Abstract
Background:
Co-administration of Angelicae gigantis radix (AGR) and Lithospermi radix (LR) has been commonly applied to patients to treat cardiac and hepatic disorders. Individual bioactivities of these herbal medicines have been widely investigated, but the hepatoprotective effects of co-treatment of AGR and LR have yet to be clarified.
Objective:
The present study investigated the protective effects of extracts of AGR and LR on carbon tetrachloride (CCl4) induced hepatic injury.
Materials and Methods:
In this study, we measured the hepatoprotective activity of individual and co-treatment of the two herbal medicines on hepatic injury induced by CCl4 by measuring different biochemical parameters such as serum aspartate aminotransaminase (AST) and serum alanine aminotransaminase (ALT). Microarray technology also used to compare ontological difference with individual and co-treatment of these two.
Results:
Combined treatment with AGR and LR (AGR + LR) decreased AST and ALT level in serum which demonstrate hepatoprotective effect of the therapy. When the effect of AGR and LR according to treatment conditions was measured, co-treatment showed the most prominent effect on hepatic injury by CCl4 rather than individual treatment condition. We further defined gene set that could be the molecular target of herbal effect on hepatic injury by CCl4 using bioinformatical analysis of interaction network. Highly recovered genes by treating AGR + LR play significant roles in response to hepatic injury induced by CCl4.
Conclusion:
Combined treatment with AGR and LR showed synergistic protective effects on the CCl4-induced rat hepatic tissue injury.
Keywords: Angelicae gigantis radix, carbon tetrachloride, combined treatment, hepatic injury, lithospermi Radix
INTRODUCTION
Herbal medicine has been widely used to treat a variety of disease in traditional Korean medicine. Co-treatment of Angelicae gigantis radix (AGR) and Lithospermi radix (LR) was frequently used in Korean Medical Hospital of Dongshin University in Republic of Korea. AGR, the root of Angelica gigas, is one of the most popular medicinal herbs and is commonly used constituents in herbal prescriptions in traditional Korean medicine. Recently, memory ameliorating effects on scopolamine-induced memory impairment[1] and anti-allergic effects in dinitrofluorobenzene-induced inflammation models[2] of AGR have been demonstrated. LR, the root of Lithospermum erythrorhizon, is not as popular as AGR but has been used as therapy for cancer, wound, and skin diseases.[3,4,5]
Not only the Korean Medical Hospital of Dongshin University but also many local traditional Korean medicine clinics prescribe co-treatment of AGR and LR to treat hepatic disease, but few studies conducted to clarify the effects of the prescription on hepatic disease. In traditional and folk medicine, some of the doctors tend to use their own prescriptions without proving their effects or toxicity. Therefore, we aimed to clarify the effects of AGR, LR, and co-treatment of these two herbal medicines on hepatic injury on rats.
Hepatic cell injury caused by carbon tetrachloride (CCl4)[6,7] and others, as well as chronic alcohol consumption,[8] is well-studied. CCl4-induced hepatic injury is one of the most investigated animal models, and in the past decades, molecular mechanisms for hepatic necrosis and steatosis induced by CCl4 were well-documented.[9]
Herbal remedies on hepatic diseases have gained popularity in recent due to their safety.[10,11,12] Herbal extract is composed of various kinds of phytochemicals, so it would be difficult to identify major components having pharmaceutical effect. Therefore, high throughput screening systems like microarray analysis is an essential process to elucidate the molecular effects of herbal extract on disease animal model.
Therefore, in this experiment, we measured hepatoprotective effects AGR and LR on by CCl4-induced hepatic injury using biochemistry and microarray technology with three treatment conditions including individual and co-treatment of these two.
MATERIALS AND METHODS
Animal
Sprague-Dawley rat (Daehan Biolink Co., Korea), weighing 200 ± 20 g, was maintained under 12 h day-night cycles for at least 2 weeks prior to the experiment. The animals were free access to food and water. All experiments were conducted under the Institutional Guidelines for Animal Experimentation at Dongshin University School of Korean Medicine.
Preparation of crude extracts from Angelicae gigantis radix, Lithospermi radix, and Angelicae gigantis radix + Lithospermi radix
AGR and LR were supplied by Gwangmyung Pharmaceutical Company (Ulsan, Korea) and identified by Dr. Gumsan Lee (College of Korean Medicine, Wonkwang University, Korea). Voucher specimens (201-AGR and 201-LR) were placed in the Herbarium of School of Korean Medicine, Pusan National University (Yangsan, Korea), and all the materials satisfy the quality control guidelines from Korean Food and Drug Administration.
The three kinds of raw materials (AGR, LR, AGR + LR) were ground into powders to 1 kg, and were extracted with 75% aqueous ethanol for two times (2 days for each) under sonication at room temperature. The ratio of AGR and LR in AGR + LR is 50:50. The extracted solutions were filtered using Whattman filter paper No. 3. The filtrate was then freeze-dried to a powder, which was stored at 4°C until use. The powder was dissolved into water prior to administration.
