Phenotypic variation of maize root system architecture highlights a genetic trade-off between small compact and large, exploratory root systems.
Abstract
The quest to determine the genetic basis of root system architecture (RSA) has been greatly facilitated by recent developments in root phenotyping techniques. Methods that are accurate, high throughput, and control for environmental factors are especially attractive for quantitative trait locus mapping. Here, we describe the adaptation of a nondestructive in vivo gel-based root imaging platform for use in maize (Zea mays). We identify a large number of contrasting RSA traits among 25 founder lines of the maize nested association mapping population and locate 102 quantitative trait loci using the B73 (compact RSA) × Ki3 (exploratory RSA) mapping population. Our results suggest that a phenotypic tradeoff exists between small, compact RSA and large, exploratory RSA.
Maize (Zea mays) serves a key role in food, feedstock, and biofuel production throughout the world. To date, maize improvement through breeding has kept pace with the increasing demand for this crop (faostat3.fao.org). This feat has been accomplished through the utilization of the tremendous genetic diversity in maize (Flint-Garcia et al., 2005; Jiao et al., 2012), but increasing environmental pressures and a growing global population will require unprecedented gains in yield in the coming years. In the last decade, researchers have begun to explore the possibility of yield improvements through the manipulation of root systems, for example through breeding for roots better able to cope with drought (Uga et al., 2013) and flooding (Jackson and Armstrong, 1999), the use of plant growth-promoting rhizobacteria (Silby et al., 2009), or increasing nutrient use efficiency (Garnett et al., 2009). The potential of belowground solutions to enhanced plant productivity has driven the development of numerous methodologies for phenotyping root system architecture (RSA), which is the spatial organization of the plant’s root system.
Several methods ranging from techniques adapted from medical imaging, such as x-ray tomography (Hargreaves et al., 2008) and combined positron emission tomography-magnetic resonance imaging (Jahnke et al., 2009), to refined versions of classical methods, such as field excavations (Trachsel et al., 2010) and pouch systems (Le Marié et al., 2014), have been used in attempts to understand the phenotypic consequences of genetic and environmental variation on root traits. Each root-phenotyping method has its advantages and disadvantages. Although the medical imaging-based techniques can produce highly detailed representations of roots, they are also very time consuming and require specialized equipment. Excavations, although more easily scaled to higher throughput and not requiring special equipment, are destructive and offer only coarse measurements of RSA. An alternative method for root phenotyping based on an optically clear gel substrate strikes an effective balance between throughput and detail, using a simple digital camera while maintaining precise control over environmental conditions. This platform has been used to quantify and classify distinctive root architectures from 12 rice (Oryza sativa) genotypes (Iyer-Pascuzzi et al., 2010), conduct a quantitative trait locus (QTL) mapping study of rice root traits in three dimensions (Topp et al., 2013), study interspecific and intraspecific rice root interactions (Fang et al., 2013), and quantify contributions of different root types to overall RSA (Clark et al., 2011).
Here, we describe the adaptation of this gel imaging platform for use with the large maize root system. We used the platform to quantify the phenotypic diversity of RSA among 25 of the 26 nested association mapping (NAM) founder lines, which encompass a wide spectrum of maize genetic diversity (Yu et al., 2008; McMullen et al., 2009). We found that these lines exhibit diverse RSAs, ranging from small and compact to large and exploratory, suggesting tradeoffs between different types of architectures. In order to identify genetic loci that control maize RSA traits, we characterized a subpopulation that best represented the contrast between the compact and exploratory RSAs. We phenotyped the B73 (compact) × Ki3 (exploratory) recombinant inbred line (RIL) NAM subpopulation for 19 RSA traits at three time points (Topp et al., 2013). These data were used to map 102 QTLs that localized to nine genomic clusters. We found high heritability and large-effect QTLs for most traits, in contrast to maize flowering time QTLs (Buckler et al., 2009). Additionally, several of our QTL clusters overlapped with meta-QTLs for yield traits (Tuberosa et al., 2003; Semagn et al., 2013) as well as novel and previously unreported loci, suggesting that this system can provide a time- and cost-effective means to identify genes controlling root architecture in maize.
RESULTS
Adaptation of a Gel-Based Root Imaging System for Use with Maize
We previously developed a high-throughput root imaging and analysis pipeline to identify the genetic basis of RSA in rice (Iyer-Pascuzzi et al., 2010; Fang et al., 2013; Topp et al., 2013). The pipeline combined a dedicated imaging platform with semiautomated image processing and analysis software. To adapt this gel-based phenotyping system for maize, we made several modifications to accommodate the larger size and faster growth rate of maize roots. These modifications included the use of custom-manufactured glass containers and the construction of a more robust imaging table (Supplemental Fig. S1). The initial modification of the imaging table, used for characterization of the NAM founders, was a direct scale up of the rice platform. The second modification, used for imaging the B73 × Ki3 mapping population, integrated several improvements to better deal with the scale of those experiments. The commercial digital cameras were replaced with higher quality computer vision cameras, the fluorescent backlight was replaced with light-emitting diode panels to improve image contrast, and the plastic turntable was replaced with a heavy-duty industrial turntable to better handle the 40-pound weight of the growth containers (see “Materials and Methods”).
To facilitate germination, maize seeds were pregerminated in petri dishes and transplanted to the gel after emergence of the primary root and coleoptile. Additionally, a more stringent, two-step surface sterilization protocol using 35% (v/v) hydrogen peroxide was implemented, because we found a higher prevalence of fungal spores on maize seeds than on rice. Due to light penetration issues through the larger diameter maize containers, the transparency of the gel was increased by lowering the Gelzan concentration to 0.15% (w/v). Lastly, we used a Hoagland solution-based growth medium better suited to maize (Hoagland and Arnon, 1950).
