Natural variations in photosynthetic acclimation to different growth irradiances, and to a step-wise increase in growth irradiance, allows genetic analysis of this complex phenomenon.
Abstract
Plants are known to be able to acclimate their photosynthesis to the level of irradiance. Here, we present the analysis of natural genetic variation for photosynthetic light use efficiency (ΦPSII) in response to five light environments among 12 genetically diverse Arabidopsis (Arabidopsis thaliana) accessions. We measured the acclimation of ΦPSII to constant growth irradiances of four different levels (100, 200, 400, and 600 µmol m−2 s−1) by imaging chlorophyll fluorescence after 24 d of growth and compared these results with acclimation of ΦPSII to a step-wise change in irradiance where the growth irradiance was increased from 100 to 600 µmol m−2 s−1 after 24 d of growth. Genotypic variation for ΦPSII is shown by calculating heritability for the short-term ΦPSII response to different irradiance levels as well as for the relation of ΦPSII measured at light saturation (a measure of photosynthetic capacity) to growth irradiance level and for the kinetics of the response to a step-wise increase in irradiance from 100 to 600 µmol m−2 s−1. A genome-wide association study for ΦPSII measured 1 h after a step-wise increase in irradiance identified several new candidate genes controlling this trait. In conclusion, the different photosynthetic responses to a changing light environment displayed by different Arabidopsis accessions are due to genetic differences, and we have identified candidate genes for the photosynthetic response to an irradiance change. The genetic variation for photosynthetic acclimation to irradiance found in this study will allow future identification and analysis of the causal genes for the regulation of ΦPSII in plants.
Light is the driving force for photosynthesis, and the relation between light and photosynthesis is complex; light is essential for photosynthesis, but absorbed light naturally gives rise to reactive intermediates and by-products that can damage the photosynthetic machinery (Powles, 1984; Asada, 2006). Even at low irradiances, these damaging reactions occur, and they increase with increasing irradiance as photosynthesis becomes increasingly light saturated. Protection from these damaging processes, while at the same time permitting high photosynthetic rates under high-light conditions and a high light use efficiency under low-light conditions, seems to be the driving force behind the evolution of many of the regulatory processes of photosynthesis.
Managing the fate of absorbed light energy is regulated within a plant at the level of the chloroplast membranes. Chloroplast membranes are highly organized; the total amount of photosystems in a chloroplast, the amount of PSI in relation to PSII, and within each photosystem the amount of light-harvesting complexes in relation to the amount of reaction centers are tightly organized in response to the prevailing light condition around the leaf (Dekker and Boekema, 2005; Kouřil et al., 2012, 2013; Tikkanen et al., 2012). A high growth irradiance will lead to relatively more photosystem-core protein complexes, electron transport complexes, ATP synthase, and enzymes in the Calvin-Benson cycle, whereas low growth irradiances will lead to relatively more light-harvesting complexes and stacking of the thylakoid membranes (Bailey et al., 2001).
With increasing irradiance, the rate of excitation of PSII and PSI exceeds the capacity of photosynthetic electron transport or metabolic capacity, resulting in excess irradiance. In the short term, this provokes a physiological, regulatory response, while in the longer term, a sufficiently large excess irradiance often will result in alterations in the chloroplast proteome. Both the short-term and long-term responses have limits to their action (Foyer et al., 2012; Tikkanen et al., 2012). In the short term, excess irradiance imposes a strain on the capacity to protect the photosystems from damage (photoprotection; Demmig-Adams and Adams, 1992). PSII is particularly susceptible to photodamage and has evolved an active regulatory process to reduce the extent of damage combined with an active repair system to replace damaged PSII reaction centers. As a first line of defense, PSII to a certain extent is able to dissipate the excess irradiance as heat via regulated nonphotochemical quenching (NPQ) mechanisms reducing the rate of reaction center damage (Rabinowitch, 1951). This process is initiated within seconds after an increase in light intensity (Müller et al., 2001). The mechanism of NPQ in plants has been intensively studied, with most focused on the role of lumen pH-dependent, or energy-dependent, dissipation of excess light (also known as qE; Demmig et al., 1987) via a mechanism that involves subunit S of PSII and the xanthophyll cycle, one of the major short-term regulatory responses to excess irradiance.