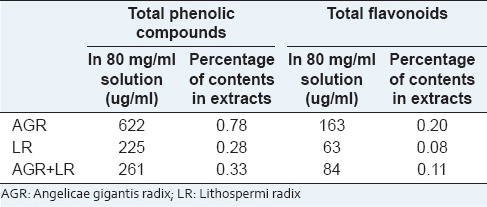
Measurements of total phenolic compounds and flavonoids contained in Angelicae gigantis radix, Lithospermi radix, and Angelicae gigantis radix + Lithospermi radix
80 ul of aqueous solutions (80 mg/ml) of each extracts was mixed with 20 ul of 50% Folin-ciocalteu reagent, and 5 min add 100 ul of 2% sodium carbonate solution and incubate for 30 min at room temperature. The absorbance was read after the absorbance of the reaction mixture was measured at 725 nm with ultraviolet (UV) spectrometer. All determination was performed in triplicate. The calibration curve was prepared by preparing tannic acid standard solutions at concentration 4–20 ug/ml in water.
Diethylene glycol colorimetric method was used for flavonoids determination. 100 ul of aqueous solutions (80 mg/ml) of each extract was mixed with 1 ml of diethylene glycol and 100 ul of 1 N NaOH. It remained at 37°C for 1 h. The absorbance of the reaction mixture was measured at 420 nm with UV spectrometer. All determination was performed in triplicate. The calibration curve was prepared by preparing naringin standard solutions at concentration 0.4–20 ug/ml in water. Total content (%) of flavonoids in each extract was calculated as naringin equivalent.
Total contents (%) of phenolic compound and flavonoids in each extract were calculated as tannic acid and naringin equivalent (TAE and NE), respectively.
TAE = ([C×V] / M) ×100
NE = ([C × V] / M) × 100
C = the concentration of Naringin established from calibration curve mg/ml
V = Volume of extract (ml)
M = the weight of herbal extract (mg)
Folin-ciocalteu method for total phenolic compounds clears that AGR, LR and AGR + LR extracts contain small amount of phenolic compound, that is, 0.78%, 0.28%, and 0.33%, respectively, and diethylene glycol colorimetric method shows 0.20%, 0.08%, and 0.11% amount of flavonoids in each extract [Table 1].
Table 1.
Total phenolic compounds and flavonoids contents in the herbal medicines
High-performance thin layer chromatography-based fingerprinting
It is difficult to maintain consistent qualities of herbal medicines because they are natural resource-derived materials such as AGR and LR, therefore, quality control or standardization of herbal medicine is very important. A Camag high performance thin layer chromatography system was used for the fingerprinting of the herbal extracts, and the linear ascending development was carried out in a Camag horizontal development chamber (10 cm × 120 cm) which was presaturated with mobile phase chloroform: Methanol (10:1 v/v) at room temperature. After development, derivatization was carried out in UV or anisaldehyde solution spraying followed by heating [Figure 1].
Figure 1.
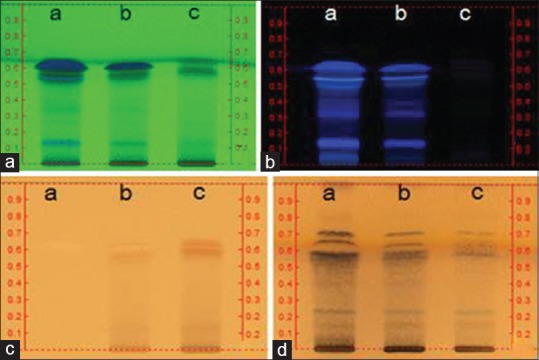
High-performance thin layer chromatography (HPTLC) images of Angelicae gigantis radix (AGR), AGR + Lithospermi radix (LR) and LR (lane a, b, and c, respectively) fingerprinting. HPTLC plate (silica gel F254; mobile phase, chloroform:Methanol/8:2) and visualizer (Camag, Swiss) were used in acquiring the images. (a) 254 nm ultraviolet (UV); (b) 366 nm UV; (c) white light; (d) p-anisaldehyde sprayed followed by white light detected
Chemical treatment
Hepatic injury was induced by intraperitoneal injection of CCl4 at 1 ml/kg (10%, diluted in corn oil). Control group received only corn oil intraperitoneally. For oral administration of herbal extracts, 1% (w/v) of each of the extracts was freely supplied in water to seven rats for five consecutive days after the induction of hepatic injury.
Blood collection and estimation of serum aspartate aminotransaminase and alanine aminotransaminase
Before sacrificing the animals, blood was collected from cardiac puncture under anesthesia without the use of anticoagulant. The blood was allowed to stand for 20 min before centrifugation at 5000 ×g for 10 min to obtain serum. The obtained serum was used for analysis of aspartate aminotransaminase (AST) and alanine aminotransaminase (ALT). Serum levels of AST and ALT were assayed from kits (Asan Pharma. Co., Ltd., Hwasung, Korea).
Estimation of lipid peroxidation
Estimation of lipid peroxidation level in hepatic tissue homogenate was conducted by measuring malondialdehyde (MDA) content (pmol/mg protein), a measure of lipid peroxidation in the form of thiobarbituric acid reacting substances by the method of Gutteridge.[13]
Histopathological examination of the liver
After blood collection from rats, livers from all of the rats were removed quickly, and were stored separately in a deep freezer. Some pieces were fixed in phosphate-buffered formalin (10%) for histopathological examination stained hematoxylin and eosin (Hematoxylin and Eosin stain), and the representative features were examined under light microscope.