Imaging of NAM Founder Lines Reveals the Phenotypic Diversity of Maize Root Architecture
To characterize the extent of root trait diversity in maize, we imaged roots of 25 of the 26 NAM founder lines. We chose these lines for two reasons: (1) the NAM founders encompass a large proportion of the genetic diversity available in maize and, therefore, should encompass a wide range of RSA traits (Yu et al., 2008; McMullen et al., 2009; Hansey et al., 2012); and (2) the NAM population is composed of 25 individual RIL subpopulations, providing many options for subsequent QTL mapping. We produced a comprehensive survey of early root development by imaging each founder line at 3, 6, 9, and 12 d after planting (dap) for a total of 564 image sets representing 225 individual plants. All images were analyzed for 19 two-dimensional traits (Galkovskyi et al., 2012). A representative image of each founder (Fig. 1; Supplemental Fig. S2) and a description of all traits used (Supplemental Table S1) are provided.
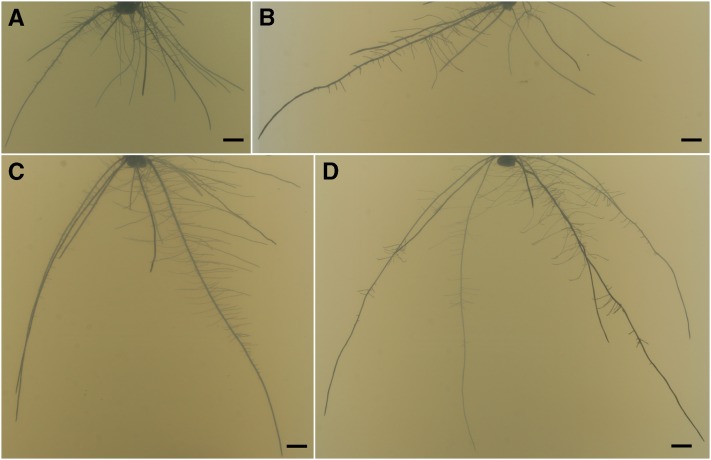
Figure 1.
RSA of several NAM founder lines grown in gellan gum. Images are from day 9 plants and represent several types of RSA. The founders include B73 (A), NC358 (B), OH43 (C), and Ki3 (D). Supplemental Figure S2 shows a representative image for each founder line. Bars = 10 mm.
Among the NAM founders, we observed RSAs that ranged from large and expansive to small and compact. At one extreme were Tx303 and Ki3 (Fig. 1; Supplemental Fig. S2), which displayed the greatest depths, maximum widths, and convex hull areas but the smallest solidities and bushiness. This combination of traits resulted in root systems that encompassed a large area/volume but did so superficially. A characteristic feature of these root systems was the presence of large pockets of unexplored space. At the other extreme were B73 (the common founder) and Ki11 (Fig. 1), which had much denser root systems, with small values for depths, maximum widths, and convex hull areas but large values for solidities and bushiness. These root systems were more tightly packed into a small area, forgoing initial expansive exploration in favor of thoroughness. This compact RSA form could be especially useful in a densely planted (Hammer, 1999) and nutrient-rich (Jobbágy and Jackson, 2001) environment, such as a well-fertilized field.
The B73 and Ki3 lines were of special interest as they represented extremes in RSA. To further explore the differences between their compact and exploratory RSAs, we attempted to determine if the differences were due to biomass allocation tradeoffs or represented intrinsic phenotypic tradeoffs. We performed Student’s t test to compare these varieties for several key traits that epitomize the contrasting RSAs, including solidity, depth, and convex hull area. To control for biomass differences between the plants, we normalized the traits using root volume as a biomass proxy. In all cases, for both transformed and untransformed data, there were significant differences between the means (Student’s t test, P < 0.0003), suggesting that the different RSA types are likely the result of phenotypic tradeoffs between thorough soil exploration and occupation and do not result merely from limiting biomass. These results indicate that root growth decisions result in architectures that determine the range of soil exploration versus occupation. A similar pattern was reported previously in rice using the same gel imaging system (Topp et al., 2013).
The remaining founders could be described as having intermediate but distinct phenotypes. For example, OH43 (Fig. 1) had a deep root system, similar to Ki3 and Tx303, but a much smaller maximum width and average solidities and convex hull areas. In contrast, NC358 (Fig. 1) had a shallow root system, similar to B73, but large maximum width similar to Ki3. Interestingly, there were no varieties with roots that both encompassed a large area and also explored it thoroughly, further supporting the idea of phenotypic tradeoffs.
Principal Component Analysis Demonstrates a Need for Multiple Traits to Describe RSA
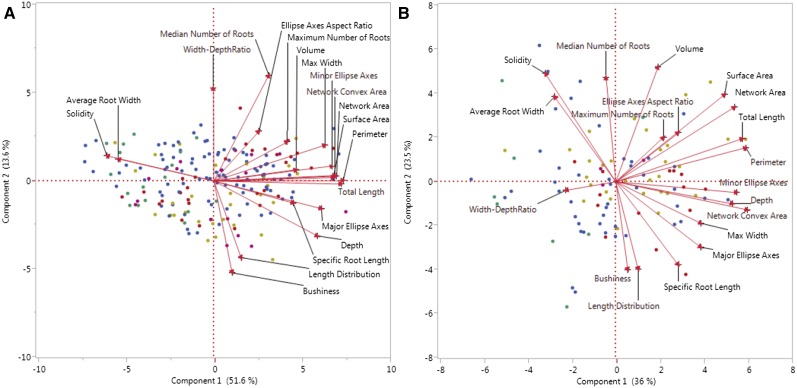
To explore the relationship between different traits, we performed principal component analysis (PCA) for each analysis day. In all cases, over 90% of the variation between the founder lines could be explained by the first five principal components. Although for the youngest plants (day 3), the first principal component was able to explain 51.6% of the variation, that number decreased with each successive day, to 44% on day 6, 36.4% on day 9, and 36% on day 12 (Supplemental Table S2). This indicated that there was less variation between the RSA of younger plants, which was not surprising given that the RSAs of the younger plants appeared very similar to each other. As the plants grew, their unique RSAs began to appear, resulting in partitioning of the variation across more principal components.
