A Ca2+ signal mediated by a glutamate receptor homolog promotes seed germination, opposing the effect of abscisic acid.
Abstract
Seed germination is a critical step in a plant’s life cycle that allows successful propagation and is therefore strictly controlled by endogenous and environmental signals. However, the molecular mechanisms underlying germination control remain elusive. Here, we report that the Arabidopsis (Arabidopsis thaliana) glutamate receptor homolog3.5 (AtGLR3.5) is predominantly expressed in germinating seeds and increases cytosolic Ca2+ concentration that counteracts the effect of abscisic acid (ABA) to promote germination. Repression of AtGLR3.5 impairs cytosolic Ca2+ concentration elevation, significantly delays germination, and enhances ABA sensitivity in seeds, whereas overexpression of AtGLR3.5 results in earlier germination and reduced seed sensitivity to ABA. Furthermore, we show that Ca2+ suppresses the expression of ABSCISIC ACID INSENSITIVE4 (ABI4), a key transcription factor involved in ABA response in seeds, and that ABI4 plays a fundamental role in modulation of Ca2+-dependent germination. Taken together, our results provide molecular genetic evidence that AtGLR3.5-mediated Ca2+ influx stimulates seed germination by antagonizing the inhibitory effects of ABA through suppression of ABI4. These findings establish, to our knowledge, a new and pivotal role of the plant glutamate receptor homolog and Ca2+ signaling in germination control and uncover the orchestrated modulation of the AtGLR3.5-mediated Ca2+ signal and ABA signaling via ABI4 to fine-tune the crucial developmental process, germination, in Arabidopsis.
Germination converts a seed from a quiescent embryonic state to a highly active phase leading to seedling establishment, which is considered to be the most critical step in the life cycle of seed plants and represents the entry of the plant into the ecosystem (Weitbrecht et al., 2011; Rajjou et al., 2012). Germination begins with water uptake by dry nondormant seeds (i.e. imbibition) and terminates with two visible sequential events: testa (seed coat) rupture due to the expansion of the embryo and endosperm and radicle (embryonic root) protrusion through the endosperm (Nonogaki et al., 2010; Weitbrecht et al., 2011). This progression is controlled by various complex intrinsic signals, such as phytohormones, and environmental factors that include water, temperature, and light (Weitbrecht et al., 2011; Rajjou et al., 2012).
Plant hormones play crucial roles in germination regulation, among which abscisic acid (ABA) has been best investigated (Schopfer and Plachy, 1985; Finkelstein et al., 2002). ABA exerts its influence in germination by suppressing water uptake by the seed to reduce embryo growth potential at early stages (Schopfer and Plachy, 1985) and by inhibiting endosperm rupture to delay germination (Müller et al., 2006). Genetic and molecular studies seeking mutants with altered ABA sensitivity have revealed that transcriptional regulation is important in ABA responses in seeds (Finkelstein et al., 2002; Kucera et al., 2005; Holdsworth et al., 2008), which is mediated chiefly by three seed-expressed ABSCISIC ACID-insensitive (ABI) transcription factors, ABI3, ABI4, and ABI5 (Giraudat et al., 1992; Finkelstein et al., 1998; Finkelstein and Lynch, 2000; Penfield et al., 2006). ABI3 is essential for seed maturation and desiccation tolerance during embryogenesis (Nambara et al., 1995), and both ABI4 and ABI5 act in the ABA inhibition of seed germination (Finkelstein, 1994; Lopez-Molina et al., 2001; Penfield et al., 2006). In contrast to the negative regulation by ABA, another plant hormone GA promotes germination (Ogawa et al., 2003) through weakening the mechanical restraint of tissues that surround the embryo and through increasing the embryo growth potential (Ogawa et al., 2003). During the germination process, the endogenous ABA content of seeds declines, while bioactive GA levels accumulate prior to radicle emergence (Ogawa et al., 2003; Weitbrecht et al., 2011). This GA-to-ABA ratio in the seeds is the primary control of germination vigor (Kucera et al., 2005; Holdsworth et al., 2008; Rajjou et al., 2012). Emerging evidence shows that other signals are also involved in seed germination. For example, reactive oxygen species act as second messengers required for germination (Bailly et al., 2008) and interact with plant hormones to coordinate germination (Müller et al., 2009). Overall, germination control, a sophisticated process, is determined by concerted actions among various endogenous signals including some yet to be discovered.
Not only is calcium (Ca2+) an indispensable nutrient for plants (White and Broadley, 2003), it also acts as a versatile second messenger through changes in free cytosolic Ca2+ level ([Ca2+]cyt) to participate in numerous developmental and adaption processes in plants, such as root hair elongation, pollen tube growth, stomatal movements, plant-microbe interactions, light signaling, and hormone responses (McAinsh and Pittman, 2009; Dodd et al., 2010; Kudla et al., 2010). The [Ca2+]cyt elevations, relative to their low levels in the resting conditions (McAinsh and Pittman, 2009), are caused by extracellular Ca2+ influx or internal Ca2+ release from the stores, both of which may encode specific cellular Ca2+ signatures to further initiate downstream gene expressions and biological events (McAinsh and Pittman, 2009; Dodd et al., 2010; Kudla et al., 2010). The spatiotemporal changes of [Ca2+]cyt in living cells are accurately controlled and sensed by an efficient Ca2+ signaling regulatory network, among which Ca2+ channels play a large role (Dodd et al., 2010; Kudla et al., 2010). In plants, several gene families are thought to encode Ca2+-permeable channels (Ward et al., 2009; Hedrich, 2012), among them Arabidopsis (Arabidopsis thaliana) glutamate receptor homologs (AtGLRs) share high structural similarity to ionotropic Glu receptor Ca2+ channels in animals (Lam et al., 1998), and consist of 20 gene family members (Lacombe et al., 2001). Studies have shown that these plant GLRs participate in diverse physiological and developmental procedures, including pollen tube morphogenesis (Michard et al., 2011), root development (Kang et al., 2004; Li et al., 2006; Vincill et al., 2013), stomatal closure regulation (Cho et al., 2009), and various stress responses (Kim et al., 2001; Meyerhoff et al., 2005; Li et al., 2013; Mousavi et al., 2013), which are mostly dependent on Ca2+. Despite the importance of AtGLR-related Ca2+ modulation in these processes, the function and molecular mechanism of Ca2+ signaling and AtGLRs in seed germination, the early plant developmental event that greatly impacts the plant’s entire life and agricultural performance (Rajjou et al., 2012), is largely unknown.
Physiological studies have shown that Ca2+ plays a protective role in salt stress responses (Liu and Zhu, 1997; Bonilla et al., 2004) and that more Ca2+ can alleviate the effects of salinity toxicity on seed germination in agricultural crops (Bonilla et al., 2004). However, the modulation of germination by Ca2+ and the underlying molecular mechanism remain elusive. In this study, we investigated and characterized the importance of the cytosolic Ca2+ signal, mediated by Arabiodopsis glutamate receptor homolog3.5 (AtGLR3.5), in germination. AtGLR3.5 regulates [Ca2+]cyt, its expression levels correlate with the onset and completion of the germination process, and it is required for normal germination. Furthermore, we show that the AtGLR3.5-mediated Ca2+ signal antagonizes the effect of ABA in seeds through inhibiting the expression of ABI4 and that ABI4 plays a fundamental role in Ca2+ responses in germination. Our results demonstrate and uncover the mechanism by which AtGLR3.5 regulates Ca2+ signaling in germination, thus providing a molecular explanation for the observation that Ca2+ alleviates decreasing in germination caused by stress. Therefore, the results highlight the significance of Ca2+ in this developmental process, particularly under stress conditions.
RESULTS AND DISCUSSION
Extracellular Calcium Enhances Seed Germination
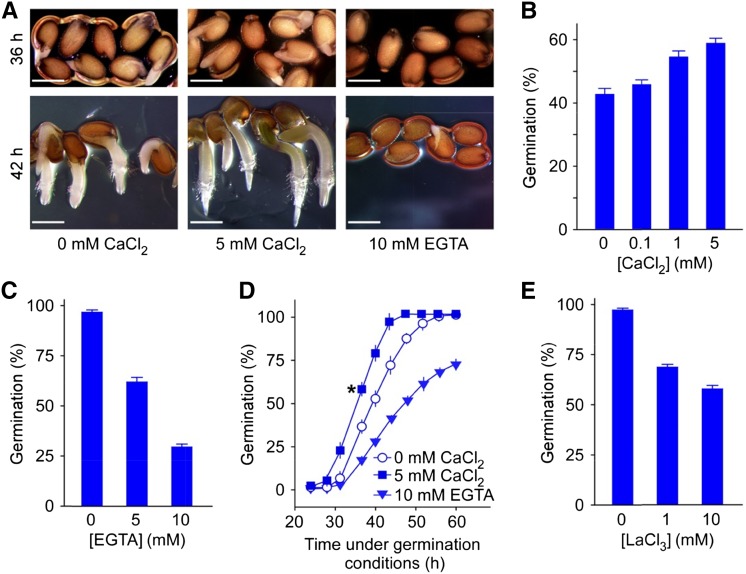
To gain insight into the role of Ca2+ in germination, we investigated whether and how Ca2+ modulates seed germination in Arabidopsis. As shown in Figure 1A, the application of CaCl2-stimulated germination (enhanced radicle emergence and subsequent root growth) compared with the control, whereas chelating Ca2+ using EGTA severely inhibited these processes. External Ca2+ at physiological levels promoted germination in a dose-dependent manner (Fig. 1B; P < 0.01 at 0.1 mm, P < 0.001 at 1 and 5 mm), although high concentrations of external Ca2+ (20–80 mm) delayed germination (Supplemental Fig. S1, A–C), presumably due to a cytotoxic effect of too much Ca2+ (White and Broadley, 2003). Moreover, EGTA drastically reduced germination, to as low as 30% of the control at 10 mm (Fig. 1C; P < 0.001 at 5 and 10 mm). These data show that adequate external Ca2+ is required for seeds to germinate, which points to an important role of Ca2+ in the earliest phase of the plant life cycle.
