Abstract
Phosphatase and tensin homolog (PTEN), which negatively regulates tumorigenic PIP3 signaling, is a commonly mutated tumor suppressor. The majority of cancer-associated PTEN mutations block its essential PIP3 phosphatase activity. However, there is a group of clinically identified PTEN mutations that maintain enzymatic activity, and it is unknown how these mutations contribute to tumor pathogenesis. Here, we show that these enzymatically competent PTEN mutants fail to translocate to the plasma membrane where PTEN converts PIP3 to PI(4,5)P2. Artificial membrane tethering of the PTEN mutants effectively restores tumor suppressor activity and represses excess PIP3 signaling in cells. Thus, our findings reveal a novel mechanism of tumorigenic PTEN deficiency.
Keywords: PTEN, Membrane localization, PIP3, Dictyostelium
Introduction
More than 1,500 different mutations in the PTEN gene have been identified in different cancers (http://cancer.sanger.ac.uk) (1–6). Approximately 40% of these are substitution missense mutations. These cancer-associated amino acid substitutions are distributed along the entire length of PTEN with the highest frequency in the lipid phosphatase catalytic residues, mutations of which abolish lipid phosphatase activity. However, there is a group of PTEN substitutions that maintain PTEN enzymatic activity (7–9), and it is currently unknown how these mutations disrupt the tumor suppressor function of PTEN.
The membrane recruitment of PTEN is important for its lipid phosphatase function in PIP3 signaling (10–15). Membrane association of PTEN is negatively regulated by phosphorylation of the inhibitory C-terminal tail domain. When phosphorylated at four serine/threonine residues, the C-terminal tail binds to the membrane-binding regulatory interface and masks the membrane binding domain containing the CBR3 loop of the C2 domain (10, 11, 16). When the phosphorylation sites are mutated to alanine (termed PTENA4), the C-terminal tail dissociates from the membrane-binding regulatory interface and promotes membrane recruitment of PTEN (10, 16). Therefore, we hypothesized that the cancer-associated mutations that sustain enzymatic activity might affect the membrane recruitment of PTEN. To test our hypothesis and determine whether the enzymatically competent mutants represent a new class of tumorigenic PTEN deficiency, we took advantage of PTENA4. Although functionally important, PTEN membrane association is difficult to analyze because a large number of PTEN molecules are present in the cytosol (10, 11). The use of PTENA4 dramatically decreases the cytosolic pool of PTEN and allows us to visualize membrane localization.
Results and Discussion
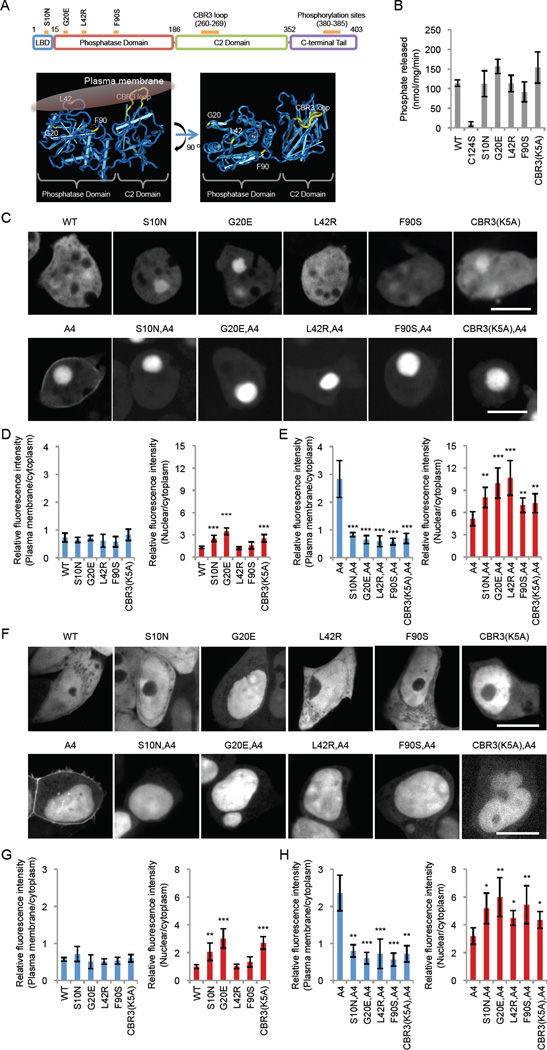
We focused on a group of PTEN missense mutants selected by two criteria. We first chose mutations that do not inhibit phosphatase activity (7–9). Among these, we further narrowed the choice down to four mutations, S10N, G20E, L42R, and F90S, that are located in or near the membrane-binding regulatory interface (Fig. 1A and Table S1) (10). We first biochemically confirmed intact PIP3 phosphatase activity of these mutants using purified PTEN as we described previously (10). WT and mutant forms of human PTEN fused with GFP were expressed and immunopurified from Dictyostelium cells using agarose bead-coupled anti-GFP antibodies. PTENS10N, PTENG20E, PTENL42R, and PTENF90S showed enzymatic activities indistinguishable from that of WT PTEN (Fig. 1B). Activity was completely abolished in the negative control PTENC124S, which carries a substitution in the catalytic residue C124. Fluorescence microscopy of Dictyostelium cells (Fig. 1C and D) and the human cell line HEK293T (Fig. 1F and G) expressing PTEN mutants fused to GFP showed that PTEN-GFP, PTENS10N-GFP, PTENG20E-GFP, PTENL42R-GFP, and PTENF90S-GFP were mainly located in the cytosol and nucleus.
