SUMMARY
Evidence from animal studies and human famines suggests that starvation may affect the health of the progeny of famished individuals. However, it is not clear whether starvation affects only immediate offspring or has lasting effects; it is also unclear how such epigenetic information is inherited. Small RNA-induced gene silencing can persist over several generations via transgenerationally inherited small RNA molecules in C. elegans, but all known transgenerational silencing responses are directed against foreign DNA introduced into the organism. We found that starvation-induced developmental arrest, a natural and drastic environmental change, leads to the generation of small RNAs that are inherited through at least three consecutive generations. These small, endogenous, transgenerationally transmitted RNAs target genes with roles in nutrition. We defined genes that are essential for this multigenerational effect. Moreover, we show that the F3 offspring of starved animals show an increased lifespan, corroborating the notion of a transgenerational memory of past conditions.
INTRODUCTION
The environment’s ability to influence inherited traits has been a subject of heated debate for over a century (Koonin and Wolf, 2009). Although the underlying mechanism has been elusive, multiple environmentally induced transgenerational epigenetic effects have been described (Jablonka and Lamb, 2008). In particular, evidence collected from diverse animal species, and broad epidemiologic surveys of different famines in the Netherlands (1944–1945) (Painter et al., 2008), China (1959–1961) (Song et al., 2009), and Russia (1941–1944) (Stanner et al., 1997) have demonstrated that susceptibility to disease is correlated with malnutritioned ancestors. In particular, it has been shown that there is an increased capacity to transmit epigenetic information about food availability during development before the prepubertal peak in growth speed (Bygren et al., 2001; Kaati et al., 2007). In all the above-mentioned studies the effects were restricted to the immediate offspring, leaving the possibility open that rather than being transgenerationally inherited, the effects were directly exerted onto the germ cells of the exposed animals (Heard and Martienssen, 2014). Thus, a truly epigenetic effect that could transmit the somatic response to satiety or famine to the generations beyond the immediate next generation remains to be found.
The model organism Caenorhabditis elegans has been successfully used for the study of transgenerational epigenetic effects (Greer et al., 2011; Rechavi et al., 2011), in part because of its short generation time, C. elegans is particularly suited for studying transgenerational inheritance of dietary history, since worms are often faced with scarcity of nutrients in the wild. In fact, a dedicated genetic program allows worms to reversibly arrest postembryonic development in the first larval stage (L1) in the absence of food (Baugh, 2013). Similar to the situation in humans, it was shown that the C. elegans F1 progeny is affected by the maternal diet (Harvey and Orbidans, 2011); again, a genetic effect that extends beyond one generation was not demonstrated to act in response to nutrient availability in C. elegans. Nevertheless, it was shown that an association between odor and food (olfactory imprinting), which can only be encoded during the L1 stage (Remy and Hobert, 2005), is memorized for multiple generations (Remy, 2010).
L1 starvation leads to a particularly drastic change in gene expression. The expression of 27% of protein-coding genes is altered, more than the number of genes which are differentially expressed throughout the entire course of larval development (Maxwell et al., 2012). Survival during and recovery from L1 arrest is in part regulated by RNA interference (RNAi), and also depends on microRNA repression of insulin pathway genes (Zhang et al., 2011). Global regulation of small RNA levels during L1 starvation, could be achieved, for example, by the long noncoding RNA rncs-1, which was shown to be induced during L1 starvation, and to in vitro “block” DICER, reducing its capacity to process other small RNAs (Hellwig and Bass, 2008).
C. elegans possesses a dedicated RNAi inheritance mechanism that could, in theory, allow memorization of dietary history-dependent small RNA response for multiple generations. Double-strand RNA (dsRNA) spreads systemically and transmits from the soma to the germline in C. elegans (Fire et al., 1998). Experimental silencing of certain genes by administration of dsRNA has been demonstrated to persist for more than 80 generations (Vastenhouw et al., 2006). The same inheritance mechanism that acts in response to artificial dsRNA was later shown to also play a role in antiviral and transposon immunity (Rechavi, 2013; Rechavi et al., 2011; Sterken et al., 2014). While the different biogenesis mechanisms are not yet fully understood, endogenous small RNAs (endo-siRNAs) align to thousands of genes across the genome (Grishok, 2013). Recently a number of groups proposed that endo-siRNAs survey all germline-expressed genes, to silence invading elements and license the expression of autogenous sequences (Ashe et al., 2012; Buckley et al., 2012; Shirayama et al., 2012). Specifically, it was suggested that two argonaute proteins, HRDE-1 (heritable RNAi deficient or WAGO-9) and CSR-1 (chromosome-segregation and RNAi deficient), carry small RNAs that compete over binding to cognate germline-expressed target mRNAs, and the result of this antagonism determines whether a gene would be transcriptionally silenced (HRDE-1 targets) or licensed (CSR-1 targets) for expression in the germline (Seth et al., 2013). The inherited regulation of germline-expressed genes persists across multiple generations by the action of small RNAs, which are amplified by RNA-dependent RNA Polymerases (RdRPs) (Gu et al., 2012).
Importantly, while it is now clear that RNAi inheritance mechanisms are induced by different exogenous genetic elements, it has not been demonstrated that an environmental change is capable of eliciting a heritable response that involves endogenous small RNAs and endogenous small RNA targets. In all the previous studies, the transgenerational silencing responses were directed against foreign DNA (transposons, viruses, transgenes) (Ashe et al., 2012; Buckley et al., 2012; Lee et al., 2012; Rechavi et al., 2011; Shirayama et al., 2012). Therefore, it is still unknown whether an endogenous, biologically relevant response, which is induced under natural circumstances, could elicit a small RNA response that would be passed on to ensuing generations.
In this paper, we describe a set of small RNAs that are induced by starvation. We show that these starvation-induced small RNAs are transmitted transgenerationally, providing a mean for starved worms to control the expression of relevant genes in consecutive generations.
RESULTS
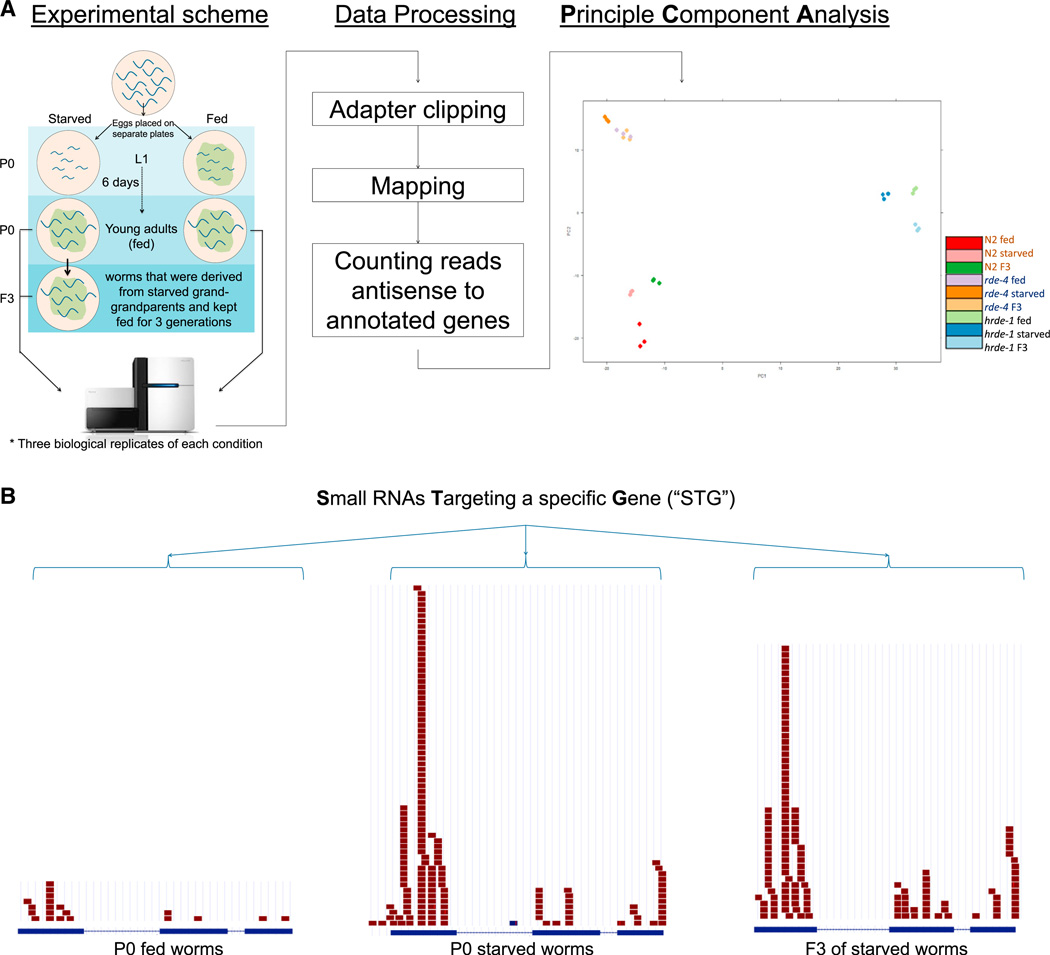
We first sequenced small RNAs from young adult worms that were either continuously fed or severely starved for 6 days as L1s, and from their F3 progeny (see scheme in Figure 1A). We used a protocol that enables cloning of “secondary” endo-small RNAs (See Extended Experimental Procedures). These are 22 nt long transcripts that possess a 5′ guanosine, which are amplified by RdRPs and constitute the most abundant type of endogenous small RNAs (Gu et al., 2009; Pak and Fire, 2007).