Figure 1.
Seed germination is enhanced by external calcium. A, Effect of calcium on germination. Arabidopsis wild-type seeds sown on modified MS medium containing 0 or 5 mm CaCl2 or 0 mm CaCl2 supplemented with 10 mm EGTA were incubated at 4°C (in the dark) for 3 d and transferred to the growth chamber (22°C) for 36 (top) or 42 h (bottom) prior to analysis. Three biological replicates were performed, and representative images are shown. B, Quantification of germination percentage at various concentrations of external Ca2+. Seeds were sown on modified MS medium containing indicated amounts of CaCl2 and scored for germination 36 h after incubation under the germination conditions. C, Effect of EGTA on germination. Seeds were sown on modified MS medium containing no CaCl2 supplemented with 5 or 10 mm EGTA and scored for germination 42 h after incubation under the germination conditions. D, Time course quantification of germination at various amounts of external Ca2+. Seeds were incubated on the same medium as in A at 22°C for 24 to 60 h and scored for germination. Asterisk indicates the germination percentage at 36 h after incubation of the seeds under the germination conditions. E, Effect of Ca2+ channel blocker LaCl3 on germination. Seeds were incubated on MS medium supplemented with indicated concentrations of LaCl3 and scored for germination 42 h after incubation under the germination conditions. For the germination analysis in B to E, data from three independent replicates are shown; error bars indicate se of the mean. Bar = 0.5 mm.
Next, we carried out a time course analysis of seed germination at 0 and 5 mm external Ca2+ concentration and found a rapid increase in the germination between 30 and 40 h after the transfer of stratified seeds to a growth chamber, with 36 h being approximately one-half of the maximum germination (Fig. 1D). This observation is comparable with previous reports showing that germination is completed between 31 and 45 h after sowing (Dekkers et al., 2013) or 36 h after imbibition under optimal conditions (Holdsworth et al., 2008; Piskurewicz et al., 2008). This supports the appropriateness of our experimental conditions and reliability of our analysis. The rapid germination increase was dramatically diminished by the removal of the Ca2+ using EGTA (Fig. 1D), indicating that Ca2+ is required for this developmental process. It is noteworthy that at approximately 60 h, when most seeds had germinated even without a Ca2+ supply, EGTA continued to inhibit germination (Fig. 1D). As the concentration of EGTA used in the experiment was high enough to chelate all the Ca2+ ions in the media, it appears likely that Ca2+, other than that in the medium, also contributes to germination. Because high amounts of Ca2+ exist in the seed coat (Punshon et al., 2012), cell wall, and apoplast (Clarkson, 1984; Hepler and Wayne, 1985) and Ca2+-permeable channels function in Ca2+ influx to the cytoplasm (Dodd et al., 2010; Kudla et al., 2010), we hypothesized that extracellular Ca2+ enters the cytosol through plasma membrane Ca2+-permeable channels to positively regulate germination. Our hypothesis was supported by pharmacological assays that showed LaCl3, a commonly used external Ca2+ channel blocker (Knight et al., 1996), significantly reduced germination (Fig. 1E; P < 0.001 at 1 and 10 mm). Together, these results suggest that extracellular Ca2+ influx by Ca2+-permeable channels in the plasma membrane plays a large role in germination.
AtGLR3.5 Mediates [Ca2+]cyt Elevation and Is Required for Seed Germination
Because our results indicate the involvement of Ca2+ channels in germination (Fig. 1), we searched publicly available gene expression databases for candidate Arabidopsis GLR Ca2+ channels that modulate seed germination. Using the eFP Browser that presents gene expression data throughout Arabidopsis development (http://bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi; Winter et al., 2007), we found that among the 20 members of the AtGLR family, AtGLR3.5 (At2g32390) and AtGLR3.7 (At2g32400) show clear transcriptional increases during the transition from dry seeds to imbibed seeds. We focused on AtGLR3.5 in this study because its expression increases much more markedly during the germination transition than AtGLR3.7, and AtGLR3.5 is most highly expressed in imbibed seeds but much less in dry seeds and vegetative tissues compared with the ubiquitous, high expression of AtGLR3.7 in various tissues. AtGLR3.7 knockout seeds showed a weak germination phenotype (Supplemental Fig. S2; P < 0.05).
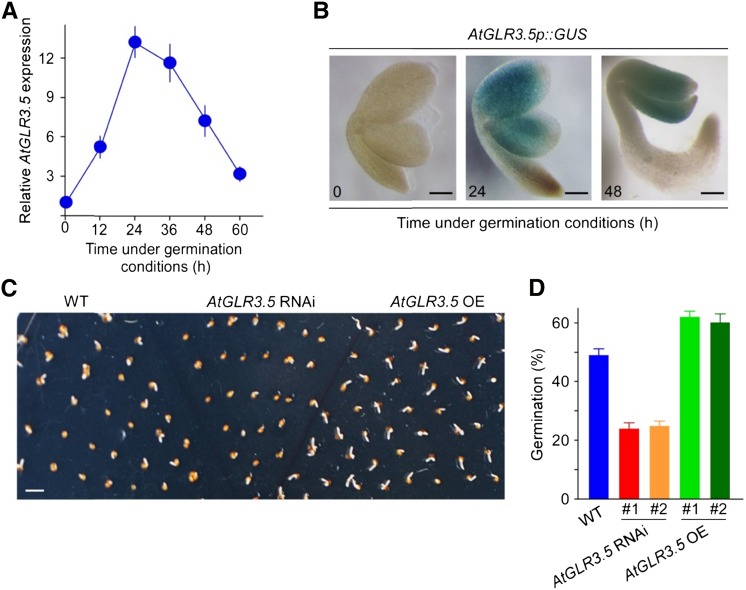
To verify the connection of AtGLR3.5 with germination, we performed quantitative reverse transcription (qRT)-PCR analysis that showed clear up-regulation of AtGLR3.5 mRNA transcript in 24-h-imbibed seeds (Supplemental Fig. S3A). Then, to obtain a more detailed understanding of the AtGLR3.5 expression pattern, we analyzed AtGLR3.5 transcript levels during the entire germination process. This revealed an immediate steep increase in the transcript level at the outset and a gradual decrease upon completion of germination, peaking at 24 h (approximately 12-fold; Fig. 2A). These data show that AtGLR3.5 is predominantly expressed in germinating seeds but not in dry or fully germinated seeds. To determine the site of AtGLR3.5 expression in germinating seeds, we created transgenic plants expressing the GUS reporter gene under a 2.1-kb promoter sequence of AtGLR3.5 and found that AtGLR3.5 was mainly expressed in embryonic cotyledons (Fig. 2B). At the seedling stage, AtGLR3.5 transcript was detected in both roots and shoots (Supplemental Fig. S3B). Compared with no detectable GUS signal at 0 h, AtGLR3.5 expression was highly up-regulated at 24 h and then decreased to a lower level at 48 h after the onset of germination (Fig. 2B), consistent with the qRT-PCR results (Fig. 2A). Intriguingly, the dynamic changes (increase and decrease) in AtGLR3.5 expression level in seeds (Fig. 2A) correlate with the time points of the onset of the rapid germination (30–40 h; Fig. 1D) and the completion of germination (60 h; Fig. 1D). The spatiotemporal expression pattern of AtGLR3.5 strongly implies a possible role for the gene in this critical developmental process.
Figure 2.
AtGLR3.5 is required for seed germination. A, Time course analysis of AtGLR3.5 expression during germination by qRT-PCR. Wild-type (WT) seeds incubated in water were stratified and collected at the indicated time points after incubation under the germination conditions. Data shown indicate means ± se of the mean (n = 3). B, AtGLR3.5p::GUS reporter analysis in seed embryo during germination. Seeds were stratified and collected for staining at the indicated time points after incubation under the germination conditions. Images were taken after removal of the seed coat. C, Germination phenotype of wild-type, AtGLR3.5 RNAi, and AtGLR3.5 OE seeds on MS medium 40 h after incubation under the germination conditions. Three biological replicates were performed, and a representative image is shown. D, Germination analyses of wild-type, AtGLR3.5 RNAi, and AtGLR3.5 OE seeds 36 h after incubation under germination conditions. Bars = 100 µm (B) and 2 mm (D).
To assess the function of AtGLR3.5 in germination, we produced transgenic plants with altered AtGLR3.5 expressions and tested their germination phenotypes. A 35S::AtGLR3.5 RNA interference (RNAi) construct and a 35S::AtGLR3.5 overexpression (OE) construct were introduced into wild-type Arabidopsis, and a large reduction or increase in the AtGLR3.5 transcript level was found in the resulting AtGLR3.5 RNAi or AtGLR3.5 OE lines, respectively (Supplemental Fig. S4, A and B). Significantly, the transcript levels of AtGLR3.4, the closest homolog to AtGLR3.5 (Chiu et al., 2002), and AtGLR3.1 were unaffected in the two AtGLR3.5 RNAi lines (Supplemental Fig. S5). The AtGLR3.5 RNAi seeds and AtGLR3.5 OE seeds displayed opposing patterns in germination in Murashige and Skoog (MS) media, with the former germinating more slowly and the latter faster than the wild type (Fig. 2, C and D; P < 0.01), suggesting that AtGLR3.5 has an essential positive effect on germination.