Figure 1. Cancer-associated PTEN mutations inhibit membrane association.
(A) The domain structure of PTEN. S10N is located in the lipid binding domain (LBD) whereas G20E, L42R, and F90S are present in the phosphatase domain. The same mutations are indicated in the 3-D structure of PTEN with the phosphatase and C2 domains (R14-V351) (27) (http://www.ncbi.nlm.nih.gov/Structure/mmdb/mmdbsrv.cgi?uid=11638). (B) The S10N, G20E, L42R, and F90S mutations do not decrease the PIP3 phosphatase activity of purified PTEN in vitro. (C–H) The indicated PTEN-GFP constructs were expressed in Dictyostelium cells (C–E) and HEK293T cells (F–H). Bar, 10 µm. Fluorescence intensity in the plasma membrane (blue bars) and the nucleus (red bars) was quantified relative to that in the cytosol (n = 8).
To determine whether the S10N, G20E, L42R, and F90S mutations affect membrane association, we combined them with the A4 mutations. PTENA4-GFP was located at the plasma membrane in addition to the nucleus in Dictyostelium cells (Fig. 1C and E) and HEK293T cells (Fig. 1F and H), as previously reported (10, 11). In contrast, PTENS10N,A4-GFP, PTENG20E,A4-GFP, PTENL42R,A4-GFP, and PTENF90S,A4-GFP were unable to localize at the plasma membrane in both cell types (Fig. 1C and E for Dictyostelium, F and G for HEK293T). Their nuclear localization was increased, as expected from our previous study showing that dissociation of PTEN from the plasma membrane leads to accumulation in the nucleus (10). The negative control PTENCBR3(K5A)-GFP, which carries mutations in the CBR3 loop of the membrane binding C2 domain (10), showed cytosolic localization (Fig. 1C–H).
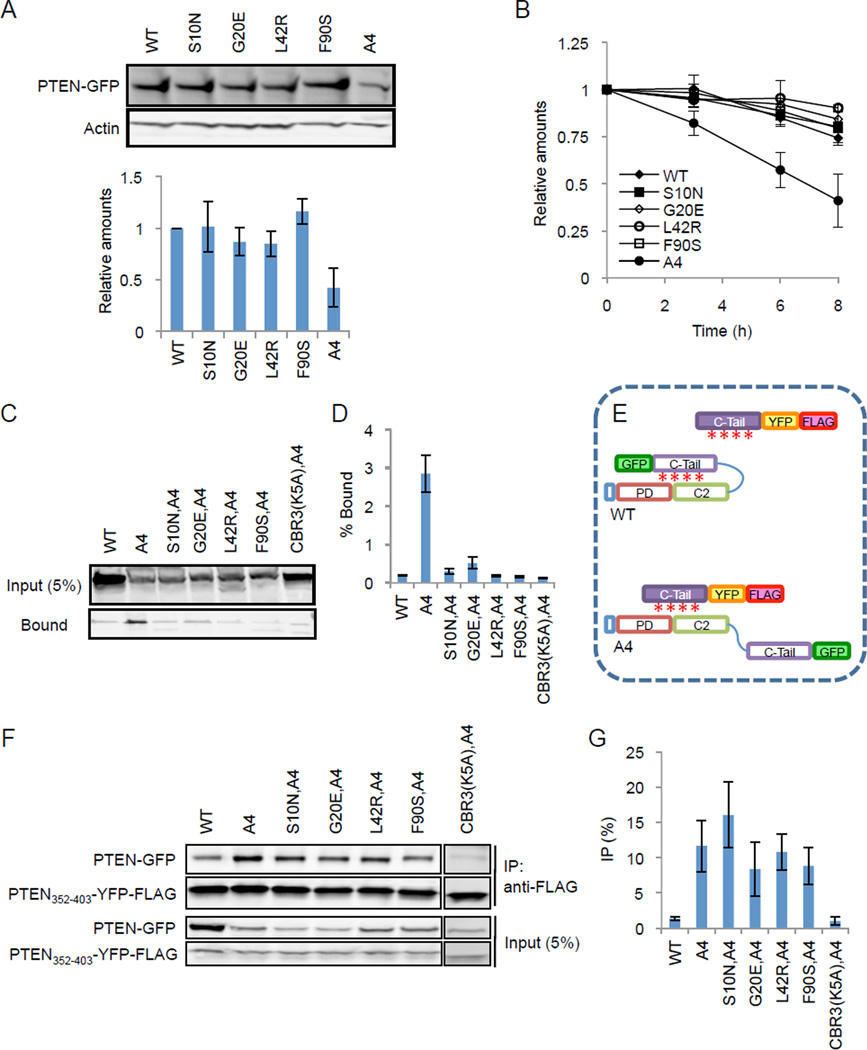
Because decreased stability of PTEN contributes to tumorigenesis (17), we tested whether the S10N, G20E, L42R, and F90S mutations affect the turnover of PTEN. Immunoblotting revealed similar levels of PTEN-GFP, PTENS10N-GFP, PTENG20E-GFP, PTENL42R-GFP, and PTENF90S-GFP in HEK293T cells (Fig. 2A). In contrast, PTENA4-GFP, which has decreased stability, showed lower steady-state levels (Fig. 2A). Consistent with these results, PTEN-GFP, PTENS10N-GFP, PTENG20E-GFP, PTENL42R-GFP, and PTENF90S-GFP had comparable half-lives in the presence of the protein synthesis inhibitor cycloheximide, whereas PTENA4-GFP showed a shorter half-life (Fig. 2B). PTEN binds to another tumor suppressor, p53 (18, 19). We tested whether the four mutations defective in membrane localization affected this protein-protein interaction using co-immunoprecipitation. The results showed similar binding for WT and mutant PTEN to p53 (Fig. S1).
Figure 2. S10N, G20E, L42R, and F90S mutations block interaction of PTEN with membrane lipids.
