Abstract
Post-synthesis DNA modification is a very useful method for DNA functionalization. This is achieved by using a modified NTP, which has a handle for further modifications, replacing the corresponding natural NTP in polymerase-catalyzed DNA synthesis. Subsequently, the handle can be used for further functionalization after PCR, preferably through a very fast reaction. Herein we describe polymerase-mediated incorporation of trans-cyclooctene modified thymidine triphosphate (TCO-TTP). Subsequently, the trans-cyclooctene group was reacted with a tetrazine tethered to other functional groups through a very fast click reaction. The utility of this DNA functionalization method was demonstrated with the incorporation of a boronic acid group and a fluorophore. The same approach was also successfully used in modifying a known aptamer for fluorescent labelling applications.
Introduction
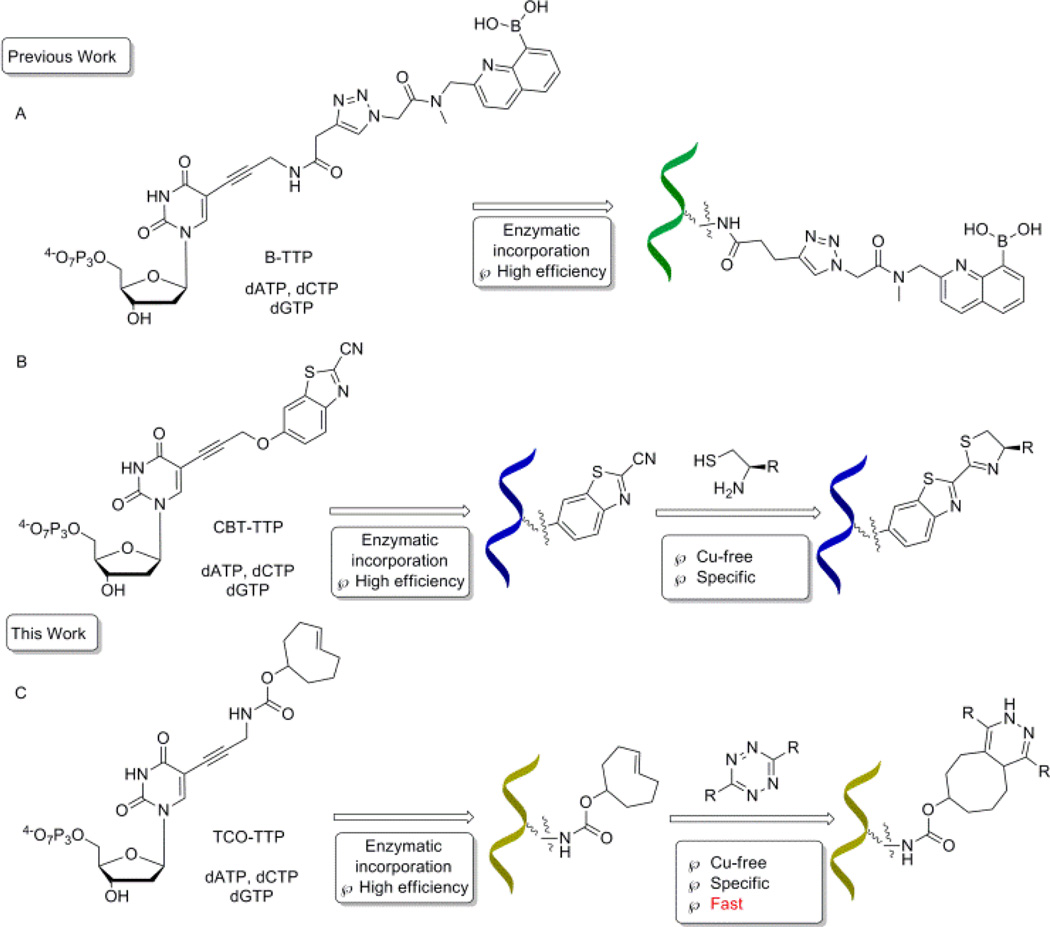
It has long been recognized that DNA has versatile functions with possible applications in sensing,1, 2 serving as new building blocks,3 developing new therapeutics,4–6 and in vitro aptamer selections.7–9 The incorporation of functional groups endows DNA molecules with additional properties and broadens application potentials.10, 11 In 2006, Seela12 and Carell13 first reported modifications of deoxynucleotides through copper(I)-catalyzed azide-alkyne cycloaddition (CuAAC) reaction developed by Sharpless14 and Maldel.15 Later on, copper-free click chemistry was widely used in chemical biology applications,16–18 in order to avoid issues related to copper.19 Especially in DNA modification, several pieces of recent work including crossing-linking,20, 21 biomolecules labeling,22–26 and cellular imaging27 have been published in this area. Our group has had a long-standing interest in using the boronic acid moiety for various sensing applications.28–31 Along this line, we have been working on the incorporation of the boronic acid functional group into DNA for aptamer selection32 and other applications.33–36 In incorporating the boronic acid group into DNA, there are special considerations because of the hygroscopic properties and Lewis acidity of this functional group. We have recently demonstrated the feasibility of direct PCR incorporation of boronic acid-modified TTP (Scheme 1, Route A)32 and post-PCR incorporation of the boronic acid group into DNA (Scheme 1, Routes B).34, 36 In post PCR DNA modification work, it is critical that the reaction goes to completion within a short period of time for various practical reasons. Thus it would be desirable to use click reactions that are fast enough so that completion of the reaction is assured on the scale of minutes.
Scheme 1.