Statistical analysis
The values are expressed as mean ± standard error, and the results were analyzed using the Statistical Package for the Social Sciences (version 18.0, SPSS Inc. Chicago, USA) and Sigmaplot (version 12, Systat Software Inc. San Jose, USA) software and statistical significance was evaluated by one-way analysis of variance followed by Duncan's multiple comparison test. A value of P < 0.05 was considered statistically significant.
RNA isolation from hepatic tissue
The rats were sacrificed by intraperitoneal injection of sodium pentobarbital. The liver was then surgically removed. After immediate snap frozen, hepatic tissue was stored in liquid nitrogen. The total RNA was then isolated from the frozen tissue using the Qiagen RNeasy Kit following the manufacturer's instructions (Qiagen Korea Ltd.). For the evaluation of quality of RNA, the ratio of 28S/18S ribosomal RNA was measured to be over 1.6.
Microarray experiment
RNA isolated from the 7 rats in each group was pooled to eliminate individual variability. An Agilent microarray system (Agilent Technologies Co.) was used, which contains approximately 45,000 oligo-spots representing ~17,000 genes. Initially, 20 μg of RNA was labeled fluorescently and hybridized with reference RNA using 3 DNA array detection system according to the manufacturer's protocol (Genisphere, PA). RNA from normal rats was used as a reference. Microarray was scanned using a ScanArray scanner (Perkin-Elmer, Boston, MA) to produce raw image file.
Microarray data analysis
Microarray data were normalized using the Lowess method as previously described.[14] Only spots with the intensity level >1.4 times to that of the local background were selected for analysis and only genes that were well-measured in all sample were included in the analysis. The hierarchical clustering was performed using CLUSTER and TREEVIEW program (M.B. Eisen, http://rana.lbl.gov). We considered 1.5 fold of expression change as baseline of up- or down-regulation. OntoExpress program was applied for ontological analysis,[15] and cytoscape program was applied for interaction network analysis[16] in which databases of BOND (http://bond.unleashedinformatics.com) and the BioGrid (http://www.thebiogrid.org/) were used. Common genes present in rat and human were obtained from the database of The Jackson Laboratory (http://www.informatics.jax.org).
RESULTS
Effects of Angelicae gigantis radix, Lithospermi radix and Angelicae gigantis radix + Lithospermi radix extracts on hepatic injury markers
Hepatoprotective effects of AG, LR, and AGR + LR on hepatic injury induced by CCl4 administration was observed by evaluating serum level of AST and ALT in treatment groups. These enzymes are cytoplasmic in nature but marked increased release of AST and ALT indicates a severe damage to hepatic tissue. As shown in Figure 2a and b, CCl4 treatment significantly increased the serum levels of AST and ALT from 120.69 ± 11.16 and 30.35 ± 11.16 to 224.06 ± 13.73 and 101.82 ± 10.67, respectively. However, pretreatment with AGR + LR before the injection of CCl4 prevented the elevation of the serum levels of AST and ALT (P < 0.05).
Figure 2.
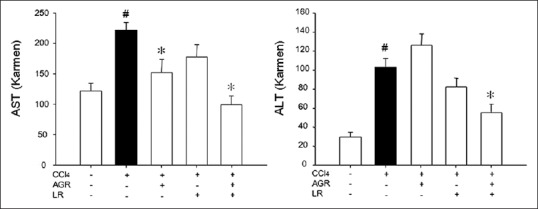
Effects of herbal medicines on serum aspartate aminotransaminase and alanine aminotransaminase activities in rats treated with carbon tetrachloride (CCl4). Rats were orally administered with Angelicae gigantis radix (AGR), AGR + Lithospermi radix (LR), and LR for five consecutive days, respectively, and then blood samples were obtained. Data are mean ± standard error of seven experiments. #P < 0.05 when compared to normal rats; *P < 0.05 when compared to CCl4 group
Exposures to CCl4 showed a significant increase in lipid peroxidation in hepatic tissue homogenate from 23.03 ± 8.37 to 168.38 ± 15.74, however, oral administration of AGR and AGR + LR extract decreased the liver MDA level to 134.06 ± 20.13 and 80.93 ± 10.52, respectively [Figure 3].
Figure 3.
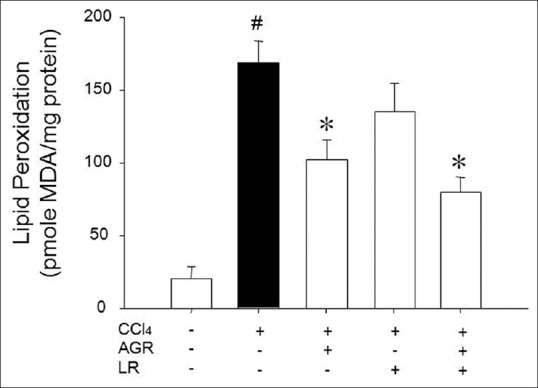
Effects of herbal medicines on liver homogenate lipid peroxidation of carbon tetrachloride (CCl4) treated rats. The results represent mean ± standard error of seven animals in each group. #P < 0.05 when compared to normal rats; *P < 0.05 when compared to CCl4 group
As depicted in Figures 2 and 3, CCl4 administration induced severe hepatic injury as shown as elevation in tissue level of MDA and serum levels of AST and ALT. Histological examination also confirmed above data by showing the inflammatory features of hepatic tissues [Figure 4], whereas pretreatment with AGR significantly reduced the hepatotoxicity of CCl4.