(A) HEK293T cells were transfected with the indicated PTEN-GFP constructs. Whole-cell lysates were analyzed by immunoblotting with antibodies to PTEN and actin. The band intensity was quantified relative to WT PTEN-GFP. Values represent the mean ± SD (n = 3). (B) HEK293T cells were transfected with expression constructs containing GFP fused to the indicated forms of PTEN and were incubated with 100 µg/ml cycloheximide. Whole-cell lysates were analyzed by immunoblotting with anti-GFP antibodies at the indicated time points. Band intensity was quantified relative to a 0-hour sample (n = 3). (C and D) Liposome binding assay. Liposomes (PC:PS:PI(4,5)P2:PE-biotin = 70:20:5:5) were incubated with whole cell lysates of Dictyostelium expressing the indicated PTEN-GFP proteins. Liposomes were collected using streptavidin-conjugated magnetic beads. The cell lysate (input) and bound fractions were analyzed by SDS-PAGE and immunoblotting using anti-GFP antibodies. Quantification of band intensity is shown in (D). Values represent the mean ± SD (n = 3). (E) Association of the C-terminal tail of PTEN with full-length PTEN was examined by an “in trans” interaction assay. Associations between the core and tail domains of the same PTEN molecule (close conformation) are maintained through phosphorylation of the tail (indicated by red asterisks). In contrast, the core region of PTENA4 dissociates from the tail (open conformation) and binds to an exogenously added tail domain. (F) Whole cell lysates expressing the indicated PTEN-GFP proteins were incubated with PTEN352–403-YFP-FLAG. PTEN352–403-YFP-FLAG was immunoprecipitated with beads coupled to anti-FLAG antibodies. Bound fractions (IP) were analyzed with antibodies to GFP and FLAG. (G) Quantification of band intensity (n = 3).
To probe the mechanism by which the S10N, G20E, L42R, and F90S mutations inhibited membrane localization of PTEN, we performed an in vitro liposome binding assay. Liposomes containing PC (70%), PS (20%), PI(4,5)P2 (5%), and PE-biotin (5%) were incubated with lysates of Dictyostelium cells expressing different PTEN-GFP proteins (Fig. 2C and D). Consistent with its cytoplasmic localization, WT PTEN-GFP bound only weakly to the liposomes. In contrast, PTENA4, which was enriched at the plasma membrane, showed approximately10-fold higher association with the liposomes (Fig. 2C and D). The PTENA4-liposome interactions were reduced to the WT level by the mutations S10N, G20E, L42R, and F90S, thus PTENS10N,A4-GFP, PTENG20E,A4-GFP, PTENL42R,A4-GFP, and PTENF90S,A4-GFP only weakly interacted with the liposomes. Therefore, the decreased localization of the PTEN mutants at the plasma membrane likely results from their inability to bind to membrane phospholipids.