Figure 1. A Protocol for Analyzing the Starvation-Induced Transgenerational Small RNA Response.
(A) Aschematic description of the experimental protocol: secondary (RdRP-amplified) small RNAs were cloned and sequenced from three biological replicates of young adult worms that were either continuously fed or severely starved for 6 daysasL1s and from theirF3 progeny. Following adapters trimming, the reads were mapped to the WS220 version of the C. elegans genome. Reads which mapped antisense to annotated gene were counted and then analyzed by principle component analysis to reveal the similarities/differences between the experimental conditions (see Figure S1 for an additional PCA of reads which were mapped to the entire genome).
(B) Visualization in the UCSC browser of the vhl-1 gene. An STG (small rnas targeting a given gene) is defined as the sum of the small RNAs that are antisense to a certain gene (see Table S1 for the fold-change and p value ranking of all the STGs).
We examined the small RNA pools of wild-type (N2), rde-4 (RNAi Deficient, a DICER interactor), and hrde-1 worms using principal components analysis (PCA). PCA reduces multidimensional data into two dimensions, so that the relative distance between samples could be determined. The PCA revealed that the three biological replicates cluster together, while the experimental conditions and the different mutants are clearly separated (see processing steps and PCA in Figure 1A, and Figure S1 available online; additional details are provided in the Extended Experimental Procedures).
The most dramatic changes between the experimental conditions were found to arise from small RNAs that align antisense to gene-coding regions. Past work has shown that endo-siRNAs generally align in the antisense orientation and almost exclusively to exons (Grishok, 2013). This typical pattern was clearly apparent in the differentially expressed small RNAs (Figure 1B). We thus analyzed gene-targeting small RNAs that were differentially expressed between the experimental conditions, and considered a gene as a “putative target” of small RNAs-mediated regulation if multiple small RNAs align to it in the antisense orientation. For short, we dubbed clusters of small RNAs that align in the antisense orientation to particular genes, as “STGs” (small RNAs targeting a given gene, see Figure 1B).
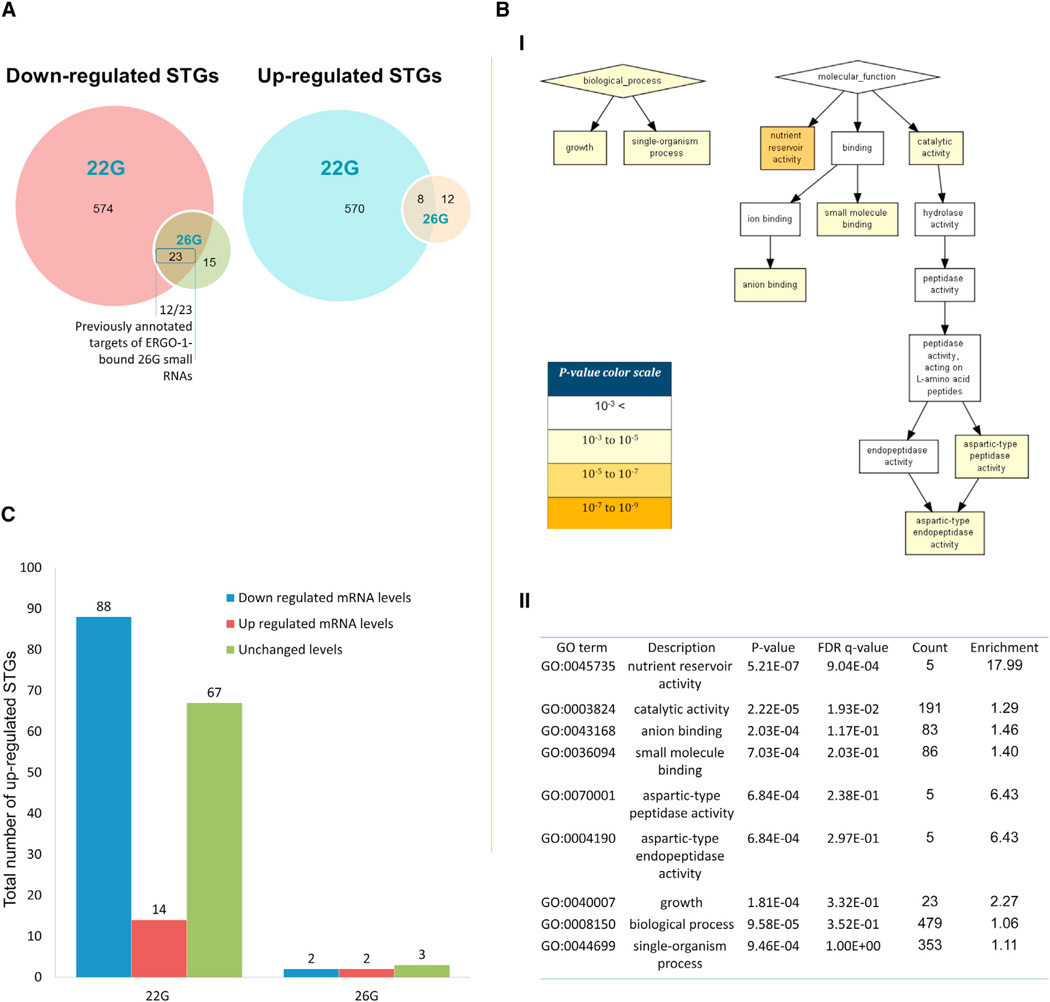
To examine whether there were changes in the small RNA pools between the P0 fed animals and the animals that were starved as L1s (hereby, P0 “starved”), we compared the samples using the DESeq2 method (FDR < 0.1) and distinguished between 22Gs and 26Gs sequences, which are the two different species of endo-siRNAs (Gent et al., 2010; Vasale et al., 2010). In the P0 adults, the 22G STGs (secondary endo-siRNAs) exhibited the most dramatic changes following L1 starvation (578 upregulated, 597 downregulated, see Table S1). However, a statistically significant differential expression in 26G STGs (primary endo-siRNAs) was also observed (20 upregulated,38 downregulated, see Figures 2A and S2). Although the exact mechanism is not yet clear, it has been shown that this primary small RNA species can induce the biogenesis of secondary 22Gs small RNAs in cis or in trans (Gent et al., 2010; Han et al., 2009; Montgomery et al., 2012; Vasale et al., 2010). This process probably entails primary small RNA-mediated guiding of RdRPs to the mature mRNAs target, and it is hypothesized that the amplified 22Gs can be sequentially loaded onto either HRDE-1 or CSR-1 (Seth et al., 2013). It is currently very difficult to reliably predict whether primary small RNAs would initiate secondary small RNA production because primary small RNAs were shown to also trigger amplification using very permissive and imperfect base pairing (Ashe et al., 2012; Montgomery et al., 2012; Shirayama et al., 2012). Nevertheless, even when allowing only high degree of complementarity (see Extended Experimental Procedures), 31 putative targets of secondary differentially expressed 22G STGs were also targeted by primary differentially expressed 26G small RNAs (8 upregulated and 23 downregulated) following L1 starvation (Figure 2A). We noticed that 12 out of the 23 STGs that were downregulated in both the 22G and the 26G analysis (Figure 2A) were also found in a previous study that identified a small number (48) of genes that are targeted by ERGO-1-bound and 3′-modified 26G small RNAs (Vasale et al., 2010). These ERGO-1 pathway 26G STGs, that we find to be downregulated upon L1 starvation, were subsequently shown to be stabilized by HENN-1-mediated 3′ methylation (Billi et al., 2012; Kamminga et al., 2012; Montgomery et al., 2012).