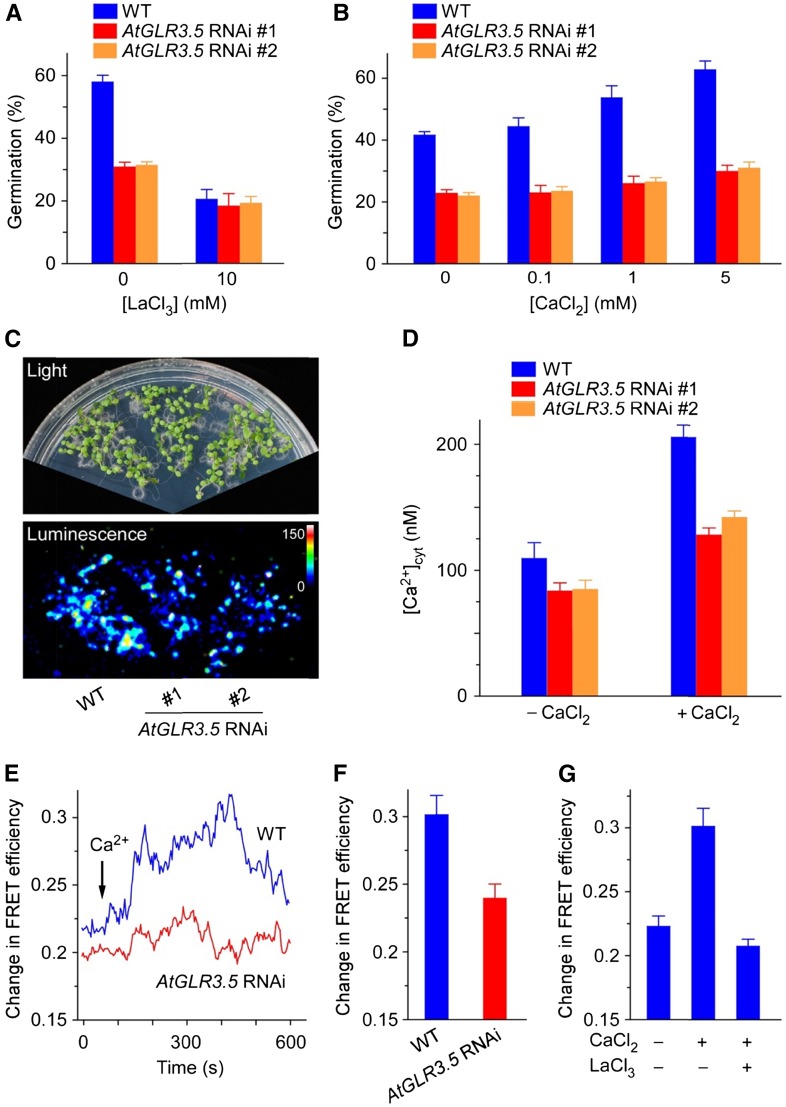
We next checked the connection of AtGLR3.5 with calcium in seed germination. It was found that the AtGLR3.5 RNAi seeds were much less sensitive to LaCl3 than the wild type (Fig. 3A), implying the Ca2+ channel function of AtGLR3.5 in germination. As external Ca2+ concentration increases, wild-type germination increased, an effect that was largely abolished in the AtGLR3.5 RNAi seeds (Fig. 3B; P < 0.001 at all Ca2+ concentrations examined), indicating the importance of AtGLR3.5 in mediating Ca2+ influx in germination. External Ca2+ treatment had no clear effect on the expression of AtGLR3.5 gene under the germination conditions (Supplemental Fig. S6), suggesting a posttranscriptional and/or posttranslational regulation of AtGLR3.5 by Ca2+. These results together indicate that the essential role of AtGLR3.5 in germination is presumably achieved by mediating Ca2+ influx.
Figure 3.
AtGLR3.5 modulates [Ca2+]cyt fluctuation. A, Germination analysis of AtGLR3.5 RNAi seeds in the absence and presence of LaCl3. Seeds sown on MS medium supplemented with indicated concentrations of LaCl3 were scored for germination 36 h after incubation under the germination conditions. B, Germination analysis of AtGLR3.5 RNAi seeds at various concentrations of CaCl2. Seeds sown on modified MS medium containing indicated amounts of CaCl2 were scored for germination 36 h after incubation under the germination conditions. C, Ca2+-sensitive photoprotein aequorin imaging analyses of [Ca2+]cyt in 8-d-old wild-type (WT) and AtGLR3.5 RNAi seedlings. D, Aequorin luminescence-based [Ca2+]cyt quantification in 8-d-old seedlings before and after 10 mm CaCl2 treatment. Data shown are means ± se of the mean (n = 30 seedlings). E, [Ca2+]cyt-dependent FRET efficiency changes in wild-type and AtGLR3.5 RNAi seedlings in response to CaCl2 (10 mm). The primary roots of 4-d-old Arabidopsis plants expressing YC3.60 were used for the analysis. Arrows indicate the time point of CaCl2 addition. F, Quantification of [Ca2+]cyt changes in the wild type and AtGLR3.5 RNAi mutant 300 s after CaCl2 (10 mm) application, based on FRET efficiency changes as in E. Data shown are means ± se of the mean (n = 10 seedlings). G, Effects of CaCl2 (10 mm) and LaCl3 (10 mm) on FRET efficiency changes in wild-type plants. Data shown are means ± se of the mean (n = 16 seedlings). In A and B, data shown indicate means ± se of the mean (n = 3).
We further examined whether AtGLR3.5 directly modulates [Ca2+]cyt by using calcium indicator proteins aequorin (Knight et al., 1991) and yellow cameleon3.60 (YC3.60; Rincón-Zachary et al., 2010). Aequorin luminescence-based Ca2+ imaging analyses showed that knockdown of AtGLR3.5 lowered the steady-state [Ca2+]cyt in young seedlings (Fig. 3, C and D). Importantly, the reduction in [Ca2+]cyt in the AtGLR3.5 RNAi seedlings was more pronounced upon external Ca2+ treatment by which Ca2+-induced [Ca2+]cyt increase (CICI) occurred (Fig. 3D; P < 0.01). These results suggest that AtGLR3.5 controls the resting [Ca2+]cyt as well as CICI response. We further conducted fluorescence resonance energy transfer (FRET)-sensitized emission imaging analysis of Arabidopsis plants expressing YC3.60, as previously described (Rincón-Zachary et al., 2010). We observed the expected CICI in the wild type and strongly impaired response in AtGLR3.5 RNAi plants (Fig. 3, E and F; P < 0.01). By contrast, the hydrogen peroxide (H2O2)-induced [Ca2+]cyt increase (Demidchik et al., 2007) appeared normal in both wild-type and AtGLR3.5 RNAi plants (Supplemental Fig. S7). Moreover, preincubation of wild-type seedlings with LaCl3 largely abolished the CICI (Fig. 3G), giving further evidence that the CICI is mediated primarily by Ca2+-permeable channels on the plasma membrane. Thus, the findings that AtGLR3.5 RNAi lines had lower [Ca2+]cyt than the wild type (Fig. 3, C and D) as well as impaired CICI response (Fig. 3, D and E) are in agreement with and also explain our prior observations that AtGLR3.5 RNAi seeds geminated more slowly than wild-type controls (Fig. 2, C and D) and were less sensitive to various amounts of external Ca2+ in germination (Fig. 3B). It indicates that external Ca2+ may activate AtGLR3.5 at the protein level, rather than up-regulation of the gene expression to promote seed germination (Supplemental Fig. S6). In addition, the dynamic expression pattern of AtGLR3.5 during seed germination (Fig. 2A) may indicate a transient induction of AtGLR3.5 and an increase in [Ca2+]cyt in germinating seeds. Collectively, these results imply that AtGLR3.5 modulates Ca2+ influx, and thus [Ca2+]cyt increases during the germination process, to ensure proper seed germination.
AtGLR3.5-Mediated Ca2+ Signal Alleviates the Inhibitory Effects of ABA on Germination
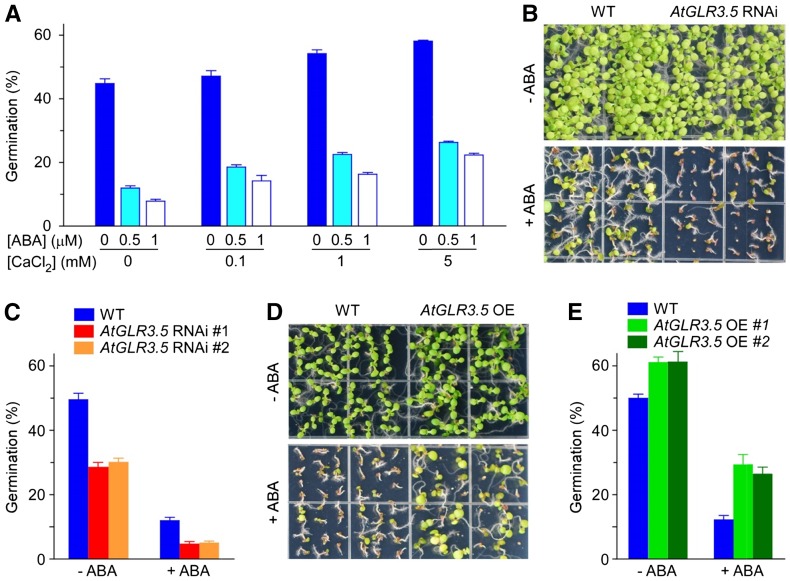
The phytohormone ABA has a prominent role in germination inhibition (Schopfer and Plachy, 1985; Finkelstein et al., 2002). Because the effect of Ca2+ in promoting radicle emergence and germination was observed (Fig. 1), we were curious as to whether the effects of Ca2+ and ABA, two signaling molecules that have opposite influences in germination regulation, are correlated. To test this idea, we scored seed germination at various concentrations of Ca2+ and ABA and found that with an increase in external Ca2+ amount, the inhibitory effect of exogenous ABA on germination was mitigated (Fig. 4A). Supporting this finding, AtGLR3.5 RNAi lines, which had lower [Ca2+]cyt and were less responsive to external Ca2+ than the wild type (Fig. 3, C–F), were hypersensitive to ABA in seed germination compared with the wild type (Fig. 4, B and C; P < 0.001). Also consistent, AtGLR3.5 OE seeds were more resistant to ABA than the wild type (Fig. 4, D and E; P < 0.001). qRT-PCR analysis shows that ABA had no clear effect on the AtGLR3.5 expression level under germination conditions (Supplemental Fig. S8). Taken together, these results indicate that the Ca2+ signal, mediated by AtGLR3.5, antagonizes the effect of ABA in suppressing seed germination.
Figure 4.