Interestingly, there was no obvious linkage between the genetic relatedness of the founders and their RSAs, as the PCA showed no grouping of varieties based on their phylogenetic relatedness (Fig. 2; Flint-Garcia et al., 2005). As a whole, this suite of traits was able to distinguish among NAM founder genotypes. The fact that they did not correlate with the genetic relatedness of demographic groups of the founders suggests that other selective factors, such as management practices or environmental influences (i.e. water and nutrient availability [Lynch and Brown, 2012], planting density [Fang et al., 2013], and soil quality [Gamuyao et al., 2012]), were important in RSA trait selection during breeding improvement.
Figure 2.
PCA for NAM founder lines for day 3 (A) and day 12 (B). The color of the dots indicates the subgroup of each data point (Flint-Garcia et al., 2005): red, mixed; yellow, nonstiff stalk; green, other; pink, stiff stalk; and blue, tropical.
Logistic Regression Identifies Key Traits for Distinguishing Founders
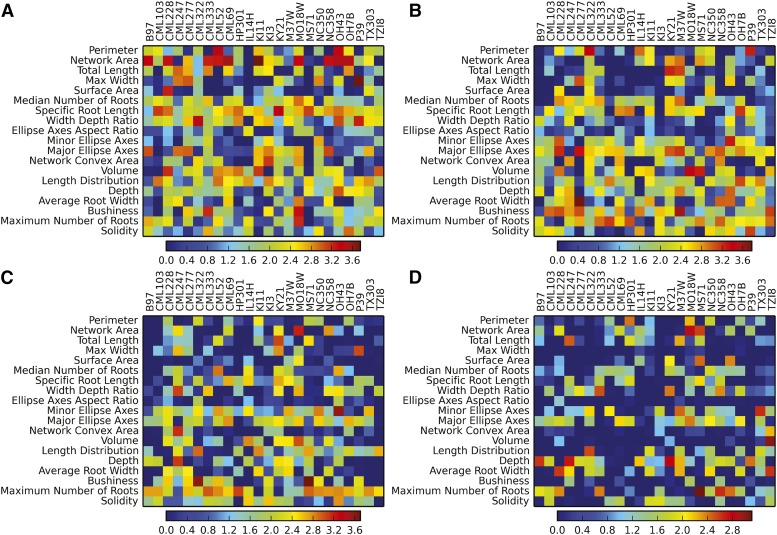
To determine the specific RSA traits that best differentiate the founders, we used a machine-learning approach, logistic regression, which is both accurate in its predictions and produces coefficient terms that reflect the importance of each individual trait in identifying different founders (Fan et al., 2008). The analysis was performed separately for each B73 by other founder pairs. To validate the predictions, a control analysis was performed by randomizing all of the data prior to the analysis (Supplemental Fig. S3). Accuracy values were calculated for classifications for both true and randomized data (Supplemental Table S3). For any given day and pair comparison, we found at least a few key traits that were best suited for that particular classification (Fig. 3). A few of the traits played important roles in a large number of classifications (such as maximum number of roots or depth); however, no single trait was key for all classifications. Which traits were key, as well as the number of informative traits, were highly dependent on the differences between the RSAs of the tested pair and the imaging day. In general, the younger the plants and the more similar the RSAs, the more traits had to be used to correctly identify the founder.
Figure 3.
Pairwise classification of the B73 founder versus all other NAM founders. Logistic regression was used to obtain coefficient values for each trait for each classification. The data were normalized by taking the natural log of the absolute value of each coefficient. Higher values of coefficients indicate especially informative traits for differentiating between B73 and other founders. A, Day 3. B, Day 6. C, Day 9. D, Day 12. Controls based on randomized data and classifier accuracies are provided in Supplemental Figure S3 and Supplemental Table S3.
For example, all of the day 3 classifications (Fig. 3) depended on a large number of traits, with network area, width-depth ratio, and length distribution being especially prominent. This was because the RSAs of the plants at 3 dap were still quite small, so a large mix of traits had to be used to distinguish between the varieties. With each subsequent imaging day, fewer and fewer traits needed to be used. This was especially true for day 12 (Fig. 3), as each pairwise classification included only a few key traits. On that day, a set of different traits, such as depth and maximum number of roots, took on more prominent roles in the classification process. Altogether, this illustrates the complexity of RSA both between varieties and over time, reinforcing the necessity of assessing a large number of traits to distinguish between different varieties as well as individual varieties at different ages (Iyer-Pascuzzi et al., 2010; Topp et al., 2013).
Selection and Phenotyping of an NAM RIL Subpopulation
We chose the B73 × Ki3 RIL population for QTL mapping because of the large phenotypic differences between their parent RSAs (Fig. 1) and because of their many genetic differences (Yu et al., 2008). Of the 200 lines in this RIL population, we omitted 14 due to the lack of seed availability or poor germination. Additionally, 11 lines were removed from further analysis due to the lack of genetic marker data. A total of 774 plants were imaged and phenotyped at 4, 6, and 8 dap for a total of 80,080 individual images across 2,020 image sets. As expected, individual RIL trait values fell between the parental values, although a number of transgressive phenotypes also were observed (Supplemental Fig. S5).
Since each plant was imaged on 3 separate days, we were able to observe RSA changes over time. Of the 19 traits, 11 showed a progressive increase between the three time points, seven remained relatively static, and a single trait decreased in value (Fig. 4; Supplemental Fig. S4). Traits such as network area showed a tremendous increase over just 4 d of growth, increasing by 120% between days 4 and 6 and by 55% between days 6 and 8. Additionally, the range between the smallest and largest values also increased significantly, from 7.29 cm2 on day 4 to 15.35 cm2 on day 6 to 24.28 cm2 on day 8. This not only demonstrates the rapid rate of growth of maize roots but also the rapid development of many root traits over a small window of time.