A schematic representation of strategies for DNA modifications with a boronic acid: A). Enzymatic incorporation of boronic acid-modified nucleotides; B). Enzymatic incorporation of CBT modified nucleotides, followed by post-PCR modification; C). Enzymatic incorporation of trans-cyclooctene modified nucleotides, followed by Cu-free post-PCR modification
With most of the DNA click-modification reactions reported, one fast condensation reaction of 1, 2-aminothiol with 2-cyanobenzothiazole (CBT) has a second-order rate constant of 22 M−1·s−1.25, 36 This means that at 1 µM, the reaction’s first half-life (t1/2) would be more than 12 h. One would have to force the reaction by increasing the concentration of the non-DNA click partner agent because the concentrations of the DNA product from PCR amplifications are limited by PCR efficiency at high concentrations and primer concentrations, and are generally in the low µM range. Even under forcing conditions, it would still take hours for the reaction to go to completion. Thus, we set out to develop modification chemistry, which would give a shorter half-life. Tetrazine-trans-cyclooctene cycloaddition is recognized as one of the fastest click reactions, with second-order rate constant being up to 22000 M−1·s−1, which would give a first half-life of 45 second at 1 µM concentration.17, 33, 37–39 Thus in this study, we examined the feasibility of using the tetrazine-trans-cyclooctene chemistry for the post-PCR labelling of DNA with the boronic acid moiety (Route C, Scheme 1).
Results and Discussion
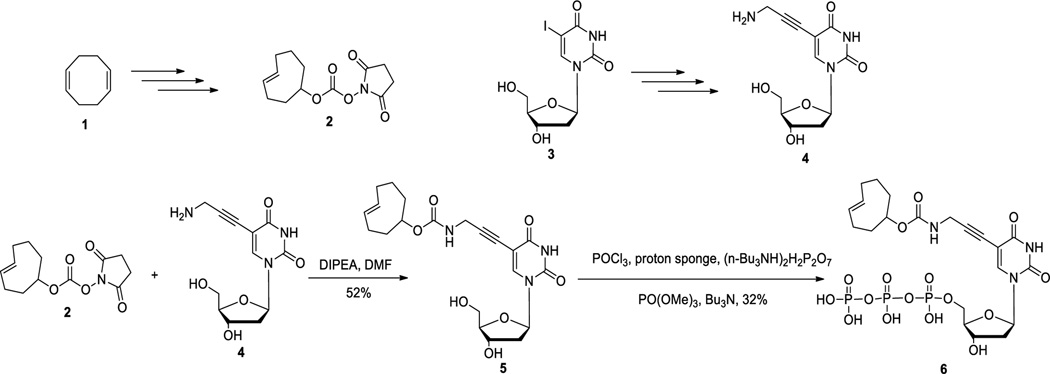
It has long been demonstrated that modification at the C5-position of deoxyuridine can be tolerated by various polymerases and such modified nucleotide triphosphates can be incorporated as a thymidine surrogate into DNA.40, 41 Thus we chose to install a click handle at the C5-position of deoxyuridine for further modifications. The trans-cyclooctene moiety can be introduced into compound 5 using amidation chemistry.42 Then subsequent triphosphorylation followed a one-pot three-step method43 to generate the final compound trans-cyclooctene triphosphate (6) in 32 % yield (Scheme 2).
Scheme 2.
Synthesis of trans-cyclooctene-modified deoxyuridine/thymidine triphosphate
Before PCR incorporation studies, we examined the stability of trans-cyclooctene modified nucleotide 6 at high temperature using NMR. It was found that there was no noticeable difference after heating the trans-cyclooctene at 90 °C for 20 min (Figure S1 and S2). Thus trans-cyclooctene is generally stable under PCR conditions because normally it takes 30 cycles (denaturing at 90 °C for 20 s each cycle) to complete the PCR step, which would subject the sample to 90 °C for no more than 15 min.
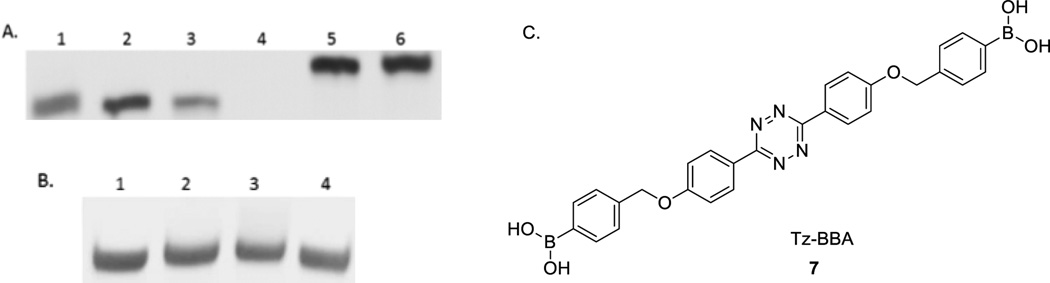
To study whether the modified nucleotide 6 can be recognized by a polymerase, we first incorporated trans-cyclooctene thymidine triphosphate (TCO-TTP) into a short DNA via primer extension using the Klenow fragment-catalyzed reaction. Based on previous successful incorporation studies,33, 36 we used a 21-mer oligonucleotide (nt) template-1 (5’-GGTTCCACCAGCAACCCG-CTA-3’), a 14-mer primer (5’-TAGCGGGTTGCTGG-3’) for the primer extension work (See experimental section for details). The template and primer were designed in such a way that the first incorporated nucleotide would be a T. Thus if TCO-TTP cannot be successfully recognized by the polymerase, there would be no primer extension. The extension products were examined by polyacrylamide gel electrophoresis (PAGE) (Figure 1). The experiments included several negative controls: (1) primer extension with all the necessary components except TTP (Lane 1, Figure 1A) (This is to make sure that enzyme fidelity would not be an issue and mis-matched incorporation does not happen), (2) primer extension with all the necessary components except the enzyme (Lane 2, Figure 1A), (3) primer extension only in the presence of the template and primer without the dNTPs and the enzyme (Lane 3, Figure 1A), and (4) primer only without the other necessary components (enzyme, template, and dNTPs) (Lane 4, Figure 1A). Positive control was Klenow fragment catalyzed primer extension using natural dNTPs (Lane 5, Figure 1A). TCO-TTP incorporation product was studied under Klenow fragment catalysis (Lane 6, Figure 1A)., It is clear that primer extension with all natural dNTPs (Lane 5) and TCO-TTP (Lane 6) showed similar full-length double stranded DNA bands, demonstrating successful incorporation of TCO-TTP. Next, we wanted to explore the feasibility of post-PCR modification with the incorporation of a boronic acid. Thus, tetrazine-bisboronic acid (Tz-BBA) was synthesized (See SI for details) for click-modification of the DNA product. In doing the modification work, we ran the reaction for 15 min. As shown in Figure 1B, similar full-length 21-bp DNA bands were observed from primer extension reaction (Figure 1B) using natural dNTPs (Lane 1), TCO-TTP (Lane 2), TCO-TTP labelled with Tz-BBA (Lane 3), and dNTP primer extension product reacting with Tz-BBA (Lane 4). There were some mobility differences among Lanes 1–3 with the click product (Lane 3) having the lowest mobility indicating increased molecular weight and the presence of the boronic acid group, which is a Lewis acid and can put a drag on mobility. However, the difference was small. Thus we sought additional evidence in support of successful incorporation of TCO-TTP and successful click modification.
