | 0.001189 | |
| VIP PATHWAY | 19 | 0.686932 | 1.951678 | 0 | 0.007198 | yes |
| NTHI PATHWAY | 19 | 0.654488 | 1.869974 | 0 | 0.019713 | |
| HSA05216_THYROID_CANCER | 21 | 0.631448 | 1.862768 | 0.001319 | 0.018062 | yes |
| P38MAPK PATHWAY | 27 | 0.591922 | 1.819187 | 0.001267 | 0.029064 | yes |
| HSA00530_AMINOSUGARS_METABOLISM | 24 | 0.598269 | 1.809769 | 0 | 0.028299 | |
| KREBS_TCA_CYCLE | 27 | 0.561241 | 1.722913 | 0.00246 | 0.087624 | |
| GSK3 PATHWAY | 17 | 0.613683 | 1.717902 | 0.006658 | 0.082197 | |
| FMLP PATHWAY | 28 | 0.549093 | 1.700237 | 0.003686 | 0.093788 | |
| RAS PATHWAY | 21 | 0.580599 | 1.690651 | 0.002581 | 0.093672 | |
| HSA03050_PROTEASOME | 21 | 0.57235 | 1.666294 | 0.007762 | 0.115172 | |
| HSA05221_ACUTE_MYELOID_LEUKEMIA | 44 | 0.490264 | 1.656008 | 0.004739 | 0.119373 | yes |
| MRNA_PROCESSING_REACTOME | 90 | 0.434236 | 1.643548 | 0 | 0.126395 | |
| B. Pathways Upregulated in Y640F Mutants Versus D661 Mutants. | ||||||
|---|---|---|---|---|---|---|
| PATHWAY NAME | SIZE | ES | NES | NOM p-val | FDR q-val | |
| HSA00190_OXIDATIVE_PHOSPHORYLATION | 94 | 0.567646 | 2.042502 | 0 | 0.001062 | |
| OXIDATIVE_PHOSPHORYLATION | 46 | 0.536051 | 1.757709 | 0 | 0.167077 | |
| PTDINSPATHWAY | 18 | 0.637419 | 1.744706 | 0.001264 | 0.13437 | |
| MRNA_PROCESSING_REACTOME | 90 | 0.485078 | 1.741168 | 0 | 0.106182 | |
| RIBOSOMAL_PROTEINS | 78 | 0.482901 | 1.703704 | 0 | 0.13044 | |
| HSA03050_PROTEASOME | 21 | 0.602159 | 1.687073 | 0.002478 | 0.135277 | |
| HSA00530_AMINOSUGARS_METABOLISM | 24 | 0.569262 | 1.661507 | 0.00885 | 0.15462 | |
| PURINE_METABOLISM | 79 | 0.472723 | 1.66001 | 0.002125 | 0.13758 | |
| PROTEASOME | 16 | 0.627454 | 1.654199 | 0.005013 | 0.131303 | |
| HSA04940_TYPE_I_DIABETES_MELLITUS | 31 | 0.538567 | 1.640318 | 0.004717 | 0.138087 | |
| ERK PATHWAY | 21 | 0.570421 | 1.622 | 0.015971 | 0.155567 | |
Pathways from KEGG, GenMAPP and BioCarta as identified in GSEA. Size= gene set size ES = Enrichment signal, NES=Normalized enrichment signal, NOM p-val = nominal p-value, FDR q-val = False discovery rate q-value.
The pathway indicated as VIP is a gene set that contributes to the inhibition of Activation Induced Cell Death (AICD) by Vasoactive Inhibitory Protein(VIP). Inhibition of AICD, not necessarily through VIP involvement, has been shown previously to be involved in the pathogenesis of LGL leukemia.(4)
The upregulation of genes related to oxidative phosphorylation could indicate increased energy demands by the cell or the replication of mitochondria in preparation for division. In support of the latter, one of the most consistently elevated genes in responders is Dynamin 1-like a gene with an established role in mitochondrial fission.(20) This is consistent with the observed ability of Stat3 to increase the electron transport chain in support of Ras transformation,(21) which also shows pathway upregulation in the responder phenotype. The cellular location and mechanism of interaction that brings this about continues to be controversial.(22) Dynamin-1-like has also been shown to interact with Glycogen Synthase Kinase 3 (GSK3)(23) which also has demonstrated gene set enrichment in the responder phenotype.
