Abstract
Aberrant activation of Notch signaling plays an essential role in colorectal cancer (CRC) progression. Amplified in breast cancer 1 (AIB1), also known as SRC-3 or NCOA3 is a transcriptional coactivator that promotes cancer cell proliferation and invasiveness. However AIB1 implication in CRC progression through enhancing Notch signaling is unknown. In this study we found that several CRC cell lines expressed high levels of AIB1, and knockdown of AIB1 decreased cell proliferation, colony formation and tumorigenesis of these CRC cells. Specifically, knockdown of AIB1 inhibited cell cycle progression at G1 phase by decreasing the mRNA levels of Cyclin A2, Cyclin B1, Cyclin E2 and Hes1. Furthermore, AIB1 interacted with Notch intracellular domain (NICD) and Mastermind-like 1 (MAMAL1) and was recruited to the Hes1 promoter to enhance Notch signaling. Downregulation of AIB1 also decreased CRC cell invasiveness in vitro and lung metastasis in vivo. Besides that, knockout of AIB1 in mice inhibited colon carcinogenesis induced by AOM/DSS treatment. The mRNA levels of Cyclin B1 and Hes5 were downregulated, but p27, ATOH1, and MUC2 were upregulated in the colon tumors from AIB1-deficient mice compared with those from wild-type mice. Thus our results signify the importance of AIB1 in CRC and demonstrate that AIB1 promotes CRC progression at least in part through enhancing Notch signaling, suggesting that AIB1 is a potential molecular target for CRC treatment.
Keywords: Amplified in breast cancer 1, Colorectal cancer, Cancer progression, Notch signaling
INTRODUCTION
Colorectal cancer (CRC) is the second leading cause of cancer deaths with a mortality rate of 33% in developed countries1. Despite much advances in screening, diagnosis and treatment of CRC in recent decades, which have led to increased cure and response rates, the etiology of CRC is still unknown. Extensive investigations have uncovered several critical genes and signaling pathways important for CRC initiation and progression which include WNT, Notch, RAS-MAPK, PI3K, TGF-β, p53 and DNA mismatch–repair pathways. However further exploration of molecular mechanism of CRC is required to develop novel therapeutic targets for cure of CRC.
Increasing evidence has shown that Notch signaling regulates many aspects of intestinal development and epithelial renewal2. Aberrant expression of Notch is found in most types of cancers and the activation of notch signaling is related to human colon cancers3. The activation of notch signaling is dependent on the interactions of ligand-receptor between neighboring cells and followed by gamma-secretase-cleaved release of the active form of Notch intracellular domain (NICD). The released NICD translocates into the nucleus to form a ternary protein complex with CSL and MAML1 to transactivate their target genes, such as those in the hairy and enhancer of split and hes related with YRPW motif (hey) families4, 5.
Amplified in breast cancer 1 (AIB1), also known as steroid receptor coactivator 3 (SRC-3, ACTR, RAC3, TRAM2, pCIP, NCOA3), is a member of the SRC/p160 coactivator family that also includes SRC-1 (NCOA1) and SRC-2 (GRIP1, TIF2, NCOA2)6. AIB1 is highly expressed in several human cancers such as breast cancer7, prostate cancer8, and liver cancer9-10, and has been demonstrated to be a key regulator for tumor initiation, progression, metastasis and survival9-16. AIB1 can interact with nuclear receptors and other transcription factors to regulate the expression of their target genes involved in many signaling pathways, including ERα, EGFR, Akt, MAPK, E2F1, C/EBPβ, NFκB, HER2/neu and PEA317-27. It has been reported that AIB1 is overexpressed in 35% of human CRC samples28, but the role of AIB1 in CRC progression is still unknown.
In this study we demonstrate that the expression of AIB1 is significantly increased in CRC cell lines as compared to normal colon epithelial cells and its downregulation reduces cell proliferation, invasion and tumor formation. We also demonstrate that AIB1 can interact with NICD to enhance Notch signaling and AIB1-deficient mice are resistant to AOM/DSS-induced CRC formation.
RESULTS
AIB1 is overexpressed in CRC cell lines
To evaluate the expression of AIB1 in CRC cell lines, Western blot analysis was performed to determine the protein levels of AIB1 in several CRC cell lines. In comparison with normal colon epithelial cells, all five human CRC cell lines (RKO, Caco-2, HCT-116, SW620 and SW480) and the CT26, a mouse CRC cell line, expressed high levels of AIB1, suggesting a plausible role of AIB1 in CRC cells (Figure 1a).
Figure 1. AIB1 is overexpressed in CRC cell lines and promotes CRC cell proliferation.
(a) Western blot analysis of expression of AIB1 protein in normal colon epithelium cells and 6 CRC cell lines. (b,c,d) Proliferation of CRC cell lines RKO, HCT116, and CT26 transiently transfected with AIB1 siRNA or control siRNA was measured by MTT assay. (e,f,g) Proliferation of CRC cell lines RKO, HCT116, and CT26 stably transfected with AIB1 shRNA or control shRNA was measured by MTT assay. The knockdown efficiency of AIB1 was measured by Western blot analysis. All experiments were performed at least twice with similar results. All data are the means +s.d. (n=3) at each time point. Statistically significant difference: *P<0.05 and **P<0.01(t-test).
Downregulation of AIB1 suppresses CRC cell proliferation, but does not affect cell survival
To determine the role of AIB1 in CRC cell proliferation, two different siRNAs against human AIB1 and two different siRNAs against mouse AIB1 were used to knock down the expression of AIB1 in two human CRC cell lines, RKO and HCT116, and one mouse CRC cell line CT26, respectively, and then cell proliferation was measured by MTT assay. As shown in Figures 1b, c and d, all AIB1-specific siRNAs efficiently reduced the levels of endogenous AIB1 protein and significantly decreased cell proliferation. Furthermore, RKO, HCT116, and CT26 cells were stably transfected with control plasmid (pSUPER-shCtrl/pll3.7-shCtrl) or human/mouse AIB1 knockdown plasmids (pSUPER-sh-hAIB1/pll3.7-sh-mAIB1)9 and cell proliferation was measured by MTT assay. Stable knockdown of AIB1 in these cell lines also significantly decreased cell proliferation (Figures 1e, f and g). These results indicate that AIB1 is vital for the proliferation of CRC cells.
It has been reported that downregulation of AIB1 could reduce prostate cancer cell survival8. To determine whether downregulation of AIB1 could affect CRC cell survival, the extent of cell death was compared between control and AIB1-knockdown CRC cells under normal growth conditions by using flow cytometric analysis. As shown in Supplementary Figure S1, downregulation of AIB1 did not affect CRC cell survival under normal growth conditions.