Figure 1.
A). Primer extension using dNTP or TCO-TTP replacing TTP catalyzed by the Klenow fragment, 20% PAGE analysis: 1. primer extension with all the other necessary components except TTP, 2. primer extension with all the necessary components except the enzyme, 3. primer extension in presence of template-1 and primer only, 4. primer extension in presence of primer only without the other components, 5. primer extension with dNTPs, 6. primer extension using TCO-TTP instead of dTTP; B). Post-synthesis boronic acid incorporation using TCO-TTP primer extension product and Tz-BBA, 20% PAGE analysis: 1. primer extension product using dNTPs, 2.primer extension product using TCO-TTP instead of dTTP, 3. primer extension product using TCO-TTP after reaction with Tz-BBA, 4. primer extension product using dNTPs after reacting with Tz-BBA; C). Structure of Tz-BBA.
The primer extension products were studied using MALDI mass spectrometry. The TCO-TTP primer extension product (Lane 2, Figures 1B and S2) showed a molecular ion of m/z 6710.2 (calculated: 6710.4, [M+ H+]). Tz-BBA click labelling product (Lane 3, Figures 1B and S3) showed a molecular ion of m/z 7237.6 (calculated: 7238.6, [M+ Na+]). The primer extension product using dNTPs was also reacted with Tz-BBA (Lane 4, Figure 1B). The product gave a molecular ion of m/z of 6520.0 (Figure S4, calculated: 6519.3, [M+ H+]), which is the same as the original extension product, suggesting no reaction with Tz-BBA, as one would have expected. The mass spectrometric results further support the successful incorporation of one TCO-TTP moiety into DNA molecule and boronic acid labelling of DNA.
To further explore the feasibility of incorporating multiple TCO-TTP into DNA, we designed two more 21-mer templates complementary to the same 14-mer primer, template-2 (5’-TCAGTCACCAGCAA-CCCGCTA-3’) and template-3 (5’-CACGACACCAGCAACCCG-CTA-3’). These templates contained 2 or 3 A in the sequence, respectively, that can incorporate 2 or 3 TCO-TTP moieties into DNA. Based on previous optimization work using the Klenow fragment,36 incorporation of additional modified TTP required higher temperature and longer incubation time than incorporating a single modified TTP. Under harsh conditions, Klenow fragment catalysis would give incomplete extension products and several blunt bands on gel. As a result, we chose the KOD-XL DNA polymerase, a family B polymerase from thermococcus kodakaraensis, which is more tolerant of unnatural TTP.44 Analysis using 20% PAGE indicated successful primer extensions with the incorporation of both 2 and 3 Ts (Figure 2). Template-1 primer extension using dNTPs (Lane 1, Figure 2A) was conducted as positive control. Primer extension products using TCO-TTP in place of dTTP were obtained with template-1 (Lane 2, Figure 2A), template-2 (Lane 3, Figure 2A) and template-3 (Lane 4, Figure 2A). As seen in Figure 2A, the electrophoretic mobility of primer extension products decreased progressively with increasing number of TCO-TTP moieties, indicating successful incorporation of multiple TCO-TTP units into DNA. To further confirm TCO-TTP’s incorporation and successful boronic acid labelling work, the extension product using template-3 was further studied through reaction with Tz-BBA in a similar fashion as the results described in Figure 1. As shown in Figure 2B, Tz-BBA-labelled primer extension products led to a slow-moving band (Lane 5). Such results are consistent with previous findings in similar studies.36 It is well known that H2O2 can oxidatively cleave the boronic acid moiety leaving a phenol group behind.45 Thus we also treated the boronic acid labelled primer extension product with H2O2 (Lane 6, Figure 2B). The faster moving band after H2O2 treatment was presumably due to decreased molecular weight as well as the lack of the boronic acid group, which tends to interact with Lewis bases in the matrix and thus provide a drag on mobility.
Figure 2.
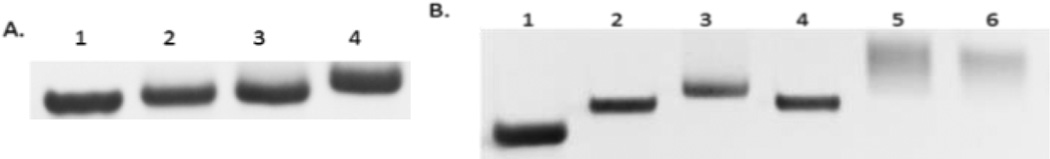
A). Primer extension incorporating multiple TCO-TTP catalyzed by KOD-XL DNA polymerase, 20% PAGE analysis: 1. primer extension using template-1 with dNTPs, 2. primer extension using template-1 with TCO-TTP in place of TTP, 3. primer extension using template-2 with TCO-TTP in place of TTP, 4. primer extension using template-3 with TCO-TTP in place of TTP; B). Post-synthesis boronic acid labelling of primer extension product using template-3 with TCO-TTP incorporation, 20% PAGE analysis: 1. primer extension with all the necessary components except TTP, 2. primer extension with dNTPs, 3. primer extension using TCO-TTP in place of TTP; 4. primer extension product using dNTPs reacted with Tz-BBA, 5. primer extension product using TCO-TTP reacted with Tz-BBA, 6. Tz-BBA labelled primer extension product (from lane 5) treated with H2O2.