Pathway enrichment by major mutation types
As noted before, the Y640F mutated genotype strongly correlated with response to therapy with MTX, whereas the D661 mutated genotype did not. Gene set analysis identical to that performed between responder and nonresponder was conducted on seven, Y640F mutant samples and eight samples with D661 mutation, predominantly D661Y. Intriguingly, both Oxidative Phosphorylation and Aminosugars Metabolism pathways, previously deemed to be upregulated in MTX responders, were differentially upregulated in Y640F mutants (Table 3B). Another related, key pathway upregulated by Y640F mutations was that of Purine Metabolism which may explain differential MTX response, as inhibition of purine biosynthesis is a primary mechanism of action of low-dose MTX.17
STAT3 Y640F mutant has strongest transcriptional activity
We speculated that expression of gene sets enriched in Y640F mutants might be the result of stronger transcriptional activity of the Y640F mutants versus the D661 mutant. Both the D661V and Y640F mutations of STAT3 have been shown to increase the transcriptional activity of STAT3, with the Y640F mutation being considerably stronger.(15) The tra nscriptional activity of the major D661 mutation observed in this study, D661Y, had not been described. We performed luciferase assays to determine the transcriptional strength of this mutation. D661Y had increased transcriptional activity when compared to an equally expressed wildtype, but much less than that observed for the Y640F mutation, 15-fold versus 131-fold respectively. We additionally report the activity of an S614R mutation from exon 20 that was found in two patients from this study (Figure 3A).
Figure 3. Evidence for Increased Transcriptional Activity of the Y640F STAT3 Mutation.
A) Y640F mutant greatly increased STAT3 transcriptional activity compared to STAT3 wild type and other mutants. HEK293T cells were co-transfected with Cignal STAT3 reporter harboring an SIE response element upstream of luciferase reporter and vector alone (vector), wild type STAT3 (WT), or STAT3 mutants (S614R, Y640F, D661Y or D661V). (Bottom) Western blot to detect the expression of the different STAT3 variants using human STAT3 antibody and using antibody β-actin as loading control.
B) Quantitative RT-PCR indicating a reduction of the mir-223 precursor transcript in patient cells harboring the Y640F mutation. Relative expression is normalized to the mean of the Y640F sample group.
C) Western blot of MYC and E2F1 in leukemic LGL and normal unactivated control CD8+ (NCD8, negative control) and activated CD8+ cells (AcCD8, positive control). Two baseline patient samples from each mutation class are di splayed. Bar graphs depicts the average level of expression of E2F1 and MYC as a proportion of GAPDH, normalized to unactivated control CD8+ samples.
D) Response rates to Step 1 by mutation type as percent of total responding.
Additional but indirect evidence of strong STAT3 activity is our finding that the mir-223 precursor transcript was the most significantly downregulated transcript in the Y640F mutation group (2.2 fold downregulated with a t-test p-value of 0.0004). We confirmed this array finding with quantitative RT-PCR (Figure 3B). Mir-223 has been shown to repress E2F1 translation by binding the 3' UTR of its transcript and E2F1 has been shown to reciprocally repress the transcript for mir-223 through promoter binding and inhibition of transcription.(24) Potentially, very high E2F1 levels such as those that are present prior to replication, could be achieved that would silence mir-223. An increase in E2F1 in leukemic LGL could be the result of increased Myc levels driven by STAT3(18) (diagrammed in Figure S1, online only). Indeed, we observe increased Myc and E2F1 protein levels in LGL cells compared to normal CD8+ controls (Figure 3C). LGL counts of 3280/mm3 observed for patients with STAT3 mutation, compared to 1576/ mm3 for patients not harboring a mutation provides additional support for a more proliferative state in mutated patients (Wilcoxon p-value of 0.051). Intriguingly, ranking the percent response in the mutational groups follow the same trend as the transcriptional strength of that mutant (Figure 3D).