Figure 4.
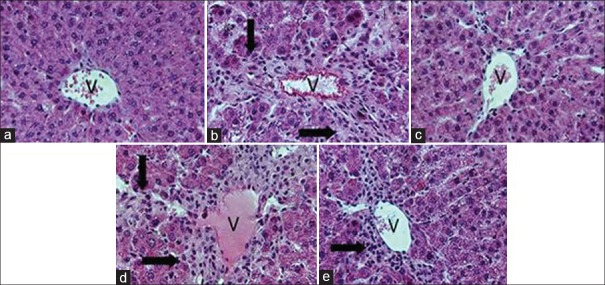
Histological examination of rat acute hepatic injury induced by carbon tetrachloride (CCl4). (a) naïve rats; (b) CCl4 treated; (c) CCl4 and Angelicae gigantis radix (AGR) treated; (d), CCl4 and Lithospermi radix (LR) treated; (e) CCl4 and AGR + LR treated group, respectively. V, central vein. Arrows indicate inflammation cells. Histological examinations were performed under a light microscope (×100)
Clustering pattern of gene expression
The effects of pretreatment of AGR, LR, and AGR + LR oral administration on gene expression in the rat hepatic tissue injured by CCl4 treatment were measured by microarray analysis. Of a total of approximately 17,000 genes on the microarray, about 2,400 genes that were up- or down-regulated in at least one sample were filtered to be clustered based on gene expression level [Figure 5a]. AGR, LR and AGR + LR administration induced and repressed different genes, respectively. The effect of herbal medicines according to treatment conditions on up (194 genes) and down-regulated genes (513 genes) induced by hepatic injury was also measured. As shown in Figure 5b and c, many up- or down-regulated genes by hepatic damage were restored to a normal level by AGR, LR, and AGR + LR, respectively.
Figure 5.
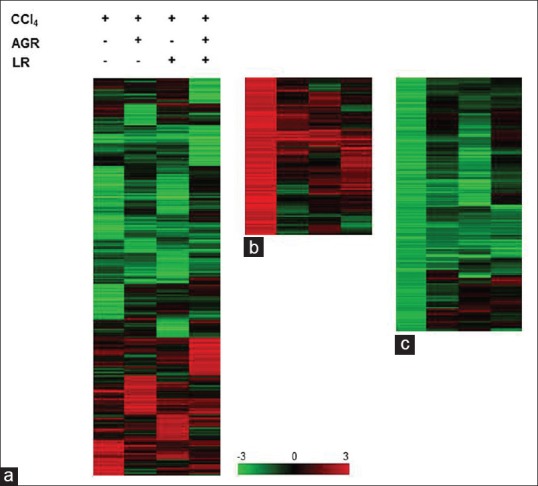
Expression profiles of genes regulated by carbon tetrachloride (CCl4) and herbal medicines. Normalized microarray data were hierarchically clustered according to the level of gene expression. Columns represent each experimental group, and rows represent genes. Red and green indicates up- and down-regulation, respectively. Scale bar represents the color intensity corresponding to the expression ratio (logarithm of base 2). Cluster (a) shows expression profile of total genes (2,423 genes) according to treatment of herbal medicines followed by CCl4 treatment. A detailed view of the individual gene expression is shown in b and c. Cluster b (194 genes) consists primarily of genes that were up-regulated while cluster (c) (513 genes) consists primarily of genes that were down-regulated in response to CCl4 treatment