The long-term response of plants to excess irradiance occurs over the time scale of hours or days and results in acclimation to the excess irradiance environment (Walters, 2005). This leads to increased capacities for electron and proton transport coupled with increased photosynthetic metabolic capacity, often combined with alterations in the organization of the photosystems. These changes in capacity are due to changes in the amounts of soluble enzymes of photosynthesis, in electron transport components and pathways, and in pigment-protein complexes (Murchie and Niyogi, 2011). Regarding the photosystem subunit stoichiometry, PSII in response to high irradiance decreases its antenna size by decreasing the amount of light-harvesting complex II (LHCII) proteins associated with the PSII supercomplex (Kouřil et al., 2013). Although the ratio of LHCI to PSI is not altered by irradiance, the antenna size of PSI decreases with increasing irradiance due to a decreased association of LHCII with PSI (Ballottari et al., 2007; Wientjes et al., 2013; i.e. decreases in the amount of LHCII seem to alter the antenna size of both photosystems). In addition to this long-term adjustment of PSI cross section, a short-term adjustment of the relative cross sections of PSII and PSI can be brought about by state transitions, driven by phosphorylation of LHCII, with phosphorylation being associated with a decrease in the cross section of PSII and an increase in that of PSI (Allen, 1992; Tikkanen et al., 2010).
In the event that the sum of the short- and long-term regulatory responses is insufficient, then more persistent damage to the photosynthetic apparatus (photoinhibition), especially to PSII, results. Damage to PSII, which is conveniently measured as a reduction of the parameter maximum photochemical efficiency of PSII in the dark-adapted state (Fv/Fm) derived from chlorophyll fluorescence measurements (Baker, 2008), is commonly used as an indicator of decreased photosynthetic performance arising from excess irradiance, although other parameters, such as decreased leaf chlorophyll content, also are used (Björkman and Demmig, 1987). The irradiance at which this damage will become apparent is difficult to predict.
Natural genetic variation in plant photosynthesis is a valuable resource (Flood et al., 2011). Naturally occurring variation in the photosynthetic acclimation response to high light has been observed in different plant species, with different studies using different light regimes and focusing on different acclimation responses (Sims and Pearcy, 1993; Valladares et al., 1997; Balaguer et al., 2001; Leakey et al., 2003; Portes et al., 2008). However, research combining different light regimes and looking at the acclimation responses of different aspects of photosynthesis in different genotypes in one experiment is lacking. Arabidopsis (Arabidopsis thaliana) is the model species for plant genetic research, a choice that was motivated partly because of the considerable variation found in this species for many traits. The number and variety of natural genotypes of Arabidopsis make it increasingly valuable as a physiological model (Alonso-Blanco et al., 2009). The variability of photosynthetic acclimation in naturally occurring genotypes of Arabidopsis has been investigated for light use efficiency in one growth environment (El-Lithy et al., 2005), for NPQ responses to high light (Jung and Niyogi, 2009), for photosynthetic capacity in response to high light (Athanasiou et al., 2010), and for the response to short-term light flecks (Alter et al., 2012). To explore the phenotypic plasticity and genetic variation within Arabidopsis, we have investigated the variability of the acclimation of multiple photosynthesis parameters to four constant growth irradiances and to a step-wise increase in the growth irradiance among 12 genotypically diverse accessions. To assess what part of the variability is due to genetic variation, we have calculated trait heritabilities, which are a measure of the extent of trait variation that is due to genetic variation. The possible genetic basis for trait variation can be determined using a genome-wide association study (GWAS), in which genetic loci associated with a trait are identified by correlating genetic variation with trait variation (Atwell et al., 2010). To perform a GWAS, there must be a description of genetic variation and the trait must be variable and heritable. It requires a large number of genotypes, so we used a population of 344 diverse Arabidopsis accessions that all had been genotyped for approximately 215,000 single-nucleotide polymorphisms (SNPs; Kim et al., 2007; Li et al., 2010). Importantly, because the genotypic description is restricted to the nuclear genome, any phenotypic variation arising from variation in cytoplasmic genomes cannot be associated with those genomes and will be a source of noise in the analysis. A GWAS for the light use efficiency of photosystem II (ΦPSII) measured 1 h after a step-wise change in irradiance was used to identify genomic regions that are associated with the response of photosynthesis to step-wise increases in irradiance. These results highlight the genetic variation and physiological adaptability (or lack of it) of photosynthesis in Arabidopsis. The presence or absence of variability is of particular importance in relation to both the evolution of the photosynthetic properties of Arabidopsis and the future identification of those genetic factors that give rise to the photosynthetic phenotype.
