Abstract
We summarized published data on the associations of apolipoprotein E (APOE) gene ε2/ε3/ε4 polymorphism with both cancer risk and circulating lipid profiles, aiming to examine the causal relevance between lipids and cancer risk. Article identification and data abstraction were conducted in duplicate and independently by two authors. Data were analyzed by STATA software. Twenty-five articles that examined the associations of APOE gene ε2/ε3/ε4 polymorphism with either cancer risk (n = 22) or circulating lipid changes (n = 4) were eligible. The presence of ε2 and ε4 alleles showed no overall associations with overall cancer risk when compared with ε3 allele. The ε4 allele was significantly associated with 1.40-fold (odds ratio or OR = 1.40; 95% confidence interval or CI: 1.00–1.94; P = 0.047) increased risk of developing cancer in Asian populations, and the presence of heterogeneity was low (I2 = 37.6%). Carriers of ε3/ε4 genotype had a significant reduction in circulating HDL-C (WMD = −2.62; 95% CI: −4.19 to −1.04; P = 0.001) without heterogeneity (I2 = 16.6%). The predicted odds of having cancer for 1 mg/dL reduction in circulating HDL-C was 1.14 (95% CI: 1.00 to 1.89). The findings of this Mendelian randomization meta-analysis demonstrate that reduced circulating HDL-C might be a potentially causal risk factor for the development of overall cancer in Asians.
Some observational studies have revealed that people with low circulating cholesterol level tended to be more susceptible to many malignancies, such as lung cancer and breast cancer1,2. As a central regulator in cholesterol metabolism, apolipoprotein E (APOE) is increasingly recognized as playing a potent inhibitory role in angiogenesis and cancer cell growth3. It has been estimated that close to 60% of circulating cholesterol variation is under genetic control, and thereof 14% variation is attributable to APOE genetic defects4. The genomic sequence of human APOE (gene ID: 348, 19q13.2) is polymorphic at two nucleotides, which yields 3 alleles (ε2, ε3, ε4) and 6 genotypes (ε2/ε2, ε2/ε3, ε3/ε3, ε2/ε4, ε3/ε4, ε4/ε4), with diverse receptor-binding capabilities5. As evidenced, this capability was proven to be defective for the ε2 allele with its carriers exhibiting lower circulating cholesterol level and higher triglyceride level when compared with ε3 homozygotes; in contrast, circulating total cholesterol and low-density lipoprotein cholesterol appear to be higher in those with ε4 allele6. In spite of exhaustive investigations, published data on the associations between APOE gene ε2/ε3/ε4 polymorphism and cancer risk are conflicting and inconclusive5,7,8,9. A recent meta-analysis by Anand et al who examined this association in 16 studies failed to detect any positive signal except in cohort studies9. However, they did not compare the changes of circulating lipid levels across APOE gene ε2/ε3/ε4 genotypes, which would be of importance to provide background data to infer causality between circulating lipids and cancer risk. To fill this gap in knowledge and generate added information, we revisited this topic and summarized the associations of APOE gene ε2/ε3/ε4 polymorphism with both cancer risk and circulating lipid profiles in a large meta-analysis implementing Mendelian randomization technique.
Results
Eligible articles
Of 530 potentially relevant articles identified according to our search strategy, 25 articles that examined the associations of APOE gene ε2/ε3/ε4 polymorphism with either cancer risk (n = 22) or circulating lipid changes (n = 4) were eligible according to the predefined inclusion and exclusion criteria5,7,8,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31. The first article was published in 199610. The total sample size ranged from 78 in McDonald et al study31 to 74033 in Benn et al study27. For 22 APOE-cancer association articles with 26 independent studies, there were 13478 cancer patients and 77592 controls in total. For 4 APOE-lipids association studies, data provided in both cancer patients and controls were analyzed separately, resulting in 6 independent studies for TG and 7 studies respectively for TC, HDL-C and LDL-C.
Host characteristics
Baseline host characteristics of study populations for the associations of APOE gene ε2/ε3/ε4 polymorphism with cancer risk and circulating lipid changes are presented in Table 1 and Table 2, respectively.
Table 1. Baseline characteristics of all study populations.