Knockdown of AIB1 induces CRC cell cycle arrest
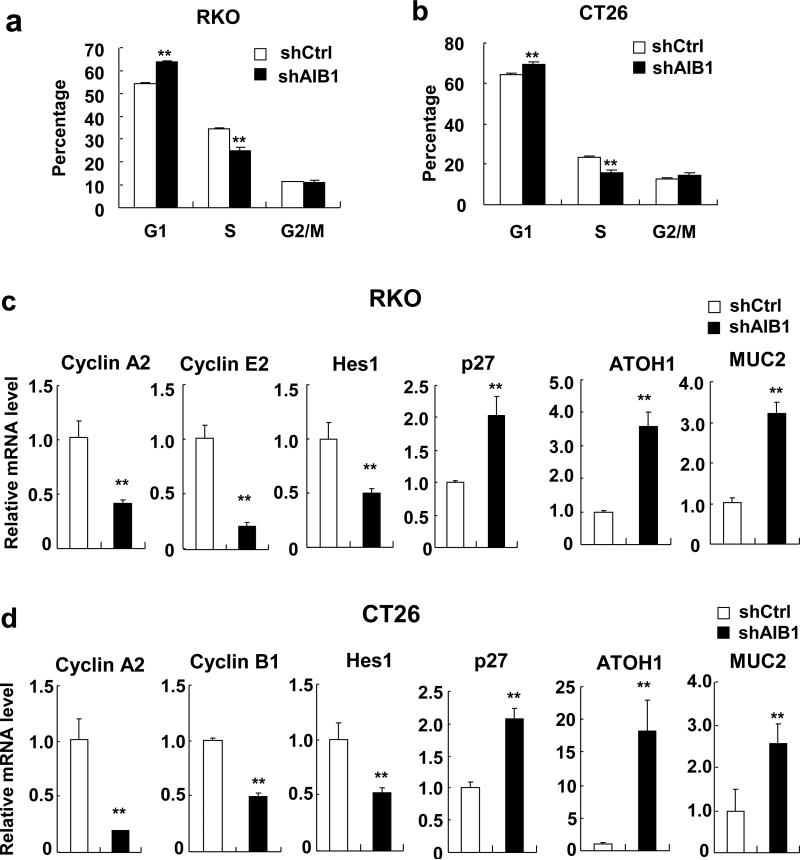
To explore the mechanism by which downregulation of AIB1 inhibits CRC cell proliferation, cell cycle analysis was performed to examine whether AIB1-knockdown cells were arrested in a specific phase of the cell cycle. Cells were synchronized by serum starvation for 24 hours and then cultured in serum-containing medium for another 24 hours before harvesting for flow cytometric analysis. As shown in Figure 2a and b, AIB1 knockdown led to an increase of cell number at the G1 phase and a concomitant decrease of cell number at the S phase as compared with control cells. These results indicate that AIB1 regulates the G1/S phase transition.
Figure 2. Knockdown of AIB1 induces CRC cell cycle arrest.
(a,b) Cell cycle progress of RKO and CT26 cells stably trasnfected with AIB1 shRNA or control shRNA was measured by flow cytometry. (c,d) The mRNA levels of Cyclin A2, Cyclin B1, cyclin E2, Hes1, p27, ATOH1, and MUC2 in AIB1-knockdown RKO and CT26 cell as well as control cells were determined by real-time PCR, respectively. Each experiment was performed at least twice with similar results. All data are the means +s.d. (n=3) at each time point. Statistically significant difference: **P<0.01(t-test).
To understand the underlying mechanisms of cell cycle arrest, the mRNA levels of several cell proliferation/cycle-related genes in control and AIB1-knockdown CRC cells were measured by real-time qPCR. As shown in Figures 2c and d, knockdown of AIB1 significantly decreased the expression of cyclin A2, cyclin E2, and Hes1 in RKO cells as well as cyclin A2, cyclin B1, and Hes1 in CT26 cells. Hes1 is a repressor of cell cycle inhibitors and a downstream target of Notch29. As a consequence of Hes1 downregulation, the mRNA levels of p27, ATOH1, and MUC2 which have been reported to be transcriptionally repressed by Hes130-31, were significantly increased in AIB1-knockdown CRC cells (Figures 2c and d). These results suggest that AIB1 promotes cell cycle progression through regulating the expression of several cell cycle-related genes.
AIB1 enhances Notch signaling
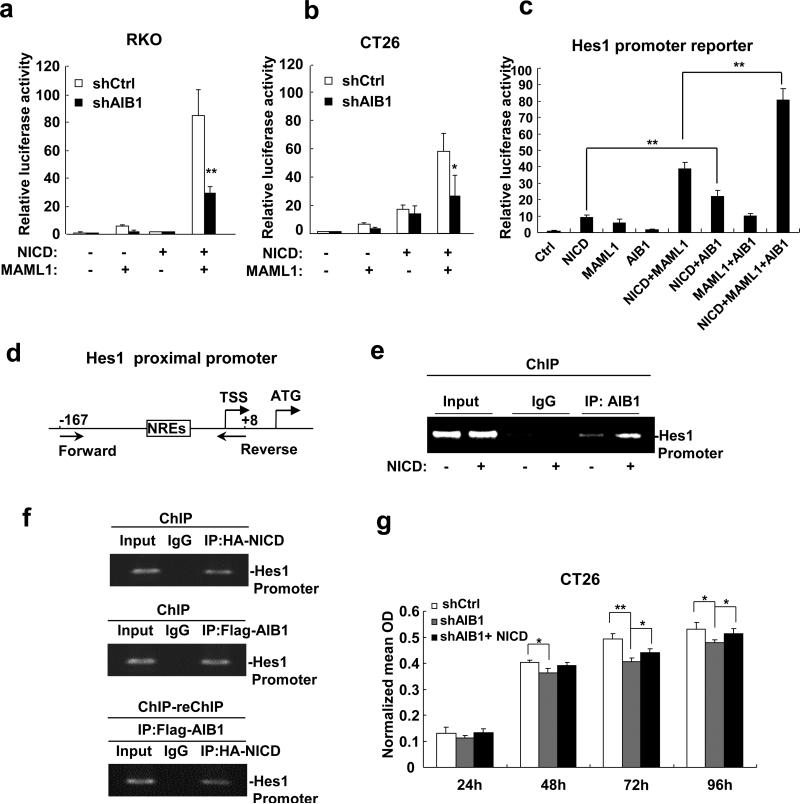
Hes1 is a typical Notch downstream target, therefore we hypothesized that AIB1 may serve as a coactivator for Notch signaling to regulate Hes1 transcription and subsequent cell proliferation. To test this hypothesis, AIB1-knockdown and control cells were co-transfected with the Hes1 promoter luciferase reporter, NICD, and MAML1. As shown in Figures 3a and b, knockdown of AIB1 decreased Hes1 promoter activity as compared with control cells. Simultaneously overexpression of AIB1, MAML1, and NICD could significantly increase Hes1 promoter activity (Figure 3c). These results suggest that AIB1 regulates Hes1 expression at transcriptional level.
Figure 3. AIB1 enhances Notch signaling.
(a, b) Knockdown AIB1 significantly decreases Hes1 promoter activity in RKO and CT26 cells as determined by luciferase reporter assay. (c) AIB1 cooperated with NICD and MAML1 to enhance Hes1 promoter activity. (d) Schematic of the Hes1 proximal promoter. Forward and Reverse: Forward and Reverse primers used for ChIP assay. NREs: Notch response elements. TSS: Transcriptional start site. (e) AIB1 was recruited to the Hes1 proximal promoter in the absence and presence of ectopic NICD as measured by ChIP assay. (f) Simultaneous recruitment of AIB1 and NICD to the Hes1 proximal promoter was measured by ChIP-reChIP assay. Cells were transfected with HA-NICD and Flag-AIB1 expression plasmids and then lysed for ChIP and ChIP-reChIP assays using control IgG, anti-HA antibody, and anti-Flag antibody for IP. (g) NICD rescued the proliferation of AIB1-knockdown CT26 cells. Each experiment was performed at least twice with similar results. All data are the means + s.d. (n=3) at each time point. Statistically significant difference: *P<0.05 and **P<0.01(t-test).
To determine whether AIB1 can be recruited to the Notch response elements (NREs) located in the Hes1 proximal promoter (Figure 3d), the chromatin-immunoprecipitation (ChIP) assay was performed. ChIP results showed that AIB1 was recruited to the Hes1 proximal promoter and the recruitment was increased in the presence of exogenous NICD (Figure 3e). Furthermore, ChIP-reChIP results showed that AIB1 and NICD were simultaneously recruited to Hes1 proximal promoter (Figure 3f).