RESULTS
Phenotypic and Genotypic Variation in Light Response Curves of Various PSII Parameters among 12 Arabidopsis Accessions Grown at 100 µmol m−2 s−1
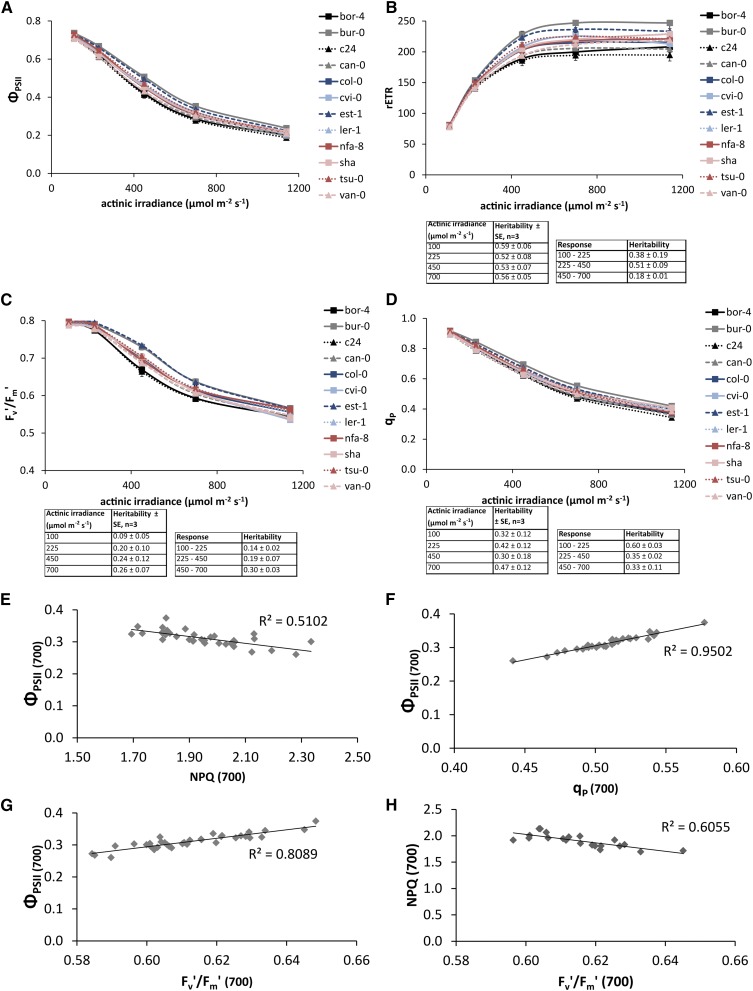
To assess genotypic variation for the irradiance response of photosynthesis in 12 genotypically diverse Arabidopsis accessions grown under constant growth irradiance of 100 µmol m−2 s−1, the operating ΦPSII was measured under a range of steady-state actinic irradiances. ΦPSII measures the proportion of the light absorbed by PSII that is used in photochemistry; it is widely used as a proxy for the quantum yield of linear electron transport and for photosynthesis in general (Maxwell and Johnson, 2000). Figure 1A shows the light response curves of ΦPSII for the 12 accessions used in this study, and Figure 1B shows the light response curves of the values for the relative linear electron transport rate (rETR; the product of ΦPSII and irradiance) derived from the ΦPSII values in Figure 1A. The electron transport rate is considered relative because we do not account for leaf light absorptance or the distribution of excitation energy between the two photosystems, nor do we apply any correction to the apparent quantum yield of PSII photochemistry provided by chlorophyll fluorescence measurements to give actual quantum yields for PSII electron transport. We will use the rETR at light saturation as a proxy for the maximum photosynthetic capacity in vivo (Genty et al., 1989). Figure 1B shows that, when grown at 100 µmol m−2 s−1, light saturation of photosynthesis occurs between 550 and 650 µmol m−2 s−1 for all 12 accessions. While the amount of phenotypic variance among the 12 accessions increases with the increase of the actinic irradiance level, so does the genetic variance, resulting in similar heritabilities (Fig. 1B).