DISCUSSION
We describe here t he results of the first large prospective study of immunosuppressive agents for the treatment of LGL leukemia. Overall, the response rate of the first line of treatment with MTX was 38%. This compares to a previously reported response rate of 56% when combining several small retrospective studies.(14) A design feature of the study was to stratify patients according to the indication for treatment, i.e. anemia versus neutropenia. Similar response rates to MTX were observed whether treatment indication was severe/symptomatic neutropenia or severe/symptomatic anemia. For those patients not responding to MTX, Cy proved to be an effective second line of treatment with 64% of patients achieving at least PR. Given the high rate of response of the second line of treatment, it can be inferred that prior failed treatment with MTX does not negatively influence future response to Cy. Our correlative laboratory studies confirm and extend previous observations indicating that production of proinflammatory cytokines is characteristic of LGL leukemia. We previously showed high serum levels of soluble Fas, Fas ligand, IFNgamma, IFN-alpha2, IL-6, IL-8 and IL-18 in patients with LGL leukemia.(16) A fundamental pathogenic mechanism in LGL leukemia is resistance to Fas-mediated death despite high expression of both Fas and Fas ligand in leukemic LGL. Blockade of Fas signaling by soluble Fas is one potential mechanism leading to apoptotic resistance.(25) Of interest, serum levels of TRAIL, a pro-apoptotic molecule similar to Fas ligand were also markedly elevated. Inhibition of TRAIL signaling by TRAIL decoy receptors and subsequent resistance to TRAIL induced apoptosis has been observed in a number of cancers. We note that LGL leukemic sera have high levels of such decoy receptors (unpublished observation). For the first time in LGL leukemia, we report the presence of elevated soluble VCAM and ICAM which have been implicated in the pathogenesis of rheumatoid arthritis(26) and the subject of potential therapeutic targeting.
An intriguing finding of this study was the identification of a gene signature that correlates with response. A gene signature similar to that associated with response was further enriched in patients with STAT3 Y640F mutated genotype. Indeed, the presence of this strongest activating mutation, Y640F, alone predicted therapeutic response to MTX. All patients with this mutation that completed at least 4 cycles of MTX responded to therapy. This finding appears counterintuitive as STAT3 activation has been associated with drug-resistant phenotype.(27) Therefore, it will be most important to validate our preliminary finding in a larger cohort of patients, particularly since the relatively small sample size did not allow the traditional strategy of biomarker evaluation utilizing a validation cohort. Nevertheless, such a putative role of STAT3 mutation in leukemic LGL biology is provocative. We speculate that purine depletion by MTX in the presence of E2F1 expression leads to apoptosis of leukemic LGL as they attempt to enter cell cycle (Figure S1). This model suggests that increased transcriptional activity of STAT3 may lead to increased sensitivity to MTX. Indeed, we showed that the Y640F mutation associated with clinical response had much stronger transcriptional activity than the other most common STAT3 mutation observed in this study (D661Y). Also supportive of this model is our finding that Y640F mutants displayed 2.2 fold lower expression of the precursor for mir-223. E2F1 and mir-223 have been shown to negatively regulate each other.(24) Activation of this regulatory loop would lead to sustained high levels of E2F1 and low levels of mir-223, as demonstrated in leukemic LGL with a mutated Y640F genotype (Figure 3). It is also of interest that not all of the patients with a responder gene signature harbored mutations in STAT3. Our previous work showed that STAT3 was a key hub in the LGL leukemia survival network.(4) It is conceivable that there may be mutations in other genes that would allow them to activate STAT3.
In summary, we report the results of the first prospective trial of immunosuppressive therapy for the treatment of LGL leukemia. Correlative studies suggest a gene signature and mutated STAT3 Y640F genotype as potential predictors of response to MTX, which need validation in larger studies. The two-step design of the protocol did not allow for direct analysis of efficacy of MTX versus Cy. A recent retrospective study showed a 72% overall response rate to first-line Cy therapy.(28) A prospective trial comparing MTX versus Cy as initial treatment of LGL leukemia has just been activated in France. Nevertheless, the somewhat low response rates to immunosuppressive agents highlight the need for development of new therapeutics for LGL leukemia.
Supplementary Material
ACKNOWLEDGEMENTS
We acknowledge Rob Brucklacher of Penn State Hershey Functional Genomics for performing microarrays. We also thank Arjan van Adrichem for the creation of the Y640F STAT3 plasmid. Su-Fern Tan provided assistance with the conversion of figures.
This study was conducted by the Eastern Cooperative Oncology Group (Robert L. Comis, M.D., Chair) and supported in part by Public Health Service Grants CA23318, CA66636, CA21115, CA73590, CA11083, CA14548, CA13650, CA17145, CA78724, CA90633, CA94872 and CA133525 from the NCI, NIH and the DHHS. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NCI.
Footnotes
The authors have no conflicts of interest to disclose.
Supplementary material is included at Leukemia’s website.