Figure 2. Transcriptome Analysis of P0 Adults that Experienced Starvation at the L1 Stage.
(A) Overlaps of up/downregulated STGs of the different endo-siRNA species (see Figure S2 for an MA Plot visualization of the differentially expressed STGs of each small RNA species).
(B) A functional annotation of the 22G STGs (secondary small RNAs which are 22 nt long and possess a 5′ guanosine) that are differentially expressed following starvation in wild-type animals. Annotation was performed using the GOrilla web-server. (I) Functional enrichment map. (II) A table of the specific GO terms, statistical significances, the number of genes that share each specific GO term, and their relative enrichments. An additional GO analysis, which also takes into account the fold-change ranking of the differentially expressed STGs is presented in Figure S3.
(C) Changes in the mRNA levels that correspond to changes in STGs. The x axis represents the two species of small RNAs that were examined: 22Gs (secondary small RNAs) and 26Gs (primary small RNAs which are 26 nt long and possess a 5′ guanosine). The y axis represents the total number of STGs which were upregulated following L1-starvation, in correspondence to the changes in mRNA levels (see Table S3 for fold-change ranking of the genes).
22G RNAs are the most abundant differentially expressed small RNAs, and since these RNA molecules are amplifiable, and are known to be carried by HRDE-1 and CSR-1, the argonaute proteins that transmit inherited small RNAs across generations (Rechavi, 2013), we hypothesized that this RNA species can be inherited following L1 starvation, and thus focused our analysis on it.
The putative target genes of the differentially expressed 22G STGs were enriched for a number of biological functions (GOrilla analysis, Figures 2B and Figure S3), and most significantly for nutrient reservoir activity (FDR < 8.61 × 10−4). Regulation of nutrient reservoir activity could in theory affect the worm’s ability to handle starvation-induced stress. Some of these genes (37/1,175, 1.5-fold enrichment, p value < 0.011) were previously found to be regulated by small RNAs following recovery from dauer arrest, an alternative developmental stage, during which worms are food deprived (Hall et al., 2013).
To test the potential of the differentially expressed STGs to regulate the levels of their cognate target genes, we sequenced mRNAs from the same samples. It is currently not clear to what extent, or depending on which conditions, endo-siRNAs affect their targets. We observed a downregulation in the mRNA levels of 88 genes that were each predicted to be a target of a specific 22G STG, that was upregulated in young adults that experienced starvation as L1s (Figure 2C and Table S3). The observed reduction in mRNA levels is consistent with the canonical mechanism of small RNA-mediated gene silencing. Upregulation of a small number of mRNAs (14), which were similarly predicted to be targets of the upregulated STGs, was also observed. Upregulation of mRNA that are targeted by small RNAs could be the result of indirect interactions, or may be a reflection of the recently described “RNA licensing” or “RNA activation” (RNAa) mechanism (Seth et al., 2013; Wedeles et al., 2013).
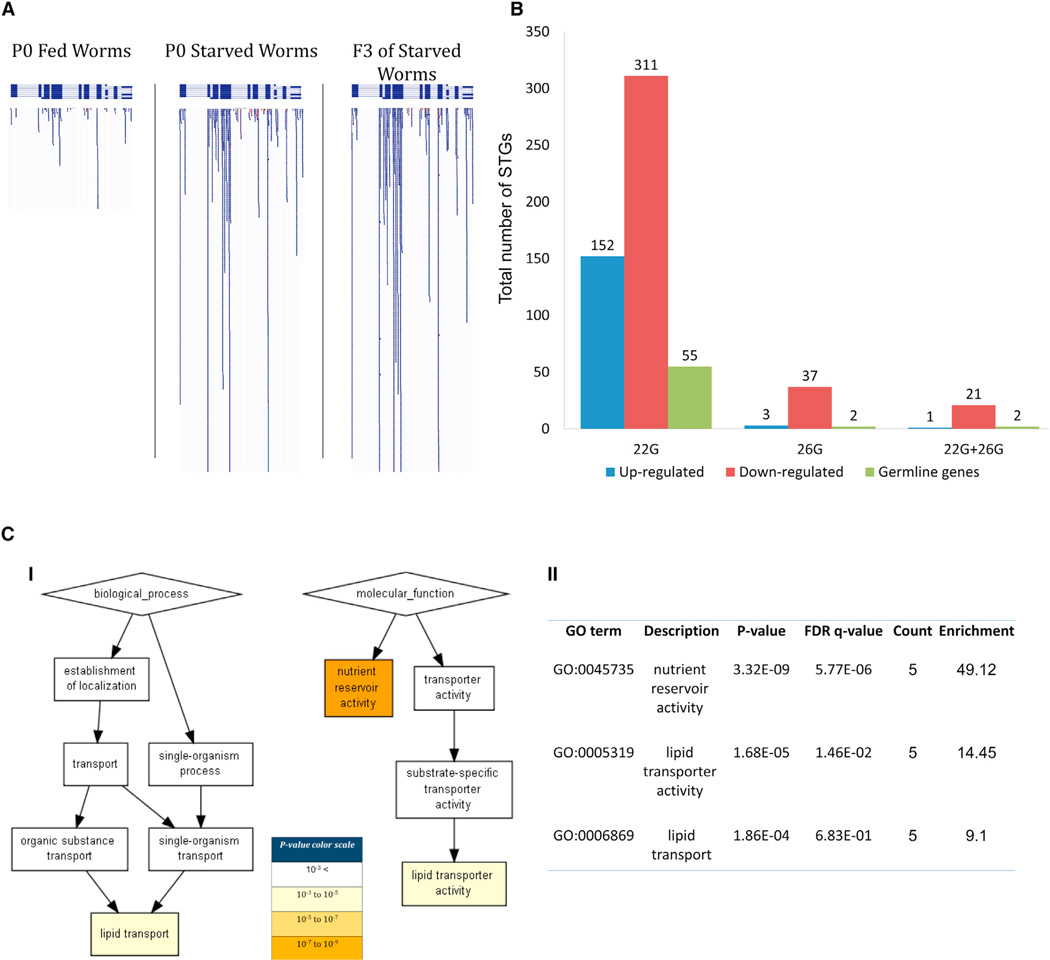
To examine whether the small RNA response could be transmitted across generations, we sequenced small RNAs from the fed F3 progeny of the worms that were starved as L1s (hereby referred to as “F3 of starved worms”). Surprisingly, although the “F3 of starved worms” were fed for three generations, clustering of the STGs that were differentially expressed following starvation in the P0 generation revealed that the F3 worms resemble the “starved” parents more than the “fed” parents (Figure S5). We examined which small RNAs were responsible for this effect and found that 26.3% (152/578) of the 22G STGs that were upregulated in their ancestors following L1 starvation were also upregulated in a statistically significant manner in the F3 progeny in comparison to the fed ancestors (hypergeometric test, p value < 1.399 × 10−71, for a typical gene see Figure 3A). In addition to inherited upregulation of STGs, we found that 52% (311/597) of the STGs that were depleted following starvation were depleted also in the F3 progeny (p value < 1.256 × 10−292, Table S2). Thus, the small RNA pools of the F3 worms reflect the changes that occurred following starvation in the P0 generation. As mentioned above, in the P0 generations 31/1,175 differentially expressed STGs which are targeted by secondary 22Gs are also targeted by differentially expressed primary 26G small RNAs STGs, which are predicted by the current model to trigger their biogenesis. We found that levels of 91% of the 22Gs that overlap with 26G STGs were transgenerationally downregulated in the F3 samples (p value < 5.629 × 10−29, fold-enrichment = 26.1, Figure 3B, Tables S2 and S5).
Figure 3. Transgenerational Inheritance of Small RNAs Following L1 Starvation.
(A) An example of a gene (hlh-30) targeted by heritable 22Gs. See Table S2 for the fold-change and p value ranking of all the inherited STGs.
(B) The total number of inherited STGs of different small RNA species and their overlapping targets.
(C) A functional annotation of STGs that are differentially expressed in an inherited manner following L1-starvation in WT animals. (I) GOrilla functional enrichment map. (II) A table of the specific GO terms, statistical significances, the number of genes that share each specific GO term, and their relative enrichments (see Figure S4 for genomic visualization of the nutrient reservoir activity genes).