Figure 4.
QTLs controlling the RSA in the B73 × Ki3 mapping population. The outer whiskers of each bar indicate the 2-log of the odds (LOD) confidence intervals, while the extent of the inner box indicates the 1-LOD confidence intervals. Closed boxes, QTL significance at α = 0.01; open boxes, QTL significance at α = 0.05. Each trait has been coded with a different color.
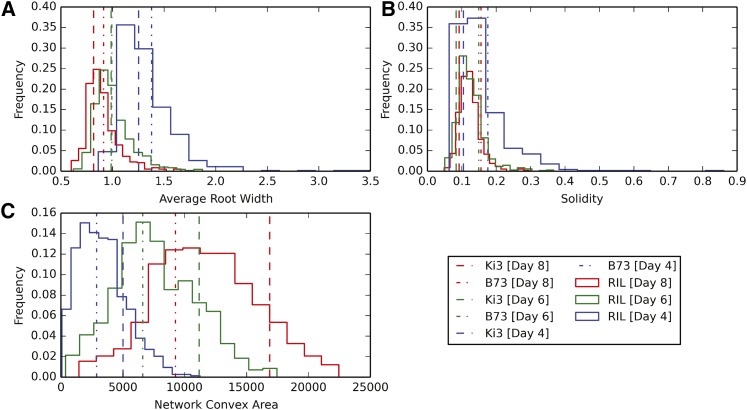
On the other hand, traits that reflected overall growth patterns, such as solidity, tended to remain static (Fig. 5) following large changes due to the initial growth burst. For example, average solidity changed by −16.5% between days 4 and 6 but only by −3.9% between days 6 and 8. The same pattern was seen across all of the NAM founder lines, with large changes in solidity at the outset of growth followed by relative stasis only a few days after planting (Supplemental Fig. S4). This is especially notable as solidity is a ratio of the number of network pixels, which reflects the size of the root, and network convex area, which reflects the expansive nature of the RSA. The fact that solidity tended to remain constant suggests a means of controlling the density of a root system through maintaining the ratio of root mass to root size. A similar pattern was reported for two rice varieties (Topp et al., 2013), reinforcing the idea that a global RSA, for example compact versus exploratory, of a given genotype or plant species can be maintained over time, irrespective of the size of the root system, and can be captured using an appropriate combination of different metrics.
Figure 5.
Distribution of traits across all imaging days for network convex area, solidity, and average root width. Histograms are based on the means of the RIL family, while horizontal lines indicate the means for the parent lines.
A single trait, average root width, showed a progressive decrease in average trait values (Fig. 5). This was due to the proportional shift from thick to thin roots over time. At day 4, a majority of the root mass is composed of thicker primary and seminal roots. At days 6 and 8, the thinner lateral roots begin to emerge, which results in a decrease of average root width. Although this makes this trait an unexpected proxy for the ratio of root type (e.g. lateral versus seminal) distributions, it also highlights the need for the further development of algorithms able to analyze specific root types. It is worth mentioning that the generally thicker maize lateral root as compared with rice roots resulted in much higher contrast between these roots and background, which in turn allowed us to more easily capture and include them in the analysis.
The Gel Platform Reduces Environmental Effects and Increases Apparent Trait Heritability
We calculated broad-sense heritability using a random-effect ANOVA to ensure that there was enough power to reliably detect QTLs. The majority of traits showed moderately high heritabilities. This was especially true for older plants, with a large number of the traits showing heritabilities higher than 40% (Table I), indicating that the observed variation has a genetic basis rather than arising from environmental factors. Additionally, key traits that defined the differences between the B73 and Ki3 founders, such as depth, network convex area, solidity, and minor ellipse axes (Table I), had especially high heritabilities. The high heritabilities for a number of RSA traits highlight the robustness of the gel platform for phenotyping. By standardizing growth conditions, confounding environmental effects are reduced, which greatly facilitates the observation of RSA differences due to genotype while minimizing the number of replicates needed. In a few rare cases, there was a slight effect due to RILs being grown in different growth chambers. Least-square means were used to transform the data to factor in and eliminate this effect. These transformed data were used for all subsequent analyses.
Table I. Broad-sense heritability and number of QTLs found.
The heritability was calculated using an ANOVA after least square means transformation.