AUTHORSHIP CONTRIBUTIONS
TL designed and was principal investigator of the study, determined clinical responses of participants and wrote the manuscript. LZ statistically analyzed clinical response and laboratory correlates and wrote the manuscript. TO designed and performed serum correlate experiments, designed and analyzed microarray experiments, designed confirmatory experiments, and wrote the manuscript. DZ designed and performed STAT3 activation assays. HR and SM designed and performed experiments measuring STAT3 mutations in the patient cohort and contributed to the manuscript. ZH designed and performed experiments measuring molecules downstream of STAT3. JB, ML, AE and MT conceptualized and administered the clinical trial and provided critical review of the manuscript.
REFERENCES
- 1.Loughran TP., Jr Clonal diseases of large granular lymphocytes. Blood. 1993;82(1):1–14. [PubMed] [Google Scholar]
- 2.Loughran TP, Jr, Kadin ME, Starkebaum G, Abkowitz JL, Clark EA, Disteche C, et al. Leukemia of large granular lymphocytes: association with clonal chromosomal abnormalities and autoimmune neutropenia, thrombocytopenia, and hemolytic anemia. Ann Intern Med. 1985;102(2):169–175. doi: 10.7326/0003-4819-102-2-169. [DOI] [PubMed] [Google Scholar]
- 3.Yang J, Epling-Burnette PK, Painter JS, Zou J, Bai F, Wei S, et al. Antigen activation and impaired Fas-induced death-inducing signaling complex formation in T-large-granular lymphocyte leukemia. Blood. 2008;111(3):1610–1616. doi: 10.1182/blood-2007-06-093823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang R, Shah MV, Yang J, Nyland SB, Liu X, Yun JK, et al. Network model of survival signaling in large granular lymphocyte leukemia. Proc Natl Acad Sci U S A. 2008;105(42):16308–16313. doi: 10.1073/pnas.0806447105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shah MV, Zhang R, Irby R, Kothapalli R, Liu X, Arrington T, et al. Molecular profiling of LGL leukemia reveals role of sphingolipid signaling in survival of cytotoxic lymphocytes. Blood. 2008;112(3):770–781. doi: 10.1182/blood-2007-11-121871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Epling-Burnette PK, Liu JH, Catlett-Falcone R, Turkson J, Oshiro M, Kothapalli R, et al. Inhibition of STAT3 signaling leads to apoptosis of leukemic large granular lymphocytes and decreased Mcl-1 expression. J Clin Invest. 2001;107(3):351–362. doi: 10.1172/JCI9940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schade AE, Powers JJ, Wlodarski MW, Maciejewski JP. Phosphatidylinositol-3-phosphate kinase pathway activation protects leukemic large granular lymphocytes from undergoing homeostatic apoptosis. Blood. 2006;107(12):4834–4840. doi: 10.1182/blood-2005-08-3076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang J, Liu X, Nyland SB, Zhang R, Ryland LK, Broeg K, et al. Platelet-derived growth factor mediates survival of leukemic large granular lymphocytes via an autocrine regulatory pathway. Blood. 2010;115(1):51–60. doi: 10.1182/blood-2009-06-223719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pandolfi F, Loughran TP, Jr, Starkebaum G, Chisesi T, Barbui T, Chan WC, et al. Clinical course and prognosis of the lymphoproliferative disease of granular lymphocytes. A multicenter study. Cancer. 1990;65(2):341–348. doi: 10.1002/1097-0142(19900115)65:2<341::aid-cncr2820650227>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 10.Semenzato G, Zambello R, Starkebaum G, Oshimi K, Loughran TP., Jr The lymphoproliferative disease of granular lymphocytes: updated criteria for diagnosis. Blood. 1997;89(1):256–260. [PubMed] [Google Scholar]
- 11.Bareau B, Rey J, Hamidou M, Donadieu J, Morcet J, Reman O, et al. Analysis of a French cohort of patients with large granular lymphocyte leukemia: a report on 229 cases. Haematologica. 2010;95(9):1534–1541. doi: 10.3324/haematol.2009.018481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dhodapkar MV, Li CY, Lust JA, Tefferi A, Phyliky RL. Clinical spectrum of clonal proliferations of T-large granular lymphocytes: a T-cell clonopathy of undetermined significance? Blood. 1994;84(5):1620–1627. [PubMed] [Google Scholar]