Ontological analysis of genes altered in liver by carbon tetrachloride
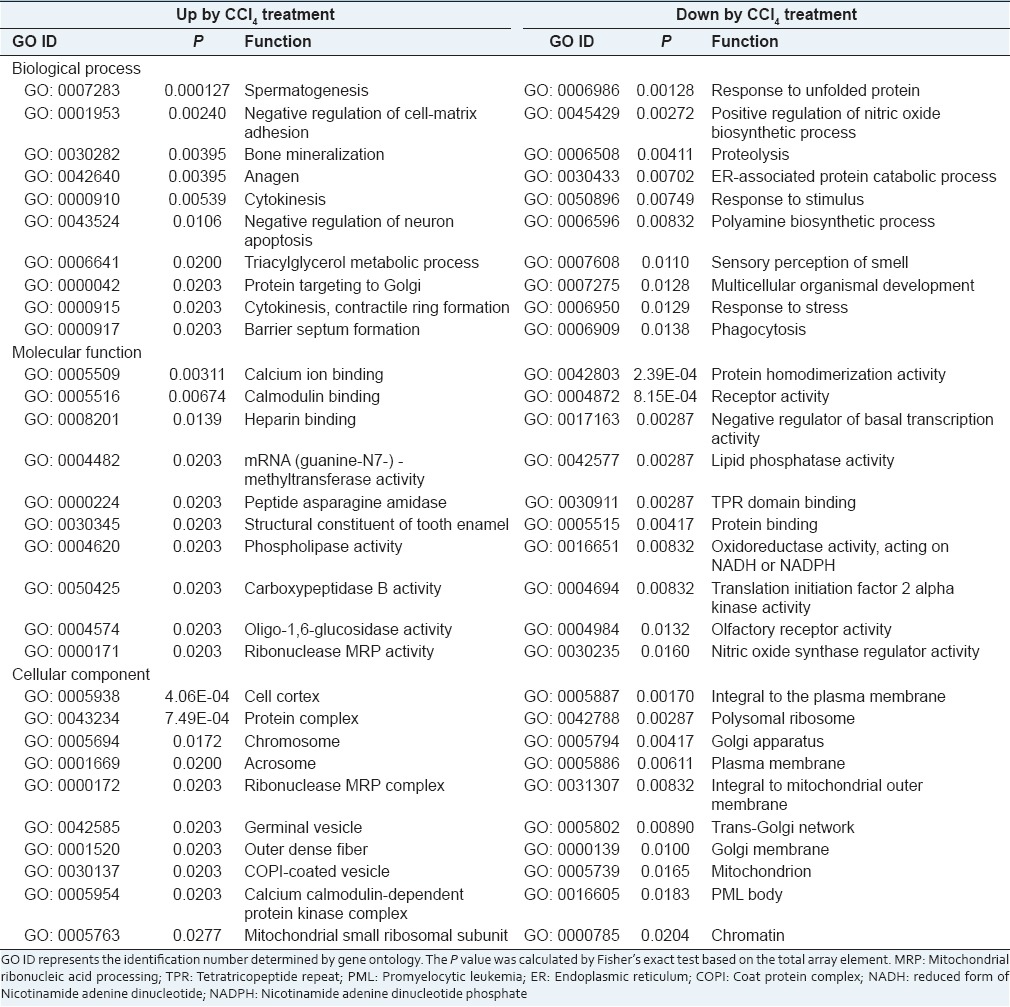
To elucidate the molecular effects of CCl4 treatment on liver, we performed an ontological analysis for genes that were up- or down-regulated in response to hepatic damage induced by CCl4. As shown in Table 2, various biological processes were involved in hepatic injury induced by CCl4, which means that treatment of CCl4 caused suppression or induction of various cellular functions in the liver.
Table 2.
Ontological analysis of genes altered by CCl4 treatment
Interaction network analysis of genes altered in liver by carbon tetrachloride
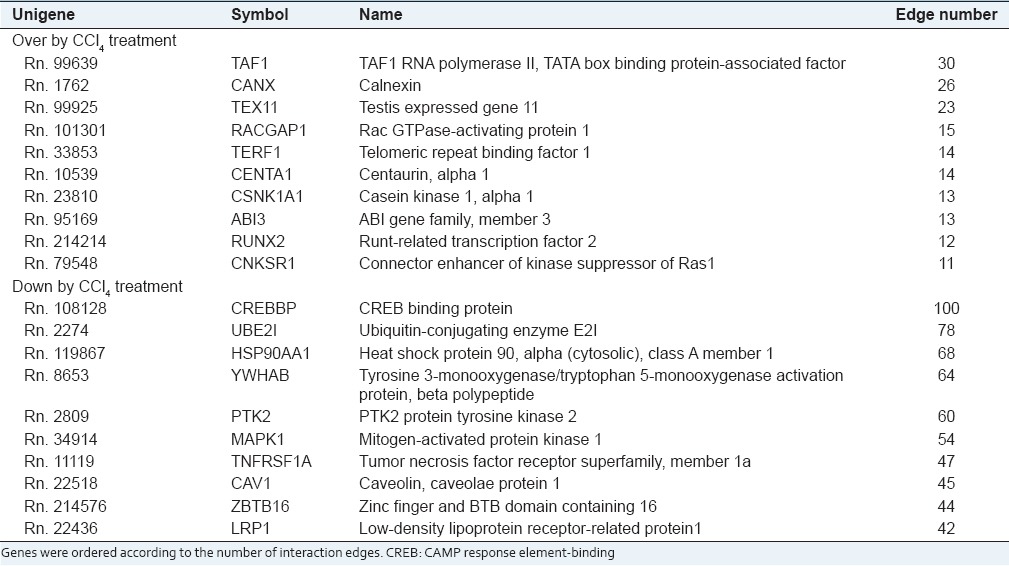
Because it was postulated that key components would interact with many other counterparts, we evaluated the interaction significance using interaction network information (Bond database, http://bond.unleashedinformatics.com BioGrid database, http://www.thebiogrid.org/). Figure 6 shows the interaction network structure of the first neighbors of proteins corresponding to up- or down-regulated genes in liver injured by CCl4. Because amount of information on interaction in rat system was much smaller than that in human system, we transformed rat genes into human genes based on gene symbol using orthology database provided by The Jaxon Laboratory (http://www.informatics.jax.org). Five hundred and thirty-seven nodes of the first neighborhood were selected for up-regulated genes [Figure 6a], whereas 1,501 nodes were selected for down-regulated genes [Figure 6b]. Of these first neighbor nodes, core nodes which have at least 10 interaction edges were selected as significant. Of core nodes, 19 genes were up-regulated, and 51 genes were down-regulated in response to hepatic injury by CCl4. Table 3 shows top 10 core node genes.