| Author (year) | Cancer type | Race | Source | Design | Match | Sample size | Age (years) | Males | BMI (kg/m2) | Smoking | Family history | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cases | Cont's | Cases | Cont's | Cases | Cont's | Cases | Cont's | Cases | Cont's | Cases | Cont's | ||||||
| Cibeira (2014) | Breast | Latinos | PB | PS | YES | 47 | 165 | 57.6 | 56.1 | 0.00 | 0.00 | 28.2 | 28.9 | NA | NA | NA | NA |
| McDonald (2013) | Breast | White | HB | RS | NA | 54 | 24 | 51.2 | 47.0 | 0.00 | 0.00 | NA | NA | NA | NA | NA | NA |
| Ahn (2012) | Hepatocellular | Asian | PB | RS | NO | 59 | 47 | 53.9 | 45.3 | 0.76 | 0.66 | NA | NA | NA | NA | NA | NA |
| De Feo (2012) | Gastric | White | HB | RS | NO | 156 | 444 | 67.1 | 59.0 | 0.53 | 0.59 | NA | NA | 0.49 | 0.46 | 0.38 | 0.29 |
| Wu (2012) | Breast | Asian | HB | RS | NO | 306 | 300 | 48.5 | 41.3 | 0.00 | 0.00 | NA | NA | NA | NA | NA | NA |
| Benn (2011) | All | White | PB | PS | YES | 6816 | 67217 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| Kulminski (2011) | All | Mixed | PB | PS | NA | 701 | 895 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| De Feo (2010) | Head and neck | White | HB | RS | NO | 417 | 436 | 63.1 | 59.3 | 0.81 | 0.58 | NA | NA | 0.86 | 0.43 | 0.31 | 0.21 |
| Porrata-Doria (2010) | Breast | Latinos | HB | RS | YES | 63 | 106 | 43.0 | 41.7 | 0.00 | 0.00 | 26.6 | 26.5 | 0.14 | 0.12 | 0.43 | 0.49 |
| Porrata-Doria (2010) | Breast | Latinos | HB | RS | YES | 142 | 123 | 63.8 | 61.8 | 0.00 | 0.00 | 27.8 | 27.9 | 0.16 | 0.13 | 0.35 | 0.40 |
| Souza (2009) | Colorectal | Latinos | HB | RS | YES | 87 | 73 | 60.6 | 61.6 | 0.47 | 0.44 | NA | NA | 0.49 | 0.48 | NA | NA |
| Kulminski (2008) | Colorectal | White | PB | RS | YES | 77 | 1644 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| Chang (2006) | Breast | Asian | HB | RS | YES | 291 | 148 | 49.5 | 50.9 | 0.00 | 0.00 | NA | NA | NA | NA | NA | NA |
| Slattery (2005) | Colorectal | Mixed | PB | RS | YES | 1556 | 1948 | NA | NA | 0.56 | 0.53 | 27.8 | 26.8 | NA | NA | 0.16 | 0.09 |
| Slattery (2005) | Colorectal | Mixed | PB | RS | YES | 777 | 988 | NA | NA | 0.59 | 0.57 | 27.8 | 27.4 | NA | NA | 0.11 | 0.08 |
| Chang (2005) | Breast | Asian | HB | RS | NO | 290 | 232 | 47.4 | 40.2 | 0.00 | 0.00 | NA | NA | NA | NA | NA | NA |
| Menzel (2004) | Breast | White | PB | RS | NO | 220 | 400 | 56.0 | 39.0 | 0.00 | 0.00 | NA | NA | NA | NA | NA | NA |
| Menzel (2004) | Breast | White | PB | RS | NA | 190 | 231 | 58.0 | 60.0 | 0.00 | 0.00 | NA | NA | NA | NA | NA | NA |
| Watson (2003) | Colorectal | White | HB | RS | NO | 206 | 353 | 68.7 | 60.6 | 0.60 | 0.67 | NA | NA | 0.41 | 0.50 | NA | NA |
| Butler (2001) | Colorectal | White | HB | RS | NO | 167 | 200 | 70.0 | 51.0 | 0.52 | 0.52 | NA | NA | NA | NA | NA | NA |
| Wessel (2001) | Prostate | White | HB | RS | NA | 230 | 798 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| Moysich (2000) | Breast | White | PB | RS | YES | 260 | 332 | 56.9 | 58.0 | 0.00 | 0.00 | 25.5 | 25.5 | NA | NA | 0.16 | 0.08 |
| Liestol (2000) | All | Mixed | HB | RS | NA | 71 | 126 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| Haapala (2000) | Prostate | White | HB | RS | NA | 38 | 163 | NA | NA | 1.00 | 1.00 | NA | NA | NA | NA | NA | NA |
| Kervinen (1996) | Colorectal | White | HB | RS | NO | 122 | 199 | 67.2 | 57.8 | 0.44 | 0.88 | NA | NA | NA | NA | NA | NA |
| Kervinen (1996) | Colorectal | White | HB | RS | NO | 135 | 199 | 62.9 | 57.8 | 0.62 | 0.88 | NA | NA | NA | NA | NA | NA |
Abbreviations: PB, population-based; HB, hospital-based; PS, prospective; RS, retrospective; BMI, body mass index; NA, not available.
Table 2. Distributions of circulating lipids across APOE gene ε2/ε3/ε4 genotypes in all qualified studies.