To corroborate that AIB1 could enhance Notch signaling, rescue experiment for proliferation of AIB1-knockdown cells was performed. AIB1-knockdown cells were transfected with NICD expression constructs, and cell proliferation was measured by MTT assay. The results showed that transfection of NICD expression constructs could indeed rescue cell proliferation of AIB1-knockdown cells (Figure 3g), suggesting that AIB1 promotes CRC cell proliferation at least in part through enhancing Notch signaling.
AIB1 directly binds to NICD and MAML1
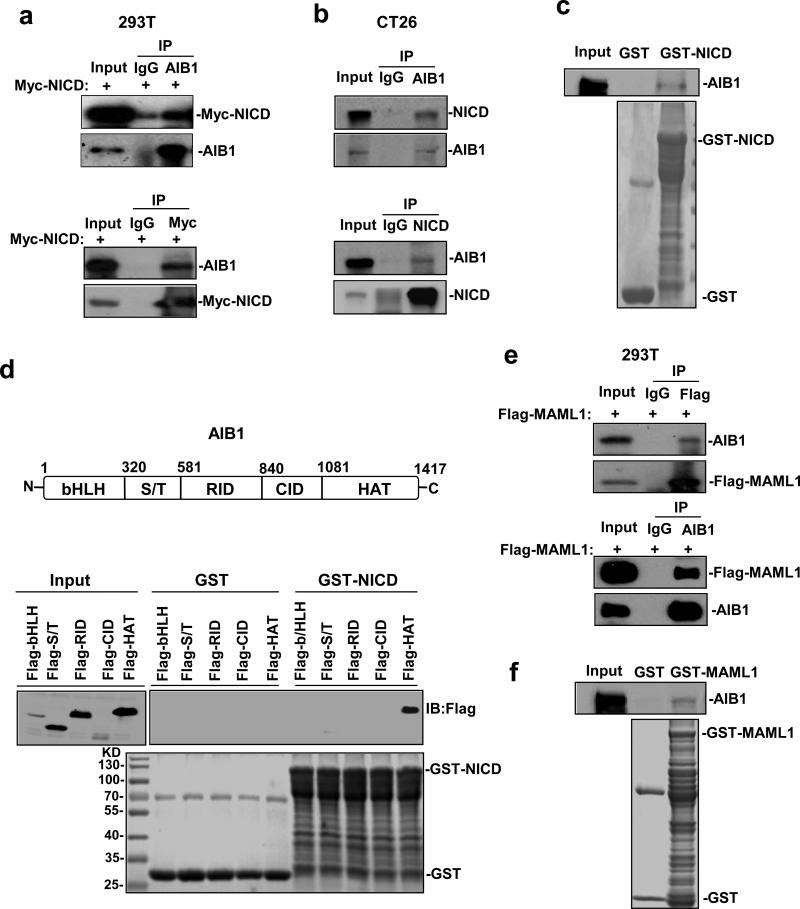
Since AIB1 is a transcriptional coactivator and we have shown that AIB1 and NICD can be simultaneously recruited to Hes1 proximal promoter to enhance its activity, it is likely that AIB1 could interact with NICD. To test this hypothesis, we transfected Myc-NICD expression construct into 293T cells and then performed Co-IP assay to determine the interactions between Myc-NICD and endogenous AIB1. The results showed that the AIB1 antibody, but not control IgG, could precipitate endogenous AIB1 and Myc-NICD (Figure 4a, upper panel). Reciprocally, AIB1 and Myc-NICD could be pulled down by Myc antibody (Figure 4a, lower panel). These results suggest that AIB1 could interact with NICD in cells. In addition, the interaction of endogenous AIB1 and NICD could also be detected in CT26 cells by Co-IP assays using anti-AIB1 and anti-NICD antibodies (Figure 4b). To determine whether AIB1 can directly bind to NICD, E. coli-produced GST-NICD protein was incubated with AIB1 protein produced by an E. coli extract-based cell free protein synthesis system for GST pull-down assays. The results showed that the GST-NICD protein, but not GST, was able to pull down AIB1 (Figure 4c), indicating that AIB1 can directly bind to NICD.
Figure 4. AIB1 directly binds to NICD and MAML1.
(a) Cells were transfected with Myc-NICD expression plasmids and then lysed for Co-IP assays using control IgG, AIB1 antibody, and anti-Myc antibody. Precipitated proteins were subjected to immunoblotting to detect AIB1 and Myc-NICD. (b) Co-IP analysis of the interaction of endogenous AIB1 and NICD in CT26 cells. (c) GST pull-down analysis of the interaction of AIB1 and NICD in vitro. E.coli-produced GST or GST-NICD protein was incubated with AIB1 protein produced by an E. coli extract-based cell free protein synthesis system for GST pull-down assays. (d) Schematic of the AIB1 protein and the interaction of AIB1 with NICD through its HAT domain. Immobilized GST-NICD or GST proteins were incubated with 5 different AIB1 domain proteins overexpressed in 293T cells for GST pull-down assays. (e) Cells were transfected with Flag-MAML1 expression plasmids and then lysed for Co-IP assays using control IgG, AIB1 antibody, and anti-Flag antibody. Precipitated proteins were subjected to immunoblotting to detect AIB1 and Flag-MAML1. (f) GST pull-down analysis of the interaction of AIB1 and MAML1 in vitro. E.coli-produced GST or GST-MAML1 protein was incubated with AIB1 protein produced by an E. coli extract-based cell free protein synthesis system for GST pull-down assays. Each experiment was performed at least twice with similar results.
AIB1 is a multidomain protein containing bHLH/Per/ARNT/Sim homologous (bHLH/PAS) domain, serine/threonine-rich(S/T) domain, receptor interaction domain (RID), CBP/p300 interaction domain (CID), and histone acetyltransferase domain (HAT) (Figure 4d, upper panel). To determine which domains of AIB1 could bind to NICD, different AIB1 domain proteins were expressed in 293T cells and GST-pull down assays were performed. Our result showed that HAT domain of AIB1 was responsible for the interaction between AIB1 and NICD (Figure 4d, lower panel).
MAML1 is a key transcriptional coactivator for Notch signaling. MAML1 binds to NICD, forms a ternary protein complex with CSL and NICD, and amplifies Notch-induced Hes1 transcription32. To determine whether AIB1 could interact with MAML1, we transfected Flag-MAML1 expression construct into 293T cells and then performed Co-IP assay. The results showed that the AIB1 antibody could precipitate endogenous AIB1 and Flag-MAML1 (Figure 4e, upper panel). Reciprocally, AIB1 and Flag-MAML1 could be pulled down by Flag antibody (Figure 4e, lower panel). These results suggest that AIB1 could interact with MAML1 in cells. To determine whether AIB1 can directly bind to MAML1, E.coli-produced GST-MAML1 protein was incubated with AIB1 protein produced by an E. coli extract-based cell free protein synthesis system for GST pull-down assays. The results showed that the GST-MAML1 protein, but not GST, was able to pull down AIB1 (Figure 4f), indicating that AIB1 can directly bind to MAML1.