| Trait | Day 4 | Day 6 | Day 8 | No. of QTLs |
|---|---|---|---|---|
| Median no. of roots | 18.85 | 13.47 | 22.16 | 2 |
| Volume | 34.13 | 31.51 | 16.67 | 2 |
| Bushiness | 8.11 | 6.8 | 15.66 | 3 |
| Maximum width | 30.53 | 34.14 | 36.65 | 3 |
| Length distribution | 13 | 13.46 | 26.86 | 3 |
| Ellipses axes aspect ratio | 19.7 | 29.89 | 48.78 | 3 |
| Specific root length | 32.31 | 33.63 | 36.93 | 3 |
| Perimeter | 31.12 | 33.27 | 44.13 | 4 |
| Surface area | 30.54 | 31.25 | 31.77 | 5 |
| Network area | 30.94 | 32.38 | 34.8 | 6 |
| Total length | 29.45 | 32.82 | 43.1 | 6 |
| Width-depth ratio | 21.79 | 28.8 | 40.49 | 6 |
| Maximum no. of roots | 17.6 | 30.21 | 40.84 | 6 |
| Average root width | 27.59 | 38.69 | 49.84 | 7 |
| Solidity | 30.62 | 43.1 | 43.21 | 7 |
| Minor ellipse axes | 32.22 | 44.27 | 54.99 | 8 |
| Depth | 29.55 | 42.22 | 49.85 | 8 |
| Network convex area | 31.81 | 42.64 | 49.79 | 9 |
| Major ellipse axes | 29.86 | 41.57 | 43.75 | 12 |
Mapping of QTLs Identifies Colocalized Clusters
Using QTL Cartographer, we identified 102 RSA QTLs across all 19 traits and three time points (Fig. 4; Basten et al., 1994, 2004). Sixty-nine were significant at α = 0.01, and an additional 33 were significant at α = 0.05. The effect size of these ranged from 5.5% to 23.8%, which was higher than expected based on previously mapped flowering time QTLs from the whole NAM population (Buckler et al., 2009). Although the smaller population size of our RIL population may have produced QTL effects with some upward bias, this would not have played a large role due to the high heritability values for our traits (Xu, 2003). The QTLs were unevenly distributed across the time points, with only 20 QTLs on day 4 but 40 on day 6 and 42 on day 8. As noted previously, this was not surprising, as the roots of different genotypes were less distinct and traits are less heritable on day 4 (Table I). Individual QTLs also were unevenly distributed across the genome, with chromosomes 2, 3, 7, 9, and 10 being especially QTL rich. Chromosome 10 was notable for its 29 QTLs, eight of which also ranked among the top most significant and having the strongest effects (Supplemental Table S4).
We used overlapping 2-LOD confidence intervals to group 94 of the QTLs into nine clusters (Mangin and Goffinet, 1997). These were distributed across the genome with single clusters on chromosomes 1 to 4, 6, 9, and 10 and a pair of clusters on chromosome 7. The remaining eight QTLs formed two clusters of two QTLs or remained unclustered. As seen previously in rice (Topp et al., 2013), in a large proportion of the clusters, overlapping QTLs spanned multiple days for the same trait, indicating that the allelic differences of genes underlying RSA QTLs have durable effects despite the rapid growth of the whole root system. Further comparison of the QTL clusters, their individual trait makeup, and the direction of the additive effects of each individual trait illustrate tradeoffs between overarching RSAs. The architectural extremes of large solidity for compact RSA and large network convex area for exploratory RSA seen in the NAM founder analysis were reflected in the individual QTL clusters.
For example, in the chromosome 10 cluster, most of the individual QTLs, such as depth and network convex area, had large additive effects due to the Ki3 allele, while solidity and width-depth ratio had large effects due to the B73 allele (Supplemental Table S4). The same pattern was seen for the QTL cluster on chromosome 2, with most QTLs in that cluster having large effects due to the Ki3 allele, with the exception of solidity and average root widths. The opposite was true for the clusters of QTLs on chromosome 9, with the additive effect of average root width being due to the Ki3 allele and the additive effect of the remaining QTLs being due to the B73 alleles.
Traits could be clearly partitioned into two opposing groups, with the smaller group composed of average root width, solidity, median and maximum number of roots, and width-depth ratio and the larger group composed of the remaining traits. In those cases where significant QTLs from different groups were located in the same cluster, they had additive effects due to different parents, emulating the same tradeoffs seen in the parental lines. A single exception was the cluster on chromosome 6, which was composed of only two traits, bushiness and length distribution, for days 6 and 8, both of which resulted in larger values for those traits due to B73 alleles.
DISCUSSION
By scaling up the various components of the gel RSA imaging platform, we adapted it for maize roots. We used this system to phenotype a number of genetically diverse maize lines and observed a large number of variable root architectures bounded on the extremes by two distinct ideotypes. One is a larger, more exploratory RSA that encompasses a large amount of space but occupies that space sparsely, resulting in empty pockets of space surrounding individual roots. This type of strategy could allow for rapid expansion of the primary and seminal roots, serving to locate resource-rich areas before committing to denser growth via lateral branching. Additionally, it has been suggested that this type of RSA would be much better at reaching deep subsurface water sources (Hammer, 1999), giving it a potential advantage in drought-prone conditions. Due to the reductionist nature of our system and the ability of the nutrients to rapidly and uniformly diffuse through the medium (Fang et al., 2013), we believe that the phenotypic differences we observed between the RSAs of the different founders were due to genetic differences and not to root plasticity or scale foraging (Campbell et al., 1991).
On the other extreme, we observed genotypes with small and compact RSAs that could thoroughly explore sites proximal to the soil surface. This growth pattern might favor varieties that grow in phosphorous-poor soils (Jobbágy and Jackson, 2001; Ho et al., 2005). Although shorter roots would not allow access to deep water resources, management practices could mitigate this need, and a more compact RSA may contribute to increased tolerance to planting density.
Between these two extremes, we observed a spectrum of diverse architectures. Despite this, it is striking that no NAM founder line had an RSA with an extensive root system that also thoroughly explores all of the encompassing space. Although the underlying mechanism is unknown, it suggests constraints on the pattern of root distribution and highlights the tremendous amount of phenotypic variability in maize RSA. Additionally, although these experiments only followed root growth for 12 d, the growth rate and direction suggest that these patterns could continue into adulthood, such that plants with compact young RSAs produce compact adult RSAs and plants with exploratory young RSAs produce large and wide-ranging adult RSAs.
To find loci controlling RSA, we phenotyped the B73 × Ki3 RIL population and performed QTL mapping. Interestingly, the effect sizes of the QTLs were substantially higher than those found for flowering time (Buckler et al., 2009), suggesting that there are fewer and stronger effect loci that control RSA. QTLs from different imaging days for the same trait tended to cluster together, suggesting that the causative alleles have persistent effects on RSA development. Clustering of different traits to the same region of the genome indicates that one locus, either a single gene or a number of linked ones, has multiple effects on many aspects of RSA.