After successful incorporation of TCO-TTP into short DNA sequences using primer extensions, we conducted PCR amplifications of a 90-mer DNA template (See experimental section for details). Before the incorporation work, we investigated amplification efficiencies of three commercially available polymerases, KOD-XL, Taq and Deep VentR exo−. PCR amplifications were examined under the same conditions for three polymerases. The screening results showed the highest efficiency by KOD-XL (Lane 2, Figure 3A). Such results are consistent with previous work on using other 5-modified deoxyuridine-5’-triphosphates.32, 36 As a result, further PCR amplifications were conducted with the KOD-XL polymerase. According to the TCO-TTP thermo-stability test, optimized PCR amplification method was built based on an improvement of the CBT-TTP incorporation work.36 With TCO-TTP modified DNA, 30 thermal cycles were run with 20 s melting at 90 °C, 20 s annealing at 48 °C, and 30 s extension at 72 °C, which gave 10-min exposure at 90 °C. Under this PCR condition, TCO-TTP can be introduced into DNA sequences in place of TTP. Following similar conditions as in the boronic acid labelling of primer extension products, PCR-amplified product was also reacted with Tz-BBA for boronic acid labelling. As shown in Figure 3B, TCO-TTP was successfully incorporated into 90-nt PCR product (Lane 3). Experiments using dATP, dCTP, and dGTP, without dTTP for PCR amplification were used as a negative control (Lane 1) and dNTPs PCR amplification as a positive control (Lane 2). PCR-amplified TCO-TTP product was reacted with Tz-BBA (Lane 4), leading to a slow mobility band. Boronic acid labelling control experiment was performed with the dNTPs PCR amplification product by reacting with Tz-BBA (Lane 5). As expected, dNTP-PCR product didn’t react with Tz-BBA and gave the same mobility as dNTP-PCR amplification product (Lane 2). The results demonstrate successful TCO-TTP incorporation during PCR and successful boronic acid labelling by reacting with Tz-BBA.
Figure 3.

A). Polymerase screening for TCO-TTP incorporation, 15% PAGE analysis: 1. KOD-XL catalyzed PCR product using dNTPs, 2. KOD-XL catalyzed PCR product using TCO-TTP in place of TTP, 3. Taq catalyzed PCR product using dNTPs, 4. Taq catalyzed PCR product using TCO-TTP in place of TTP; 5. Deep VentR catalyzed PCR product using dNTPs, 6. Deep VentR catalyzed PCR product using TCO-TTP in place of TTP. B). Post-PCR boronic acid labelling of amplified DNA product with TCO-TTP incorporation, 15% PAGE analysis: 1. PCR amplification in absence of TTP, 2. PCR amplification with dNTPs, 3. PCR amplification using TCO-TTP in place of TTP; 4. PCR amplification product using TCO-TTP reacted with Tz-BBA, 5. PCR amplification product using dNTPs reacted with Tz-BBA
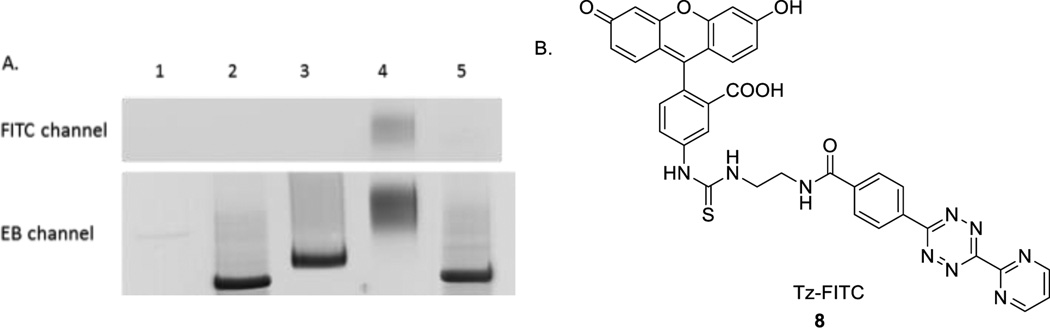
Encouraged by the results from TCO-TTP incorporation into a 90-nt DNA sequence and successful post-PCR boronic acid labelling, we further explored incorporating TCO-TTP into a published aptamer to see whether we can use the TCO-TTP handle for labelling DNA sequences with other structural moieties, such as fluorophore. N37 is a known aptamer selectively targeting the thrombospondin-1 (TSP-1) protein on the surface of inflamed endothelial cells with high potential of atherosclerosis.46, 47 Using the same PCR amplification conditions described above, N37 was used as new template for amplifications. Tetrazine-FITC (fluorescein isothiocyanate reacting with an amino handle of tetrazine, Tz-FITC, Figure 4B, see SI for details) was synthesized for post-PCR fluorescent labelling of TCO-TTP modified DNA. We studied the incorporation of fluorescein into a known aptamer46 N37 and the applicability of using such a fluorophore-labelled DNA for binding applications. TCO-TTP modified N37 (TCO37) (Lane 3, Figure 4A) was obtained and analyzed with 15% PAGE after KOD-XL catalyzed PCR. This amplification product generated fluorescein-labelled N37 aptamer (Lane 4, Figure 4A) after reacting with Tz-FITC. Natural N37 aptamer (Lane 2, Figure 4A) was used as positive control. The product of N37 (generated using dNTPs) reacting with Tz-FITC (Lane 5, Figure 4A) was used as negative control for the fluorescein labelling experiments. As shown in Figure 4A, only the PCR product with the trans-cyclooctene moiety reacted with Tz-FITC as indicated by the fluorescent band (FITC channel, λem: 520 nm, Lane 4, Figure 4A). No fluorescence was detected in other lanes. Ethidium bromide (EB) staining results were consistent with successful incorporation of TCO-TTP and boronic acid labelling with 90-mer DNA template.