Knockdown of AIB1 inhibits tumorigenesis of CRC cells
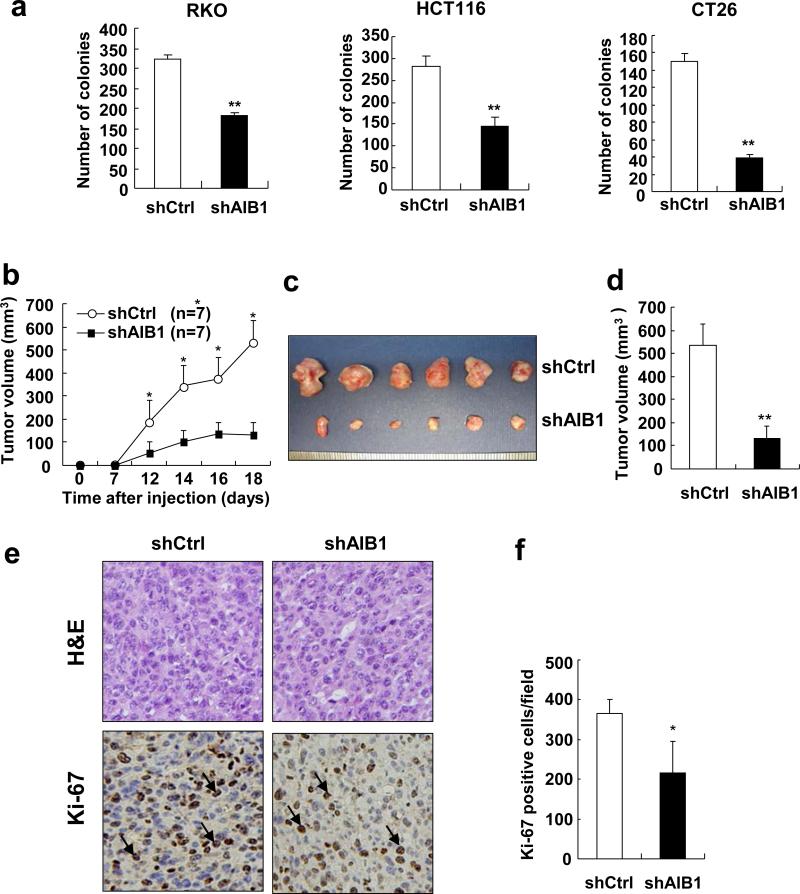
Having shown that knockdown of AIB1 decreased CRC cell proliferation in vitro, it was next determined whether AIB1 affects tumorigenesis of CRC cells. Colony formation assay was performed with AIB1-knockdown and control cells. The results showed that the number of colonies in AIB1-knockdown cells were significantly less as compared to control cells (Figure 5a).
Figure 5. Knockdown of AIB1 reduces the tumor formation.
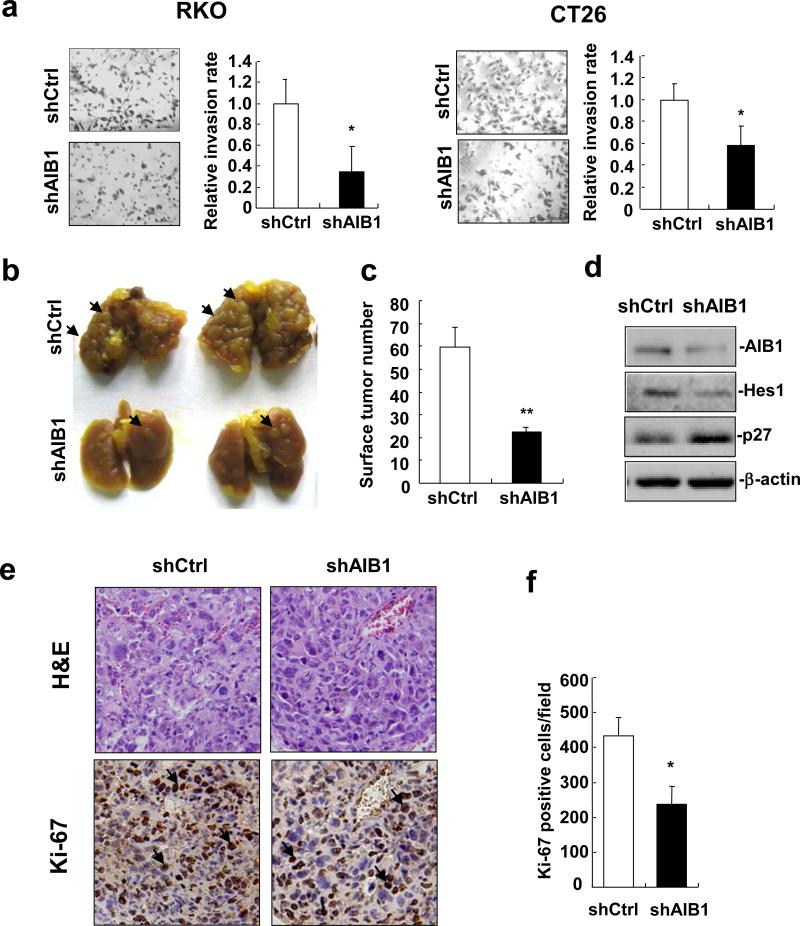
(a) The foci numbers in colony formation of AIB1-knockdown RKO, HCT116 and CT26 cells and control cells was measured by crystal violet staining. Each experiment was performed twice with similar results. All data are the means + s.d. (n=3). Statistically significant difference: **P<0.01(t-test). (b) AIB1-knockdown CT26 and control cells were injected subcutaneously into the dorsal flanks of BALB/c mice, respectively. Tumor growth was monitored for 18 days. The experiment was performed once. (c,d) Eighteen days after injection, mice were euthanized and tumors were harvested for imaging and volume measuring (**P<0.01, N=6 tumors). (e) Hematoxylin and eosin (H&E) and Ki-67 staining of tissue sections from control and AIB1-knockdown CT26 tumors. (f) Quantitation of Ki-67-positive tumor cells (*P<0.05, N=6 tumors).
As aberrant Notch signaling activation could be involved in the pathogenesis of colorectal tumors2-3, 33-35, we hypothesized that downregulation of AIB1 and subsequently reduced Notch signaling would inhibit CRC growth. Therefore, AIB1-knockdown CT26 and control cells were subcutaneously injected into BALB/c mice and tumor growth was monitored. Knockdown of AIB1 significantly decreased tumor growth (Figure 5b), and the mean volume of AIB1-knockdown tumors was about 4-fold smaller than that from control tumors (Figures 5c and d). Furthermore, Ki-67 and TUNEL staining for tissue sections from AIB1-knockdown CT26 and control tumors were performed to determine the effects of AIB1 knockdown on CT26 cell proliferation and survival in vivo. As shown in Figures 5e and f and Supplementary Figure S2, knockdown of AIB1 significantly reduced the number of Ki-67-positive cells, but had no effect on the number of TUNEL-positive cells. These results indicate that AIB1 promotes CRC growth by enhancing cell proliferation but not cell survival.
Downregulation of AIB1 decreases cell invasion and tumor metastasis
It has been reported that AIB1 is involved in the invasion of several types of cancers9, 16, 27. Therefore, we presumed that downregulation of AIB1 might decrease the invasive ability of CRC cells. To test this hypothesis, cell invasion was assessed using a transwell matrigel invasion assay. Knockdown of AIB1 significantly decreased the ability of cells to penetrate through the matrigel-coated membrane (Figure 6a), suggesting that downregulation of AIB1 decreases CRC cell invasion. To further investigate the potential role of AIB1 in CRC cell invasion in vivo, tumor metastasis in BALB/c mice was preformed. 14 days after tail vein injection of AIB1-knockdown or control CT26 cells, lungs were harvested for metastasis analysis. The number and size of tumors on the lung surface from AIB1-knockdown group was significantly less than that on control group (Figures 6b and c). Western blot analysis of AIB1-knockdown and control tumors confirmed the downregulation of AIB1 and Hes1 in AIB1-knockdown tumors (Figure 6d). Furthermore, Ki-67 and TUNEL staining for tissue sections from AIB1-knockdown and control tumors showed that knockdown of AIB1 significantly reduced the number of Ki-67-positive cells (Figures 6e and f), but had no effect on the number of TUNEL-positive cells (Supplementary Figure S3). These results indicate that AIB1 promotes CRC metastasis by enhancing cell invasion and proliferation, but not cell survival.