We detected the same kind of phenotypic tradeoffs in the maize NAM founders as in rice (Topp et al., 2013). The RSA traits can be divided roughly into two groups: those that support a compact RSA and those that support an exploratory RSA. For most QTL clusters, traits that have strong additive effects due to the parent exhibiting the first group of traits would have strong additive effects in the other group due to the other parent. Prime examples of this are the clusters found on chromosomes 9 and 10, with the former having mostly strong overall additive effects for compact architecture and the latter having strong overall additive effects for exploratory root architecture. With the exception of the chromosome 6 cluster, which was composed of four QTLs with additive effects due to B73, all clusters contained some QTLs that favored either compact or expansive RSA.
We show that some profound quantitative differences in RSA can be controlled by a few large-effect loci, consistent with what has been observed with the two crop root QTLs for which genes have been identified, Deep Rooting1 and Phosphorus-Starvation Tolerance1 (Gamuyao et al., 2012; Uga et al., 2013). It is possible that the maize loci we characterized are the result of single strongly acting genes or groups of tightly linked genes. Since the effect size of our QTLs was larger than those found previously in flowering studies (Buckler et al., 2009), there is greater potential for their use in marker-assisted breeding. It also should be noted that previous QTLs mapped in the NAM population were associated with the entire NAM population with QTL segregating across multiple families, which likely would result in smaller effect sizes. Since, in our work, we focused on a single RIL subpopulation, the QTLs may segregate only in this population, resulting in larger effect sizes.
A valid concern in the use of artificial medium such as gellan gum is the question of the transferability of findings to soil-grown plants. Although there is no definitive study, there is some evidence for positive correlations between controlled environment and field traits. The rice QTLs mapped in the gel system had a strong correlation to QTLs mapped for the same population in soil (Topp et al., 2013). Additionally, a number of the QTL clusters reported here have good overlap with meta-QTLs found in several studies for yield and root traits (Lynch, 1995; Tuberosa et al., 2003; Wang et al., 2012; Semagn et al., 2013), despite those studies being based on plants of different ages, different growth conditions, and different traits. To date, large numbers of QTLs have been reported. However, due to the difficulties of fine-mapping, few causal alleles have been found (Gamuyao et al., 2012; Uga et al., 2013). This is especially true for maize root traits, for which some QTLs have been fine-mapped but none has been cloned. The rapid phenotyping made possible by the gel platform, as well as other controlled environment systems, offers an efficient method for fine-mapping and cloning RSA QTLs.
MATERIALS AND METHODS
Plants and Growth Conditions
Maize (Zea mays) seeds were sterilized with 35% (v/v) hydrogen peroxide for 20 min on a shaker, followed by three washes with sterile water. Seeds were then incubated partially submerged in water at 28°C for 8 h, resterilized, and transferred to individual 30-mm petri dishes filled with enough water to keep them partially submerged (approximately 25 mL). The plates were then incubated in dark conditions at 28°C until germination (2 or 3 d). Developmentally similar seedlings (emerged coleoptile and approximately 1-inch-long primary root) were planted in 28-L custom glass cylinders filled with 14 L of one-half-strength Hoagland solution, pH 6 [3.01 mm KNO3, 2.53 mm Ca(NO3)2∙4H2O, 2.10 mm MgSO4∙7H2O, 1.03 mm NH4H2PO4, 84.61 μm KCl, 23.45 μm H3BO3, 3.52 μm ZnSO4∙7H2O, 2.31 μm MnSO4∙H2O, 0.51 μm NH4NO3, 0.17 μm CuSO4∙5H2O, 0.1 μm Na2MoO3∙2H2O, and 7.5 mg L−1 diethylaminoethyl acid iron(III) disodium salt hydrate (Sequestrene 330; phytotechlab.com)]. Plants were grown on a 16-h-day/8-h-night cycle, 28°C day and 25°C night, with 800 μmol m−2 s−1 photosynthetically active radiation. NAM founders were imaged at 3, 6, 9, and 12 dap, while the B73 × Ki3 RILs were imaged at 4, 6, and 8 dap.
Imaging
Imaging was done using the maize imaging platform described above. A 40-image rotational series of each plant was taken and uploaded to a server for further processing and analysis using the previously described pipeline (Topp et al., 2013). Six replicates were performed for most NAM founders. M162W was not included in the NAM founder analysis due to its poor germination and performance in the gel system. Three or more replicates were performed for the 175 RILs that had available seeds and marker data. A total of 25,560 images for the NAM data set and 80,800 images for the B73 × Ki3 data set were used.
Image Processing and Phenotyping
All of the image processing and RSA phenotyping were done as described previously (Galkovskyi et al., 2012; Topp et al., 2013). Each image set was cropped and segmented using double adaptive thresholding with a set of maize-specific parameters. Each set underwent an iterative process of thresholding and quality control aided by a Python script until a set of clean images (an all-white root on an all-black background) was produced. These underwent two-dimensional phenotyping and scaling from pixels to appropriate International System units.
Statistical Analysis
Student’s t test and PCA were performed based on the correlation among line means as described previously (Topp et al., 2013) using JMP Pro version 11 (www.jmp.com/software/jmp11/).
Logistic Regression
A machine-learning approach using L1-regularized logistic regression using the mlpy Python library was performed to isolate the distinguishing traits of any B73 × other founder pairs (Albanese et al., 2012). The data were first normalized to a zero mean and unit variance (in order to improve computation speed) and then randomly partitioned into two subsets. Half of the data were labeled as the training set, while the other half were labeled as the validation set. A classification model was constructed using the training set and used to determine its accuracy by determining the classes of the data in the validation set. This model was a linear combination of terms composed of a learned coefficient multiplied by trait value. As such, traits modified by large coefficient values (orange and red in Fig. 3) point to those being the key traits that differentiate B73 from any other individual founder. A cross-validation was performed by permuting the above process 1,000 times and averaging the coefficients across the runs. A control was performed by randomly permuting the data labels and running the same analysis (Supplemental Fig. S3).