| Author (year) | Cancer type | Race | Status | Lipids (mg/dL) | ε2/3 | ε3/3 | ε3/4 | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Mean | SD | Mean | SD | Mean | SD | |||||
| Cibeira (2014) | Breast | Latinos | Cases | HDL | 51.00 | 14.40 | 48.40 | 12.10 | 50.20 | 15.20 |
| Cibeira (2014) | Breast | Latinos | Controls | HDL | 57.20 | 22.70 | 53.60 | 11.10 | 50.10 | 13.60 |
| Trompet (2009) | All types | White | Both | HDL | 50.66 | 14.09 | 49.50 | 16.26 | 47.95 | 9.74 |
| Souza (2009) | Colorectal | Latinos | Cases | HDL | 33.30 | 8.30 | 41.90 | 16.10 | 34.40 | 13.50 |
| Souza (2009) | Colorectal | Latinos | Controls | HDL | 52.10 | 15.40 | 43.40 | 13.70 | 45.10 | 14.90 |
| Moysich (2000) | Breast | White | Cases | HDL | 53.00 | 14.00 | 54.00 | 15.00 | 49.00 | 12.00 |
| Moysich (2000) | Breast | White | Controls | HDL | 57.00 | 18.00 | 54.00 | 18.00 | 50.00 | 13.00 |
| Cibeira (2014) | Breast | Latinos | Cases | LDL | 166.30 | 56.40 | 109.70 | 51.40 | 106.50 | 56.40 |
| Cibeira (2014) | Breast | Latinos | Controls | LDL | 110.60 | 35.40 | 100.70 | 31.30 | 135.70 | 50.90 |
| Trompet (2009) | All types | White | Both | LDL | 128.77 | 28.18 | 146.95 | 32.52 | 154.68 | 29.23 |
| Souza (2009) | Colorectal | Latinos | Cases | LDL | 122.60 | 0.57 | 118.90 | 48.50 | 109.10 | 32.40 |
| Souza (2009) | Colorectal | Latinos | Controls | LDL | 93.20 | 23.60 | 143.90 | 54.10 | 121.00 | 25.70 |
| Moysich (2000) | Breast | White | Cases | LDL | 124.00 | 43.00 | 143.00 | 41.00 | 159.00 | 58.00 |
| Moysich (2000) | Breast | White | Controls | LDL | 126.00 | 39.00 | 153.00 | 40.00 | 152.00 | 49.00 |
| Cibeira (2014) | Breast | Latinos | Cases | TC | 253.50 | 72.50 | 189.90 | 55.20 | 194.70 | 50.30 |
| Cibeira (2014) | Breast | Latinos | Controls | TC | 199.50 | 35.20 | 204.80 | 42.30 | 223.60 | 51.80 |
| Trompet (2009) | All types | White | Both | TC | 206.50 | 35.23 | 218.87 | 32.52 | 226.99 | 38.98 |
| Souza (2009) | Colorectal | Latinos | Cases | TC | 179.60 | 13.00 | 182.30 | 56.00 | 176.50 | 36.50 |
| Souza (2009) | Colorectal | Latinos | Controls | TC | 161.20 | 25.80 | 213.70 | 59.40 | 185.60 | 36.90 |
| Moysich (2000) | Breast | White | Cases | TC | 204.00 | 44.00 | 227.00 | 42.00 | 241.00 | 58.00 |
| Moysich (2000) | Breast | White | Controls | TC | 213.00 | 45.00 | 236.00 | 42.00 | 232.00 | 45.00 |
| Cibeira (2014) | Breast | Latinos | Cases | TG | 181.00 | 159.30 | 159.10 | 61.80 | 190.30 | 241.10 |
| Cibeira (2014) | Breast | Latinos | Controls | TG | 158.10 | 91.30 | 153.20 | 83.10 | 188.80 | 122.80 |
| Souza (2009) | Colorectal | Latinos | Cases | TG | 134.00 | 10.50 | 107.60 | 55.80 | 131.90 | 63.40 |
| Souza (2009) | Colorectal | Latinos | Controls | TG | 106.00 | 67.40 | 132.60 | 61.60 | 109.80 | 24.80 |
| Moysich (2000) | Breast | White | Cases | TG | 136.00 | 77.00 | 146.00 | 95.00 | 161.00 | 129.00 |
| Moysich (2000) | Breast | White | Controls | TG | 154.00 | 90.00 | 147.00 | 113.00 | 153.00 | 143.00 |
Abbreviations: HDL, high-density lipoprotein; LDL, low-density lipoprotein; TC, total cholesterol; TG, triglyceride; SD, standard deviation.
For 26 APOE-cancer association studies, breast cancer was reported in 10 studies, colorectal cancer in 8 studies, multiple cancers in 3 studies, prostate cancer in 2 studies, gastric, head and neck, hepatocellular cancers respectively in 1 study. 14 of 26 studies were conducted in White populations, and 4 respectively in Asian, Latinos and mixed populations. As for source of controls, 10 studies enrolled population-based controls, and 16 studies enrolled hospital-based studies. The majority of 26 studies were retrospective in design (n = 23) with the rest being prospective (n = 3). Cancer patients and controls were reported to be matched in 10 studies, unmatched in 10 studies and unreported in 6 studies. The mean age was significantly higher in cancer patients than in controls (58.07 years versus 52.69 years, P = 0.001). No significance was observed in gender, BMI, smoking and family history of cancer between the two groups (P > 0.05).