Figure 4.
A). Post-PCR FITC labelling of TCO-TTP incorporation products using N37 template, 15% PAGE analysis: 1. PCR amplification with all the essential components except TTP, 2. PCR amplification with dNTPs, 3. PCR amplification using TCO-TTP in place of dTTP; 4. PCR amplification product using TCO-TTP reacted with Tz-FITC, 5. PCR amplification product using dNTPs reacted with Tz-FITC; B). Structure of Tz-FITC.
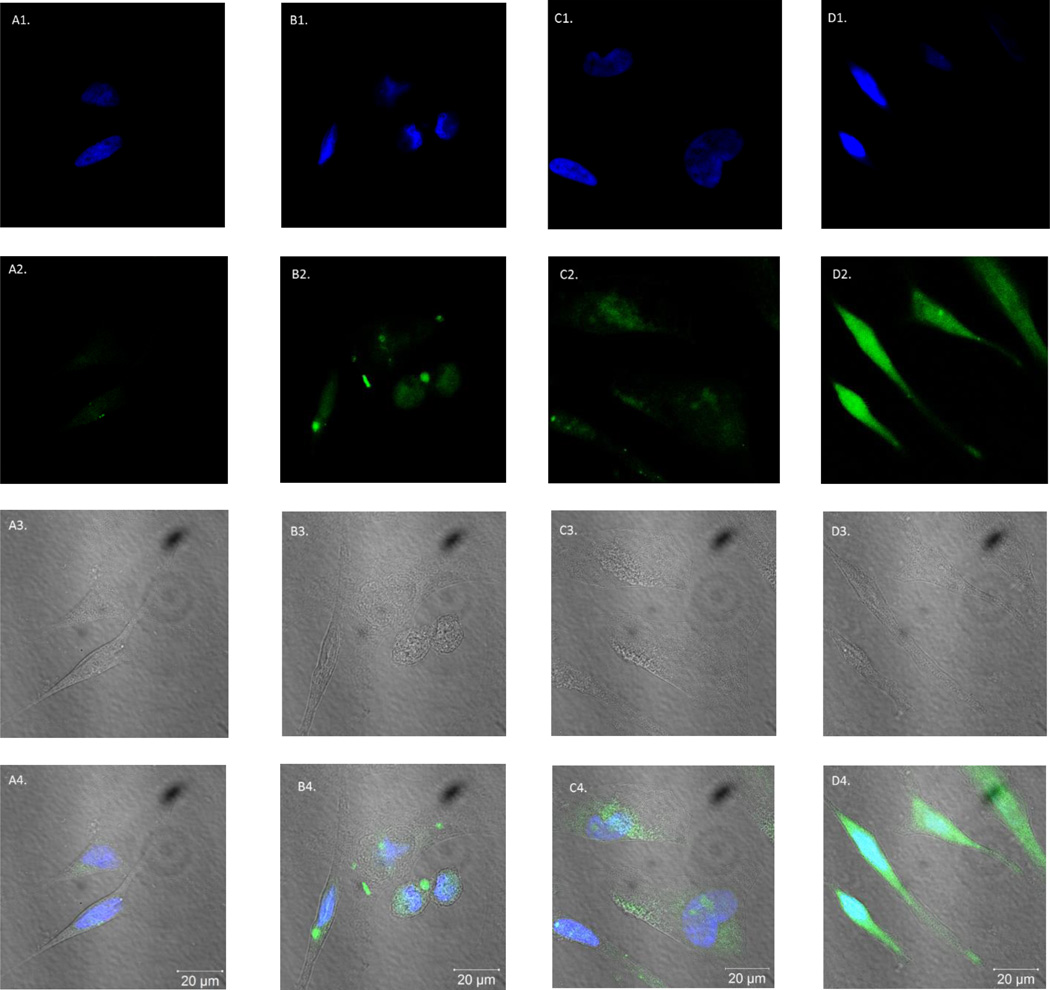
Based on the successful incorporation of a fluorophore into a known N37 aptamer, we wanted to explore its in vitro cell fluorescent imaging applications. In natural N37 aptamer studies, the binding affinity with TSP-1 was determined with 5’-FAM labelled primer, which has a single fluorophore attached in each aptamer sequence.46 However, with the ability to incorporate a fluorophore using the tetrazine-trans-alkene chemistry, it was possible to introduce a fluorophore in multiple positions of the TCO-modified aptamer N37 (TCO37). Such labelled aptamers could have higher fluorescent intensity without losing binding affinity. Thus fluorescent labelling studies were conducted using Hela cells, which express TSP-1 on the surface.48 After incubation with 1 µM of 5’-FAM N37 aptamer (Figure 5B) and FITC-labelled TCO37 aptamer (Figure 5D) at 37 °C for 30 min, the cells were examined under a fluorescent microscope (see experimental section for details). Two control experiments were conducted with cell only (Figure 5A) and N37 (no click handle) reacted with Tz-FITC (Figure 5C). 4',6-Diamidino-2-phenylindole (DAPI) staining (for nuclei) was also used. Cells were examined using three channels (Figure 5): (1) DAPI channel (λem: 461 nm; first row) for nuclear staining; (2) FITC channel (λem: 520 nm, second row) for detecting the fluorescein signal; (3) phase contrast channel for morphological observations (third row); and (4) Merged channel, with images of DAPI, FITC and phase merged into one (fourth row). As one can see, all cells stained normally with DAPI and have normal morphology under phase contrast, indicating normal cell growth. However, only the cells treated with FITC-labelled TCO37 aptamer showed significant fluorescent labelling in the fluorescein channel. Interestingly, the FAM-N37 labelled cells did not show the same significant fluorescent labelling. The results so far indicate that replacing T with TCO-T in aptamer N37 didn’t affect its binding affinity to TSP-1 and allowed for fluorescent labelling of cells after the incorporation of a fluorophore. The ability to incorporate multiple fluorophores could provide enhanced sensitivity for labelling. Conceivably, the same approach can be used in other known aptamers for binding studies.