The C-terminal tail of PTEN binds to the membrane-binding regulatory interface and closes the protein conformation, physically limiting access of the membrane-binding site to the membrane (Fig. 2E) (10, 16). Mutations in the C-terminal phosphorylation sites in PTENA4 release this inhibitory interaction (Fig. 2E) (11). To determine whether the S10N, G20E, L42R, and F90S mutations restore this interaction in PTENA4 we performed an “in trans” interaction assay and examined the binding of PTEN-GFP to a separate tail domain fused to FLAG after immunoprecipitation with anti-FLAG antibodies. As previously reported (10, 11), although PTEN-GFP did not interact with PTEN352–403-FLAG, PTENA4-GFP showed a strong interaction (Fig. 2F and G). The S10N, G20E, L42R, and F90S mutations did not affect the interaction of PTENA4-GFP with PTEN352–403-FLAG (Fig. 2F and G). A negative control, PTENCBR3(K5A),A4-GFP which carries mutation in the tail-binding CBR3 loop (10), did not interact with PTEN352–403-FLAG. Therefore, S10N, G20E, L42R, and F90S mutations do not stimulate interaction of the membrane-binding regulatory interface with the C-terminal tail.
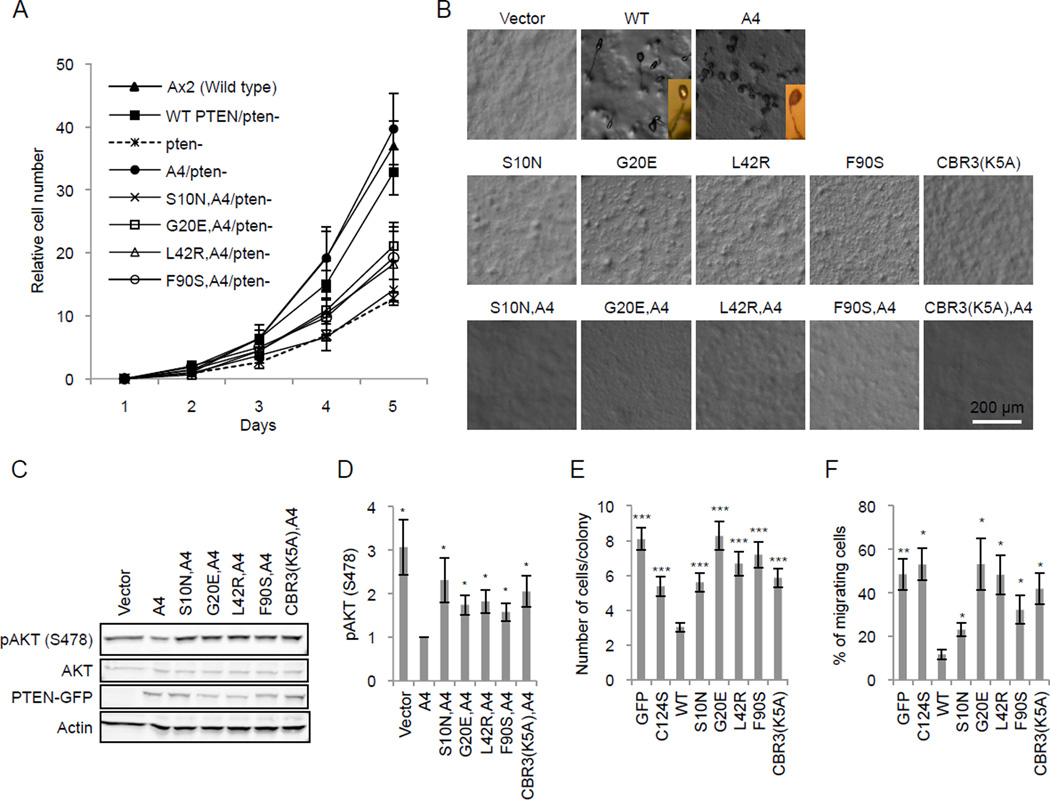
To determine the function of PTENS10N, PTENG20E, PTENL42R, and PTENF90S, we first investigated cell proliferation and starvation-induced differentiation of Dictyostelium cells. PTEN function is evolutionarily conserved in Dictyostelium and human cells (10, 20, 21). The loss of PTEN in Dictyostelium causes defects in cell proliferation and differentiation into stress-resistant fruiting bodies under starvation conditions (20). Expression of WT PTEN and PTENA4 restored these phenotypes in Dictyostelium PTEN-null cells (Fig. 3A and B). However, PTENS10N, PTENG20E, PTENL42R, and PTENF90S were unable to rescue the defects in the presence or absence of the A4 mutation (Fig. 3A and B). We then examined the role of PTEN in PIP3 signaling in HEK293T cells, in which PIP3 is overproduced, by measuring phosphorylation of AKT, a major downstream event of PIP3 signaling (22). PTENA4 suppressed AKT phosphorylation by approximately 2.5 fold, whereas addition of the mutations (S10N, G20E, L42R, or F90S) blocked the suppressive activity of PTENA4 (Fig. 3C and D). Furthermore, we examined the effect of these cancer mutations on PIP3-dependent cell proliferation and migration in the MCF-10A PIK3CA cell line, which carries a constitutively active form of PI3Kα and has increased PIP3 signaling (23, 24). When WT PTEN-GFP was expressed in MCF-10A PIK3CA cells, cell proliferation was significantly decreased approximately 2.5 fold compared with expression of the negative controls of GFP alone or the catalytic mutant PTENC124S (Fig. 3E). S10N, G20E, L42R, and F90S mutations completely blocked the ability of PTEN to decrease proliferation rates. Similarly, the function of PTEN in cell migration was significantly impaired by these four cancer mutations (Fig. 3F).