Heritabilities and Data Transformation
A random-effect model ANOVA (using JMP Pro version 11) for the RIL families and chambers used was constructed separately for each trait. Broad-sense heritability was reported as the proportion of the genotypic variance over the total phenotypic variance of individual plants. The model included plant averages as well as growth chambers used and the r2 due to the family-estimated individual heritabilities. Due do the small sample number, this resulted in smaller, but still robust, heritabilities than those calculated for whole families. Due to the few analyses showing marginally significant chamber effects, least square means-adjusted data provided by the ANOVA (which corrected for chamber effects) were used in subsequent QTL mapping analyses.
Composite Interval QTL Mapping
QTL analysis was performed using the Linux version of QTL Cartographer, version 1.17 (Basten et al., 1994, 2004). Composite interval mapping (model 6, 1-centimorgan [cM] walk speed, 10-cM window) was performed using ranked markers from forward and backward step-wise regression as covariates. The α = 0.05 and α = 0.01 significance thresholds were obtained through permutation (1,000), while the confidence intervals were set at 2- and 1-LOD distance from the key marker. Due to the high density of markers used here, a gap of 5 cM was allowed for confidence interval calculations.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Imaging table used for photographing the root systems of maize.
Supplemental Figure S2. Representative image of each NAM founder.
Supplemental Figure S3. Logistic regression control.
Supplemental Figure S4. Mean solidity of NAM founder lines versus imaging day.
Supplemental Figure S5. Distribution of individual trait values in the B73 × Ki3 mapping population.
Supplemental Table S1. Trait descriptions.
Supplemental Table S2. Breakdown of the components for the first five principal components for the PCA analysis on NAM founder lines.
Supplemental Table S3. Logistic regression accuracies.
Supplemental Table S4. List of QTLs.
Supplementary Material
Acknowledgments
We thank Jim Holland for providing seeds for the B73 × Ki3 RIL population and the members of the Benfey laboratory and Tom Mitchell-Olds for support and critical reading of the article.
Glossary
- RSA
root system architecture
- QTL
quantitative trait locus
- NAM
nested association mapping
- RIL
recombinant inbred line
- dap
days after planting
- PCA
principal component analysis
- LOD
log of the odds
- cM
centimorgan
Footnotes
This work was supported by the U.S. Department of Agriculture Food Research Initiative (grant no. 2011–67012–30773 to C.N.T.), the National Science Foundation Division of Biological Infrastructure (grant no. 0820624 to P.N.B.), the Howard Hughes Medical Institute, and the Gordon and Betty Moore Foundation (grant no. GBMF3405 to P.N.B.).
Articles can be viewed without a subscription.
References
- Albanese D, Visintainer R, Merler S, Riccadonna S, Jurman G, Furlanello C (2012) mlpy: Machine Learning Python. http://arxiv.org/abs/1202.6548v2 (March 12, 2015)
- Basten CJ, Weir BS, Zeng ZB (1994) Zmap: a QTL cartographer. In C Smith, JS Gavora, B Benkel, J Chesnais, W Fairfull, JP Gibson, BW Kennedy, EB Burnside, eds, Proceedings of the 5th World Congress on Genetics Applied to Livestock Production: Computing Strategies and Software, Vol 22. Organizing Committee, 5th World Congress on Genetics Applied to Livestock Production, Guelph, Ontario, Canada, pp 65–66 [Google Scholar]
- Basten CJ, Weir BS, Zeng ZB (2004) QTL Cartographer, Version 1.17. Department of Statistics, North Carolina State University, Raleigh [Google Scholar]
- Buckler ES, Holland JB, Bradbury PJ, Acharya CB, Brown PJ, Browne C, Ersoz E, Flint-Garcia S, Garcia A, Glaubitz JC, et al. (2009) The genetic architecture of maize flowering time. Science 325: 714–718 [DOI] [PubMed] [Google Scholar]
- Campbell BD, Grime JP, Mackey JML (1991) A trade-off between scale and precision in resource foraging. Oecologia 87: 532–538 [DOI] [PubMed] [Google Scholar]
- Clark RT, MacCurdy RB, Jung JK, Shaff JE, McCouch SR, Aneshansley DJ, Kochian LV (2011) Three-dimensional root phenotyping with a novel imaging and software platform. Plant Physiol 156: 455–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan RE, Chang KW, Hsieh CJ, Wang XR, Lin CJ (2008) LIBLINEAR: a library for large linear classification. J Mach Learn Res 9: 1871–1874 [Google Scholar]
- Fang S, Clark RT, Zheng Y, Iyer-Pascuzzi AS, Weitz JS, Kochian LV, Edelsbrunner H, Liao H, Benfey PN (2013) Genotypic recognition and spatial responses by rice roots. Proc Natl Acad Sci USA 110: 2670–2675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint-Garcia SA, Thuillet AC, Yu J, Pressoir G, Romero SM, Mitchell SE, Doebley J, Kresovich S, Goodman MM, Buckler ES (2005) Maize association population: a high-resolution platform for quantitative trait locus dissection. Plant J 44: 1054–1064 [DOI] [PubMed] [Google Scholar]
- Galkovskyi T, Mileyko Y, Bucksch A, Moore B, Symonova O, Price CA, Topp CN, Iyer-Pascuzzi AS, Zurek PR, Fang S, et al. (2012) GiA Roots: software for the high throughput analysis of plant root system architecture. BMC Plant Biol 12: 116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamuyao R, Chin JH, Pariasca-Tanaka J, Pesaresi P, Catausan S, Dalid C, Slamet-Loedin I, Tecson-Mendoza EM, Wissuwa M, Heuer S (2012) The protein kinase Pstol1 from traditional rice confers tolerance of phosphorus deficiency. Nature 488: 535–539 [DOI] [PubMed] [Google Scholar]