It was previously hypothesized that germline transcription of the mRNA target provides a template on which heritable small RNAs can amplify (Buckley et al., 2012; Burton et al., 2011). We thus asked whether the putative targets of the heritable STGs are enriched for germline expression. We found that 21.7% of the putative targets of the upregulated inherited 22G STGs were germline expressed (based on data from (Reinke et al., 2004), fold-enrichment = 1.5, p value < 0.012, Figure 3B). This enrichment suggests that inherited small RNAs may indeed preferentially target germline-expressed genes. On the other hand, only 8% of the putative targets of downregulated inherited STGs are germline-expressed genes (fold-depletion = 1.85, p value < 1.13 × 10−4).
Examination of the list of 463 potential targets (see Table S6) of the inherited STGs using GOrilla revealed functional enrichments that resembled the enrichments that were detected in the P0 generation, and most notably where again enriched for nutrient reservoir activity (Figure 3C). Interestingly, differentially expressed heritable STGs align to the entire family of the vitellogenins (6/6, 38.7-fold enrichment, p value < 2.950 × 10−10), worm yolk proteins that provision the egg, and are encoded in different locations in the genome (DePina et al., 2011). Biogenesis of the secondary 22G STGs that target the vitellogenins could be initiated by dsRNA precursors. On the complementary strands of all the vitellogenins, there are genes which are transcribed in the opposite direction, suggesting that cotranscription of these genes and the vitellogenins could lead to the formation of dsRNA (Figure S4). 20 F box proteins (5-fold enrichment, p value < 5.057 × 10−9) were also found to be targeted by heritable STGs. While the function of these proteins in the starvation response of C. elegans is unknown, in plants homologous proteins monitor protein turnover following stress (Meloche and Roux, 2012).
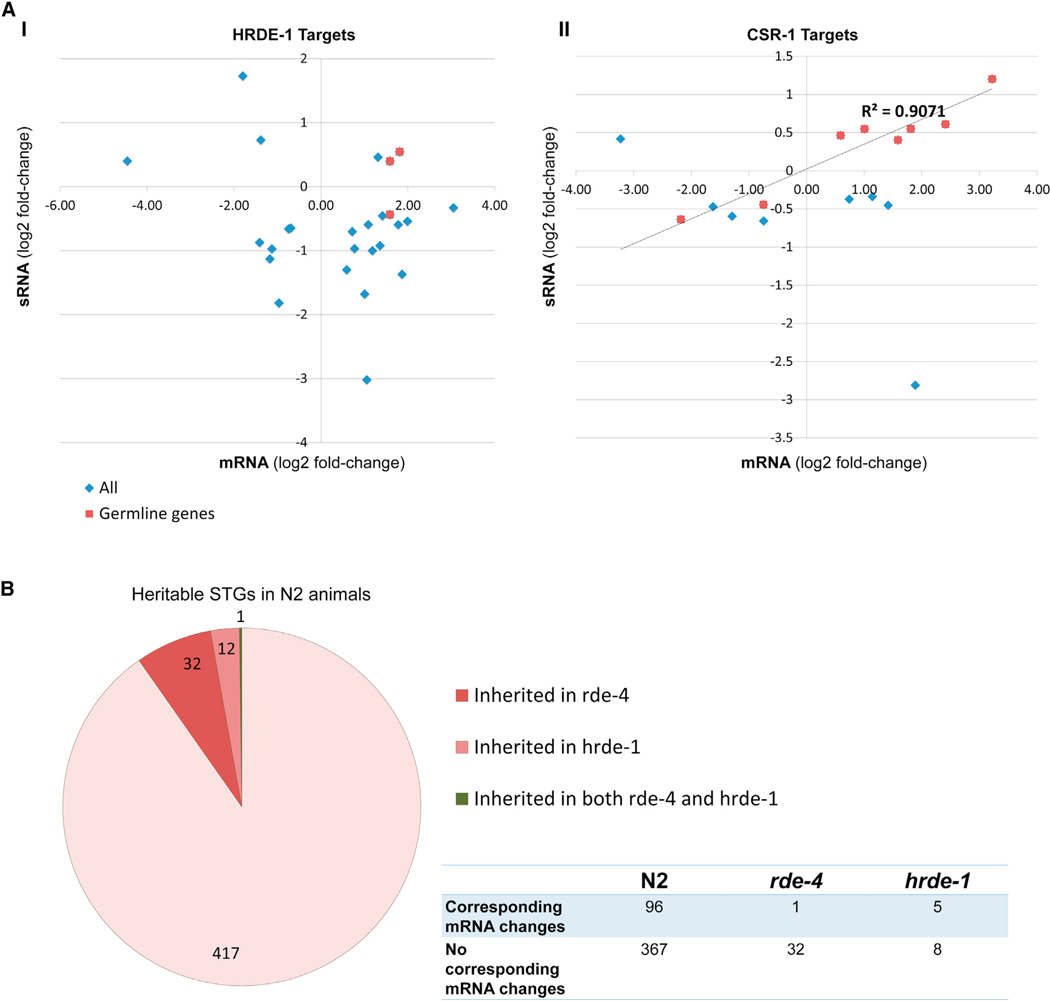
To examine whether inherited small RNAs could regulate these enriched genetic pathways across multiple generations, we analyzed the mRNA expression pattern of the genes that were targeted by inherited STGs using transcriptome sequencing. The expression levels of 96/463 genes that are targeted by differentially expressed heritable small RNAs were changed between the F3 progeny of starved worms and the fed P0s (Table S4). We could not detect any distinct pattern of up or downregulation when we plotted the inherited small RNAs against their cognate mRNAs. However, separate examination of the mRNA targets of the heritable small RNAs that were previously shown to bind HRDE-1 or CSR-1 (Buckley et al., 2012; Claycomb et al., 2009), revealed opposite trends of regulation. We observed a downregulation of 65.3% (17/26) of the mRNAs that were targeted by HRDE-1-bound small RNAs, and upregulation of 68.7% (11/16) of the mRNAs that were targeted by CSR-1-bound small RNAs. Interestingly, a strong correlation (R2 = 0.907, p value < 0.00026) was detected between the levels of inherited CSR-1 STGs and germline expressed, but not somatically expressed, cognate mRNA targets (Figure 4A).
Figure 4. Transgenerationally Inherited Small RNAs Differentially Regulate Their Cognate mRNAs and Are RDE-4 and HRDE-1-Dependent.
(A) Plotting of the log2-fold changes of the inherited STGs, versus the mRNA log2-fold changes of their cognate targets. Shown separately are (I) HRDE-1 (II) CSR-1 targets (Buckley et al., 2012; Claycomb et al., 2009). Somatic genes are represented by blue dots and germline-expressed genes are represented by red dots. Shown is a linear correlation line (R2 = 0.907) between the inherited STGs that were previously shown to bind CSR-1, and their germline-expressed mRNA targets.
(B) The majority of the inherited STGs are depleted in rde-4 and hrde-1 mutants. Shown are the numbers of the small minority of STGs which are nevertheless inherited in the RNAi mutants and their corresponding mRNA changes.
Previous studies described that levels of DICER-dependent small RNAs (both endo-siRNAs and microRNAs) are altered by L1 starvation (Hellwig and Bass, 2008; Reinke et al., 2004; Zhang et al., 2011), and therefore in theory the levels of these small RNAs, if inherited, could be affected in future nonstarved generations. The dsRNA-binding protein RDE-4 is a specific component in the DICER endo-siRNA pathway (Lee et al., 2006). Conveniently, while RDE-4 was shown to play a role in response to different stresses (Mansisidor et al., 2011), multiple generations of rde-4 mutants can be followed (unlike DICER mutants, which are sterile), since these mutants are surprisingly fertile and lack obvious phenotypes under normal growth conditions. Moreover, RDE-4 operates upstream to ERGO-1, and as mentioned above, we found that a large fraction of the differentially expressed 26G STGs were previously found to bind this argonaute (Vasale et al., 2010). We therefore examined which small RNAs are induced by L1 starvation and persist until adulthood in rde-4 mutants and whether small RNA inheritance following L1-starvation is abrogated in these animals. We found that inheritance of most (93%) of the STGs is dependent on RDE-4. Only 33 out of the 463 (Table S5) STGs that were found to be inherited following L1-starvation in N2 were also found to be inherited following L1-starvation in rde-4 animals (Figure 4B). In addition to the significant reduction in the inherited small RNAs response, while in N2 expression of 96 out of 463 genes (20%) that were found to be targeted by heritable STGs were changed, the mRNA levels of only 33 putative targets (7%) were changed in rde-4 animals. RDE-4 was previously hypothesized to function specifically in response to stress. We observed that more STGs are reduced in rde-4 animals in comparison to N2 animals when comparing the starved conditions (1,496 genes) or the F3 of starved worms (2,686 genes), relative to the smaller difference which is observed between the fed parents (1,024 genes).
hrde-1 hermaphrodites, but not csr-1 hermaphrodites, are fertile at nonrestrictive temperature (Buckley et al., 2012; Conine et al., 2013), and so the progeny of starved hrde-1 mutants could be examined. We found in additional sequencing experiments that 96.8% of the heritable STGs were no longer inherited in hrde-1 mutants (13/463) (Figure 4B and Table S5), in line with HRDE-1’s role in transgenerational shuttling of heritable small RNAs. 25.9% of these inherited STGs were previously shown to physically bind HRDE-1 (3.7-fold enrichment, p value < 1.139 × 10−40) (Buckley et al., 2012). In contrast to the clustering pattern of the STGs that was observed in N2 animals, in hrde-1 animals the “fed” parents clustered with the “F3 of starved worms,” separately from the “starved” parents. These patterns are consistent with the reduced small RNA inheritance in hrde-1 mutants. In rde-4 animals, no clear clustering pattern was observed, possibly because of RDE-4’s upstream role in biogenesis of small RNAs (Figure S5) (Gent et al., 2010; Vasale et al., 2010).