Overall comparisons for cancer risk
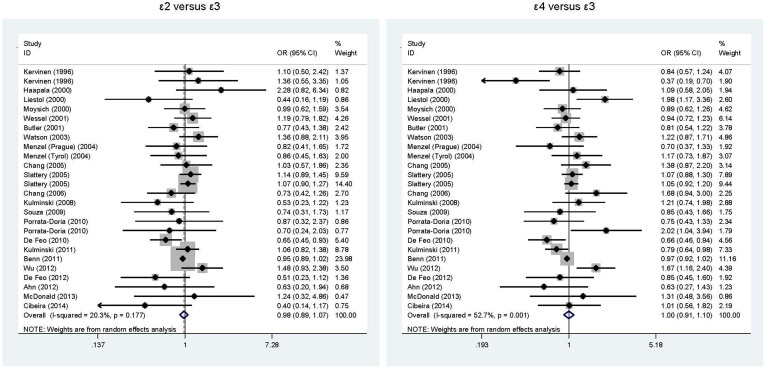
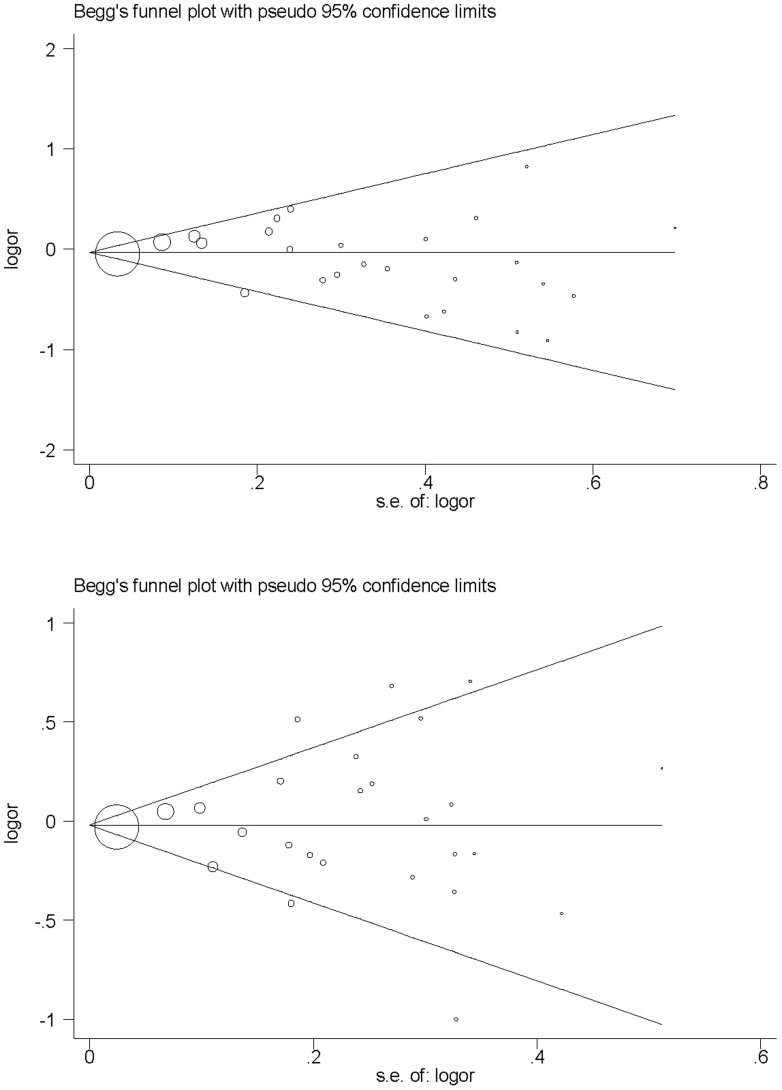
Considering the low numbers of APOE gene ε2/ε2, ε2/ε4, ε4/ε4 genotypes, only allelic comparisons (ε2 versus ε3 and ε4 versus ε3) were computed. As shown in Figure 1, the presence of ε2 and ε4 alleles showed no overall associations with overall cancer risk when compared with ε3 allele. There was no evidence of heterogeneity for the comparison of ε2 with ε3 (I2 = 20.3%), but significant heterogeneity for the comparison of ε4 with ε3 (I2 = 20.3%). The low probabilities of publication bias for both comparisons were reflected by the Begg's funnel plots (Figure 2) and Egger's tests (P = 0.512 for ε2 with ε3 and 0.662 for ε4 with ε3). The trim and fill method indicated that only one missing study was required for the comparison of ε4 with ε3 to make the Filled funnel plot symmetrical (Supplementary Figure S1).
Figure 1. Overall comparisons of APOE gene ε2 versus ε3 (the left) and ε4 versus ε3 (the right) in association with cancer risk.
Figure 2. Begg's funnel plots for the comparisons of APOE gene ε2 versus ε3 (the upper) and ε4 versus ε3 (the lower).
Sensitivity analysis
The direction and magnitude of pooled effect estimates regarding the comparisons of APOE gene ε2 and ε4 alleles with ε3 allele were confirmed in our sensitivity analysis, respectively.
Stratified comparisons for cancer risk
In an attempt to examine whether risk prediction was heterogeneous between different subgroups, several subgroup analyses were conducted according to cancer type, ethnicity, source of controls, study design, matched status and sample size, respectively (Table 3). There was no indicative of significant associations for the comparisons of ε2 versus ε3 and ε4 versus ε3 across all subgroups except for the latter comparison in Asians. The ε4 allele was significantly associated with 1.40-fold (OR = 1.40; 95% CI: 1.00–1.94; P = 0.047) increased risk of developing cancer in Asian populations, and the presence of heterogeneity was low (I2 = 37.6%), as compared with 8% reduced risk in Caucasian populations (OR = 0.92; 95% CI: 0.81–1.03; P = 0.135).
Table 3. Subgroup analysis of APOE gene ε2/ε3/ε4 polymorphism with cancer risk.