AtGLR3.5 and calcium affect ABA sensitivity in seed germination. A, Calcium alleviates the inhibitory effect of ABA on germination. B, Phenotypes of 15-d-old wild-type (WT) and AtGLR3.5 RNAi seedlings treated with or without ABA (1 µm). C, Germination analysis of wild-type and AtGLR3.5 RNAi seeds treated with or without ABA (1 µm). D, Phenotypes of 9-d-old wild-type and AtGLR3.5 OE seedlings in the absence and presence of ABA (1 µm). E, Germination analysis of wild-type and AtGLR3.5 OE seeds in the presence and absence of ABA (1 µm). For the germination analysis in A, C, and E, seeds were scored for germination 36 h after incubation under the germination conditions, and data shown are means ± se of the mean (n = 3). In B and D, three independent experiments were performed and representative images are shown.
Cytosolic Ca2+ Signal Mediated by AtGLR3.5 Acts against ABI4 to Foster Seed Germination
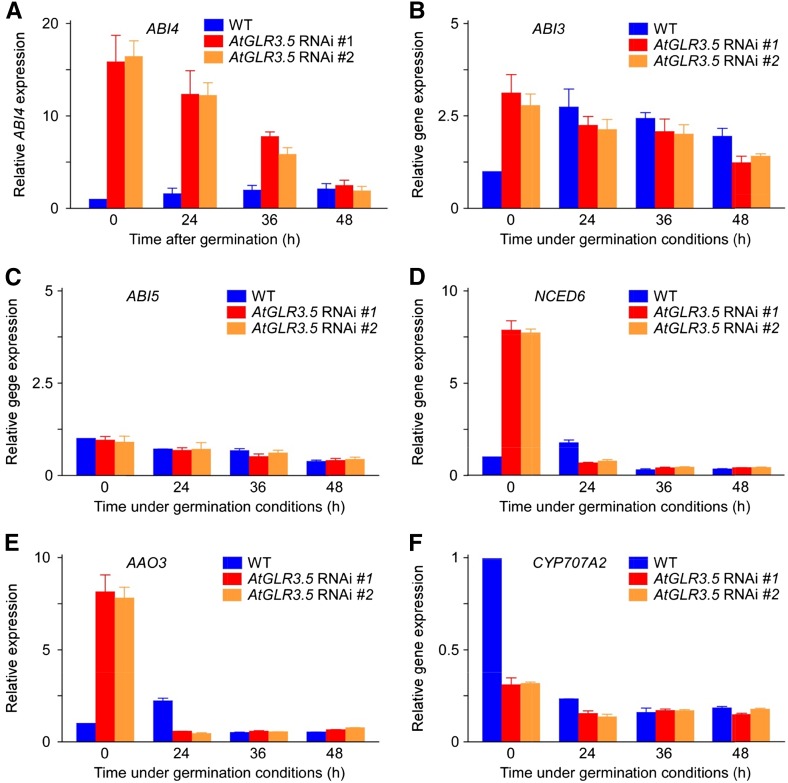
To understand the molecular mechanism by which Ca2+ counteracts the effect of ABA on seed germination, we examined the expression of some key ABA regulators involved in germination, including ABI3, ABI4, and ABI5 from the ABA signaling network, 9-CIS-EPOXYCAROTENOID DIOXYGENASE6 (NCED6) and ABSCISIC ALDEHYDE OXIDASE3 (AAO3) from the ABA biosynthesis pathway, and CYTOCHROME P450, FAMILY 707, SUBFAMILY A, POLYPEPTIDE2 (CYP7072A2) in ABA catabolism, in wild-type and AtGLR3.5 RNAi seeds. As shown in Figure 5A, a drastic up-regulation (approximately 15-fold) of ABI4 expression was observed in stratified AtGLR3.5 RNAi seeds compared with the wild type. This elevation began to decrease and gradually dropped to the wild-type level at approximately 48 h after incubation of the seeds under germination conditions. A 3-fold increase in the ABI3 transcript level was found in the stratified AtGLR3.5 RNAi seeds compared with the wild type, but there was no difference in the ABI3 transcript level between wild-type and AtGLR3.5 RNAi seeds after 24-h incubation under the germination conditions (Fig. 5B). In addition, a previous study has shown that ABI3 expression does not regulate seed germination (Bassel et al., 2006). No apparent difference was found in the expression level of ABI5 during the entire germination period (Fig. 5C). In contrast to the higher expression of ABI4 in the germinating AtGLR3.5 RNAi seeds (Fig. 5A), the stratified AtGLR3.5 OE seeds had lower ABI4 expression than the wild type (Supplemental Fig. S9; P < 0.01).
Figure 5.
AtGLR3.5 influences the expression of key ABA regulators in seed germination. Expression analyses of genes involved in ABA signaling (A–C) and metabolic pathways (d–F) in wild-type (WT) and AtGLR3.5 RNAi seeds at indicated time points after incubation under germination conditions. Seeds were stratified at 4°C for 3 d, transferred to a growth chamber for indicated time periods, and collected for analysis. Data shown are means ± se of the mean (n = 3).
A recent finding has shown that ABI4 positively regulates ABA biogenesis during seed germination and directly represses the expression of the ABA catabolic gene CYP707A2 (Shu et al., 2013). In line with this report, our results showed that the expression levels of two crucial ABA biosynthesis genes NCED6 and AAO3 were significantly higher in stratified AtGLR3.5 RNAi seeds and then rapidly decreased to levels lower than the wild type 24 h after incubation under germination conditions (Fig. 5, D and E). We also found that the transcript of CYP707A2 was down-regulated in the AtGLR3.5 RNAi seeds prior to germination and recovered to the wild-type level 24 h after incubation under germination conditions (Fig. 5F). Consistent with the expression patterns of these ABA biogenesis and catabolic genes, the ABA content in the stratified AtGLR3.5 RNAi seeds was higher than the wild type, and this difference did not persist 24 h after incubation of the seeds (Supplemental Fig. S10; P < 0.01 at 0-h time point). The expression levels of NCED6 and AAO3 were lower in the stratified AtGLR3.5 OE seeds (Supplemental Fig. S9; P < 0.01), and the ABA content in the stratified AtGLR3.5 OE seeds was clearly lower than that in the wild type (Supplemental Fig. S10; P < 0.05 at 0 h). Thus, we conclude that the expression level of AtGLR3.5 affects both ABA biogenesis and ABA signaling in seed germination and that the expression of ABI4 is significantly increased at early germination stages in the AtGLR3.5 RNAi seeds compared with the wild type.
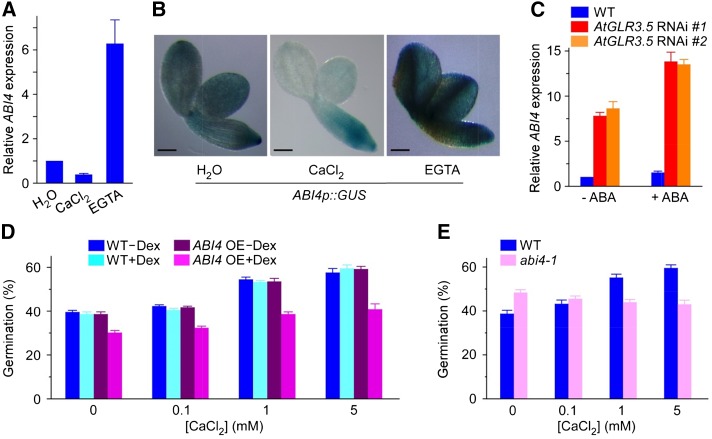
To further determine whether the increase in ABI4 expression level in the AtGLR3.5 RNAi seeds is relevant to the impaired elevation in [Ca2+]cyt (Fig. 3, C–F), we next analyzed the effect of external Ca2+ or EGTA on ABI4 expression in wild-type seeds. As shown in Figure 6A, the ABI4 expression level was clearly suppressed by CaCl2 but greatly induced by EGTA after 24-h incubation under germination conditions. In addition, histochemical analysis of GUS activity in ABI4p::GUS transgenic seeds displayed a very similar pattern. Compared with the control, the addition of Ca2+ caused a decrease in ABI4 expression, while adding EGTA had the opposite effect (Fig. 6B). It is worth noting that the suppression of ABI4 expression by Ca2+ occurred preferentially in the embryonic cotyledons (Fig. 6B), the same region in seeds where AtGLR3.5 is predominantly expressed (Fig. 2B). Together, these results provide strong evidence that AtGLR3.5-mediated [Ca2+]cyt increase causes a decrease in ABI4 expression in germinating seeds.
Figure 6.
Interplay between calcium and ABA converges at ABI4. A, Relative expression of ABI4 in response to CaCl2 (1 mm) or EGTA (5 mm) in wild-type (WT) seeds. B, Histochemical staining of the GUS reporter driven by the ABI4 promoter (ABI4p::GUS) in response to CaCl2 (1 mm) or EGTA (5 mm) treatment. Images were taken after removal of the seed coat. C, Expression analysis of ABI4 in wild-type or AtGLR3.5 RNAi lines in response to ABA (20 µm). D, Germination analysis of wild-type and Dex-inducible ABI4 OE seeds at various concentrations of CaCl2 in the absence or presence of Dex (2 µm). E, Germination analysis of abi4-1 mutant at various concentrations of CaCl2. In A to C, seeds were stratified in indicated solution and transferred to the growth chamber for 24 h before analysis. In D and E, seeds were scored for germination 36 h after incubation under the germination conditions. In A and C, data shown are means ± se of the mean (n = 3). Bar = 100 µm.
ABI4 regulates the ABA response in seeds (Finkelstein et al., 2002; Penfield et al., 2006), and its expression is induced by ABA (Penfield et al., 2006). Because Ca2+ represses ABI4 expression during germination (Fig. 6, A and B) and antagonizes the effects of ABA (Fig. 4A), we examined whether the influence of Ca2+ on ABI4 expression is effective in the presence of ABA. As shown in Figure 6C, compared with the control (no ABA treatment), the increase in the level of ABI4 transcript was much larger in AtGLR3.5 RNAi seeds than the wild type in the presence of ABA, indicating that AtGLR3.5 plays a crucial role in repression of ABA-induced ABI4 expression during germination. Together, these results provide unequivocal molecular evidence illustrating the importance of the change in the [Ca2+]cyt mediated by AtGLR3.5 in ABA response during seed germination, which is consistent with our former observations that AtGLR3.5 and Ca2+ affect ABA sensitivity in seeds (Fig. 4, A, C, and E). Furthermore, our results also suggest that AtGLR3.5 may impede ABA signaling, at least in part, through modulating ABI4 expression to promote seed germination.