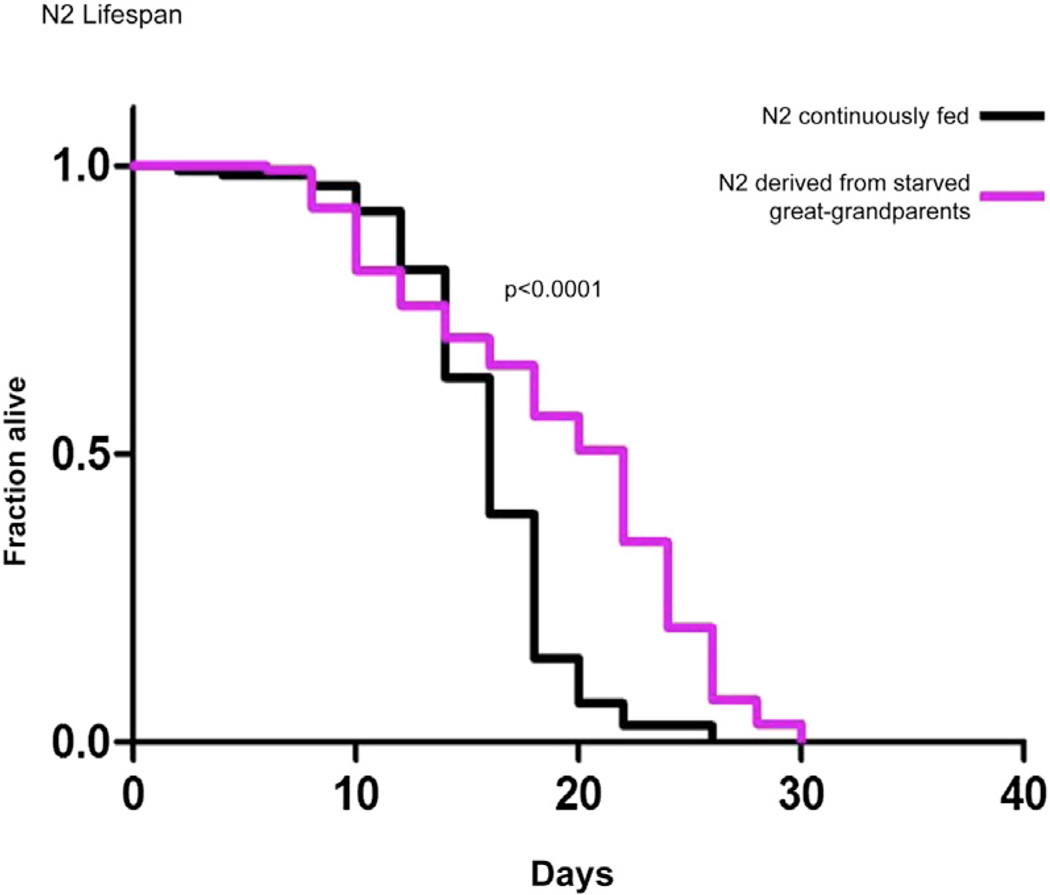
To assess whether starvation also alters the physiology of the progeny of starved parents, we performed lifespan assays. We compared the lifespan of animals whose great-grandparents had experienced starvation at the first larval stage with animals whose great-grandparents were continuously fed. In three independent experiments, we observed a significant increase in the lifespan of animals whose ancestors had experienced starvation (37% increase, p value < 0.0001; 70% increase, p value < 0.0001; 22% increase, p value = 0.004, Figure 5 and Figure S6). At this point, we do not know whether inherited RNA molecules are responsible for this lifespan effect, but the lifespan effect independently corroborates the notion that there is a transgenerational memory of dietary history.
Figure 5. Animals with a Starvation History Show an Extended Lifespan.
A comparison of the lifespan of N2 worms whose great-grandparents had experienced starvation at the first larval stage with animals whose great-grandparents were continuously fed. The x axis shows lifespan in days of adulthood. The y axis shows the fraction of worms alive. N2 derived from starved great-grandparents show extended lifespan in comparison to N2 that were continuously fed over generations (live 37% longer, log-rank, p < 0.0001). Each of the lifespan experiments was performed three times (data from two additional experiments are shown in Figure S6).
DISCUSSION
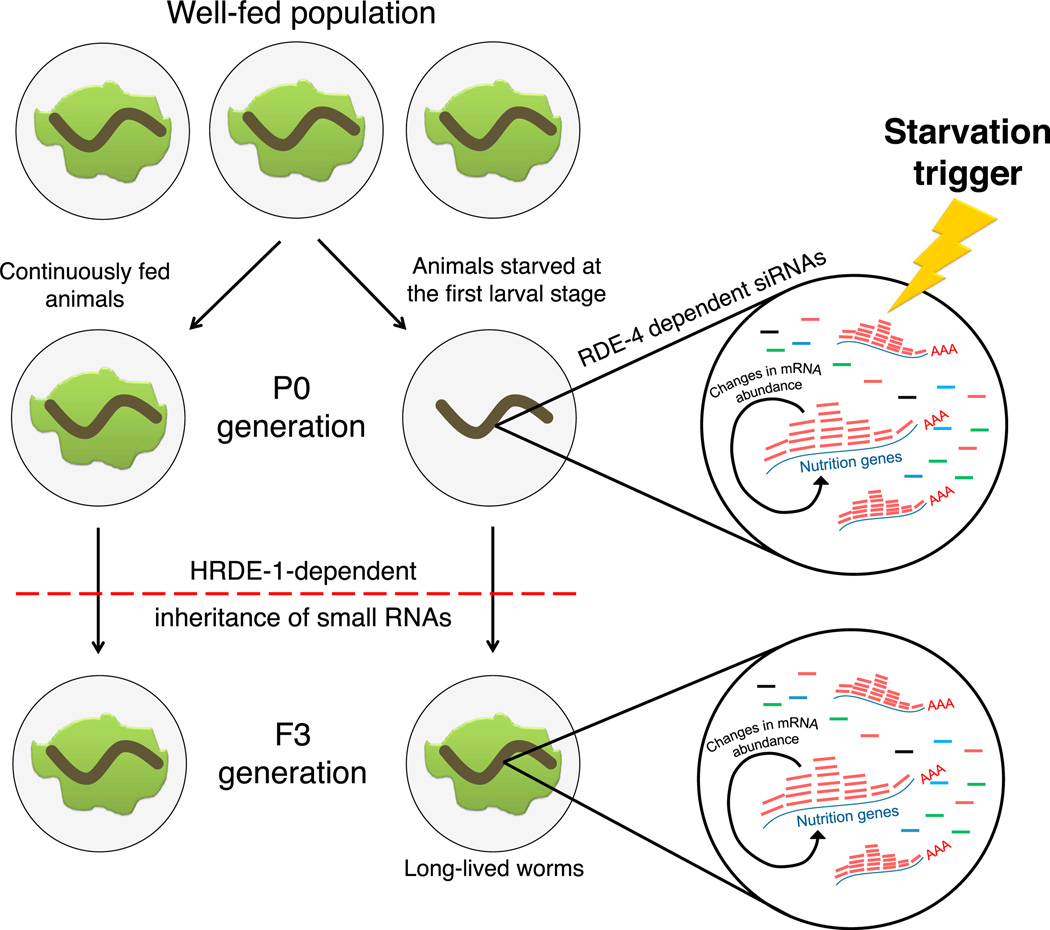
We discovered that changes in small RNA (22Gs) levels that follow L1-starvation-induced developmental arrest are passed on for at least three generations (Figure 6). While it was previously shown that RNAi could act transgenerationally, in all the previous studies heritable silencing was directed against foreign DNA introduced into the worm. This is the first study that demonstrates that an endogenous response could trigger small RNA inheritance. The targets of these inherited small RNAs include sets of genes involved in nutrition. This epigenetic response is compromised in rde-4 mutants, and transgenerational transmission of the starvation-induced small RNAs depends on the germ-line-expressed nuclear argonaute HRDE-1.