| Subgroup | No. of studies | ε2 versus ε3 | ε4 versus ε3 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| OR | 95% CI | P | I2 | OR | 95% CI | P | I2 | ||
| Cancer type | |||||||||
| Breast cancer | 10 | 0.95 | 0.78–1.17 | 0.654 | 0.0% | 1.18 | 0.95–1.48 | 0.132 | 42.8% |
| Colorectal cancer | 8 | 1.05 | 0.95–1.21 | 0.277 | 0.0% | 0.96 | 0.82–1.14 | 0.667 | 50.4% |
| All cancers | 3 | 0.96 | 0.81–1.13 | 0.617 | 32.9% | 1.02 | 0.77–1.36 | 0.088 | 80.5% |
| Ethnicity | |||||||||
| Caucasian | 14 | 0.94 | 0.81–1.08 | 0.389 | 22.8% | 0.92 | 0.81–1.03 | 0.135 | 33.1% |
| Asian | 4 | 1.01 | 0.69–1.47 | 0.969 | 34.7% | 1.40 | 1.00–1.94 | 0.047 | 37.6% |
| Latinos | 4 | 0.67 | 0.41–1.09 | 0.109 | 0.0% | 1.05 | 0.69–1.60 | 0.819 | 44.8% |
| Mixed | 4 | 1.07 | 0.93–1.22 | 0.339 | 9.7% | 1.05 | 0.85–1.31 | 0.649 | 74.2% |
| Source of controls | |||||||||
| HB | 16 | 0.95 | 0.78–1.16 | 0.629 | 32.2% | 1.05 | 0.86–1.28 | 0.661 | 64.8% |
| Population-based | 10 | 0.98 | 0.91–1.04 | 0.434 | 2.4% | 0.97 | 0.93–1.02 | 0.297 | 3.0% |
| Study design | |||||||||
| Retrospective design | 23 | 0.97 | 0.86–1.11 | 0.679 | 20.0% | 1.03 | 0.90–1.16 | 0.704 | 53.2% |
| Prospective design | 3 | 0.96 | 0.81–1.14 | 0.639 | 36.7% | 0.92 | 0.80–1.06 | 0.239 | 36.6% |
| Matched status | |||||||||
| Yes | 10 | 0.97 | 0.90–1.05 | 0.455 | 6.5% | 1.02 | 0.94–1.11 | 0.669 | 23.5% |
| No | 10 | 0.95 | 0.74–1.21 | 0.648 | 38.9% | 0.92 | 0.72–1.20 | 0.549 | 69.0% |
| NA | 6 | 1.06 | 0.82–1.37 | 0.667 | 17.6% | 1.01 | 0.77–1.32 | 0.971 | 55.8% |
| Total sample size | |||||||||
| <500 | 13 | 0.82 | 0.65–1.04 | 0.097 | 0.0% | 0.97 | 0.75–1.25 | 0.808 | 58.0% |
| ≥500 | 13 | 1.01 | 0.90–1.13 | 0.897 | 38.7% | 1.01 | 0.92–1.11 | 0.863 | 50.5% |
Abbreviations: OR, odds ratio; 95% CI, 95% confidence interval; NA, not available.
Meta-regression analysis
As age, gender, BMI, smoking and family history of cancer were continuous, several meta-regression models were constructed by including them as covariates separately, and still no significance was attained.
Overall comparisons for lipid changes
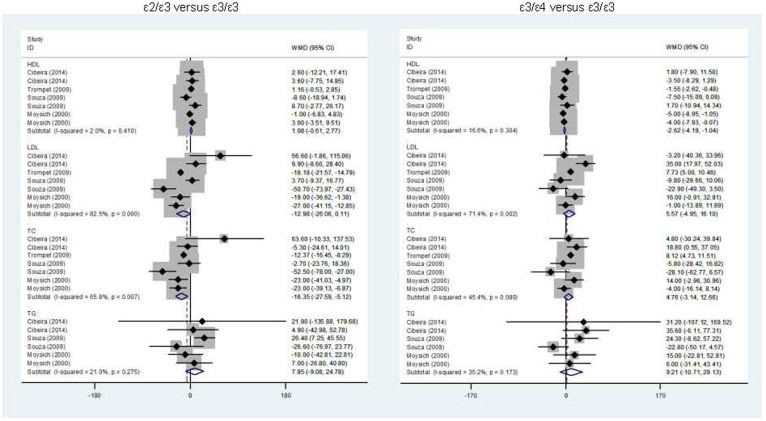
In view of limited data on APOE gene ε2/ε2, ε2/ε4, ε4/ε4 genotypes, mean lipid changes were only compared for genotype ε2/ε3 versus ε3/ε3 and ε3/ε4 and ε3/ε3 (Figure 3). Out of four lipids (TG, TC, HDL-C and LDL-C) examined, carriers of ε2/ε3 genotype had a significant reduction in circulating TC (WMD = −16.35; 95% CI: −27.59 to −5.12; P = 0.004) when compared with those with ε3/ε3 genotype, yet with strong evidence of heterogeneity (I2 = 65.8%). In contrast, carriers of ε3/ε4 genotype had a significant reduction in circulating HDL-C (WMD = −2.62; 95% CI: −4.19 to −1.04; P = 0.001) without heterogeneity (I2 = 16.6%). No statistical significance was observed for the other comparisons.
Figure 3. Overall lipid changes for the comparisons of APOE gene ε2/ε3 versus ε3/ε3 (the left) and ε3/ε4 versus ε3/ε3 (the right).
Causal prediction of circulating lipids for cancer
At the requirements of Mendelian randomization technique, causal relevance between circulating lipids and cancer risk was only calculated based on the association between APOE gene ε4 allele and cancer risk in Asians and the relationship between ε3/ε4 genotype and circulating HDL-C reduction. The predicted odds of overall cancer for 1 mg/dL reduction in circulating HDL-C was 1.14 (95% CI: 1.00 to 1.89), and this estimate was significant at a significance level of 5% as the null hypothesis value of 1 was not included by the estimated 95% CI.
Discussion
Extending the findings of a recent meta-analysis by Anand et al,9 we through a larger Mendelian randomization meta-analysis of the data from 25 articles and on 91070 participants, found that reduced circulating HDL-C might be a potentially causal risk factor for the development of overall cancer in Asians by using APOE gene ε2/ε3/ε4 polymorphism as a surrogate marker. This meta-analysis is unique to our knowledge, as it is to date the first to address the causal relevance between circulating lipids and cancer risk in medical literature.