Our results show that the effects of the AtGLR3.5-mediated Ca2+ signal on germination may be largely determined by changes in ABI4 expression (Fig. 6, A–C). Thus, we further tested the Ca2+ responses in ABI4 OE and abi4-1 mutant plants. The germination of ABI4 OE seeds in which ABI4 expression is driven by the dexamethasone (Dex)-inducible promoter (Shkolnik-Inbar and Bar-Zvi, 2010) was indistinguishable from the wild type when Dex was absent (Fig. 6D). In the presence of Dex, however, the germination of ABI4 OE seeds was significantly lower than the wild type, irrespective of external Ca2+ concentrations (Fig. 6D; P < 0.001), indicating the inhibitory effect of ABI4 on seed germination. Thus, the phenotype of AtGLR3.5 RNAi seeds, which show highly up-regulated ABI4 expression, late germination, and less responsiveness to external Ca2+ compared with the wild type (Figs. 2D, 3B, and 5A), bears a strong resemblance to that of the ABI4 OE plants (Fig. 6D; Shkolnik-Inbar and Bar-Zvi, 2010). By contrast, the abi4-1 mutants that contain no functional ABI4 (Finkelstein, 1994) showed unimpaired or slightly better germination on medium containing 0 or 0.1 mm Ca2+ and reduced germination at 1 and 5 mm Ca2+ compared with wild-type seeds (Fig. 6E). These results suggest that ABI4 plays not only negative but also positive roles in germination when Ca2+ is present. Interestingly, a previous report has shown both positive and negative roles of ABI4 in root growth (Cui et al., 2012). Together, these results indicate a fundamental role of ABI4 in modulation of Ca2+-dependent germination and further support that the elevated expression of ABI4 is a major cause of the delayed germination of AtGLR3.5 RNAi seeds.
The ability of the seed embryo to resume its biochemical and molecular activities determines germination vigor (Rajjou et al., 2012). ABI4 (Finkelstein et al., 1998), an APETALA2/ETHYLENE-RESPONSIVE ELEMENT BINDING FACTOR (AP2/ERF) transcription factor involved in ABA responses in seeds, is specifically transcribed in the embryo, where it represses the breakdown of seed lipids (Penfield et al., 2006), thereby contributing to ABA sensitivity in the seed embryo (Penfield et al., 2006; Weitbrecht et al., 2011). Studies have shown that transcriptional regulation plays a critical role in the ABA response in germinating seeds (Finkelstein et al., 2002; Kucera et al., 2005; Holdsworth et al., 2008). Thus, the AtGLR3.5-mediated Ca2+ repression of ABI4 expression (Figs. 5A and 6, A–C) may indicate more lipid breakdown and better embryonic growth potential, which is supported by our observations that AtGLR3.5 OE seeds germinated faster than the wild type (Fig. 2, C and D) and that Ca2+ stimulates enhanced germination both in the presence and absence of ABA (Figs. 1 and 4A).
The completion of seed germination requires coordination between the embryo and the surrounding endosperm and testa layers (Penfield et al., 2006). In Arabidopsis, ABA represses embryo growth potential in seeds (Schopfer and Plachy, 1985) and delays endosperm rupture but not testa rupture (Müller et al., 2006). The ABI4 transcript is confined to the embryo (Penfield et al., 2006), while ABI5 is expressed in both embryo and micropylar endosperm, a barrier for the radicle protrusion (Lopez-Molina et al., 2001; Penfield et al., 2006). These two ABI transcriptional factors play a synergistic role in regulating downstream gene expression and physiological responses (Reeves et al., 2011). Our results demonstrate that the [Ca2+]cyt signal generated by AtGLR3.5 specifically affects ABI4 but not ABI5 expression in germinating seeds (Fig. 5, A and C), and the ABI4 regulation by Ca2+ also occurs when ABA is present (Fig. 6C), suggesting a specific role of Ca2+ in the embryo tissue during germination. Studies have shown that an early signal originating from the embryo is required to induce endosperm weakening to facilitate germination (Müller et al., 2006). Our results suggest that Ca2+ regulation of ABI4 expression occurs at early germination stages, 24 h before germination onset (Fig. 6, A and B). Thus, it will be interesting to investigate whether Ca2+ modulation of ABI4 in the embryo tissue also plays a role in the early embryo-endosperm interaction and endosperm rupture during seed germination.
Beyond ABA, germination is largely modulated by plant hormone GA (Ogawa et al., 2003; Kucera et al., 2005). A recent report shows that ABI4 positively regulates ABA biogenesis and negatively regulates GA biosynthesis during seed germination (Shu et al., 2013). Because we have found that AtGLR3.5-mediated Ca2+ signal represses ABI4 expression and counteracts the ABA effect on germination, interesting future work might be to investigate whether Ca2+ is also a part of GA signaling system in germination control.
AtGLR3.5-Modulated Seed Germination May Involve Cellular Ca2+-Sensing Molecules
As a crucial cellular signaling molecule, Ca2+ functions by eliciting characteristic transient [Ca2+]cyt fluctuations at various developmental stages or upon specific stimuli (Dodd et al., 2010; Kudla et al., 2010). These Ca2+ signatures are detected and decoded by downstream cellular Ca2+-sensing components that contain Ca2+-binding domains, including calmodulin (CaM), Ca2+-dependent protein kinase (CPK), and calcineurin B-like protein, which convert the information of the Ca2+ transients into biochemical events and physiological responses (Luan, 2009; Dodd et al., 2010; Kudla et al., 2010). Interestingly, studies have shown that disturbing the function of some Ca2+-sensing proteins cause germination problems (Rivetta et al., 1997; Pandey et al., 2004; Zhao et al., 2011). The finding that AtGLR3.5 modulates [Ca2+]cyt and germination prompted us to examine the expression of Ca2+-sensing genes in AtGLR3.5 RNAi seeds. It was found that consistent with the up-regulation of AtGLR3.5 at early germination stages (Fig. 2A), the expression of CaM1, CaM7, and CPK3 in wild-type seeds was significantly up-regulated as germination begins; however, these increases were largely abolished in germinating AtGLR3.5 RNAi seeds (Fig. 7). Germinating AtGLR3.5 OE seeds had a higher level of CaM1, CaM7, and CPK3 transcripts compared with the wild type (Supplemental Fig. S11, P < 0.01 in A and C and P < 0.05 in B). These results indicate the involvement of the Ca2+-decoding system triggered by the AtGLR3.5-mediated [Ca2+]cyt signal in germination control. Ca2+-sensing proteins either combine the calcium-sensing function mediated by the Ca2+-binding domains with an enzymatic activity (e.g. CPKs) or have calcium-binding abilities but lack other effector domains (e.g. CaMs), which therefore need to interact with target proteins to further transmit the Ca2+ signals (Luan, 2009; Dodd et al., 2010; Kudla et al., 2010). A recent report has shown that CPK5 directly phosphorylates NADPH/respiratory burst oxidase protein D, which is a prerequisite for the downstream signal propagation (Dubiella et al., 2013). While it has been established that CaM7 directly interacts with the promoters of several light-inducible genes and regulates their expression (Kushwaha et al., 2008), other CaMs appear to modulate gene expression through their interactions with Ca2+/calmodulin-binding transcription activators that function as transcriptional (co)regulators (Finkler et al., 2007). Thus, our finding of the altered expression of CaM1, CaM7, and CPK3 in germinating AtGLR3.5 RNAi and OE seeds may indicate a complex Ca2+-decoding network involving protein phosphorylation, protein-protein interaction, and even protein-DNA interaction, which acts downstream of the AtGLR3.5-mediated Ca2+ signal prior to ABI4. Future investigation into the Ca2+-decoding network will more precisely elucidate the signaling mechanism as to how AtGLR3.5 and Ca2+ signal to ABI4 in germination control.
Figure 7.
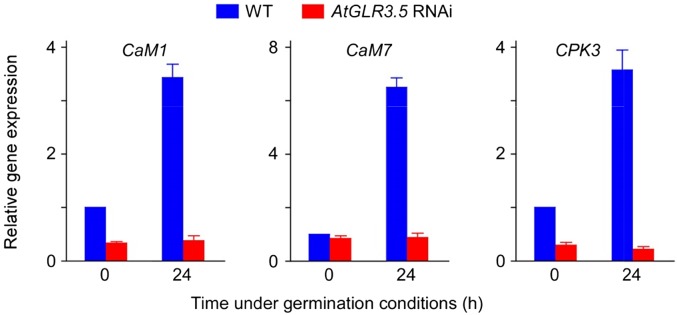
AtGLR3.5 affects the expression of cellular Ca2+-sensing molecules. Expression analyses of CaM1, CaM7, and CPK3 in wild-type (WT) and AtGLR3.5 RNAi seeds at indicated time points after incubation under the germination conditions. Seeds were stratified and collected at 0 or 24 h after transfer to a growth chamber. Data shown are means ± se of the mean (n = 3).