Figure 6. Knockdown of AIB1 decreases CRC cell invasion in vitro and tumor metastasis.
in vivo. (a) Images and quantitation of AIB1-knockdown RKO and CT26 cells and control cells penetrating through the matrigel-coated membrane Each experiment was performed twice with similar results. All data are the means + s.d. (n=3). Statistically significant difference: **P<0.01(t-test). (b) Images of the lungs at 14 days after injection of AIB1-knockdown CT26 and control cells. Arrow indicated tumors on the surface of the lungs. The experiment of tumor metastasis in vivo was performed once. (c) Quantitation of tumor number on the surface of the lungs (**P<0.01, N=5 lungs). (d) Western blot analysis of the expression of AIB1, Hes1 and p27 in control and AIB1-knockdown CT26 tumors. (e) H&E and Ki-67 staining of tissue sections from control and AIB1-knockdown CT26 tumors. Arrow indicated Ki-67-positive tumor cells. (f) Quantitation of Ki-67-positive tumor cells (* P<0.05, N=5 tumors).
AIB1-deficient mice are resistant to AOM/DSS-induced CRC formation
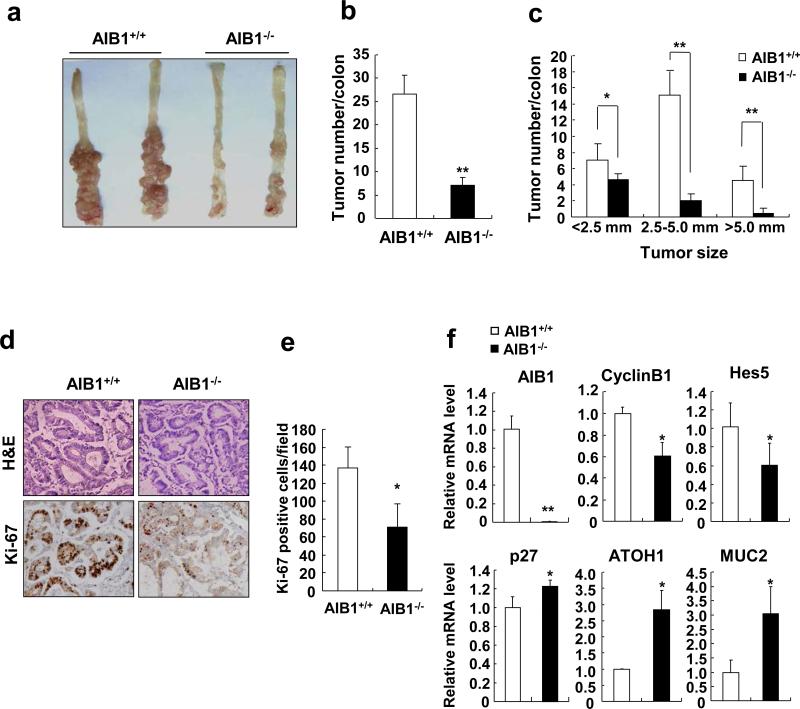
To determine whether AIB1 ablation in mice reduced the development and growth of CRC, we used AOM/DSS-induced CRC model for in vivo study36. After 12 weeks of AOM/DSS treatment, wild-type mice developed severe colorectal tumors, whereas AIB1-deficient mice had very few colorectal tumors (Figures 7a and b). The diameter of colon tumors formed in AIB1-deficient mice also showed a significant decrease compared with those in wild-type mice (Figure 7c). Consistently, AIB1-deficient tumors exhibited significantly reduced proliferation compared to wild-type tumor as determined by Ki-67 staining (Figures 7d and e). The mRNA levels of cyclin B1 and Hes5 were reduced in AIB1-deficient mice compared to wild-type mice (Figure 7f), whereas the mRNA levels of p27, ATOH1, and MUC2 were upregulated in the colorectal tumors from AIB1-deficient mice (Figure 7f). These in vivo results strongly suggest that AIB1 deficiency reduces AOM/DSS-induced CRC at least in part through attenuating Notch signaling.
Figure 7. AIB1-deficient mice are resistant to AOM/DSS-induced colorectal cancer.
(a) Images of colorectal tumors on the colons from AIB1-deficient and wild-type mice after 12-week AOM/DSS treatment. The experiment of AOM/DSS-induced colorectal cancer was performed twice with similar results. (b,c) Quantitation of the number and size of colorectal tumors (**P<0.01, N=5 colons). (d) H&E and Ki-67 staining of tissue sections of colorectal tumors from AIB1-deficient and wild-type mice(*P<0.05, N=5 tumors). (e) Quantitation of Ki-67-positive tumor cells (* P<0.05, N=5 tumors). (f) The mRNA levels of AIB1, cyclin B1, Hes5, ATOH1, and MUC2 were measured in colorectal tumors from AIB1-deficient and wild-type mice, respectively (*P<0.05, N=5 tumors).
DISCUSSION
Accumulating evidence indicates that aberrant AIB1 expression is a common phenomenon in human cancers6. Although previous studies showed that AIB1 was overexpressed in more than 35% of CRCs28, the exact role of AIB1 in colorectal tumorigenesis remains elusive. The present study clearly demonstrates an important role of AIB1 in CRC growth and progression: (1) AIB1 was overexpressed in several CRC cell lines and knockdown of AIB1 significantly reduced cell proliferation and invasiveness (2) knockdown of AIB1 significantly reduced CRC growth and metastasis in vivo; (3) AIB1 ablation in mice dramatically reduced AOM/DSS-induced CRC formation.
Consistent with reduced cell proliferation, downregulation of AIB1 caused CRC cell cycle arrest at G1 phase and affected the expression of several proliferation and cell cycle related genes such as cyclin A2, cyclin B1/ E2, Hes1, and Hes1's downstream targets p27, ATOH1, and MUC2. Since Hes1 is a typical target gene of Notch signaling that plays an important role in CRC progression, it prompted us to examine whether AIB1 is involved in the activation of Notch signaling to promote CRC progression. Our present results demonstrated that AIB1 directly interacted with NICD and recruited to Hes1 promoter to enhance Hes1 transcription and subsequent CRC cell proliferation. More importantly, AIB1-deficient colorectal tumors grew much slower and expressed less Notch target gene Hes5 than wild-type colorectal tumors, providing genetic evidence that AIB1 is indeed required for activation of Notch signaling pathway for CRC growth.
A simple model for the activation of Notch signaling could be that the NICD translocates into the nucleus to form a functional core activation complex with the transcription factor CSL and transcriptional coactivator MAML (NICD-CSL-MAML) to activate transcriptional events that are specific to Notch signaling. However, this model does not illustrate the whole picture of the regulation of NICD-CSL-MAML transactivation. Emerging evidence implicates several nuclear factors such as p300, PCAF, GCN5, and DDX5 can interact with the NICD-CSL-MAML complex to regulate Notch signaling37-40. In addition, the Notch1 nuclear interactome contains some key regulators such as LSD1 and PHF8 acting through their demethylase activity to promote epigenetic modifications at Notch-target genes41, highlighting the importance of coactivators in the regulation of Notch signaling. In this study, we found that AIB1 could enhance Notch signaling and promote proliferation, invasive and metastatic potential of CRC in vitro and in vivo. To the best of our knowledge, this is the first time that AIB1 is reported to serve as a critical coactivator for Notch signaling. Although the exact mechanism by which AIB1 enhance notch signaling is yet to be identified. Our study showed that AIB1 interacted with NICD by its HAT domain, but not CBP/p300 interaction domain (CID), implicating that after binding to NICD, AIB1 might recruit p300 via its CID to NICD-CSL-MAML complex to enhance Notch signaling. Further study is required to reveal the molecular mechanism by which AIB1 enhances NICD-CSL-MAML transactivation.