Several observational and clinical studies have demonstrated an inverse association between circulating HDL-C and cancer risk; however, this association is currently subject to an ongoing debate, as the issues of confounding and reverse causation are intractable in classic epidemiology. Fortunately, Mendelian randomization has been introduced as a viable technique to overcome drawbacks of observational studies and obtain robust causal estimates32. Recently, a large-scale prospective study that examined the association of HDL-C with cancer incidence in patients with type II diabetes demonstrated that this significant association might be attributable to confounding and reverse causation33. Another prospective study by Kucharska-Newton et al identified a relatively weak inverse association between HDL-C and lung cancer, and this association was dependent on smoking status1. It is widely believed that circulating HDL-C is under considerable genetic control with heritability estimates of up to 60%4,34. Several lines of evidence supported a close relation between APOE genetic alterations and circulating HDL profiles7,35,36,37, which was mirrored in the current meta-analysis revealing that the presence of APOE gene ε4 allele was associated with significantly reduced HDL-C in circulation, reinforcing the soundness of selecting ε2/ε3/ε4 polymorphism as a surrogate marker. Besides, we observed that the ε4 allele was particularly overrepresented in Asian cancer patients relative to controls. Based on these observations, it is reasonably expected that low circulating HDL-C conferred by APOE gene ε4 allele is causally related with an increased risk of cancer in Asians. Nevertheless, given the inadequate statistical power of this meta-analysis in subgroup analyses, far larger sample sizes than examined here will be required to produce sufficient power to evaluate the causality between circulating HDL-C and cancer risk.
Several limitations of the present meta-analysis need to be acknowledged. Firstly, we restricted our search scope to published articles written in only English language, and we cannot totally rule out the likelihood of selective publication bias. Secondly, almost all involved studies had circulating lipids measured only once, which cannot reflect its long-term profile in the development of cancer. Thirdly, this meta-analysis was based on summarized data, rather than individual participant data, precluding further gene-to-environment interactions. Fourthly, only APOE gene ε2/ε3/ε4 polymorphism was selected in this study, and investigations on other candidate genes or polymorphisms involved in HDL-C regulation were highly encouraged, leaving a challengeable task to test whether this polymorphism integrated with other risk determinants will enhance cancer risk prediction. Fifthly, one key assumption of Mendelian randomization is that the genetic polymorphism under study should not exhibit a pleiotropic effect, which is beyond our capability in this meta-analysis to eliminate this effect. Nevertheless, the present meta-analysis enriched our understandings of circulating HDL-C in molecular carcinogenesis, which would facilitate the identification of at-risk individuals who would develop cancer later in future clinical screening.
Taken together, the findings of this Mendelian randomization meta-analysis demonstrate that reduced circulating HDL-C might be a potentially causal risk factor for the development of overall cancer in Asians. For practical reasons, it is encouraging to deem this study as a beginning instead of an endpoint of investigations to establish and optimize the background data to understanding the causal relevance of circulating HDL-C to carcinogenesis of multiple solid tumors.
Methods
The present meta-analysis was carried out in accordance with the guidelines formulated in the PRISMA (Preferred Reporting Items for Systematic Reviews and Meta-analyses) statement (see the Supporting Checklist)38.
Search strategy
To identify all relevant articles that assessed the associations of APOE gene ε2/ε3/ε4 polymorphism with cancer risk or circulating lipid changes, we systematically searched PubMed and Embase electronic databases as of December 20, 2014 using the following subject terms, ‘apolipoprotein E or apo E or APOE or apo-E′, in combination with ‘cancer or carcinoma or neoplasia or tumor or adenoma or neoplasm or myeloma or melanoma or lymphoma or leukaemia or leiomyoma’ and ‘polymorphism or variant or variation or mutation or genotype or allele or SNP’. We also manually checked the reference lists of major original articles and reviews for the missing citations of relevance.
The titles and abstracts of all retrieved articles were independently read by two authors of this meta-analysis (Chunhua Yang and Xuri Li) to assess their eligibility. If we cannot reject an article with certainty, its full text was reviewed to ascertain whether relevant data were provided and if necessary we contacted study authors by emails to request additional information. We extracted data from the most recent or complete article if a same study group was reported by more than one publications. This process was run in duplicate and independently by the same two authors, and any uncertainty over the eligibility was adjudicated by a discussion or further joint inspection of original articles.
Inclusion/exclusion criteria
All studies that met the following criteria were included: (a) regarding cancer risk, data on associations between APOE gene ε2/ε3/ε4 polymorphism and all sites of cancer except for skin were provided; (b) regarding circulating lipid changes, the mean or medium values and the corresponding standard deviation of circulating lipids including triglyceride (TG), total cholesterol (TC), high-density lipoprotein cholesterol (HDL-C) and low-density lipoprotein cholesterol (LDL-C) were provided across APOE gene ε2/ε3/ε4 alleles or genotypes; (c) study design should be either prospective or retrospective; (d) detailed genotype or allele counts of APOE gene ε2/ε3/ε4 polymorphism were tractable between cancer patients and controls.
Conference abstracts or proceedings that did not specifically address the topic of our analysis were excluded from full-text review. Case reports or series, editorials, narrative or systematic reviews, or non-English articles were also not covered. Also this meta-analysis did not involve studies that examined the progression, severity or response to treatment or survival of cancer in association with APOE gene ε2/ε3/ε4 polymorphism or that were lack of cancer-free controls.