Figure 3. S10N, G20E, L42R, and F90S mutations interfere with PTEN function in the regulation of PIP3 signaling.
(A) The proliferation of PTEN-null Dictyostelium cells expressing different PTEN-GFP constructs is shown. The numbers of cells were normalized relative to those at day 1. Values represent the mean ± SD (n = 3). (B) PTEN-null Dictyostelium cells expressing different PTEN-GFP constructs were starved to induce differentiation into fruiting bodies. Pictures were taken at 36 hours after the onset of starvation. PTEN-null cells expressing WT PTEN or PTENA4 formed fruiting bodies whereas PTEN-null cells expressing vector alone did not aggregate and remained undifferentiated. PTEN and PTENA4 molecules carrying the cancer mutations did not differentiate into fruiting bodies. Inserts show side views of fruiting bodies. (C) HEK293T cell lysates containing the indicated PTEN-GFP constructs were analyzed by immunoblotting with antibodies to phospho-AKT, AKT, GFP (PTEN-GFP), and actin. (D) Quantification of band intensity (n = 3). (E and F) MCF-10A PIK3CA cells were infected with lentiviruses carrying the indicated PTEN constructs and analyzed for cell proliferation by counting the number of cells in individual colonies (E) and for cell migration using transwell plates (F). Values represent the mean ± SD (n = 4).
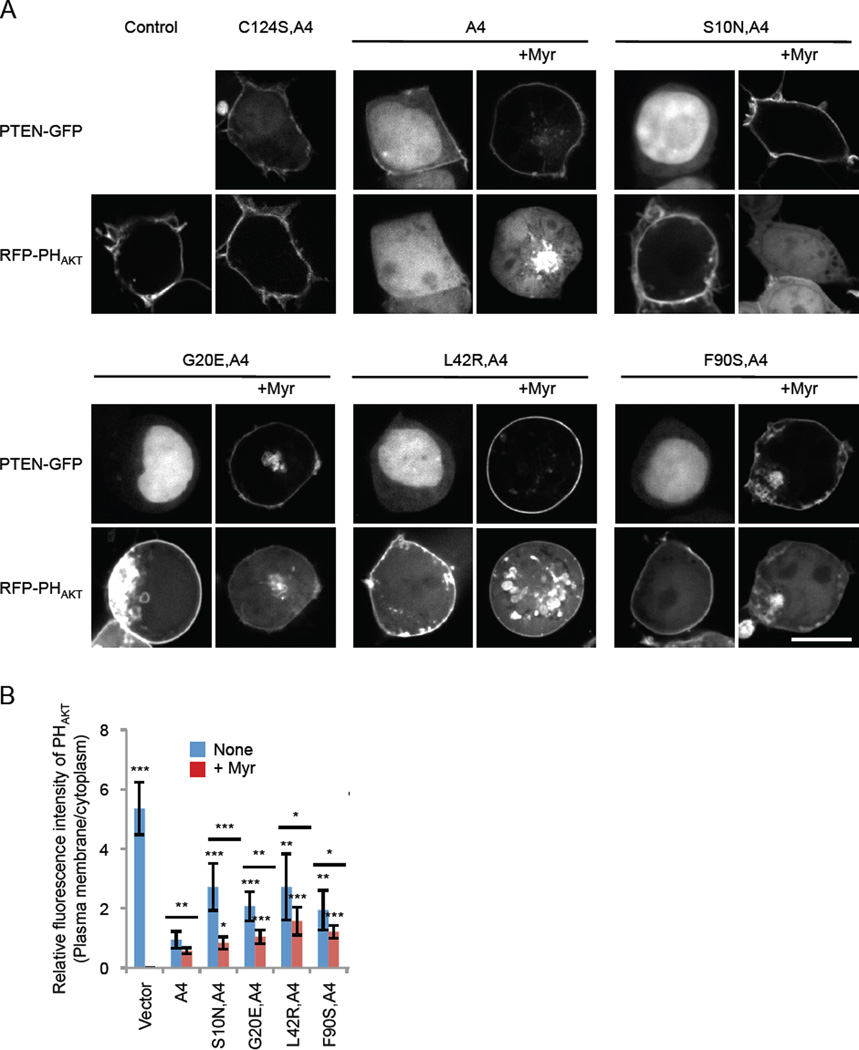
Finally, we tested whether artificial tethering of PTEN to the plasma membrane restored the function of PTENS10N, PTENG20E, PTENL42R, and PTENF90S. For this, we added a myristoylation motif derived from the plasma membrane protein PKBR1 (25) to the N-terminus of PTEN. The lipid anchor motif effectively recruited WT and mutant PTEN to the plasma membrane as revealed by fluorescence microscopy (Fig. 4A). To assess the effect of membrane recruitment on the amount of PIP3 in the plasma membrane, we co-expressed a biosensor for PIP3, RFP fused to the PIP3-binding PH domain from AKT1 (26). Bringing PTEN to the plasma membrane significantly improved the ability of PTENS10N,A4, PTENG20E,A4, PTENL42R,A4, and PTENF90S,A4 to remove PIP3 in the plasma membrane. These data further show that the primary defect of the PTEN cancer mutations can be traced to the defects in their membrane association.
Figure 4. Artificial tethering to the plasma membrane neutralizes the effect of S10N, G20E, L42R, and F90S on PIP3 signaling and PIP3-regulated cancer cell behavior.