- 13.Neben MA, Morice WG, Tefferi A. Clinical features in T-cell vs. natural killer-cell variants of large granular lymphocyte leukemia. Eur J Haematol. 2003;71(4):263–265. doi: 10.1034/j.1600-0609.2003.00136.x. [DOI] [PubMed] [Google Scholar]
- 14.Lamy T, Loughran TP., Jr How I treat LGL leukemia. Blood. 2011;117(10):2764–2774. doi: 10.1182/blood-2010-07-296962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koskela HL, Eldfors S, Ellonen P, van Adrichem AJ, Kuusanmaki H, Andersson EI, et al. Somatic STAT3 mutations in large granular lymphocytic leukemia. N Engl J Med. 366(20):1905–1913. doi: 10.1056/NEJMoa1114885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kothapalli R, Nyland SB, Kusmartseva I, Bailey RD, McKeown TM, Loughran TP., Jr Constitutive production of proinflammatory cytokines RANTES, MIP-1beta and IL-18 characterizes LGL leukemia. Int J Oncol. 2005;26(2):529–535. [PubMed] [Google Scholar]
- 17.Liu JH, Wei S, Lamy T, Epling-Burnette PK, Starkebaum G, Djeu JY, et al. Chronic neutropenia mediated by fas ligand. Blood. 2000;95(10):3219–3222. [PubMed] [Google Scholar]
- 18.Shirogane T, Fukada T, Muller JM, Shima DT, Hibi M, Hirano T. Synergistic roles for Pim-1 and c-Myc in STAT3-mediated cell cycle progression and antiapoptosis. Immunity. 1999;11(6):709–719. doi: 10.1016/s1074-7613(00)80145-4. [DOI] [PubMed] [Google Scholar]
- 19.Gronowski AM, Zhong Z, Wen Z, Thomas MJ, Darnell JE, Jr, Rotwein P. In vivo growth hormone treatment rapidly stimulates the tyrosine phosphorylation and activation of Stat3. Mol Endocrinol. 1995;9(2):171–177. doi: 10.1210/mend.9.2.7776967. [DOI] [PubMed] [Google Scholar]
- 20.Smirnova E, Griparic L, Shurland DL, van der Bliek AM. Dynamin-related protein Drp1 is required for mitochondrial division in mammalian cells. Mol Biol Cell. 2001;12(8):2245–2256. doi: 10.1091/mbc.12.8.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gough DJ, Corlett A, Schlessinger K, Wegrzyn J, Larner AC, Levy DE. Mitochondrial STAT3 supports Ras-dependent oncogenic transformation. Science. 2009;324(5935):1713–1716. doi: 10.1126/science.1171721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Phillips D, Reilley MJ, Aponte AM, Wang G, Boja E, Gucek M, et al. Stoichiometry of STAT3 and mitochondrial proteins: Implications for the regulation of oxidative phosphorylation by protein- protein interactions. J Biol Chem. 285(31):23532–23536. doi: 10.1074/jbc.C110.152652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hong YR, Chen CH, Cheng DS, Howng SL, Chow CC. Human dynamin-like protein interacts with the glycogen synthase kinase 3beta. Biochem Biophys Res Commun. 1998;249(3):697–703. doi: 10.1006/bbrc.1998.9253. [DOI] [PubMed] [Google Scholar]
- 24.Pulikkan JA, Dengler V, Peramangalam PS, Peer Zada AA, Muller-Tidow C, Bohlander SK, et al. Cell-cycle regulator E2F1 and microRNA-223 comprise an autoregulatory negative feedback loop in acute myeloid leukemia. Blood. 115(9):1768–1778. doi: 10.1182/blood-2009-08-240101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu JH, Wei S, Lamy T, Li Y, Epling-Burnette PK, Djeu JY, et al. Blockade of Fas-dependent apoptosis by soluble Fas in LGL leukemia. Blood. 2002;100(4):1449–1453. [PubMed] [Google Scholar]
- 26.Littler AJ, Buckley CD, Wordsworth P, Collins I, Martinson J, Simmons DL. A distinct profile of six soluble adhesion molecules (ICAM-1, ICAM-3, VCAM-1, E-selectin, L-selectin and P-selectin) in rheumatoid arthritis. Br J Rheumatol. 1997;36(2):164–169. doi: 10.1093/rheumatology/36.2.164. [DOI] [PubMed] [Google Scholar]
- 27.Alas S, Bonavida B. Inhibition of constitutive STAT3 activity sensitizes resistant non-Hodgkin's lymphoma and multiple myeloma to chemotherapeutic drug-mediated apoptosis. Clin Cancer Res. 2003;9(1):316–326. [PubMed] [Google Scholar]
- 28.Moignet A, Hasanali Z, Zambello R, Pavan L, Bareau B, Tournilhac O, et al. Cyclophosphamide as a first-line therapy in LGL leukemia. Leukemia. 2014;28(5):1134–1136. doi: 10.1038/leu.2013.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.