Figure 5.
Images of Hela cells were taken using a confocal microscope. Fluorescent images of four samples were shown: A column: blank, cell only; B column: FAM-N37 aptamer; C column: product of N37 aptamer reacting with Tz-FITC (negative control); D column: FITC-labelled TCO37 sequence. Images were taken with 3 channels: 1. DAPI channel (λem: 461 nm, first row); 2. FITC channel (λem: 520 nm, second row); 3. Phase contrast channel (third row); 4. Images from the DAPI channel merged that of the FITC channel and phase channel (fourth row)
Conclusions
A fast and efficient approach of post-PCR DNA modification is described. The modified TTP (TCO-TTP) has a click handle for reaction with tetrazine, which can be used for tethering other functional groups. Compared to previously reported methods, the approach described uses a very fast click reaction and has the advantage of significantly improved reaction kinetics. The incorporation of a boronic acid functional group or a fluorophore has been described after the successful incorporation of TCO-TTP.
The boronic acid-labelled DNA can be used for aptamer selection against carbohydrates and glycoproteins, while the fluorophore-labelled DNA can be used for cell labelling studies among other applications. In addition, the trans-alkene functional group in the modified DNA can also be used to tether other functional group for various purposes.
Experimental Section
Experimental Procedure for Klenow Fragment Catalyzed Primer Extension
The 50 µL reaction mixtures of contained 21-nt template (5’-GGTTCCACCAGCAACCCG-CTA-3’ (20 µM)), 14-nt primer (5’- TAGCGGGTTGCTGG-3’ (20 µM)), Tris-HCl (10 mM), NaCl (50 mM), MgCl2 (10 mM), dithiothreitol (1 mM) at pH 7.9, Klenow fragment (0.5 U µL−1), dATP, dCTP, dGTP, and dTTP (or TCO-TTP) (200 µM). Reactions were performed by incubating the prepared solutions at 25 °C for 30 min. The primer extension products were analyzed by 20 % PAGE.
Experimental Procedure for KOD-XL Catalyzed Primer Extension
The 50 µL reaction mixture contained 21-nt template (Template-2: 5’-TCAGTCACCAGCAACCCGCTA-3’, Template-3: 5’-CACGACACCAG-CAACCCGCTA-3’ (20 µM)), 14-nt primer (5’-TAGCGGGTTGCTGG-3’ (20 µM)), Tris-HCl (10 mM), NaCl (50 mM), MgCl2 (10 mM), dithiothreitol (1 mM) at pH 7.9, KOD-XL (0.5 U µL−1), dATP, dCTP, dGTP, dTTP, or TCO-TTP (200 µM). Reactions were performed by incubating the prepared solutions at 90 °C for 1 min, 20 °C for 1 min, and 66 °C for 20 min. The primer extension products were analyzed by 20 % PAGE.
Experimental Procedure for Post-synthesis Labelling of Primer Extension Products with Tz-BBA
The primer extension product TCO-DNA21 was purified using Millipore Amicon 3 kDa spin column. 2 µL of 10 mM Tz-BBA in DMSO was added into pre-purified TCO-DNA21 (20 µL). The reaction was allowed to vortex at r.t. for 15 min. The negative control experiment was performed following the same procedure except using dNTPs-DNA21, the primer extension product using dNTPs and other reagents. The resulting DNA products after post-synthesis modification were purified with Millipore Amicon 3 kDa spin column and analyzed by 20 % PAGE.
Experimental Procedure for PCR Incorporation and Post-PCR Labelling of TCO-TTP
The 50 µL PCR mixture contained DNA 90-nt template (PBA, 5’-CCTTCGTTGTCTGCCTTCGTGAGCGGAGTCAGACGCACGCTCGTACCTGTGCGCAAGCACTATGACGGACACCCTTCAGAATTCGCACCA-3’, 10 nM), 20-nt primer 1 (5’-TGGTGCGAATTCTGAAGGGT-3’, 1 µM), 20-nt primer 2 (5’-CCTTCGTTGTCTGCCTTCGT-3’, 1 µM), dATP, dCTP, dGTP, dTTP or TCO-TTP at a concentration of 200 µM, 0.5 U µL−1 KOD-XL DNA polymerase and 1× reaction buffer as provided by vender. Thirty thermal cycles were conducted with melting at 90 °C for 20 s, annealing at 48 °C for 20 s, and extending at 72 °C for 30 s with initial denaturing at 90 °C for 2 min and final extension at 72 °C for 5 min. The PCR products were then analyzed by 15% PAGE.
Post-synthesis labelling of the PCR products was performed using similar procedures as those for primer extension. Specifically, TCO-DNA90 prepared from PCR was purified using Millipore Amicon 10 kDa spin column. Pre-purified TCO-DNA90 (20 µL) was then added to the 2 µL of 10 mM Tz-BBA DMSO solution, which was further reacted at r.t. for 30 min. The negative control experiment was performed following the same procedure except using dNTPs-DNA90, the PCR product using dNTPs and other reagents. The resulting DNA product after post-synthesis modification was purified with Millipore Amicon 10 kDa spin column and analyzed by 15% PAGE.