Notch signaling not only plays a critical role in CRC progression but also plays an essential role in intestinal development and homeostasis. Gut-specific inactivation of the Notch effectors Hes1, Hes3 and Hes5 in mice leads to impaired intestinal homeostasis by reducing cell proliferation, increasing goblet cell formation, and altering intestinal structures42. These results indicate that although Notch signaling is a promising target for anti-CRC therapy43, direct inhibition of Notch signaling could cause severe side effects by inducing stem/progenitor cells in healthy intestinal regions to differentiate into goblet cells. Thus, a strategy to inhibit Notch signaling only in CRC cells without affecting healthy cells is desirable. AIB1-deficient mice exhibit relatively normal intestinal structures, enterocyte/goblet cell ratio, and colon epithelial cell proliferation (Supplementary Figure S4), indicating that AIB1 deficiency does not affect intestinal development and homeostasis in general. Given that AIB1 is dispensable in normal intestinal homeostasis, but plays an essential role in CRC progression at least in part through enhancing Notch signaling, AIB1 emerges as a potential drug target for anti-CRC therapy through interference of Notch signaling without severe side effects on normal intestine. Recently, Wang et al. reported that a natural polyphenol gossypol and a cardiac glycoside bufalin were capable of binding to AIB1 protein to promote its degradation44-45, providing the proof-of-principle basis for the development of AIB1 targeting drugs for CRC treatment.
It has been shown that AIB1 promotes cancer progression by enhancing several oncogenic pathways such as ER, AR, EGFR, Akt, AP1, NRF2, NF-κB, and PEA36. Some of these oncogenic pathways have been shown to be involved in CRC progression. Therefore, AIB1 may simultaneously activate Notch and other oncogenic pathways to promote CRC progression. It is possible that therapeutically targeting AIB1 dampens not only Notch signaling pathway but also other oncogenic pathways to inhibit CRC progression.
Collectively our study demonstrates that AIB1 plays an essential role in CRC progression. AIB1 promotes CRC progression by directly interacting with NICD and MAMAL1 to enhance Notch signaling. Therefore pharmacological targeting of AIB1 may be a promising approach for treating CRC.
MATERIALS AND METHODS
Tissue culture
RKO, Caco-2, SW620, SW480 and HEK293T were maintained in DMEM supplemented with 10% FBS and antibiotics. HCT116 was cultured in MyCoy5A's medium and mouse CRC cell CT26 were cultured in RPMI1640 supplemented with 10% fetal bovine serum (FBS) and antibiotics.
AIB1-knockdown experiments
Two different small interfering RNAs (siRNAs) were synthesized against human AIB1 and two different siRNAs were synthesized against mouse AIB1. Human AIB1-specific targeting sequences of each siAIB1 were as follows: siAIB1-1, 5’-AGACTCCTTAGGACCGCTT-3’; siAIB1-2, 5’-TCGAGACGGAAAACATTGTA-3’. Mouse AIB1-specific targeting sequences of each siAIB1 were as follows: siAIB1-1’, 5’-GAACACGATTGTCGTTTGT-3’; siAIB1-2’, 5’-GTGTGTCAGTCAAACAGCA-3’. Human AIB1-specific targeting sequence 5’-AGACTCCTTAGGACCGCTT-3’ and mouse AIB1-specific targeting sequence 5’-GAACACGATTGTCGTTTGT-3’ were inserted into pSUPER/pll3.7 to generate pSUPER-sh-hAIB1 and pll3.7-sh-mAIB1 plasmids, respectively. All siRNAs and plasmids were transfected into different cell lines using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) following the manufacturer's instructions.
Cell proliferation and cell cycle analysis
Cell proliferation was analyzed by MTT assay and cell cycle was measured by flow cytometry as described in our previous study9, 46.
Cell viablility/death assay
The cell viablility/death assay was analyzed by propidium iodide (PI) staining. Briefly, the cells were harvested and collected in phosphate-buffered saline (PBS). After washing with PBS, the cells were resuspended in 1 ml PBS containing 5μg PI. PI incorporation and cell size were quantified by flow cytometry. All cells were divided into three regions. PI-negative cells of normal size were considered viable cells , PI-positive and smaller size cells were considered apoptotic cells of early phase , and PI-negative cells of smaller size were consider died cells of later period and the last two regions were consider cell death.
Real-time RT–PCR
Real-time RT-PCR was performed as previously described. Total RNA was isolated and reverse trancribed into cDNA using MMLV transcriptase (ToYoBo, Shanghai, China) with random primers. Real-time PCRs were performed using the StepOne Real-time PCR system (Applied Biosystems) with the SYBR Green master mix (Applied Biosystems). The primer sequences for cyclin A2, cyclin B1, cyclin E2, Hes1, p27, ATOH1 and MUC2 will be provided on request. Primers for GAPDH has been published in our previous study46.
Antibodies and Western blot
The Western blot and anti-AIB1 antibody has been described in our previous study9. Anti-Myc (9E10) and HA (F7) antibody were purchased from Santa Cruz. FLAG beads (M2 beads) and anti-FLAG antibody was purchased from Sigma Aldrich. Super-signal west Femto Lumianal/Enhancer Solution (Thermo Fisher Scientific Inc.) was used for imaging.
Chromatin immunoprecipitation (ChIP) assay and ChIP-reChIP assay
ChIP assay and ChIP-reChIP assay were processed according to the manufacturer's instructions (Abcam). The following primers were used to amplify the DNA fragment corresponding to the sequence from -167 to +8 on Hes1 promoter: Hes1 forward: cagaccttgtgcctggcg, Hes1 reverse: tgtgatccctaggccctg.
In vitro expression of AIB1 protein and GST pull-down assays
The coding sequence of AIB1 was clonded into pCR3.1 at downstream of a T7 promoter. E. coli extract-based cell free in vitro expression of AIB1 protein was performed using the S30 T7 high yield Protein Expression System (Promega) following the manufacturer's protocol. For GST pull-down assays, 1 μg of E. coli-produced GST or GST fusion proteins were immobilized on glutathione-agarose (Roche) for 1 h at room temperature. After several washes, the agarose was resuspended and then incubated with in vitro expressed proteins or cell lysates containing indicated proteins at 4°C. After extensive washes, bound proteins were eluted, resolved with SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and visualized by immumoblotting.
Cell invasion assays
Matrigel invasion assays were performed to investigate the cell invasiveness. The method has been described in our previous study9.
Mice
6 to 8-week-old male BALB/c mice were used for tumor formation and metastasis experiments. 6 to 8-week-old male SRC-3−/− mice and wild-type littermates on a BALB/c background were used for endogenous colon tumor induction experiments. Animal experiments were performed in accordance with the Guide for the Care and Use of Laboratory Animals. All animal experimental procedures were approved by Animal Care and Use Committee of Xiamen University. Every effort was made to reduce the suffering of animals.
Tumor formation
4×106 CT26-sh-mAIB1 and control cells were subcutaneously injected into BALB/c mice, respectively. Two perpendicular axis of the tumor were measured every 3 days from 10 days after injection. The volume of the tumor was calculated according to the formula: Volume=Length × Width2 × 0.52. Four weeks after injection, the mice were sacrificed and the tumors were photographed and used for Western blot and immunohistochemical analysis.
Tumor metastasis
To test tumor metastasis, CT26-sh-mAIB1 and control cells were intravenously injected into BALB/c mice. Three weeks after injection, mice were sacrificed and lungs were fixed by picric acid-formaldehyde. Tumors in the lungs were quantitated and used for Western blot analysis, H&E staining, and immunohistochemical analysis.