(A) HEK293T cells expressing a PIP3 biosensor (RFP-PHAKT) together with different PTEN-GFP constructs were observed by fluorescence microscopy. Myristoylation motif derived from the plasma membrane protein PKBR1 was added to the N-terminus of PTEN (+Myr). Bar, 10 µm. (B) Intensity of RFP at the plasma membrane was quantified relative to that in the cytosol. Values represent the mean ± SD (n = 8).
In summary, we identified a new class of cancer-associated PTEN mutations that specifically inhibit the recruitment of PTEN to the plasma membrane as a result of impaired interactions with phospholipids. Our findings reveal the importance of membrane binding of PTEN in tumorigenic PIP3 signaling. Since this class of mutations maintains normal PIP3 phosphatase activity and can be rescued by tethering to the plasma membrane, we envision that pharmacologic promotion of membrane localization may be an effective strategy for anticancer therapy for this class of PTEN deficiency. According to the cBioportal and COSMIC databases, the four PTEN mutations analyzed in our study occur in ~1% (11/1210, COSMIC) of all human cancer sequencing studies. Thus, associations with progression and prognosis cannot currently be assessed given the relatively small number of patients in these studies and the lack of follow-up information for most sequencing studies outside of the Cancer Genome Atlas. However, the fact that these mutations recur, albeit at low frequencies, substantiates the functional relevance. In addition, as stated in our Table S1, these mutations appear to occur in a variety of cancer types. The cBioportal database suggests that these mutations are more frequent in breast, lung, brain, and uterine cancers. Although the overall frequency is small, 1% of breast cancers is equivalent to ~15,000 cases globally. This demonstrates the impact of our work; low-frequency mutations, which were previously of uncertain significance, now appear to be mutations of functional consequence. Thus, these mutations offer new insights into molecular mechanisms of cancer and possible new avenues for therapeutic intervention.
Supplementary Material
Acknowledgements
We thank Dr. Todd Waldman (Georgetown University School of Medicine) for lentiviral constructs. The mutation data was obtained from the Sanger Institute Catalogue Of Somatic Mutations In Cancer (COSMIC) web site (http://www.sanger.ac.uk/cosmic) (28). This work was supported by NIH grants to MI (GM084015), PND (GM28007 and GM34933), HS (GM089853 and NS084154) and to RP (CA082783 and CA155117).