Experimental Procedure of Fluorescence Labelling and Cell Imaging Studies
The PCR mixture of a final volume of 50 µL contained DNA 86-nt template (N37, 5’- ATACCAGCTTATTCAATTCTGCCGGGAACACCGCGTGGC-TCTCTGCAACGCCCAGGACATACCACATTAGATAGTAAGTGCAATCT-3’ 10 nM each), 18-nt primer 1 or 5’-FAM 18-nt primer 1 (5’-(FAM)-ATACCAGCTTATTCAATT-3’, 1 µM), primer 2 (5’- AGATTGCACTT-ACTATCT-3’, 1 µM),46 dATP, dCTP, dGTP, dTTP or TCO-TTP at a concentration of 200 µM, 0.5 U µL−1 KOD-XL DNA polymerase and 1× reaction buffer as provided by vender. PCR thermal cycles were conducted as previous incorporation. Post-PCR labelling using Tz-FITC was performed with similar procedures as previous incorporation. The FITC-labelled DNA product was purified with Millipore Amicon 10 kDa spin column and analyzed by 15% PAGE.
Hela cells were seeded on rectangular coverslips in a 6-well plate until it achieved 90% coverage on surface before the imaging experiment and were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 1% PNS at 37 °C with 5% CO2. The cells were then incubated with either FAM-N37 aptamer, or FITC-labelled TCO37 aptamer at the same concentration in SELEX binding buffer49 for 30 min at 37 °C. After washing with phosphate buffered saline (PBS) for 2 times, the cells were fixed with ice-cold methanol for 5 min. Thereafter, the coverslips containing the cells were mounted onto the slides and imaged with a fluorescent microscope.
Supplementary Material
Acknowledgements
Financial support from the NIH (GM084933 and GM086925), the Chinese Scholarship Council to KW and the Georgia State University Molecular Basis of Disease Program (MBD) through a fellowship to DZW and KW are gratefully acknowledged.
Footnotes
Footnotes should appear here. These might include comments relevant to but not central to the matter under discussion, limited experimental and spectral data, and crystallographic data.
Electronic Supplementary Information (ESI) available: [details of any supplementary information available should be included here]. See DOI: 10.1039/b000000x/
Notes and references
- 1.Liu J, Lu Y. Angew. Chem. Int. Ed. 2006;45:90–94. [Google Scholar]
- 2.Drummond TG, Hill MG, Barton JK. Nat. Biotechnol. 2003;21:1192–1199. doi: 10.1038/nbt873. [DOI] [PubMed] [Google Scholar]
- 3.Rothemund PW. Nature. 2006;440:297–302. doi: 10.1038/nature04586. [DOI] [PubMed] [Google Scholar]
- 4.Long X, Thomas A. J. Pharm. Sci. 2011;100:38–52. doi: 10.1002/jps.22243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hurley LH. Nat. Rev. Cancer. 2002;2:188–200. doi: 10.1038/nrc749. [DOI] [PubMed] [Google Scholar]
- 6.Keefe AD, Pai S, Ellington A. Nat. Rev. Drug Discov. 2010;9:537–550. doi: 10.1038/nrd3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kanan MW, Rozenman MM, Sakurai K, Snyder TM, Liu DR. Nature. 2004;431:545–549. doi: 10.1038/nature02920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Syed MA, Pervaiz S. Oligonucleotides. 2010;20:215–224. doi: 10.1089/oli.2010.0234. [DOI] [PubMed] [Google Scholar]
- 9.Fang X, Tan W. Accounts Chem. Res. 2009;43:48–57. doi: 10.1021/ar900101s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eaton BE, Gold L, Hicke BJ, Janjić N, Jucker FM, Sebesta DP, Tarasow TM, Willis MC, Zichi DA. Bioorg. Med. Chem. 1997;5:1087–1096. doi: 10.1016/s0968-0896(97)00044-8. [DOI] [PubMed] [Google Scholar]
- 11.Sakthivel K, Barbas CF. Angew. Chem. Int. Ed. 1998;37:2872–2875. doi: 10.1002/(SICI)1521-3773(19981102)37:20<2872::AID-ANIE2872>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 12.Seela F, Sirivolu V. Chem. Biodivers. 2006;3:509–514. doi: 10.1002/cbdv.200690054. [DOI] [PubMed] [Google Scholar]
- 13.Burley G, Gierlich J, Mofid M, Nir H, Tal S, Eichen Y, Carell T. J. Am. Chem. Soc. 2006;128:1398–1399. doi: 10.1021/ja055517v. [DOI] [PubMed] [Google Scholar]
- 14.Kolb H, Finn M, Sharpless K. Angew. Chem. Int. Ed. 2001;40:2004–2021. doi: 10.1002/1521-3773(20010601)40:11<2004::AID-ANIE2004>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 15.Tornøe CW, Christensen C, Meldal M. J. Org. Chem. 2002;67:3057–3064. doi: 10.1021/jo011148j. [DOI] [PubMed] [Google Scholar]
- 16.John CJ, Carolyn RB. Chem. Soc. Rev. 2010;39:1272–1279. [Google Scholar]
- 17.Selvaraj R, Fox JM. Curr. Opi. Chem. Bio. 2013;17:753–760. doi: 10.1016/j.cbpa.2013.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Juliane S, Manfred W, Andres Js. J. Am. Chem. Soc. 2010;132:8846–8847. [Google Scholar]