Colon tumor induction
To induce colon tumor, azoxymethane (AOM) was injected into male SRC-3−/− mice and wild-type littermates intraperitoneally, followed by administration of three 1-week cycles of 2.5% dextran sodium sulfate water, each cycle separated by 2 weeks36. After 12 weeks of treatment with AOM/DSS, mice were sacrificed and colons were isolated for photographing, quantitation, H&E staining, and immunohistochemical analysis.
Immunohistochemistry
Slides were soaked in preheated citrate buffer (pH 6.0) under microwave heating for 20 min to retrieve antigen. After cooling down, slides were washed with PBS and then incubated with Ki-67 antibody (1:500) for 1 h, followed by incubation with alkaline phosphatase-conjugated secondary antibody for 1 h. After washing, NBT/BCIP reagent was added to visualize stained proteins.
Terminal deoxynucleotidyl Transferase Fluorescein-dUTP Nick End Labeling (TUNEL) assay
The TUNEL assay was performed using a commercially available kit (in situ cell death detection kit, Roche) according to the manufacturer's instruction.
Statistical analysis
The data were collected from several independent experiments, with three replicates per experiment. All data were expressed as means+s.d. Statistically significant differences (P<0.05) were examined using t-test in SPSS 11.0 for Windows (SPSS Inc., Chicago, IL, USA).
Supplementary Material
Acknowledgement
We thank Dr. Dawang Zhou (Xiamen University) for providing HA-NICD, FLAG-NICD and Myc-NICD constructs, Dr. Lizhi Wu (University of Florida) for providing the FLAG-MAML1 and pGL3-Hes1-promoter plasmids. This work was supported by grants from the National Natural Science Foundation of China (No. 81071671 and No. 31170819 to C.Y.), the Fundamental Research Funds for the Central Universities (No. 2012121038 to C.Y.), the New Teachers Award of the Ministry of Education(No.20110121120004 to P.M.), and 111 Project B12001. J. Xu is supported by the CA112403 and the DK058242 NIH grants.
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
References
- 1.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.Qiao L, Wong BC. Role of Notch signaling in colorectal cancer. Carcinogenesis. 2009;30:1979–1986. doi: 10.1093/carcin/bgp236. [DOI] [PubMed] [Google Scholar]
- 3.Zhang Y, Li B, Ji ZZ, Zheng PS. Notch1 regulates the growth of human colon cancers. Cancer. 2010;116:5207–5218. doi: 10.1002/cncr.25449. [DOI] [PubMed] [Google Scholar]
- 4.Zanotti S, Canalis E. Notch and the skeleton. Mol Cell Biol. 2010;30:886–896. doi: 10.1128/MCB.01285-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ranganathan P, Weaver KL, Capobianco AJ. Notch signalling in solid tumours: a little bit of everything but not all the time. Nat Rev Cancer. 2011;11:338–351. doi: 10.1038/nrc3035. [DOI] [PubMed] [Google Scholar]
- 6.Xu J, Wu RC, O'Malley BW. Normal and cancer-related functions of the p160 steroid receptor co-activator (SRC) family. Nat Rev Cancer. 2009;9:615–630. doi: 10.1038/nrc2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anzick SL, Kononen J, Walker RL, Azorsa DO, Tanner MM, Guan XY, et al. AIB1, a steroid receptor coactivator amplified in breast and ovarian cancer. Science. 1997;277:965–968. doi: 10.1126/science.277.5328.965. [DOI] [PubMed] [Google Scholar]
- 8.Zhou HJ, Yan J, Luo W, Ayala G, Lin SH, Erdem H, et al. SRC-3 is required for prostate cancer cell proliferation and survival. Cancer Res. 2005;65:7976–7983. doi: 10.1158/0008-5472.CAN-04-4076. [DOI] [PubMed] [Google Scholar]
- 9.Xu Y, Chen Q, Li W, Su X, Chen T, Liu Y, et al. Overexpression of transcriptional coactivator AIB1 promotes hepatocellular carcinoma progression by enhancing cell proliferation and invasiveness. Oncogene. 2010;29:3386–3397. doi: 10.1038/onc.2010.90. [DOI] [PubMed] [Google Scholar]
- 10.Chen Q, Li W, Wan Y, Xia X, Wu Q, Chen Y, et al. Amplified in breast cancer 1 enhances human cholangiocarcinoma growth and chemoresistance by simultaneous activation of Akt and Nrf2 pathways. Hepatology. 2012;55:1820–1829. doi: 10.1002/hep.25549. [DOI] [PubMed] [Google Scholar]
- 11.Chung AC, Zhou S, Liao L, Tien JC, Greenberg NM, Xu J. Genetic ablation of the amplified-in-breast cancer 1 inhibits spontaneous prostate cancer progression in mice. Cancer Res. 2007;67:5965–5975. doi: 10.1158/0008-5472.CAN-06-3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuang SQ, Liao L, Wang S, Medina D, O'Malley BW, Xu J. Mice lacking the amplified in breast cancer 1/steroid receptor coactivator-3 are resistant to chemical carcinogen-induced mammary tumorigenesis. Cancer Res. 2005;65:7993–8002. doi: 10.1158/0008-5472.CAN-05-1179. [DOI] [PubMed] [Google Scholar]
- 13.Kuang SQ, Liao L, Zhang H, Lee AV, O'Malley BW, Xu J. AIB1/SRC-3 deficiency affects insulin-like growth factor I signaling pathway and suppresses v-Ha-ras-induced breast cancer initiation and progression in mice. Cancer Res. 2004;64:1875–1885. doi: 10.1158/0008-5472.can-03-3745. [DOI] [PubMed] [Google Scholar]
- 14.Tien JC, Zhou S, Xu J. The role of SRC-1 in murine prostate cancinogenesis is nonessential due to a possible compensation of SRC-3/AIB1 overexpression. Int J Biol Sci. 2009;5:256–264. doi: 10.7150/ijbs.5.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Park KJ, Krishnan V, O'Malley BW, Yamamoto Y, Gaynor RB. Formation of an IKKalpha-dependent transcription complex is required for estrogen receptor-mediated gene activation. Mol Cell. 2005;18:71–82. doi: 10.1016/j.molcel.2005.03.006. [DOI] [PubMed] [Google Scholar]
- 16.Yan J, Erdem H, Li R, Cai Y, Ayala G, Ittmann M, et al. Steroid receptor coactivator-3/AIB1 promotes cell migration and invasiveness through focal adhesion turnover and matrix metalloproteinase expression. Cancer Res. 2008;68:5460–5468. doi: 10.1158/0008-5472.CAN-08-0955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yan J, Tsai SY, Tsai MJ. SRC-3/AIB1: transcriptional coactivator in oncogenesis. Acta Pharmacol Sin. 2006;27:387–394. doi: 10.1111/j.1745-7254.2006.00315.x. [DOI] [PubMed] [Google Scholar]
- 18.Kishimoto H, Wang Z, Bhat-Nakshatri P, Chang D, Clarke R, Nakshatri H. The p160 family coactivators regulate breast cancer cell proliferation and invasion through autocrine/paracrine activity of SDF-1alpha/CXCL12. Carcinogenesis. 2005;26:1706–1715. doi: 10.1093/carcin/bgi137. [DOI] [PubMed] [Google Scholar]
- 19.Long W, Yi P, Amazit L, LaMarca HL, Ashcroft F, Kumar R, et al. SRC-3Delta4 mediates the interaction of EGFR with FAK to promote cell migration. Mol Cell. 2010;37:321–332. doi: 10.1016/j.molcel.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yan J, Yu CT, Ozen M, Ittmann M, Tsai SY, Tsai MJ. Steroid receptor coactivator-3 and activator protein-1 coordinately regulate the transcription of components of the insulin-like growth factor/AKT signaling pathway. Cancer Res. 2006;66:11039–11046. doi: 10.1158/0008-5472.CAN-06-2442. [DOI] [PubMed] [Google Scholar]