Data gathering
Data were gathered independently from each eligible article by two authors (Chunhua Yang and Xuri Li) according to a predefined protocol developed by all contributing authors, including the first author's last name, publication year, ethnicity, cancer subtype, case-control matched status, source of controls, study design, sample size, the genotype and/or allele counts of APOE gene ε2/ε3/ε4 polymorphism between cancer patients and controls, the mean or medium (standard deviation) values of circulating TG, TC, HDL-C and LDL-C for each APOE gene ε2/ε3/ε4 allele or genotype carriers, as well as baseline characteristics of study populations when available such as age, gender, body mass index (BMI), smoking and the family history of cancer. The units of circulating TG, TC, HDL-C and LDL-C were uniformly standardized as mg/dL for consistency.
Statistics
All statistical analyses were managed with the use of STATA software (StataCorp, Texas, USA, version 12.0) on Windows.
The association of APOE gene ε2 or ε4 allele with cancer risk was expressed as odds ratio (OR) and 95% confidence interval (95% CI) when compared with the ε3 allele. Considering the confounding effect of heterogeneity between studies, only random-effects model with the DerSimonian & Laird method39 was employed.
The probability of publication bias was assessed by visual Begg's funnel plot and the Egger's test, as well as the trim-and-fill method which can infer the existence of unpublished hidden articles from a filled funnel plot and correct the meta-analysis by imputing the presence of missing studies to yield an unbiased pooled estimate.
Heterogeneity was quantified by the inconsistency index (I2) statistic, which ranges from 0% to 100% and is defined as the percentage of the observed between-study variability that is due to heterogeneity rather than chance. In this meta-analysis, I2 > 50% is designated as a threshold to indicate significant heterogeneity39. To identify potential sources of heterogeneity, predetermined subgroup analyses and meta-regression analyses were performed to model categorical and continuous host characteristics, respectively. For meta-regression analysis, given that some host characteristics had a lot of missing values such as smoking, each characteristic was modeled separately.
To evaluate the impact of individual studies on pooled effect estimates, we performed sensitivity analysis by sequentially omitting each study one at a time and computing differential estimates for remaining studies.
Under the assumptions of Mendelian randomization as formulated by Katan MB in 198640, we calculated the risk prediction as the ratio of the coefficient of the association between APOE gene ε2/ε3/ε4 polymorphism and cancer risk to that of the relationship between this polymorphism and circulating lipid changes to reflect the possible causal relevance of these lipids on cancer.
Author Contributions
G.T., J.M. and B.W. conceived and designed the experiments; C.Y. and XuriL performed the experiments; C.Y. and G.T. analyzed the data; X.W., XuriL, XianglinL and W.W. contributed materials/analysis tools; C.Y., G.T. and B.W. wrote and revised the manuscript. All authors reviewed and approved the manuscript prior to submission.
Supplementary Material
Supplementary materials
References
- Kucharska-Newton A. M. et al. HDL-cholesterol and the incidence of lung cancer in the Atherosclerosis Risk in Communities (ARIC) study. Lung Cancer 61, 292–300 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berrino F. et al. Metabolic syndrome and breast cancer prognosis. Breast Cancer Res Treat 147, 159–165 (2014). [DOI] [PubMed] [Google Scholar]
- Vogel T. et al. Apolipoprotein E: a potent inhibitor of endothelial and tumor cell proliferation. J Cell Biochem 54, 299–308 (1994). [DOI] [PubMed] [Google Scholar]
- Davignon J., Gregg R. E. & Sing C. F. Apolipoprotein E polymorphism and atherosclerosis. Arteriosclerosis 8, 1–21 (1988). [DOI] [PubMed] [Google Scholar]
- De Feo E. et al. A case-control study on the effect of Apolipoprotein E genotypes on gastric cancer risk and progression. BMC Cancer 12, 494 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallongeville J., Lussier-Cacan S. & Davignon J. Modulation of plasma triglyceride levels by apoE phenotype: a meta-analysis. J Lipid Res 33, 447–454 (1992). [PubMed] [Google Scholar]
- Cibeira G. H. et al. Apolipoprotein E genetic polymorphism, serum lipoprotein levels and breast cancer risk: A case-control study. Mol Clin Oncol 2, 1009–1015 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souza D. R. et al. Association between apolipoprotein E genotype, serum lipids, and colorectal cancer in Brazilian individuals. Braz J Med Biol Res 42, 397–403 (2009). [DOI] [PubMed] [Google Scholar]
- Anand R., Prakash S. S., Veeramanikandan R. & Kirubakaran R. Association between apolipoprotein E genotype and cancer susceptibility: a meta-analysis. J Cancer Res Clin Oncol 140, 1075–1085 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kervinen K. et al. Is the development of adenoma and carcinoma in proximal colon related to apolipoprotein E phenotype? Gastroenterology 110, 1785–1790 (1996). [DOI] [PubMed] [Google Scholar]
- Lehrer S. Possible relationship of the apolipoprotein E (ApoE) epsilon4 allele to prostate cancer. Br J Cancer 78, 1398 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haapala K., Lehtimaki T., Ilveskoski E. & Koivisto P. A. Apolipoprotein E genotype is not linked to locally recurrent hormone-refractory prostate cancer. Prostate Cancer Prostatic Dis 3, 107–109 (2000). [DOI] [PubMed] [Google Scholar]