Figure 6. A Model for L1 Starvation-Induced Transgenerational Inheritance.
Food (bacterial lawn) is shown in dark green. Starvation-induced transcripts, which are transcribed either during or after worms experience L1 starvation, enable the production or depletion of specific RDE-4-dependent small RNAs, which regulate the levels of their mRNA targets (involved in nutrition). The production of the heritable small RNAs might be initiated in the germline, or, if endogenous small RNAs act systemically, it could originate also in somatic tissues. The changes in small RNAs persist in the absence of the trigger (starvation) to the next generations. The germline-expressed argonaute, HRDE-1, carries the heritable 22G small RNAs in the germline.
Some small RNAs in C. elegans, for example PIWI-interacting RNAs (piRNAs) and microRNAs, are independent transcriptional units regulated by dedicated promoters (Bagijn et al., 2012; Grishok, 2013). Although the biogenesis mechanism is at the moment poorly understood, endo-siRNAs in general, and RDE-4-dependent 26G and 22G endo-siRNAs in particular, are not independently transcribed but originate from mRNA transcripts (Billi et al., 2014). 26Gs and 22Gs are antisense sequences that are produced as short independent transcripts by nonprocessive RdRPs (26Gs by RRF-3 and 22Gs by RRF-1 and EGO-1) that use spliced mRNAs as a template. Some, but not all of the amplified 22G directly depend on primary 26Gs that prime their production (Grishok, 2013). Thus, it is possible that the mRNA response, either during or following severe L1 starvation, triggers the starvation-induced changes in endo-siRNAs, simply because endo-siRNAs levels are determined based on the availability of the cognate mRNA template. Alternatively, changes in small RNA production could be, in theory, initiated by starvation-induced expression or modification of particular biogenesis protein factors or by starvation-induced bidirectional transcription that would result in dsRNA formation, which could serve as substrate for small RNA production. Irrespective of the nature of the initiation mechanism, our findings suggests that a transient trigger (L1-starvation) is memorized by the generation of an ensemble of small RNAs which are then available to target mRNAs in ensuing generations in the absence of the original starvation trigger (Figure 6).
Although we observed an enrichment for heritable targeting of germline genes, in accordance with previous studies on gene targets of HRDE-1-bound 22Gs (Buckley et al., 2012), many genes that were found to be targeted by heritable small RNAs were not previously found to be expressed in the germline. We observed that a large percentage of the genes (52%) which are targeted by both 26G and 22G following starvation were previously annotated as ERGO-1 targets, and it was shown that although ERGO-1-bound small RNAs are produced during oogenesis, they mostly regulate zygotic genes that are not expressed in the germline (Han et al., 2009; Reinke et al., 2004). It is not clear whether endogenous small RNAs, similarly to exogenous ones (Fire et al., 1998), move systemically in C. elegans; if this is the case, it is also possible that certain small RNAs were transmitted from the soma to the germline.
It is not known which features in a given mRNA transcript render it suitable for serving as a template for the production of 26G or 22G small RNAs. Presumably secondary structures (perhaps simply dsRNA), induce the recruitment of small RNA biogenesis factors (Billi et al., 2014). Hence, in theory, for each starvation-regulated gene, there could be a different biogenesis pathway that produces the specific starvation-provoked small RNAs. We found that genes that are involved in nutrition, and most notably all the vitellogenin genes, are under heritable small RNA control. Since on the complementary strands of all the vitellogenin genes there are genes that are transcribed in the opposite direction, we hypothesize that cotranscription and ensuing formation of dsRNA are the trigger for small RNA formation in this case (Figure S4). Regulation of the levels of yolk proteins that provide resources to the egg (DePina et al., 2011) by cotranscription-induced small RNA production upon starvation, could be a method to monitor the amount of food that is allocated to the next generations. Similarly, heritable small RNAs were found to target multiple genes that are involved in fat regulation, protein turnover, stress resistance, longevity, and reproduction, all with the potential of affecting the survival of the progeny (Table S6).
Since heritable small RNAs are hypothesized to act as memory of “self” and “nonself” genes (Rechavi, 2013), it was previously suggested the endo-siRNAs “buffer” the genome against new or foreign genes that might be deleterious (Fischer et al., 2011; Vasale et al., 2010). In addition to upregulation of inherited endo-siRNAs, we observed, upon stress, an inherited downregulation of many endo-siRNAs. It is possible that releasing silenced genes from endo-siRNA-mediated repression is a bet-hedging strategy, which could benefit the progeny if some of these “dangerous” and otherwise silent genes prove useful in overcoming stress such as food deprivation.
In parallel to the characterization of the inherited small RNA response, we also observed that F3 progeny that were derived from L1-starved parents live longer. The putative gene targets of the transgenerationally inherited small RNAs were enriched for functions that are related to aging. The inherited increased longevity could be directly dependent on gene regulation that is enforced by the inherited small RNAs (for example, the kin-29, gcy-23, polg-1, coq-3, and aco-1 genes, were shown to affect lifespan and are targeted by heritable small RNAs), or, since different small RNA species compete over common limiting resources (for example, binding to shared proteins), it could indirectly stem from a tilt in the balance within the small RNA pool, or by a heritable reduction in the productivity of the germline. Future studies will reveal if L1-starvation-induced and inherited small RNA gene regulation is related to the observed heritable increased longevity, and whether these transgenerational effects prepare the progeny for similar famines, or are adaptive in other ways.
Over the course of evolution worms and humans specialized in different regulatory epigenetic processes. For example, C. elegans have an expanded argonaute family (Yigit et al., 2006), and RdRP-mediated small RNAs amplification (Sijen et al., 2001; Smardon et al., 2000), but no DNA methylation (Simpson et al., 1986). Thus, until the proper experiments are performed, it cannot be assumed that the mechanism for memorizing starvations that we describe here is the mean by which mammals achieve the same end. Nevertheless, it was recently found in mice that traumatic depression leaves a trace of microRNAs in sperm (Gapp et al., 2014). Therefore, it is of great importance to study whether noncoding RNAs play a role in the epigenetic effects that were observed following starvation in humans as well.
EXPERIMENTAL PROCEDURES
Small RNA Library Preparation and Sequencing
All the experiments were done three times (independent biological triplicates). Worms were lysed using the TRIzol reagent (Life Technologies), repetitive freezing, thawing, and vortexing. The total RNA samples were treated with tobacco acid pyrophosphatase (Epicenter), to ensure 5′ monophosphate-independent capturing of small RNAs. Libraries were prepared using the TruSeq Small RNA protocol (illumina) according to the manufacturer’s protocol. The resulting cDNAs were separated on a 4% agarose E-Gel (Invitrogen, Life Technologies), and the 140–160 nt length species were selected. cDNA was purified using the MinElute Gel Extraction kit (QIAGEN). Libraries were sequenced using an Illumina HighSeq2500 instrument.
mRNA Library Preparation and Sequencing
Total RNA was isolated using the TRIzol reagent (Life Technologies), and libraries were prepared using the Low Sample (LS) protocol (Illumina) according to the manufacturer’s protocol. Libraries were sequenced using an Illumina HighSeq2500 instrument.
Worm Strains and L1 Starvation Protocol
Standard culture techniques were used to maintain the nematodes on NGM plates seeded with OP50 bacteria. Strains employed in this work were as follows: The wild-type strain (Bristol N2), WM35 rde-4(ne299) unc-69(e587), and YY538: hrde-1 (tm1200). The nematodes were kept fed for at least five generations before the beginning of each experiment. Synchronization of the worms at the L1 stage was achieved using “egg prepping.” Half of the eggs were transferred to NGM plates with OP50 bacterial lawn (“fed” condition) and the other half to NGM plates without food (“starved” worms). Worms that hatched on plates without food were kept starved as L1s for 6 days. The starved worms were then collected using M9 buffer and transferred to NGM plates containing food. All worms (“fed” and “starved”) were collected for sequencing as young adults 2 days after they reached the L4 stage.