Figure 6.
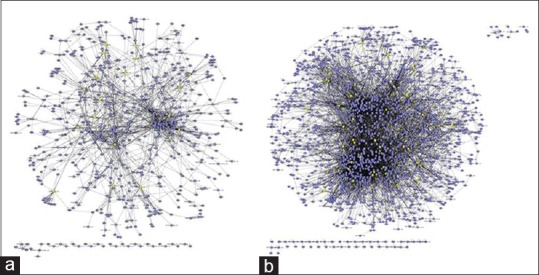
Analysis of the interaction network. The first neighborhood of genes that were altered in response to carbon tetrachloride (CCl4) treatment was identified during analysis of the interaction network. The number of first neighborhood nodes of genes that were up-regulated in response to CCl4 was 537 (a) and the number of first neighborhood nodes of genes that were down-regulated was 1,501 (b) yellow nodes indicate core node genes that were altered by CCl4 treatment and had at least 10 interaction edges with other proteins
Table 3.
Top 10 core node genes altered by CCl4 treatment
Effect of treatment of Angelicae gigantis radix, Lithospermi radix, and Angelicae gigantis radix + Lithospermi radix on genes altered in liver by carbon tetrachloride
The core node genes identified in this analysis were thought to play key roles in the response to hepatic injury by CCl4. Therefore, we measured the effect of AGR, LR, and AGR + LR on these genes. Although the overall expression pattern of the core node genes was similar to that of the expression pattern of all of the genes that were altered by hepatic injury [Figure 7], the percentage of core node genes recovered by treatment with the herbal medicines increased greatly in all treatment conditions. Specifically, of 19 core node genes up-regulated by CCl4, all 19 genes were restored to normal levels by AGR administration [Figure 7b]. However, when all of the genes that were up-regulated in response to hepatic injury were evaluated, AGR administration restored the expression level of 94.3% (183/194) of the genes [Figure 7a]. LR administration also recovered 94.7% (18/19) of genes when measured using core node genes and 93.8% (182/194) when measured using all of the genes that were up-regulated by CCl4. The most prominent effect of the herbal medicines was obtained when these two treatment conditions (AGR + LR) were co-administrated. One hundred percentage (when using core node genes) and 97.4% (when using all up-regulated genes) of recovery rates were obtained. Similar effects were also observed when the genes that were down-regulated in response to hepatic injury by CCl4 were evaluated. Specifically, by AGR administration, 89.1% (457/513) of the genes that were down-regulated in response to hepatic injury were recovered [Figure 7c], whereas this recovery rate was 72.5% (37/51) when only the core node genes were evaluated [Figure 7d]. LR administration increased recovery rate from 73.3% (376/513) to 92.2% (47/51) using core node genes rather than all down-regulated genes. As in the case of up-regulated genes, co-treatment of AGR and LR (AGR + LR) showed the highest recovery rate. 100% of recovery rate was measured with core node genes whereas 89.3% (458/513) was obtained with all down-regulated genes induced by CCl4 treatment.
Figure 7.
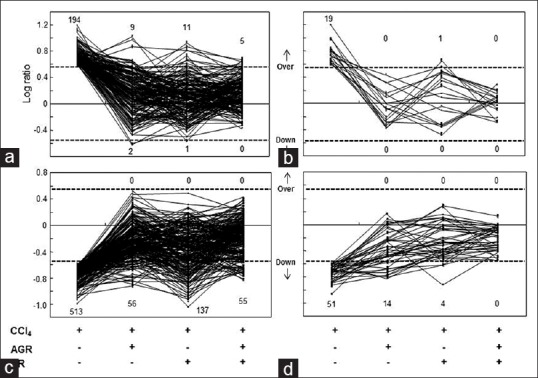
Effects of herbal medicines on the gene expression that were altered by carbon tetrachloride. The effects of herbal medicines according to treatment conditions on total up-regulated (a), up-regulated core node genes (b), total down-regulated (c), and down-regulated core node genes (d) were evaluated. The figures represent the number of up-regulated and down-regulated genes according to each treatment condition. The dotted line indicates the baseline of up-or down-regulation. Core node genes were determined as genes showing interaction edges with at least 10 neighborhood nodes
DISCUSSION
These two medicinal herbs, AGR and LR, are often prescribed together in Korean Medical Hospital of Dongshin University to treat hepatic diseases including acute hepatitis. AGR was reported to have memory ameliorating[1] and anti-allergic effects,[2] and LR was reported to have anti-cancer,[3] and wound healing[4] effects. However, the mechanism of the effect of co-treated remedy on hepatic injury has not been clearly studied. In this study, we evaluated the biochemical and overall expression profile to determine if AGR, LR, and AGR + LR exerted effects on hepatic injury induced by CCl4 treatment.