- 21.Long W, Foulds CE, Qin J, Liu J, Ding C, Lonard DM, et al. ERK3 signals through SRC-3 coactivator to promote human lung cancer cell invasion. J Clin Invest. 2012;122:1869–1880. doi: 10.1172/JCI61492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Louie MC, Zou JX, Rabinovich A, Chen HW. ACTR/AIB1 functions as an E2F1 coactivator to promote breast cancer cell proliferation and antiestrogen resistance. Mol Cell Biol. 2004;24:5157–5171. doi: 10.1128/MCB.24.12.5157-5171.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Louet JF, Coste A, Amazit L, Tannour-Louet M, Wu RC, Tsai SY, et al. Oncogenic steroid receptor coactivator-3 is a key regulator of the white adipogenic program. Proc Natl Acad Sci U S A. 2006;103:17868–17873. doi: 10.1073/pnas.0608711103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu RC, Qin J, Hashimoto Y, Wong J, Xu J, Tsai SY, et al. Regulation of SRC-3 (pCIP/ACTR/AIB-1/RAC-3/TRAM-1) Coactivator activity by I kappa B kinase. Mol Cell Biol. 2002;22:3549–3561. doi: 10.1128/MCB.22.10.3549-3561.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fereshteh MP, Tilli MT, Kim SE, Xu J, O'Malley BW, Wellstein A, et al. The nuclear receptor coactivator amplified in breast cancer-1 is required for Neu (ErbB2/HER2) activation, signaling, and mammary tumorigenesis in mice. Cancer Res. 2008;68:3697–3706. doi: 10.1158/0008-5472.CAN-07-6702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu C, York B, Wang S, Feng Q, Xu J, O'Malley BW. An essential function of the SRC-3 coactivator in suppression of cytokine mRNA translation and inflammatory response. Mol Cell. 2007;25:765–778. doi: 10.1016/j.molcel.2007.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qin L, Liao L, Redmond A, Young L, Yuan Y, Chen H, et al. The AIB1 oncogene promotes breast cancer metastasis by activation of PEA3-mediated matrix metalloproteinase 2 (MMP2) and MMP9 expression. Mol Cell Biol. 2008;28:5937–5950. doi: 10.1128/MCB.00579-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xie D, Sham JS, Zeng WF, Lin HL, Bi J, Che LH, et al. Correlation of AIB1 overexpression with advanced clinical stage of human colorectal carcinoma. Hum Pathol. 2005;36:777–783. doi: 10.1016/j.humpath.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 29.Monahan P, Rybak S, Raetzman LT. The notch target gene HES1 regulates cell cycle inhibitor expression in the developing pituitary. Endocrinology. 2009;150:4386–4394. doi: 10.1210/en.2009-0206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murata K, Hattori M, Hirai N, Shinozuka Y, Hirata H, Kageyama R, et al. Hes1 directly controls cell proliferation through the transcriptional repression of p27Kip1. Mol Cell Biol. 2005;25:4262–4271. doi: 10.1128/MCB.25.10.4262-4271.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bar Y, Russ HA, Knoller S, Ouziel-Yahalom L, Efrat S. HES-1 is involved in adaptation of adult human beta-cells to proliferation in vitro. Diabetes. 2008;57:2413–2420. doi: 10.2337/db07-1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu L, Aster JC, Blacklow SC, Lake R, Artavanis-Tsakonas S, Griffin JD. MAML1, a human homologue of Drosophila mastermind, is a transcriptional co-activator for NOTCH receptors. Nat Genet. 2000;26:484–489. doi: 10.1038/82644. [DOI] [PubMed] [Google Scholar]
- 33.Fernandez-Majada V, Aguilera C, Villanueva A, Vilardell F, Robert-Moreno A, Aytes A, et al. Nuclear IKK activity leads to dysregulated notch-dependent gene expression in colorectal cancer. Proc Natl Acad Sci U S A. 2007;104:276–281. doi: 10.1073/pnas.0606476104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sikandar SS, Pate KT, Anderson S, Dizon D, Edwards RA, Waterman ML, et al. NOTCH signaling is required for formation and self-renewal of tumor-initiating cells and for repression of secretory cell differentiation in colon cancer. Cancer Res. 2010;70:1469–1478. doi: 10.1158/0008-5472.CAN-09-2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Christofori G. Metastatic colon cancer cells negotiate the intravasation Notch. Cancer Cell. 2011;19:6–8. doi: 10.1016/j.ccr.2011.01.003. [DOI] [PubMed] [Google Scholar]
- 36.Neufert C, Becker C, Neurath MF. An inducible mouse model of colon carcinogenesis for the analysis of sporadic and inflammation-driven tumor progression. Nature protocols. 2007;2:1998–2004. doi: 10.1038/nprot.2007.279. [DOI] [PubMed] [Google Scholar]
- 37.Lin S, Tian L, Shen H, Gu Y, Li JL, Chen Z, et al. DDX5 is a positive regulator of oncogenic NOTCH1 signaling in T cell acute lymphoblastic leukemia. Oncogene. 2012;32:4845–4853. doi: 10.1038/onc.2012.482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kurooka H, Honjo T. Functional interaction between the mouse notch1 intracellular region and histone acetyltransferases PCAF and GCN5. J Biol Chem. 2000;275:17211–17220. doi: 10.1074/jbc.M000909200. [DOI] [PubMed] [Google Scholar]
- 39.Wallberg AE, Pedersen K, Lendahl U, Roeder RG. p300 and PCAF act cooperatively to mediate transcriptional activation from chromatin templates by notch intracellular domains in vitro. Mol Cell Biol. 2002;22:7812–7819. doi: 10.1128/MCB.22.22.7812-7819.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hansson ML, Popko-Scibor AE, Saint Just Ribeiro M, Dancy BM, Lindberg MJ, Cole PA, et al. The transcriptional coactivator MAML1 regulates p300 autoacetylation and HAT activity. Nucleic Acids Res. 2009;37:2996–3006. doi: 10.1093/nar/gkp163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yatim A, Benne C, Sobhian B, Laurent-Chabalier S, Deas O, Judde JG, et al. NOTCH1 Nuclear Interactome Reveals Key Regulators of Its Transcriptional Activity and Oncogenic Function. Mol Cell. 2012;48:445–458. doi: 10.1016/j.molcel.2012.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ueo T, Imayoshi I, Kobayashi T, Ohtsuka T, Seno H, Nakase H, et al. The role of Hes genes in intestinal development, homeostasis and tumor formation. Development. 2012;139:1071–1082. doi: 10.1242/dev.069070. [DOI] [PubMed] [Google Scholar]
- 43.Miyamoto S, Nakanishi M, Rosenberg DW. Suppression of colon carcinogenesis by targeting Notch signaling. Carcinogenesis. 2013;34:2415–2423. doi: 10.1093/carcin/bgt191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang Y, Lonard DM, Yu Y, Chow DC, Palzkill TG, O'Malley BW. Small molecule inhibition of the steroid receptor coactivators, SRC-3 and SRC-1. Mol Endocrinol. 2011;25:2041–2053. doi: 10.1210/me.2011-1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang Y, Lonard DM, Yu Y, Chow DC, Palzkill TG, Wang J, et al. Bufalin is a potent small molecule inhibitor of the steroid receptor coactivators SRC-3 and SRC-1. Cancer Res. 2014 doi: 10.1158/0008-5472.CAN-13-2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu Y, Tong Z, Li T, Chen Q, Zhuo L, Li W, et al. Hepatitis B virus X protein stabilizes amplified in breast cancer 1 protein and cooperates with it to promote human hepatocellular carcinoma cell invasiveness. Hepatology. 2012;56:1015–1024. doi: 10.1002/hep.25751. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.