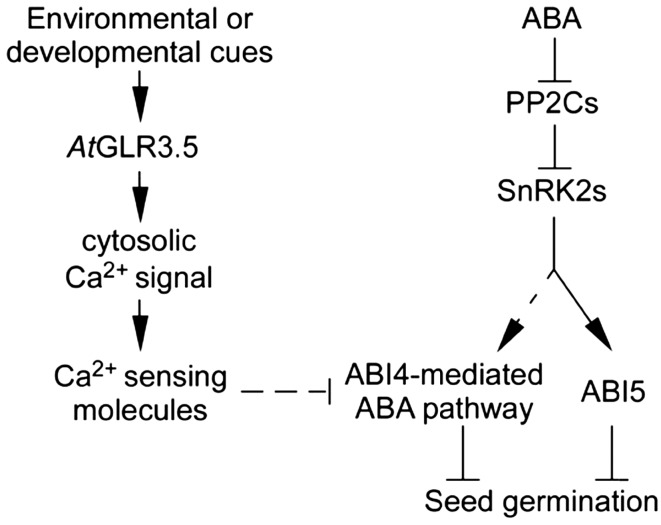
Ca2+ signals are core regulators of numerous physiological events and stress responses in plants (White and Broadley, 2003; McAinsh and Pittman, 2009; Dodd et al., 2010; Kudla et al., 2010). In this study, we investigated the role of Ca2+ signal in seed germination, the first step of plant life cycle. We show that Ca2+ signal, mediated by AtGLR3.5, positively regulates seed germination and antagonizes the inhibitory effect of ABA. AtGLR3.5 modulates steady-state [Ca2+]cyt as well as transient [Ca2+]cyt elevations upon Ca2+ stimuli. The AtGLR3.5-mediated Ca2+ signal represses ABA effects in seeds, largely through the transcription factor ABI4. Our data suggest that the signal transduction from the AtGLR3.5-mediated [Ca2+]cyt elevation to ABI4 involves cellular Ca2+-sensing molecules. A working model illustrating the AtGLR3.5-Ca2+-ABI4 regulatory network in seed germination along with the known ABA pathway that has not been approved to be Ca2+ dependent is presented in Figure 8.
Figure 8.
A working model for seed germination regulated by the AtGLR3.5-cytosolic Ca2+-ABI4 network and Ca2+-independent ABA signaling pathway. Arrows and t bars indicate activation and repression, respectively. Black lines represent direct effects, and dotted lines represent effects that have not yet been known to occur directly. PP2Cs, Type 2C protein phosphatases; SnRK2s, SNF1-related protein kinases.
Since the discovery of GLR genes in plants (Lam et al., 1998), many studies have demonstrated that Ca2+ levels mediated by AtGLRs affect myriad cellular processes (Kim et al., 2001; Meyerhoff et al., 2005; Cho et al., 2009; Michard et al., 2011; Li et al., 2013; Mousavi et al., 2013; Vincill et al., 2013). To our knowledge, this is the first report demonstrating that cytoplasmic Ca2+ dynamics modulated by a plant GLR play a pivotal role in seed germination. Recently, amino acid activation of AtGLR Ca2+ channels has been reported (Qi et al., 2006; Michard et al., 2011; Vincill et al., 2012; Tapken et al., 2013). Identification of additional AtGLR members involved in the [Ca2+]cyt modulation and germination regulation and discovering the amino acid(s) that may activate such AtGLRs would help to better describe the role of Ca2+ and AtGLRs in controlling plant physiology.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
All Arabidopsis (Arabidopsis thaliana) genotypes used in this study are in the ecotype Columbia background. Seeds of the abi4-1 mutant and transgenes expressing ABI4p::GUS, 35S::aequorin, 35S::YC3.60, or Dex-inducible ABI4 OE plants were kindly provided by Drs. Ruth Finkelstein (University of California, Santa Barbara), Marc Knight (Durham University), Magaly Rincón-Zachary and Elison B. Blancaflor (Samuel Roberts Noble Foundation), and Dudy Bar-Zvi (Ben-Gurion University), respectively. The transfer DNA insertion mutant atglr3.7 (SALK_103942) was obtained from the Arabidopsis Biological Resource Center. Arabidopsis plants were grown in soil or in petri dishes in a controlled growth chamber at 22°C under white fluorescent light (110 μmol m–2 s–1) with a 16-h-light/8-h-dark photoperiod. For the germination assay, seeds were surface sterilized with 10% (v/v) bleach (Glorix) for 20 min and rinsed five times with sterile water. Seeds were then plated on MS-based medium containing MS major salts with indicated concentrations of CaCl2, MS minor salts (Sigma), vitamins (Sigma), 1% (w/v) Suc (Sigma), and 0.8% (w/v) agar (Sigma type A) supplemented with EGTA (Sigma), LaCl3 (Sigma), or ABA (Sigma) as needed. After stratification for 3 d at 4°C in darkness, the plates were transferred to the growth chamber (22°C) for indicated time periods. Germination was scored by the first sign of radicle tip appearance based on the criteria described previously (Bewley, 1997). For the germination frequency analysis, 60 to 80 seeds per sample in each treatment were tested, and three biological replicates were performed. Seeds tested in one experiment were harvested the same day.
Plasmid Construction and Generation of Arabidopsis Transgenic Plants
As no homozygous AtGLR3.5 transfer DNA insertion lines (SALK_023880 and SALK_035264) were found, we generated AtGLR3.5 knockdown lines using the pFGC5941 RNAi vector (http://www.chromdb.org). A 279-bp fragment of the AtGLR3.5 DNA sequence was amplified by PCR using primers 5′-AATCTAGAGGCGCGCCCTCGATGTTTTCTTCAGCCAAGGC-3′ (containing XbaI and AscI restriction sites) and 5′-AAGGATCCATTTAAATCTGAGAAACCTGTATGATTCGACC-3′ (containing BamHI and SwaI restriction sites). The obtained fragment was cloned into the pFGC5941 vector first in sense orientation after cutting with AscI and SwaI and then in the opposite orientation after digesting with XbaI and BamHI, generating an AtGLR3.5 RNAi construct under the control of the 35S promoter. The AtGLR3.5 OE and AtGLR3.5p::GUS constructs were created using the Gateway cloning system (Invitrogen) by cloning the full-length AtGLR3.5 coding sequence or the 2.1-kb promoter region of AtGLR3.5 first into the pENTR/D-TOPO cloning vector and then into the binary vector pMDC32 or pMDC163 (Karimi et al., 2002), respectively. Primers used for the PCR amplifications are as follows: AtGLR3.5 full-length coding sequence, 5′-CACCATGATTCTTTCATTGGAAGAGC-3′ and 5′-TCACTGTGGAGTTTCGTGATC-3′; and AtGLR3.5 promoter region, 5′-ATTCCACTCTATAGGAAGAAAATTGTT-3′ and 5′-AAGTAAACAGAGCTCCAACGTTTACAG-3′. All constructs were verified by DNA sequencing. The resulting plasmids were introduced into Agrobacterium tumefaciens strain GV3101, which were subsequently transformed into Arabidopsis wild-type plants by the floral dip method (Clough and Bent, 1998). Transformants were selected with Basta or hygromycin. Homozygous lines carrying a single insertion in the T3 generation were used for further analysis.
Gene Expression Analysis
Total RNA was extracted using Spectrum Plant Total RNA Kit (Sigma) and reverse transcribed using iScript cDNA Synthesis Kit (Bio-Rad) after treatment with DNase I (Ambion). For the expression analyses of AtGLR3.5, AtGLR3.4, and AtGLR3.1 in the AtGLR3.5 RNAi lines, PCR amplifications were performed with 32 cycles for AtGLR3.5 and Ubiquitin21 (UBQ21) and 30 cycles for AtGLR3.4 and AtGLR3.1, using the following gene-specific primers: AtGLR3.5, 5′-ATCTCATAAGTGAAGTTGCTGCAA-3′ and 5′-TTGCTATTAAAGTGTCCATTCCTT-3′; AtGLR3.4, 5′-CAAATAGCAACTGCAAATTCCGTA-3′ and 5′-TGATTGTGAAGTCCCAGCTGCACT-3′; AtGLR3.1, 5′-TCAGATTACTGTAAGTTTGCCAT-3′ and 5′-TCATATGGGTCTTCTAGATGCAGT-3′; and UBQ21, 5′-TAGAGATGCAGGCATCAAGAGCGCGACTG-3′ and 5′-GCGGCGAGGCGTGTATACATTTGTGCCATT-3′. Quantitative PCR was performed using SYBR Green PCR Master Mix (Bio-Rad) in a CFX96 Real-Time PCR System (Bio-Rad). The expression values were normalized to that of the housekeeping gene ACTIN2. Gene-specific primers used were designed by AtRTPrimer (Han and Kim, 2006) with the following sequences: AtGLR3.5, 5′-AGGAGGCTGGTTCTTCTAGAGG-3′ and 5′-TCTCACTGTGGAGTTTCGTG-3′; and ACTIN2, 5′-GGTAACATTGTGCTCACTCGTGG-3′ and 5′-AACGACCTTAATCTTCATGCTGC-3′. Primers used for expression analyses of genes involved in ABA or GA signaling and biosynthesis pathways including ABI3, ABI4, ABI5, NCED6, AAO3, and CYP707A2 are as described (Nelson et al., 2009; Footitt et al., 2011). Primers used for the expression analyses of CaM1, CaM7, and CPK3 are as described (Liu et al., 2005; Lario et al., 2013). For each expression analysis, three biological replicates were performed with similar results, and values shown are from one experimental repetition.
Measurement of [Ca2+]cyt Level
Cytosolic free calcium concentrations were monitored using plants harboring a single functional copy of aequorin or YC3.60 as described (Knight et al., 1996; Tang et al., 2007; Rincón-Zachary et al., 2010). AtGLR3.5 RNAi lines expressing aequorin or YC3.60 were obtained by genetic crosses. For aequorin bioluminescence-based Ca2+ imaging analysis, sterilized seeds were sowed on MS plates and incubated in a growth chamber for 8 d. Seedlings were then treated with 20 µm coelenterazine (Gold Bio) 18 h prior to Ca2+ imaging. Bright-field images were taken before aequorin luminescence recording. Aequorin images were taken after 20-min recording using a PIXIS CCD camera (Princeton Instruments), both before external CaCl2 treatment to detect the resting [Ca2+]cyt level and after 10 mm CaCl2 treatment to monitor the CICI. WinView32 (Roper) software was used for the image analysis. The total amount of aequorin was estimated by treating the plants with discharging solution (0.9 m CaCl2 in 10% [v/v] ethanol). The [Ca2+]cyt was calibrated based on the aequorin luminescence intensity as described (Knight et al., 1996). Experiments were performed at room temperature.