In order to investigate the protective effects of the herbal medicines, we performed CCl4-induced hepatic injury in rats. CCl4 is frequently used to induce liver fibrosis in experiment. It is known that liver damage caused by CCl4 may be due to the free radical formed during metabolism of CCl4 in the liver.[7,17] Furthermore, in our previous study, lipid peroxidation was greatly increased in liver by the treatment with CCl4.[18,19] As depicted in Figures 2-4, CCl4 administration induced significant hepatic injury shown dramatic elevation in serum levels of AST and ALT. These results indicate that the effect of AGR + LR on AST mainly induced by AGR, but the effect of AGR + LR on ALT mainly induced by LR. Hence, we can presume that there are possibilities of having different action mechanisms of AGR and LR on hepatoprotective effects. Hepatic tissue level of MDA also supports the occurrence of acute hepatic injury. This was also confirmed by conducting histological examination as showing the typical inflammatory features, centrilobular necrosis, whereas, pretreatment with the AFR treatment significantly attenuated the hepatotoxicity induced by CCl4.
As shown in Figure 5, treatment with CCl4 induced various molecular responses in rat liver tissue. The functional annotation of altered genes reveals that the CCl4 may be related with impairment of many biological functions in the liver [Table 2]. Recent studies on expression profile also reported various functional changes, especially lipid metabolism and transport pathway in rat liver injured by CCl4.[20,21] Up-or down-regulated genes by CCl4 were restored to the normal level by the pretreatment with herbal medicines. All treatment conditions including AGR, LR, and AGR + LR were effective to recover the up-or down-regulated genes by CCl4 to the normal level. Especially, AGR + LR administration shows the most prominent effect on restoring altered genes to the normal level for both up- and down-regulated genes [Figure 5]. This result supports biochemical data depicted above in which AGR + LR administration significantly reduced hepatic injury and lipid peroxidation level compared with AGR only or sole treatment of LR. The recovery rate even increased to 100% by co-treatment of AGR and LR when measured using a subset of genes that represented core nodes of interactions with many other proteins [Figure 7]. But the restoration of the expression level in response to co-treatment does not mean the recovery of liver function itself. The variability in the recovery rate among measured biochemical parameters such as AST, ALT [Figure 2], and lipid peroxidation [Figure 3] indicates that this rate should be determined based on a combination of results, not on only one parameter. In addition, many genes were newly up-or down-regulated in rat liver by treatment with the herbal medicines. These newly expressed genes seem to play critical roles in response to hepatic injury; therefore, it is necessary to identify the functions of these newly regulated genes to elucidate the precise molecular mechanism by which Korean traditional herbal medicines can reduce hepatic injury induced by CCl4. When we take the biochemistry data and gene expression profile together, the three kinds of herbal extracts showed moderate effects on hepatic injury in biochemical analysis, on the other hand, gene expressions dramatically restored by the herbal extracts. It means that gene expression recovery can be considered as supplementary data for clinical evaluation, but not as the main.
A vulnerability of this study is inaccuracy of dosage of each extract on individual rats. We tried not to give physical stress to experimental animals, so we administered the extracts via drinking water. Nevertheless, the three kinds of extracts showed beneficial effects on hepatic injury. The hepatoprotective effect was greater when AGR and LR were administered together, and AGR was thought to have more dominant effect.
CONCLUSIONS
This is the first report of the comparative study of protective effects of AGR, LR, and AGR + LR against CCl4 induced hepatic injury in rats. The effects measured with biochemical assays are dependent on its ability of AGR, and the efficacy may have a correlation with components of AGR. The high recovery rate we obtained using the core node genes does not indicate the recovery of liver injury itself but suggests that these genes play significant roles in response to liver injury induced by CCl4. Furthermore, our study revealed that the co-treatment of AGR and LR was more effective to reduce liver injury induced by CCl4 at molecular level than only single treatment condition.
Footnotes
Source of Support: This work was generously supported by the Bio-Scientific Research Grant funded by the Pusan National University (PNU, Bio-Scientific Research Grant) (PNU-2010-101-259).
Conflict of Interest: None declared.