- Liestol K. et al. Association between apolipoprotein E genotypes and cancer risk in patients with acquired immunodeficiency syndrome. Cancer Detect Prev 24, 496–499 (2000). [PubMed] [Google Scholar]
- Moysich K. B. et al. Apolipoprotein E genetic polymorphism, serum lipoproteins, and breast cancer risk. Mol Carcinog 27, 2–9 (2000). [DOI] [PubMed] [Google Scholar]
- Butler W. J., Ryan P. & Roberts-Thomson I. C. Metabolic genotypes and risk for colorectal cancer. J Gastroenterol Hepatol 16, 631–635 (2001). [DOI] [PubMed] [Google Scholar]
- Wessel N., Liestol K., Maehlen J. & Brorson S. H. The apolipoprotein E epsilon4 allele is no risk factor for prostate cancer in the Norwegian population. Br J Cancer 85, 1418 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson M. A. et al. Apolipoprotein E gene polymorphism and colorectal cancer: gender-specific modulation of risk and prognosis. Clin Sci (Lond) 104, 537–545 (2003). [DOI] [PubMed] [Google Scholar]
- Menzel H. J. et al. Association of NQO1 polymorphism with spontaneous breast cancer in two independent populations. Br J Cancer 90, 1989–1994 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang N. W. et al. Influences of apolipoprotein E polymorphism on the risk for breast cancer and HER2/neu status in Taiwan. Breast Cancer Res Treat 90, 257–261 (2005). [DOI] [PubMed] [Google Scholar]
- Slattery M. L. et al. Associations between apoE genotype and colon and rectal cancer. Carcinogenesis 26, 1422–1429 (2005). [DOI] [PubMed] [Google Scholar]
- Chang S. J. et al. Association between the apolipoprotein E genotypes and breast cancer patients in Taiwanese. Breast Cancer Res Treat 98, 109–113 (2006). [DOI] [PubMed] [Google Scholar]
- Kulminski A. M. et al. Health-protective and adverse effects of the apolipoprotein E epsilon2 allele in older men. J Am Geriatr Soc 56, 478–483 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trompet S. et al. Apolipoprotein e genotype, plasma cholesterol, and cancer: a Mendelian randomization study. Am J Epidemiol 170, 1415–1421 (2009). [DOI] [PubMed] [Google Scholar]
- De Feo E. et al. A case-control study on the effect of apoliprotein E genotype on head and neck cancer risk. Cancer Epidemiol Biomarkers Prev 19, 2839–2846 (2010). [DOI] [PubMed] [Google Scholar]
- Kato I., Land S., Majumdar A. P., Barnholtz-Sloan J. & Severson R. K. Functional polymorphisms to modulate luminal lipid exposure and risk of colorectal cancer. Cancer Epidemiol 34, 291–297 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porrata-Doria T., Matta J. L. & Acevedo S. F. Apolipoprotein E Allelic Frequency Altered in Women with Early-onset Breast Cancer. Breast Cancer (Auckl) 4, 43–48 (2010). [PMC free article] [PubMed] [Google Scholar]
- Benn M., Tybjaerg-Hansen A., Stender S., Frikke-Schmidt R. & Nordestgaard B. G. Low-density lipoprotein cholesterol and the risk of cancer: a mendelian randomization study. J Natl Cancer Inst 103, 508–519 (2011). [DOI] [PubMed] [Google Scholar]
- Kulminski A. M. et al. Trade-off in the effects of the apolipoprotein E polymorphism on the ages at onset of CVD and cancer influences human lifespan. Aging Cell 10, 533–541 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn S. J. et al. Association between apolipoprotein E genotype, chronic liver disease, and hepatitis B virus. Clin Mol Hepatol 18, 295–301 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C. T. et al. Combined effects of peroxisome proliferator-activated receptor alpha and apolipoprotein E polymorphisms on risk of breast cancer in a Taiwanese population. J Investig Med 60, 1209–1213 (2012). [DOI] [PubMed] [Google Scholar]
- McDonald B. C., Conroy S. K., Smith D. J., West J. D. & Saykin A. J. Frontal gray matter reduction after breast cancer chemotherapy and association with executive symptoms: a replication and extension study. Brain Behav Immun 30 Suppl S117–125 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith G. D. & Ebrahim S. Mendelian randomization: prospects, potentials, and limitations. Int J Epidemiol 33, 30–42 (2004). [DOI] [PubMed] [Google Scholar]
- Morton J. et al. The association of high-density lipoprotein cholesterol with cancer incidence in type II diabetes: a case of reverse causality? Cancer Epidemiol Biomarkers Prev 22, 1628–1633 (2013). [DOI] [PubMed] [Google Scholar]
- Boes E., Coassin S., Kollerits B., Heid I. M. & Kronenberg F. Genetic-epidemiological evidence on genes associated with HDL cholesterol levels: a systematic in-depth review. Exp Gerontol 44, 136–160 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulenouar H. et al. Impact of APOE gene polymorphisms on the lipid profile in an Algerian population. Lipids Health Dis 12, 155 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu W., Zhang X. & Qi Y. Association of an apolipoprotein E polymorphism with circulating cholesterols and hypertension: a meta-based Mendelian randomization analysis. Hypertens Res 35, 434–440 (2012). [DOI] [PubMed] [Google Scholar]
- Burman D. et al. Relationship of the ApoE polymorphism to plasma lipid traits among South Asians, Chinese, and Europeans living in Canada. Atherosclerosis 203, 192–200 (2009). [DOI] [PubMed] [Google Scholar]
- Moher D., Liberati A., Tetzlaff J. & Altman D. G. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. BMJ 339, b2535 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins J. P., Thompson S. G., Deeks J. J. & Altman D. G. Measuring inconsistency in meta-analyses. BMJ 327, 557–560 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katan M. B. Apolipoprotein E isoforms, serum cholesterol, and cancer. Lancet 1, 507–508 (1986). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary materials