For YC3.60-based FRET-sensitized emission imaging analysis, sterilized seeds were sowed on MS medium and incubated in a chamber for 4 d. The FRET-sensitized emission images were taken using a Leica TCS SP5 confocal laser-scanning microscope (Leica Microsystems) as described previously (Rincón-Zachary et al., 2010). Briefly, the primary roots of the seedlings were mounted with 0.1% (w/v) low-melting agarose on glass bottom microwell dishes (MatTek) and then treated with 10 mm CaCl2 or 10 mm H2O2 to elicit CICI. Plants expressing cyan fluorescent protein or yellow fluorescent protein alone were used to obtain calibration images. [Ca2+]cyt-dependent FRET efficiency changes were calculated using the Leica software as described (Rincón-Zachary et al., 2010).
Measurement of ABA Levels in Seeds
Seeds (100 mg per sample) were stratified at 4°C for 3 d and collected at 0 and 24 h after incubation under germination conditions. ABA content was measured by liquid chromatography-tandem mass spectrometry at the Proteomics and Mass Spectrometry Facility at the Donald Danforth Plant Science Center. Three replicates were performed.
Histochemical GUS Activity Analysis
GUS activity was assayed as described (Jefferson et al., 1987). Briefly, transgenic seeds or seedlings were incubated in GUS staining buffer containing 2 mg mL–1 5-bromo-4-chloro-3-indolyl-β-d-GlcA, 100 mm sodium phosphate, pH 7.0, 1 mm EDTA, pH 8.0, 1% (v/v) Triton X-100, 5 mm potassium ferrocyanide, and 5 mm potassium ferricyanide at 37°C in the dark overnight and cleaned with 70% (v/v) ethanol to remove chlorophyll. Samples in one experiment were stained for the same amount of time for comparison. Photographs were taken using a Zeiss Stemi SV6 dissection microscope equipped with a Zeiss Axio CamICc digital camera.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. High concentrations of external calcium delay seed germination.
Supplemental Figure S2. Germination analysis of atglr3.7 mutant seeds.
Supplemental Figure S3. Expression of AtGLR3.5 in seeds (dry seeds versus imbibed seeds) and seedlings.
Supplemental Figure S4. Relative AtGLR3.5 expression in 2-week-old AtGLR3.5 RNAi and AtGLR3.5 OE transgenic lines.
Supplemental Figure S5. Reverse transcription-PCR analyses show that the AtGLR3.5 RNAi construct specifically silenced AtGLR3.5 transcript but not transcripts of AtGLR.3.4 and AtGLR3.1 in AtGLR3.5 RNAi lines.
Supplemental Figure S6. Effect of CaCl2 on the expression of AtGLR3.5 in seeds.
Supplemental Figure S7. FRET-sensitized emission analysis showing [Ca2+]cyt changes in wild-type and AtGLR3.5 RNAi seedlings in response to 10 mm H2O2 treatment.
Supplemental Figure S8. Effect of ABA on the expression of AtGLR3.5 in seeds.
Supplemental Figure S9. Relative expression of ABI4, AAO3, and NCED6 in stratified wild-type and AtGLR3.5 OE seeds.
Supplemental Figure S10. ABA contents in wild-type, AtGLR3.5 RNAi, and AtGLR3.5 OE seeds.
Supplemental Figure S11. Relative expression of CaM1, CaM7, and CPK3 in germinating wild-type and AtGLR3.5 OE seeds.
Supplementary Material
Acknowledgments
We thank Drs. Ruth Finkelstein, Magaly Rincón-Zachary, Elison B. Blancaflor, Dudy Bar-Zvi, and Marc Knight for providing seeds and plasmids; Drs. Tracy Punshon and Mary Lou Guerinot for the helpful discussion; Drs. Jian-Hua Zhu and Qingmei Guan for access to a luminescence imaging camera; Amy Beaven for confocal microscopy; Jin-Young Lee for the laboratory assistance; and Roxane Bouten and Jade Lee for critical reading of the article.
Glossary
- ABA
abscisic acid
- [Ca2+]cyt
free cytosolic Ca2+ level
- qRT
quantitative reverse transcription
- MS
Murashige and Skoog
- CICI
Ca2+-induced free cytosolic Ca2+ level increase
- FRET
fluorescence resonance energy transfer
- H2O2
hydrogen peroxide
- Dex
dexamethasone
- OE
overexpression
Footnotes
This work was supported by the National Science Foundation (grant nos. IOS–1025837 and MCB–1244303 to J.M.K. and project no. IBS–R013–G2–2014–a00 to J.M.K.).
Articles can be viewed without a subscription.
References
- Bailly C, El-Maarouf-Bouteau H, Corbineau F (2008) From intracellular signaling networks to cell death: the dual role of reactive oxygen species in seed physiology. C R Biol 331: 806–814 [DOI] [PubMed] [Google Scholar]
- Bassel GW, Mullen RT, Bewley JD (2006) ABI3 expression ceases following, but not during, germination of tomato and Arabidopsis seeds. J Exp Bot 57: 1291–1297 [DOI] [PubMed] [Google Scholar]
- Bewley JD. (1997) Seed germination and dormancy. Plant Cell 9: 1055–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonilla I, El-Hamdaoui A, Bolanos L (2004) Boron and calcium increase Pisum sativum seed germination and seedling development under salt stress. Plant Soil 267: 97–107 [Google Scholar]
- Chiu JC, Brenner ED, DeSalle R, Nitabach MN, Holmes TC, Coruzzi GM (2002) Phylogenetic and expression analysis of the glutamate-receptor-like gene family in Arabidopsis thaliana. Mol Biol Evol 19: 1066–1082 [DOI] [PubMed] [Google Scholar]
- Cho D, Kim SA, Murata Y, Lee S, Jae SK, Nam HG, Kwak JM (2009) De-regulated expression of the plant glutamate receptor homolog AtGLR3.1 impairs long-term Ca2+-programmed stomatal closure. Plant J 58: 437–449 [DOI] [PubMed] [Google Scholar]
- Clarkson DT. (1984) Calcium-transport between tissues and its distribution in the plant. Plant Cell Environ 7: 449–456 [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cui H, Hao Y, Kong D (2012) SCARECROW has a SHORT-ROOT-independent role in modulating the sugar response. Plant Physiol 158: 1769–1778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekkers BJ, Pearce S, van Bolderen-Veldkamp RP, Marshall A, Widera P, Gilbert J, Drost HG, Bassel GW, Müller K, King JR, et al. (2013) Transcriptional dynamics of two seed compartments with opposing roles in Arabidopsis seed germination. Plant Physiol 163: 205–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demidchik V, Shabala SN, Davies JM (2007) Spatial variation in H2O2 response of Arabidopsis thaliana root epidermal Ca2+ flux and plasma membrane Ca2+ channels. Plant J 49: 377–386 [DOI] [PubMed] [Google Scholar]
- Dodd AN, Kudla J, Sanders D (2010) The language of calcium signaling. Annu Rev Plant Biol 61: 593–620 [DOI] [PubMed] [Google Scholar]
- Dubiella U, Seybold H, Durian G, Komander E, Lassig R, Witte CP, Schulze WX, Romeis T (2013) A CDPK/NADPH-oxidase activation circuit is required for rapid defense signaling propagation. Proc Natl Acad Sci USA 110: 8744–8749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein R. (1994) Mutations at two new Arabidopsis ABA response loci are similar to the abi3 mutations. Plant J 5: 765–771 [Google Scholar]
- Finkelstein RR, Gampala SSL, Rock CD (2002) Abscisic acid signaling in seeds and seedlings. Plant Cell 14: S15–S45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR, Lynch TJ (2000) The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell 12: 599–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR, Wang ML, Lynch TJ, Rao S, Goodman HM (1998) The Arabidopsis abscisic acid response locus ABI4 encodes an APETALA 2 domain protein. Plant Cell 10: 1043–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkler A, Ashery-Padan R, Fromm H (2007) CAMTAs: calmodulin-binding transcription activators from plants to human. FEBS Lett 581: 3893–3898 [DOI] [PubMed] [Google Scholar]
- Footitt S, Douterelo-Soler I, Clay H, Finch-Savage WE (2011) Dormancy cycling in Arabidopsis seeds is controlled by seasonally distinct hormone-signaling pathways. Proc Natl Acad Sci USA 108: 20236–20241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraudat J, Hauge BM, Valon C, Smalle J, Parcy F, Goodman HM (1992) Isolation of the Arabidopsis ABI3 gene by positional cloning. Plant Cell 4: 1251–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S, Kim D (2006) AtRTPrimer: database for Arabidopsis genome-wide homogeneous and specific RT-PCR primer-pairs. BMC Bioinformatics 7: 179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedrich R. (2012) Ion channels in plants. Physiol Rev 92: 1777–1811 [DOI] [PubMed] [Google Scholar]
- Hepler PK, Wayne RO (1985) Calcium and plant development. Annu Rev Plant Physiol 36: 379–439 [Google Scholar]
- Holdsworth MJ, Bentsink L, Soppe WJJ (2008) Molecular networks regulating Arabidopsis seed maturation, after-ripening, dormancy and germination. New Phytol 179: 33–54 [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J, Mehta S, Turano FJ (2004) The putative glutamate receptor 1.1 (AtGLR1.1) in Arabidopsis thaliana regulates abscisic acid biosynthesis and signaling to control development and water loss. Plant Cell Physiol 45: 1380–1389 [DOI] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A (2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Kim SA, Kwak JM, Jae SK, Wang MH, Nam HG (2001) Overexpression of the AtGluR2 gene encoding an Arabidopsis homolog of mammalian glutamate receptors impairs calcium utilization and sensitivity to ionic stress in transgenic plants. Plant Cell Physiol 42: 74–84 [DOI] [PubMed] [Google Scholar]
- Knight H, Trewavas AJ, Knight MR (1996) Cold calcium signaling in Arabidopsis involves two cellular pools and a change in calcium signature after acclimation. Plant Cell 8: 489–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight MR, Campbell AK, Smith SM, Trewavas AJ (1991) Transgenic plant aequorin reports the effects of touch and cold-shock and elicitors on cytoplasmic calcium. Nature 352: 524–526 [DOI] [PubMed] [Google Scholar]