REFERENCES
- 1.Park SJ, Jung JM, Lee HE, Lee YW, Kim DH, Kim JM, et al. The memory ameliorating effects of INM-176, an ethanolic extract of Angelica gigas, against scopolamine- or Aß(1-42)-induced cognitive dysfunction in mice. J Ethnopharmacol. 2012;143:611–20. doi: 10.1016/j.jep.2012.07.019. [DOI] [PubMed] [Google Scholar]
- 2.Joo SS, Park D, Shin S, Jeon JH, Kim TK, Choi YJ, et al. Anti-allergic effects and mechanisms of action of the ethanolic extract of Angelica gigas in dinitrofluorobenzene-induced inflammation models. Environ Toxicol Pharmacol. 2010;30:127–33. doi: 10.1016/j.etap.2010.04.007. [DOI] [PubMed] [Google Scholar]
- 3.Andújar I, Recio MC, Giner RM, Ríos JL. Traditional chinese medicine remedy to jury: The pharmacological basis for the use of shikonin as an anticancer therapy. Curr Med Chem. 2013;20:2892–8. doi: 10.2174/09298673113209990008. [DOI] [PubMed] [Google Scholar]
- 4.Hsiao CY, Tsai TH, Chak KF. The molecular basis of wound healing processes induced by lithospermi radix: A proteomics and biochemical analysis. Evid Based Complement Alternat Med 2012. 2012 doi: 10.1155/2012/508972. 508972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tan YQ, Liu JL, Bai YP, Zhang LX. Literature research of Chinese medicine recipes for the treatment of psoriasis vulgaris with blood-heat syndrome type. Chin J Integr Med. 2011;17:150–3. doi: 10.1007/s11655-011-0645-y. [DOI] [PubMed] [Google Scholar]
- 6.Best DH, Coleman WB. Liver regeneration by small hepatocyte- like progenitor cells after necrotic injury by carbon tetrachloride in retrorsine-exposed rats. Exp Mol Pathol. 2010;89:92–8. doi: 10.1016/j.yexmp.2010.06.007. [DOI] [PubMed] [Google Scholar]
- 7.Weber LW, Boll M, Stampfl A. Hepatotoxicity and mechanism of action of haloalkanes: Carbon tetrachloride as a toxicological model. Crit Rev Toxicol. 2003;33:105–36. doi: 10.1080/713611034. [DOI] [PubMed] [Google Scholar]
- 8.Lee M, Kowdley KV. Alcohol's effect on other chronic liver diseases. Clin Liver Dis. 2012;16:827–37. doi: 10.1016/j.cld.2012.08.010. [DOI] [PubMed] [Google Scholar]
- 9.Fujimoto J, Iimuro Y. 9.20 – Carbon tetrachloride-induced hepatotoxicity. In: McQueen CA, editor. Comprehensive Toxicology. 2nd ed. Oxford: Elsevier; 2010. pp. 437–55. [Google Scholar]
- 10.Tiwari P, Ahirwae D, Chandy A, Ahirwar B. Evaluation of hepatoprotective activity of alcoholic and aqueous extracts of Selaginella lepidophylla. Asian Pac J Trop Dis. 2014;4(Suppl 1):S81–6. [Google Scholar]
- 11.Deng JS, Chang YC, Wen CL, Liao JC, Hou WC, Amagaya S, et al. Hepatoprotective effect of the ethanol extract of Vitis thunbergii on carbon tetrachloride-induced acute hepatotoxicity in rats through anti-oxidative activities. J Ethnopharmacol. 2012;142:795–803. doi: 10.1016/j.jep.2012.06.003. [DOI] [PubMed] [Google Scholar]
- 12.Faremi TY, Suru SM, Fafunso MA, Obioha UE. Hepatoprotective potentials of Phyllanthusamarus against ethanol-induced oxidative stress in rats. Food Chem Toxicol. 2008;46:2658–64. doi: 10.1016/j.fct.2008.04.022. [DOI] [PubMed] [Google Scholar]
- 13.Gutteridge JM. Lipid peroxidation and antioxidants as biomarkers of tissue damage. Clin Chem. 1995;41:1819–28. [PubMed] [Google Scholar]
- 14.Yang YH, Dudoit S, Luu P, Lin DM, Peng V, Ngai J, et al. Normalization for cDNA microarray data: A robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 2002;30:e15. doi: 10.1093/nar/30.4.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khatri P, Voichita C, Kattan K, Ansari N, Khatri A, Georgescu C, et al. Onto-Tools: New additions and improvements in 2006. Nucleic Acids Res. 2007;35:W206–11. doi: 10.1093/nar/gkm327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fang HL, Lin WC. Corn oil enhancing hepatic lipid peroxidation induced by CCl 4 does not aggravate liver fibrosis in rats. Food Chem Toxicol. 2008;46:2267–73. doi: 10.1016/j.fct.2008.03.005. [DOI] [PubMed] [Google Scholar]
- 18.Kim HW, Kim SW, Cho SJ, Kim BY, Yun YC, Cho SI. Effects of sayeoksanhap-pyeongweisan-gamibang (SPG) on hematological changes in animals damaged by CCl4. Korean J Herbol. 2007;22:169–74. [Google Scholar]
- 19.Kim SW, Kim HW, Cho SI, Yun YC. Effects of sayeoksanhap-pyeongweisan-gamibang (SPG) on hepatological changes in animals damaged by CCl4. J Korean Acupunct Moxibustion Soc. 2007;24:197–207. [Google Scholar]
- 20.Chung H, Kim HJ, Jang KS, Kim M, Yang J, Kang KS, et al. Comprehensive analysis of differential gene expression profiles on D-galactosamine-induced acute mouse liver injury and regeneration. Toxicology. 2006;227:136–44. doi: 10.1016/j.tox.2006.07.026. [DOI] [PubMed] [Google Scholar]
- 21.Okumura H, Katoh M, Minami K, Nakajima M, Yokoi T. Change of drug excretory pathway by CCl4-induced liver dysfunction in rat. Biochem Pharmacol. 2007;74:488–95. doi: 10.1016/j.bcp.2007.04.025. [DOI] [PubMed] [Google Scholar]