- Garnett T, Conn V, Kaiser BN (2009) Root based approaches to improving nitrogen use efficiency in plants. Plant Cell Environ 32: 1272–1283 [DOI] [PubMed] [Google Scholar]
- Hammer G. (1999) Climate change and the global harvest: potential impacts of the greenhouse effect on agriculture. Crop Sci 39: 582 [Google Scholar]
- Hansey CN, Vaillancourt B, Sekhon RS, de Leon N, Kaeppler SM, Buell CR (2012) Maize (Zea mays L.) genome diversity as revealed by RNA-sequencing. PLoS ONE 7: e33071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargreaves CE, Gregory PJ, Bengough AG (2008) Measuring root traits in barley (Hordeum vulgare ssp. vulgare and ssp. spontaneum) seedlings using gel chambers, soil sacs and x-ray microtomography. Plant Soil 316: 285–297 [Google Scholar]
- Ho M, Rosas J, Brown K, Lynch JP (2005) Root architectural tradeoffs for water and phosphorus acquisition. Funct Plant Biol 32: 737–748 [DOI] [PubMed] [Google Scholar]
- Hoagland DR, Arnon DI (1950) The water-culture method for growing plants without soil. Calif Agric Exp Stn Circ 1–32 [Google Scholar]
- Iyer-Pascuzzi AS, Symonova O, Mileyko Y, Hao Y, Belcher H, Harer J, Weitz JS, Benfey PN (2010) Imaging and analysis platform for automatic phenotyping and trait ranking of plant root systems. Plant Physiol 152: 1148–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson MB, Armstrong W (1999) Formation of aerenchyma and the processes of plant ventilation in relation to soil flooding and submergence. Plant Biol 1: 274–287 [Google Scholar]
- Jahnke S, Menzel MI, van Dusschoten D, Roeb GW, Bühler J, Minwuyelet S, Blümler P, Temperton VM, Hombach T, Streun M, et al. (2009) Combined MRI-PET dissects dynamic changes in plant structures and functions. Plant J 59: 634–644 [DOI] [PubMed] [Google Scholar]
- Jiao Y, Zhao H, Ren L, Song W, Zeng B, Guo J, Wang B, Liu Z, Chen J, Li W, et al. (2012) Genome-wide genetic changes during modern breeding of maize. Nat Genet 44: 812–815 [DOI] [PubMed] [Google Scholar]
- Jobbágy EG, Jackson RB (2001) The distribution of soil nutrients with depth: global patterns and the imprint of plants. Biogeochemistry 53: 51–77 [Google Scholar]
- Le Marié C, Kirchgessner N, Marschall D, Walter A, Hund A (2014) Rhizoslides: paper-based growth system for non-destructive, high throughput phenotyping of root development by means of image analysis. Plant Methods 10: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch J. (1995) Root architecture and plant productivity. Plant Physiol 109: 7–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch JP, Brown KM (2012) New roots for agriculture: exploiting the root phenome. Philos Trans R Soc Lond B Biol Sci 367: 1598–1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangin B, Goffinet B (1997) Comparison of several confidence intervals for QTL location. Heredity (Edinb) 78: 345–353 [Google Scholar]
- McMullen MD, Kresovich S, Villeda HS, Bradbury P, Li H, Sun Q, Flint-Garcia S, Thornsberry J, Acharya C, Bottoms C, et al. (2009) Genetic properties of the maize nested association mapping population. Science 325: 737–740 [DOI] [PubMed] [Google Scholar]
- Semagn K, Beyene Y, Warburton ML, Tarekegne A, Mugo S, Meisel B, Sehabiague P, Prasanna BM (2013) Meta-analyses of QTL for grain yield and anthesis silking interval in 18 maize populations evaluated under water-stressed and well-watered environments. BMC Genomics 14: 313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silby MW, Cerdeño-Tárraga AM, Vernikos GS, Giddens SR, Jackson RW, Preston GM, Zhang XX, Moon CD, Gehrig SM, Godfrey SAC, et al. (2009) Genomic and genetic analyses of diversity and plant interactions of Pseudomonas fluorescens. Genome Biol 10: R51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topp CN, Iyer-Pascuzzi AS, Anderson JT, Lee CR, Zurek PR, Symonova O, Zheng Y, Bucksch A, Mileyko Y, Galkovskyi T, et al. (2013) 3D phenotyping and quantitative trait locus mapping identify core regions of the rice genome controlling root architecture. Proc Natl Acad Sci USA 110: E1695–E1704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trachsel S, Kaeppler SM, Brown KM, Lynch JP (2010) Shovelomics: high throughput phenotyping of maize (Zea mays L.) root architecture in the field. Plant Soil 341: 75–87 [Google Scholar]
- Tuberosa R, Salvi S, Sanguineti MC, Maccaferri M, Giuliani S, Landi P (2003) Searching for quantitative trait loci controlling root traits in maize: a critical appraisal. Plant Soil 255: 35–54 [Google Scholar]
- Uga Y, Sugimoto K, Ogawa S, Rane J, Ishitani M, Hara N, Kitomi Y, Inukai Y, Ono K, Kanno N, et al. (2013) Control of root system architecture by DEEPER ROOTING 1 increases rice yield under drought conditions. Nat Genet 45: 1097–1102 [DOI] [PubMed] [Google Scholar]
- Wang Y, Huang Z, Deng D, Ding H, Zhang R, Wang S, Bian Y, Yin Z, Xu X (2012) Meta-analysis combined with syntenic metaQTL mining dissects candidate loci for maize yield. Mol Breed 31: 601–614 [Google Scholar]
- Xu S. (2003) Theoretical basis of the Beavis effect. Genetics 165: 2259–2268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Holland JB, McMullen MD, Buckler ES (2008) Genetic design and statistical power of nested association mapping in maize. Genetics 178: 539–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.