Small RNA-seq Analysis
For details see the Extended Experimental Procedures. In brief, the adaptor sequences were removed using CutAdapt. Clipped reads were mapped to WormBase 220 version of the genome (WS220) using Bowtie2 (v. 2.1.0). Reads which aligned antisense to genes were counted using htseq-count and Ensembl-based gff file (version 66). We used DESeq2, an R package, to determine differential expression of STGs, and considered an STG to be differentially expressed if its FDR value was less than 0.1. Hierarchical clustering was done using the pvclust package.
RNA-seq Analysis
Reads were mapped to WormBase 220 version of the genome (WS220) using Bowtie (v. 2.1.0). Reads were counted using htseq-count and Ensembl-based gff file (version 66) and normalized for RPM (reads per million). We considered a gene to be differentially expressed if its fold-changed was greater than 1.5 and it had total reads of more than 10 RPM per comparison.
Lifespan Assays
N2 worms in the late L4 stage growing at 20°C were transferred to fresh NGM plates with OP50 bacterial lawn. The first day of adulthood is day 1 in the survival curves. Animals were scored as “alive,” “dead,” or “lost” every other day. Animals that failed to display touch-provoked movement were scored as “dead.” Animals that died from causes other than aging, such as sticking to the plate walls, were scored as “lost.” Animals were transferred to fresh plates every 2 days. Each assay consisted of 8 NGM plates with 15 worms in each plate and every experiment was conducted three times. All lifespan experiments were performed at 20°C. Survival curves were plotted and statistical analyses (log-rank tests) were performed using the SPSS V 15 software.
Supplementary Material
ACKNOWLEDGMENTS
We thank Yoav Benjamini for help with the statistical methods. We thank Scott Kennedy for providing the hrde-1 strain. We also thank Assaf Gordon, Michal lineal, Shelly Mahlab, and all members of the Rechavi and Hobert lab for their helpful comments. Oded Rechavi, Leah Houri-Ze’evi, and Sarit Anava were supported by funds from the Alon, Bikura, and Yad Hanadiv fellowships, and by the CBRC, Teva NNE, ISF, and ERC grants. This work was supported by the Howard Hughes Medical Institute.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes Extended Experimental Procedures, six figures, and six tables and can be found with this article online at http://dx.doi.org/10.1016/j.cell.2014.06.020.
REFERENCES
- Billi AC, Fischer SEJ, Kim JK. Endogenous RNAi pathways in C. elegans. WormBook, ed. The C. elegans Research Community, WormBook. 2014. http://dx.doi.org/10.1895/wormbook.1.170.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashe A, Sapetschnig A, Weick E-M, Mitchell J, Bagijn MP, Cording AC, Doebley A-L, Goldstein LD, Lehrbach NJ, Le Pen J, et al. piRNAs can trigger a multigenerational epigenetic memory in the germline of C. elegans. Cell. 2012;150:88–99. doi: 10.1016/j.cell.2012.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagijn MP, Goldstein LD, Sapetschnig A, Weick E-M, Bouasker S, Lehrbach NJ, Simard MJ, Miska EA. Function, targets, and evolution of Caenorhabditis elegans piRNAs. Science. 2012;337:574–578. doi: 10.1126/science.1220952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baugh LR. To grow or not to grow: nutritional control of development during Caenorhabditis elegans L1 arrest. Genetics. 2013;194:539–555. doi: 10.1534/genetics.113.150847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billi AC, Alessi AF, Khivansara V, Han T, Freeberg M, Mitani S, Kim JK. The Caenorhabditis elegans HEN1 ortholog, HENN-1, methylates and stabilizes select subclasses of germline small RNAs. PLoS Genet. 2012;8:e1002617. doi: 10.1371/journal.pgen.1002617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley BA, Burkhart KB, Gu SG, Spracklin G, Kershner A, Fritz H, Kimble J, Fire A, Kennedy S. A nuclear Argonaute promotes multigenerational epigenetic inheritance and germline immortality. Nature. 2012;489:447–451. doi: 10.1038/nature11352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton NO, Burkhart KB, Kennedy S. Nuclear RNAi maintains heritable gene silencing in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA. 2011;108:19683–19688. doi: 10.1073/pnas.1113310108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bygren LO, Kaati G, Edvinsson S. Longevity determined by paternal ancestors’ nutrition during their slow growth period. Acta Biotheor. 2001;49:53–59. doi: 10.1023/a:1010241825519. [DOI] [PubMed] [Google Scholar]
- Claycomb JM, Batista PJ, Pang KM, Gu W, Vasale JJ, van Wolfswinkel JC, Chaves DA, Shirayama M, Mitani S, Ketting RF, et al. The Argonaute CSR-1 and its 22G-RNA cofactors are required for holocentric chromosome segregation. Cell. 2009;139:123–134. doi: 10.1016/j.cell.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conine CC, Moresco JJ, Gu W, Shirayama M, Conte D, Jr, Yates JR, 3rd, Mello CC. Argonautes promote male fertility and provide a paternal memory of germline gene expression in C. elegans. Cell. 2013;155:1532–1544. doi: 10.1016/j.cell.2013.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePina AS, Iser WB, Park S-S, Maudsley S, Wilson MA, Wolkow CA. Regulation of Caenorhabditis elegans vitellogenesis by DAF-2/IIS through separable transcriptional and posttranscriptional mechanisms. BMC Physiol. 2011;11:11. doi: 10.1186/1472-6793-11-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Fischer SEJ, Montgomery TA, Zhang C, Fahlgren N, Breen PC, Hwang A, Sullivan CM, Carrington JC, Ruvkun G. The ERI-6/7 helicase acts at the first stage of an siRNA amplification pathway that targets recent gene duplications. PLoS Genet. 2011;7:e1002369. doi: 10.1371/journal.pgen.1002369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gapp K, Jawaid A, Sarkies P, Bohacek J, Pelczar P, Prados J, Farinelli L, Miska E, Mansuy IM. Implication of sperm RNAs in transgenerational inheritance of the effects of early trauma in mice. Nat. Neurosci. 2014;17:667–669. doi: 10.1038/nn.3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gent JI, Lamm AT, Pavelec DM, Maniar JM, Parameswaran P, Tao L, Kennedy S, Fire AZ. Distinct phases of siRNA synthesis in an endogenous RNAi pathway in C. elegans soma. Mol. Cell. 2010;37:679–689. doi: 10.1016/j.molcel.2010.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer EL, Maures TJ, Ucar D, Hauswirth AG, Mancini E, Lim JP, Benayoun BA, Shi Y, Brunet A. Transgenerational epigenetic inheritance of longevity in Caenorhabditis elegans. Nature. 2011;479:365–371. doi: 10.1038/nature10572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishok A. Biology and Mechanisms of Short RNAs in Caenorhabditis elegans. Adv. Genet. 2013;83:1–69. doi: 10.1016/B978-0-12-407675-4.00001-8. [DOI] [PubMed] [Google Scholar]
- Gu W, Shirayama M, Conte D, Jr, Vasale J, Batista PJ, Claycomb JM, Moresco JJ, Youngman EM, Keys J, Stoltz MJ, et al. Distinct argonaute-mediated 22G-RNA pathways direct genome surveillance in the C. elegans germline. Mol. Cell. 2009;36:231–244. doi: 10.1016/j.molcel.2009.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu SG, Pak J, Guang S, Maniar JM, Kennedy S, Fire A. Amplification of siRNA in Caenorhabditis elegans generates a transgenerational sequence-targeted histone H3 lysine 9 methylation footprint. Nat. Genet. 2012;44:157–164. doi: 10.1038/ng.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall SE, Chirn G-W, Lau NC, Sengupta P. RNAi pathways contribute to developmental history-dependent phenotypic plasticity in C. elegans. RNA. 2013;19:306–319. doi: 10.1261/rna.036418.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han T, Manoharan AP, Harkins TT, Bouffard P, Fitzpatrick C, Chu DS, Thierry-Mieg D, Thierry-Mieg J, Kim JK. 26G endo-siRNAs regulate spermatogenic and zygotic gene expression in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA. 2009;106:18674–18679. doi: 10.1073/pnas.0906378106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey SC, Orbidans HE. All eggs are not equal: the maternal environment affects progeny reproduction and developmental fate in Caenorhabditis elegans. PLoS ONE. 2011;6:e25840. doi: 10.1371/journal.pone.0025840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heard E, Martienssen RA. Transgenerational epigenetic inheritance: myths and mechanisms. Cell. 2014;157:95–109. doi: 10.1016/j.cell.2014.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellwig S, Bass BL. A starvation-induced noncoding RNA modulates expression of Dicer-regulated genes. Proc. Natl. Acad. Sci. USA. 2008;105:12897–12902. doi: 10.1073/pnas.0805118105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jablonka E, Lamb MJ. Soft inheritance: challenging the modern synthesis. Genet. Mol. Biol. 2008;31:389–395. [Google Scholar]