- 19.Kennedy DC, McKay CS, Legault MCB, Danielson DC, Blake JA, Pegoraro AF, Stolow A, Mester Z, Pezacki JP. J. Am. Chem. Soc. 2011;133:17993–18001. doi: 10.1021/ja2083027. [DOI] [PubMed] [Google Scholar]
- 20.Varizhuk AM, Kaluzhny DN, Novikov RA, Chizhov AO, Smirnov IP, Chuvilin AN, Tatarinova ON, Fisunov GY, Pozmogova GE, Florentiev VL. J. Org. Chem. 2013;78:5964–5969. doi: 10.1021/jo400651k. [DOI] [PubMed] [Google Scholar]
- 21.Pujari SS, Ingale SA, Seela F. Bioconjugate Chem. 2014;25:1855–1870. doi: 10.1021/bc5003532. [DOI] [PubMed] [Google Scholar]
- 22.Arndt S, Wagenknecht H-A. Angew. Chem. Int. Ed. 2014 doi: 10.1002/anie.201407874. Early View. [DOI] [PubMed] [Google Scholar]
- 23.Neef AB, Luedtke NW. ChemBioChem. 2014;15:789–793. doi: 10.1002/cbic.201400037. [DOI] [PubMed] [Google Scholar]
- 24.Bußkamp H, Ellen B, Niederwieser A, Abdel-Rahman OS, Winter RF, Wittmann V, Marx A. Chem. Commun. 2014;50:10827–10829. doi: 10.1039/c4cc04332d. [DOI] [PubMed] [Google Scholar]
- 25.Stubinitzky C, Cserép GB, Bätzner E, Kele P, Wagenknecht H-A. Chem. Commun. 2014;50:11218–11221. doi: 10.1039/c4cc02855d. [DOI] [PubMed] [Google Scholar]
- 26.Pyka AM, Domnick C, Braun F. Bioconjugate Chem. 2014;25:1438–1443. doi: 10.1021/bc500302y. [DOI] [PubMed] [Google Scholar]
- 27.Rieder U, Luedtke NW. Angew. Chem. Int. Ed. 2014;126:9322–9326. [Google Scholar]
- 28.Springsteen G, Wang B. Chem. Commun. 2001:1608–1609. doi: 10.1039/b104895n. [DOI] [PubMed] [Google Scholar]
- 29.Springsteen G, Wang B. Tetrahedron. 2002;58:5291–5300. [Google Scholar]
- 30.Cheng Y, Ni N, Yang W, Wang B. Chem. Eur. J. 2010;16:13528–13538. doi: 10.1002/chem.201000637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jin S, Cheng Y, Reid S, Li M, Wang B. Med. Res. Rev. 2010;30:171–257. doi: 10.1002/med.20155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin N, Yan J, Huang Z, Altier C, Li M, Carrasco N, Suyemoto M, Johnston L, Wang S, Wang Q, Fang H, Caton-Williams J, Wang B. Nucleic Acids Res. 2006;35:1222–1229. doi: 10.1093/nar/gkl1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang X, Dai C, Molina ADC, Wang B. Chem. Commun. 2010;46:1073–1075. doi: 10.1039/b921163b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dai C, Wang L, Sheng J, Peng H, Draganov A, Huang Z, Wang B. Chem. Commun. 2011;47:3598–3600. doi: 10.1039/c0cc04546b. [DOI] [PubMed] [Google Scholar]
- 35.Cheng Y, Dai C, Peng H, Zheng S, Jin S, Wang B. Chem. Asian. J. 2011;6:2747–2752. doi: 10.1002/asia.201100229. [DOI] [PubMed] [Google Scholar]
- 36.Cheng Y, Peng H, Chen W, Ni N, Ke B, Dai C, Wang B. Chem. Eur. J. 2013;19:4036–4042. doi: 10.1002/chem.201201677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Blackman M, Royzen M, Fox J. J. Am. Chem. Soc. 2008;130:13518–13519. doi: 10.1021/ja8053805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Taylor MT, Blackman ML, Dmitrenko O, Fox JM. J. Am. Chem. Soc. 2011;133:9646–9649. doi: 10.1021/ja201844c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen W, Wang D, Dai C, Hamelberg D, Wang B. Chem. Commun. 2012;48:1736–1738. doi: 10.1039/c2cc16716f. [DOI] [PubMed] [Google Scholar]
- 40.Ötvös L, Sági J, Kovács T, Walker RT. Nucleic Acids Res. 1987;15:1763–1777. doi: 10.1093/nar/15.4.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kovács T, Ötvös L. Tetrahedron Lett. 1988;29:4525–4528. [Google Scholar]
- 42.Royzen M, Yap GP, Fox JM. J. Am. Chem. Soc. 2008;130:3760–3761. doi: 10.1021/ja8001919. [DOI] [PubMed] [Google Scholar]
- 43.Marika K, Mia H, Pasi V, Harri Ln. Bioconjugate Chem. 2010;21:748–755. [Google Scholar]
- 44.Kuwahara M, Nagashima J-i, Hasegawa M, Tamura T, Kitagata R, Hanawa K, Hososhima S-i, Kasamatsu T, Ozaki H, Sawai H. Nucleic Acids Res. 2005;34:5383–5394. doi: 10.1093/nar/gkl637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Henry GK. J. Am. Chem. Soc. 1954;76:870–874. [Google Scholar]
- 46.Ji K, Lim WS, Li SFY, Bhakoo K. Anal. Bioanal. Chem. 2013;405:6853–6861. doi: 10.1007/s00216-013-7155-z. [DOI] [PubMed] [Google Scholar]
- 47.Ji K, de Carvalho LP, Bi X, Seneviratnankn A, Bhakoo K, Chan M, Li SFY. Biosens. Bioelectron. 2014;55:405–411. doi: 10.1016/j.bios.2013.12.012. [DOI] [PubMed] [Google Scholar]
- 48.Kang J, Kim M, Chang S, Sim S, Jo Y. J. Cell. Biochem. 2008;104:1192–1203. doi: 10.1002/jcb.21697. [DOI] [PubMed] [Google Scholar]
- 49.Sefah K, Shangguan D, Xiong X, O'Donoghue MB, Tan W. Nat. Protoc. 2010;5:1169–1185. doi: 10.1038/nprot.2010.66. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.