Footnotes
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Chalhoub N, Baker SJ. PTEN and the PI3-kinase pathway in cancer. Annu Rev Pathol. 2009;4:127–150. doi: 10.1146/annurev.pathol.4.110807.092311. Epub 2008/09/05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ali IU, Schriml LM, Dean M. Mutational spectra of PTEN/MMAC1 gene: a tumor suppressor with lipid phosphatase activity. J Natl Cancer Inst. 1999;91(22):1922–1932. doi: 10.1093/jnci/91.22.1922. Epub 1999/11/24. [DOI] [PubMed] [Google Scholar]
- 3.Tan MH, Mester JL, Ngeow J, Rybicki LA, Orloff MS, Eng C. Lifetime cancer risks in individuals with germline PTEN mutations. Clin Cancer Res. 2012;18(2):400–407. doi: 10.1158/1078-0432.CCR-11-2283. Epub 2012/01/19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hollander MC, Blumenthal GM, Dennis PA. PTEN loss in the continuum of common cancers, rare syndromes and mouse models. Nat Rev Cancer. 2011;11(4):289–301. doi: 10.1038/nrc3037. Epub 2011/03/25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Song MS, Salmena L, Pandolfi PP. The functions and regulation of the PTEN tumour suppressor. Nature reviews Molecular cell biology. 2012;13(5):283–296. doi: 10.1038/nrm3330. Epub 2012/04/05. [DOI] [PubMed] [Google Scholar]
- 6.Leslie NR, Batty IH, Maccario H, Davidson L, Downes CP. Understanding PTEN regulation: PIP2, polarity and protein stability. Oncogene. 2008;27(41):5464–5476. doi: 10.1038/onc.2008.243. Epub 2008/09/17. [DOI] [PubMed] [Google Scholar]
- 7.Denning G, Jean-Joseph B, Prince C, Durden DL, Vogt PK. A short N-terminal sequence of PTEN controls cytoplasmic localization and is required for suppression of cell growth. Oncogene. 2007;26(27):3930–3940. doi: 10.1038/sj.onc.1210175. Epub 2007/01/11. [DOI] [PubMed] [Google Scholar]
- 8.Han SY, Kato H, Kato S, Suzuki T, Shibata H, Ishii S, et al. Functional evaluation of PTEN missense mutations using in vitro phosphoinositide phosphatase assay. Cancer Res. 2000;60(12):3147–3151. Epub 2000/06/24. [PubMed] [Google Scholar]
- 9.Rodriguez-Escudero I, Oliver MD, Andres-Pons A, Molina M, Cid VJ, Pulido R. A comprehensive functional analysis of PTEN mutations: implications in tumor- and autism-related syndromes. Hum Mol Genet. 2011;20(21):4132–4142. doi: 10.1093/hmg/ddr337. Epub 2011/08/11. [DOI] [PubMed] [Google Scholar]
- 10.Nguyen HN, Afkari Y, Senoo H, Sesaki H, Devreotes PN, Iijima M. Mechanism of human PTEN localization revealed by heterologous expression in Dictyostelium. Oncogene. 2013 doi: 10.1038/onc.2013.507. Epub ahead. Epub 2013/12/03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rahdar M, Inoue T, Meyer T, Zhang J, Vazquez F, Devreotes PN. A phosphorylation-dependent intramolecular interaction regulates the membrane association and activity of the tumor suppressor PTEN. Proc Natl Acad Sci U S A. 2009;106(2):480–485. doi: 10.1073/pnas.0811212106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gericke A, Leslie NR, Losche M, Ross AH. PtdIns(4,5)P2-mediated cell signaling: emerging principles and PTEN as a paradigm for regulatory mechanism. Advances in experimental medicine and biology. 2013;991:85–104. doi: 10.1007/978-94-007-6331-9_6. Epub 2013/06/19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Das S, Dixon JE, Cho W. Membrane-binding and activation mechanism of PTEN. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(13):7491–7496. doi: 10.1073/pnas.0932835100. Epub 2003/06/17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keniry M, Parsons R. The role of PTEN signaling perturbations in cancer and in targeted therapy. Oncogene. 2008;27(41):5477–5485. doi: 10.1038/onc.2008.248. Epub 2008/09/17. [DOI] [PubMed] [Google Scholar]
- 15.Vazquez F, Matsuoka S, Sellers WR, Yanagida T, Ueda M, Devreotes PN. Tumor suppressor PTEN acts through dynamic interaction with the plasma membrane. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(10):3633–3638. doi: 10.1073/pnas.0510570103. Epub 2006/03/16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nguyen HN, Yang JM, Afkari Y, Park BH, Sesaki H, Devreotes PN, et al. Engineering ePTEN, an enhanced PTEN with increased tumor suppressor activities. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(26):E2684–E2693. doi: 10.1073/pnas.1409433111. Epub 2014/07/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vazquez F, Ramaswamy S, Nakamura N, Sellers WR. Phosphorylation of the PTEN tail regulates protein stability and function. Molecular and cellular biology. 2000;20(14):5010–5018. doi: 10.1128/mcb.20.14.5010-5018.2000. Epub 2000/06/24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Freeman DJ, Li AG, Wei G, Li HH, Kertesz N, Lesche R, et al. PTEN tumor suppressor regulates p53 protein levels and activity through phosphatase-dependent and -independent mechanisms. Cancer Cell. 2003;3(2):117–130. doi: 10.1016/s1535-6108(03)00021-7. Epub 2003/03/07. [DOI] [PubMed] [Google Scholar]