- Kaati G, Bygren LO, Pembrey M, Sjöström M. Transgenerational response to nutrition, early life circumstances and longevity. Eur. J. Hum. Genet. 2007;15:784–790. doi: 10.1038/sj.ejhg.5201832. [DOI] [PubMed] [Google Scholar]
- Kamminga LM, van Wolfswinkel JC, Luteijn MJ, Kaaij LJT, Bagijn MP, Sapetschnig A, Miska EA, Berezikov E, Ketting RF. Differential impact of the HEN1 homolog HENN-1 on 21U and 26G RNAs in the germline of Caenorhabditis elegans. PLoS Genet. 2012;8:e1002702. doi: 10.1371/journal.pgen.1002702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Wolf YI. Is evolution Darwinian or/and Lamarckian? Biol. Direct. 2009;4:42. doi: 10.1186/1745-6150-4-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RC, Hammell CM, Ambros V. Interacting endogenous and exogenous RNAi pathways in Caenorhabditis elegans. RNA. 2006;12:589–597. doi: 10.1261/rna.2231506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H-C, Gu W, Shirayama M, Youngman E, Conte D, Jr, Mello CC. C. elegans piRNAs mediate the genome-wide surveillance of germline transcripts. Cell. 2012;150:78–87. doi: 10.1016/j.cell.2012.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansisidor AR, Cecere G, Hoersch S, Jensen MB, Kawli T, Kennedy LM, Chavez V, Tan M-W, Lieb JD, Grishok A. A conserved PHD finger protein and endogenous RNAi modulate insulin signaling in Caenorhabditis elegans. PLoS Genet. 2011;7:e1002299. doi: 10.1371/journal.pgen.1002299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell CS, Antoshechkin I, Kurhanewicz N, Belsky JA, Baugh LR. Nutritional control of mRNA isoform expression during developmental arrest and recovery in C. elegans. Genome Res. 2012;22:1920–1929. doi: 10.1101/gr.133587.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meloche S, Roux PP. F-box proteins elongate translation during stress recovery. Sci. Signal. 2012;5:pe25. doi: 10.1126/scisignal.2003163. [DOI] [PubMed] [Google Scholar]
- Montgomery TA, Rim Y-S, Zhang C, Dowen RH, Phillips CM, Fischer SEJ, Ruvkun G. PIWI associated siRNAs and piRNAs specifically require the Caenorhabditis elegans HEN1 ortholog henn-1. PLoS Genet. 2012;8:e1002616. doi: 10.1371/journal.pgen.1002616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Painter RC, Osmond C, Gluckman P, Hanson M, Phillips DIW, Roseboom TJ. Transgenerational effects of prenatal exposure to the Dutch famine on neonatal adiposity and health in later life. BJOG. 2008;115:1243–1249. doi: 10.1111/j.1471-0528.2008.01822.x. [DOI] [PubMed] [Google Scholar]
- Pak J, Fire A. Distinct populations of primary and secondary effectors during RNAi in C. elegans. Science. 2007;315:241–244. doi: 10.1126/science.1132839. [DOI] [PubMed] [Google Scholar]
- Rechavi O. Guest list or black list: heritable small RNAs as immunogenic memories. Trends Cell Biol. 2013;24:212–220. doi: 10.1016/j.tcb.2013.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rechavi O, Minevich G, Hobert O. Transgenerational inheritance of an acquired small RNA-based antiviral response in C. elegans. Cell. 2011;147:1248–1256. doi: 10.1016/j.cell.2011.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinke V, Gil IS, Ward S, Kazmer K. Genome-wide germline-enriched and sex-biased expression profiles in Caenorhabditis elegans. Development. 2004;131:311–323. doi: 10.1242/dev.00914. [DOI] [PubMed] [Google Scholar]
- Remy J-J. Stable inheritance of an acquired behavior in Caenorhabditis elegans. Curr. Biol. 2010;20:R877–R878. doi: 10.1016/j.cub.2010.08.013. [DOI] [PubMed] [Google Scholar]
- Remy J-J, Hobert O. An interneuronal chemoreceptor required for olfactory imprinting in C. elegans. Science. 2005;309:787–790. doi: 10.1126/science.1114209. [DOI] [PubMed] [Google Scholar]
- Seth M, Shirayama M, Gu W, Ishidate T, Conte D, Jr, Mello CC. The C. elegans CSR-1 argonaute pathway counteracts epigenetic silencing to promote germline gene expression. Dev. Cell. 2013;27:656–663. doi: 10.1016/j.devcel.2013.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirayama M, Seth M, Lee H-C, Gu W, Ishidate T, Conte D, Jr, Mello CC. piRNAs initiate an epigenetic memory of nonself RNA in the C. elegans germline. Cell. 2012;150:65–77. doi: 10.1016/j.cell.2012.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sijen T, Fleenor J, Simmer F, Thijssen KL, Parrish S, Timmons L, Plasterk RH, Fire A. On the role of RNA amplification in dsRNA-triggered gene silencing. Cell. 2001;107:465–476. doi: 10.1016/s0092-8674(01)00576-1. [DOI] [PubMed] [Google Scholar]
- Simpson VJ, Johnson TE, Hammen RF. Caenorhabditis elegans DNA does not contain 5-methylcytosine at any time during development or aging. Nucleic Acids Res. 1986;14:6711–6719. doi: 10.1093/nar/14.16.6711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smardon A, Spoerke JM, Stacey SC, Klein ME, Mackin N, Maine EM. EGO-1 is related to RNA-directed RNA polymerase and functions in germ-line development and RNA interference in C. elegans. Curr. Biol. 2000;10:169–178. doi: 10.1016/s0960-9822(00)00323-7. [DOI] [PubMed] [Google Scholar]
- Song S, Wang W, Hu P. Famine, death, and madness: schizophrenia in early adulthood after prenatal exposure to the Chinese Great Leap Forward Famine. Soc. Sci. Med. 2009;68:1315–1321. doi: 10.1016/j.socscimed.2009.01.027. [DOI] [PubMed] [Google Scholar]
- Stanner SA, Bulmer K, Andrés C, Lantseva OE, Borodina V, Poteen VV, Yudkin JS. Does malnutrition in utero determine diabetes and coronary heart disease in adulthood? Results from the Leningrad siege study, a cross sectional study. BMJ. 1997;315:1342–1348. doi: 10.1136/bmj.315.7119.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterken MG, Snoek LB, Bosman KJ, Daamen J, Riksen JAG, Bakker J, Pijlman GP, Kammenga JE. A heritable antiviral RNAi response limits Orsay virus infection in Caenorhabditis elegans N2. PLoS ONE. 2014;9:e89760. doi: 10.1371/journal.pone.0089760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasale JJ, Gu W, Thivierge C, Batista PJ, Claycomb JM, Youngman EM, Duchaine TF, Mello CC, Conte D., Jr Sequential rounds of RNA-dependent RNA transcription drive endogenous small-RNA biogenesis in the ERGO-1/Argonaute pathway. Proc. Natl. Acad. Sci. USA. 2010;107:3582–3587. doi: 10.1073/pnas.0911908107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vastenhouw NL, Brunschwig K, Okihara KL, Müller F, Tijsterman M, Plasterk RHA. Gene expression: long-term gene silencing by RNAi. Nature. 2006;442:882. doi: 10.1038/442882a. [DOI] [PubMed] [Google Scholar]
- Wedeles CJ, Wu MZ, Claycomb JM. Protection of germline gene expression by the C. elegans Argonaute CSR-1. Dev. Cell. 2013;27:664–671. doi: 10.1016/j.devcel.2013.11.016. [DOI] [PubMed] [Google Scholar]
- Yigit E, Batista PJ, Bei Y, Pang KM, Chen C-CG, Tolia NH, Joshua-Tor L, Mitani S, Simard MJ, Mello CC. Analysis of the C. elegans Argonaute family reveals that distinct Argonautes act sequentially during RNAi. Cell. 2006;127:747–757. doi: 10.1016/j.cell.2006.09.033. [DOI] [PubMed] [Google Scholar]
- Zhang X, Zabinsky R, Teng Y, Cui M, Han M. microRNAs play critical roles in the survival and recovery of Caenorhabditis elegans from starvation-induced L1 diapause. Proc. Natl. Acad. Sci. USA. 2011;108:17997–18002. doi: 10.1073/pnas.1105982108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.