- Kucera B, Cohn MA, Leubner-Metzger G (2005) Plant hormone interactions during seed dormancy release and germination. Seed Sci Res 15: 281–307 [Google Scholar]
- Kudla J, Batistic O, Hashimoto K (2010) Calcium signals: the lead currency of plant information processing. Plant Cell 22: 541–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushwaha R, Singh A, Chattopadhyay S (2008) Calmodulin7 plays an important role as transcriptional regulator in Arabidopsis seedling development. Plant Cell 20: 1747–1759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacombe B, Becker D, Hedrich R, DeSalle R, Hollmann M, Kwak JM, Schroeder JI, Le Novère N, Nam HG, Spalding EP, et al. (2001) The identity of plant glutamate receptors. Science 292: 1486–1487 [DOI] [PubMed] [Google Scholar]
- Lam HM, Chiu J, Hsieh MH, Meisel L, Oliveira IC, Shin M, Coruzzi G (1998) Glutamate-receptor genes in plants. Nature 396: 125–126 [DOI] [PubMed] [Google Scholar]
- Lario LD, Ramirez-Parra E, Gutierrez C, Spampinato CP, Casati P (2013) ANTI-SILENCING FUNCTION1 proteins are involved in ultraviolet-induced DNA damage repair and are cell cycle regulated by E2F transcription factors in Arabidopsis. Plant Physiol 162: 1164–1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Wang J, Ma C, Zhao Y, Wang Y, Hasi A, Qi Z (2013) Glutamate receptor-like channel3.3 is involved in mediating glutathione-triggered cytosolic calcium transients, transcriptional changes, and innate immunity responses in Arabidopsis. Plant Physiol 162: 1497–1509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Zhu S, Song X, Shen Y, Chen H, Yu J, Yi K, Liu Y, Karplus VJ, Wu P, et al. (2006) A rice glutamate receptor-like gene is critical for the division and survival of individual cells in the root apical meristem. Plant Cell 18: 340–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HT, Sun DY, Zhou RG (2005) Ca2+ and AtCaM3 are involved in the expression of heat shock protein gene in Arabidopsis. Plant Cell Environ 28: 1276–1284 [Google Scholar]
- Liu J, Zhu JK (1997) An Arabidopsis mutant that requires increased calcium for potassium nutrition and salt tolerance. Proc Natl Acad Sci USA 94: 14960–14964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Molina L, Mongrand S, Chua NH (2001) A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proc Natl Acad Sci USA 98: 4782–4787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan S. (2009) The CBL-CIPK network in plant calcium signaling. Trends Plant Sci 14: 37–42 [DOI] [PubMed] [Google Scholar]
- McAinsh MR, Pittman JK (2009) Shaping the calcium signature. New Phytol 181: 275–294 [DOI] [PubMed] [Google Scholar]
- Meyerhoff O, Müller K, Roelfsema MR, Latz A, Lacombe B, Hedrich R, Dietrich P, Becker D (2005) AtGLR3.4, a glutamate receptor channel-like gene is sensitive to touch and cold. Planta 222: 418–427 [DOI] [PubMed] [Google Scholar]
- Michard E, Lima PT, Borges F, Silva AC, Portes MT, Carvalho JE, Gilliham M, Liu LH, Obermeyer G, Feijó JA (2011) Glutamate receptor-like genes form Ca2+ channels in pollen tubes and are regulated by pistil D-serine. Science 332: 434–437 [DOI] [PubMed] [Google Scholar]
- Mousavi SAR, Chauvin A, Pascaud F, Kellenberger S, Farmer EE (2013) GLUTAMATE RECEPTOR-LIKE genes mediate leaf-to-leaf wound signalling. Nature 500: 422–426 [DOI] [PubMed] [Google Scholar]
- Müller K, Linkies A, Vreeburg RA, Fry SC, Krieger-Liszkay A, Leubner-Metzger G (2009) In vivo cell wall loosening by hydroxyl radicals during cress seed germination and elongation growth. Plant Physiol 150: 1855–1865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller K, Tintelnot S, Leubner-Metzger G (2006) Endosperm-limited Brassicaceae seed germination: abscisic acid inhibits embryo-induced endosperm weakening of Lepidium sativum (cress) and endosperm rupture of cress and Arabidopsis thaliana. Plant Cell Physiol 47: 864–877 [DOI] [PubMed] [Google Scholar]
- Nambara E, Keith K, Mccourt P, Naito S (1995) A regulatory role for the abi3 gene in the establishment of embryo maturation in Arabidopsis thaliana. Development 121: 629–636 [Google Scholar]
- Nelson DC, Riseborough JA, Flematti GR, Stevens J, Ghisalberti EL, Dixon KW, Smith SM (2009) Karrikins discovered in smoke trigger Arabidopsis seed germination by a mechanism requiring gibberellic acid synthesis and light. Plant Physiol 149: 863–873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonogaki H, Bassel GW, Bewley JD (2010) Germination: still a mystery. Plant Sci 179: 574–581 [Google Scholar]
- Ogawa M, Hanada A, Yamauchi Y, Kuwahara A, Kamiya Y, Yamaguchi S (2003) Gibberellin biosynthesis and response during Arabidopsis seed germination. Plant Cell 15: 1591–1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey GK, Cheong YH, Kim KN, Grant JJ, Li L, Hung W, D’Angelo C, Weinl S, Kudla J, Luan S (2004) The calcium sensor calcineurin B-like 9 modulates abscisic acid sensitivity and biosynthesis in Arabidopsis. Plant Cell 16: 1912–1924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penfield S, Li Y, Gilday AD, Graham S, Graham IA (2006) Arabidopsis ABA INSENSITIVE4 regulates lipid mobilization in the embryo and reveals repression of seed germination by the endosperm. Plant Cell 18: 1887–1899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piskurewicz U, Jikumaru Y, Kinoshita N, Nambara E, Kamiya Y, Lopez-Molina L (2008) The gibberellic acid signaling repressor RGL2 inhibits Arabidopsis seed germination by stimulating abscisic acid synthesis and ABI5 activity. Plant Cell 20: 2729–2745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punshon T, Hirschi K, Yang J, Lanzirotti A, Lai B, Guerinot ML (2012) The role of CAX1 and CAX3 in elemental distribution and abundance in Arabidopsis seed. Plant Physiol 158: 352–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Z, Stephens NR, Spalding EP (2006) Calcium entry mediated by GLR3.3, an Arabidopsis glutamate receptor with a broad agonist profile. Plant Physiol 142: 963–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajjou L, Duval M, Gallardo K, Catusse J, Bally J, Job C, Job D (2012) Seed germination and vigor. Annu Rev Plant Biol 63: 507–533 [DOI] [PubMed] [Google Scholar]
- Reeves WM, Lynch TJ, Mobin R, Finkelstein RR (2011) Direct targets of the transcription factors ABA-Insensitive (ABI) 4 and ABI5 reveal synergistic action by ABI4 and several bZIP ABA response factors. Plant Mol Biol 75: 347–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rincón-Zachary M, Teaster ND, Sparks JA, Valster AH, Motes CM, Blancaflor EB (2010) Fluorescence resonance energy transfer-sensitized emission of yellow cameleon 3.60 reveals root zone-specific calcium signatures in Arabidopsis in response to aluminum and other trivalent cations. Plant Physiol 152: 1442–1458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivetta A, Negrini N, Cocucci M (1997) Involvement of Ca2+-calmodulin in Cd2+ toxicity during the early phases of radish (Raphanus sativus L.) seed germination. Plant Cell Environ 20: 600–608 [Google Scholar]
- Schopfer P, Plachy C (1985) Control of seed-germination by abscisic-acid: III. Effect on embryo growth-potential (Minimum turgor pressure) and growth coefficient (Cell-wall extensibility) in Brassica-Napus L. Plant Physiol 77: 676–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shkolnik-Inbar D, Bar-Zvi D (2010) ABI4 mediates abscisic acid and cytokinin inhibition of lateral root formation by reducing polar auxin transport in Arabidopsis. Plant Cell 22: 3560–3573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu K, Zhang H, Wang S, Chen M, Wu Y, Tang S, Liu C, Feng Y, Cao X, Xie Q (2013) ABI4 regulates primary seed dormancy by regulating the biogenesis of abscisic acid and gibberellins in Arabidopsis. PLoS Genet 9: e1003577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang RH, Han S, Zheng H, Cook CW, Choi CS, Woerner TE, Jackson RB, Pei ZM (2007) Coupling diurnal cytosolic Ca2+ oscillations to the CAS-IP3 pathway in Arabidopsis. Science 315: 1423–1426 [DOI] [PubMed] [Google Scholar]
- Tapken D, Anschütz U, Liu LH, Huelsken T, Seebohm G, Becker D, Hollmann M (2013) A plant homolog of animal glutamate receptors is an ion channel gated by multiple hydrophobic amino acids. Sci Signal 6: ra47. [DOI] [PubMed] [Google Scholar]
- Vincill ED, Bieck AM, Spalding EP (2012) Ca2+ conduction by an amino acid-gated ion channel related to glutamate receptors. Plant Physiol 159: 40–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincill ED, Clarin AE, Molenda JN, Spalding EP (2013) Interacting glutamate receptor-like proteins in Phloem regulate lateral root initiation in Arabidopsis. Plant Cell 25: 1304–1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward JM, Mäser P, Schroeder JI (2009) Plant ion channels: gene families, physiology, and functional genomics analyses. Annu Rev Physiol 71: 59–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weitbrecht K, Müller K, Leubner-Metzger G (2011) First off the mark: early seed germination. J Exp Bot 62: 3289–3309 [DOI] [PubMed] [Google Scholar]
- White PJ, Broadley MR (2003) Calcium in plants. Ann Bot (Lond) 92: 487–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ (2007) An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao R, Sun HL, Mei C, Wang XJ, Yan L, Liu R, Zhang XF, Wang XF, Zhang DP (2011) The Arabidopsis Ca2+-dependent protein kinase CPK12 negatively regulates abscisic acid signaling in seed germination and post-germination growth. New Phytol 192: 61–73 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.