- 19.Stambolic V, MacPherson D, Sas D, Lin Y, Snow B, Jang Y, et al. Regulation of PTEN transcription by p53. Molecular cell. 2001;8(2):317–325. doi: 10.1016/s1097-2765(01)00323-9. Epub 2001/09/08. [DOI] [PubMed] [Google Scholar]
- 20.Iijima M, Devreotes P. Tumor suppressor PTEN mediates sensing of chemoattractant gradients. Cell. 2002;109(5):599–610. doi: 10.1016/s0092-8674(02)00745-6. [DOI] [PubMed] [Google Scholar]
- 21.Iijima M, Huang YE, Devreotes P. Temporal and spatial regulation of chemotaxis. Dev Cell. 2002;3(4):469–478. doi: 10.1016/s1534-5807(02)00292-7. [DOI] [PubMed] [Google Scholar]
- 22.Hawkins PT, Anderson KE, Davidson K, Stephens LR. Signalling through Class I PI3Ks in mammalian cells. Biochemical Society transactions. 2006;34(Pt 5):647–662. doi: 10.1042/BST0340647. Epub 2006/10/21. [DOI] [PubMed] [Google Scholar]
- 23.Gustin JP, Karakas B, Weiss MB, Abukhdeir AM, Lauring J, Garay JP, et al. Knockin of mutant PIK3CA activates multiple oncogenic pathways. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(8):2835–2840. doi: 10.1073/pnas.0813351106. Epub 2009/02/07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang GM, Wong HY, Konishi H, Blair BG, Abukhdeir AM, Gustin JP, et al. Single copies of mutant KRAS and mutant PIK3CA cooperate in immortalized human epithelial cells to induce tumor formation. Cancer Res. 2013;73(11):3248–3261. doi: 10.1158/0008-5472.CAN-12-1578. Epub 2013/04/13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kamimura Y, Xiong Y, Iglesias PA, Hoeller O, Bolourani P, Devreotes PN. PIP3-independent activation of TorC2 and PKB at the cell's leading edge mediates chemotaxis. Curr Biol. 2008;18(14):1034–1043. doi: 10.1016/j.cub.2008.06.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen CL, Wang Y, Sesaki H, Iijima M. Myosin I links PIP3 signaling to remodeling of the actin cytoskeleton in chemotaxis. Science signaling. 2012;5(209):ra10. doi: 10.1126/scisignal.2002446. Epub 2012/02/03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee JO, Yang H, Georgescu MM, Di Cristofano A, Maehama T, Shi Y, et al. Crystal structure of the PTEN tumor suppressor: implications for its phosphoinositide phosphatase activity and membrane association. Cell. 1999;99(3):323–334. doi: 10.1016/s0092-8674(00)81663-3. Epub 1999/11/11. [DOI] [PubMed] [Google Scholar]
- 28.Bamford S, Dawson E, Forbes S, Clements J, Pettett R, Dogan A, et al. The COSMIC (Catalogue of Somatic Mutations in Cancer) database and website. Br J Cancer. 2004;91(2):355–358. doi: 10.1038/sj.bjc.6601894. Epub 2004/06/10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Y, Senoo H, Sesaki H, Iijima M. Rho GTPases orient directional sensing in chemotaxis. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(49):E4723–E4732. doi: 10.1073/pnas.1312540110. Epub 2013/11/20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang P, Wang Y, Sesaki H, Iijima M. Proteomic identification of phosphatidylinositol (3,4,5) triphosphate-binding proteins in Dictyostelium discoideum. Proc Natl Acad Sci U S A. 2010 doi: 10.1073/pnas.1006153107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim JS, Xu X, Li H, Solomon D, Lane WS, Jin T, et al. Mechanistic analysis of a DNA damage-induced, PTEN-dependent size checkpoint in human cells. Molecular and cellular biology. 2011;31(13):2756–2771. doi: 10.1128/MCB.01323-10. Epub 2011/05/04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cai H, Huang CH, Devreotes PN, Iijima M. Analysis of chemotaxis in Dictyostelium. Methods in molecular biology. 2012;757:451–468. doi: 10.1007/978-1-61779-166-6_26. Epub 2011/09/13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Y, Steimle PA, Ren Y, Ross CA, Robinson DN, Egelhoff TT, et al. Dictyostelium huntingtin controls chemotaxis and cytokinesis through the regulation of myosin II phosphorylation. Mol Biol Cell. 2011;22(13):2270–2281. doi: 10.1091/mbc.E10-11-0926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Iijima M, Huang YE, Luo HR, Vazquez F, Devreotes PN. Novel mechanism of PTEN regulation by its phosphatidylinositol 4,5-bisphosphate binding motif is critical for chemotaxis. J Biol Chem. 2004;279(16):16606–16613. doi: 10.1074/jbc.M312098200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.