Figure 1.
A, ΦPSII of 12 Arabidopsis accessions. Error bars indicate se; n = 3. ΦPSII was measured on plants that were grown in 100 µmol m−2 s−1 for 24 d after reaching steady-state photosynthesis in five different actinic irradiances: 100, 225, 450, 700, and 1,150 µmol m−2 s−1. B, rETR in the presence of different actinic irradiances of 12 accessions of Arabidopsis, calculated from PSII operating light use efficiencies. The inset shows the average heritabilities over three independent experiments for the individual measurement points and for the response of these values to actinic irradiance level. C, Fv′/Fm′ of 12 accessions of Arabidopsis. D, qp in the presence of different actinic irradiances of 12 accessions of Arabidopsis. E, Correlation of ΦPSII and NPQ of 12 accessions each with three replicates grown in 100 µmol m−2 s−1 and measured at light saturation (700 µmol m−2 s−1). F, Correlation of ΦPSII and qp of 12 accessions each with three replicates grown in 100 µmol m−2 s−1 and measured at light saturation (700 µmol m−2 s−1). G, Correlation of ΦPSII and Fv′/Fm′ of 12 accessions, each with three replicates grown in 100 µmol m−2 s-1 1 and measured at light saturation (700 µmol m−2 s−1). H, Correlation of NPQ and Fv′/Fm′ of 12 accessions, each with three replicates grown in 100 µmol m−2 s-1 1 and measured at light saturation (700 µmol m−2 s−1).
ΦPSII is the product of the quantum efficiency of the open PSII reaction centers (Fv′/Fm′) and the PSII efficiency factor (qp; Genty et al., 1989). Therefore, the loss of ΦPSII can be accounted for by decreases in one or both of these parameters. For plants grown at a growth irradiance of 100 µmol m−2 s−1, Fv′/Fm′ remains constant at about 0.8 for actinic irradiances of 225 µmol m−2 s−1 or less, but it decreases when the actinic irradiance is increased above this (Fig. 1C). Heritability for the response of Fv′/Fm′ to increases in irradiance increases with increasing actinic irradiance (Fig. 1C). Similar to Fv′/Fm′, qp also decreases with increasing actinic irradiance for plants grown at 100 µmol m−2 s−1 growth irradiance (Fig. 1D). However, unlike Fv′/Fm′, qp shows decreases in response to actinic irradiances of less than 225 µmol m−2 s−1. In contrast to the heritability for the irradiance response of Fv′/Fm′, the heritability for the irradiance response of qp decreases with increasing actinic irradiance (Fig. 1D).
The values of NPQ calculated according to the Stern-Volmer model (Fm/Fm′ − 1, where Fm is the maximum PSII fluorescence yield in the dark-adapted state and Fm′ is the maximum PSII fluorescence yield in the light-adapted state), qp, and Fv′/Fm′ measured at an actinic irradiance of 700 µmol m−2 s−1 (i.e. at light saturation) are shown plotted against the value of ΦPSII measured at 700 µmol m−2 s−1 (Fig. 1, E–G), and the correlation between NPQ and Fv′/Fm′ is shown in Figure 1H. NPQ, qp, and Fv′/Fm′ are linearly related to ΦPSII, with the correlation being negative in the case of NPQ and positive for qp and Fv′/Fm′. The parameters Fv′/Fm′ and qp are more strongly correlated to ΦPSII than is NPQ (Fig. 1, E–G).
Light Use Efficiency Responses to Different, Constant Growth Irradiances
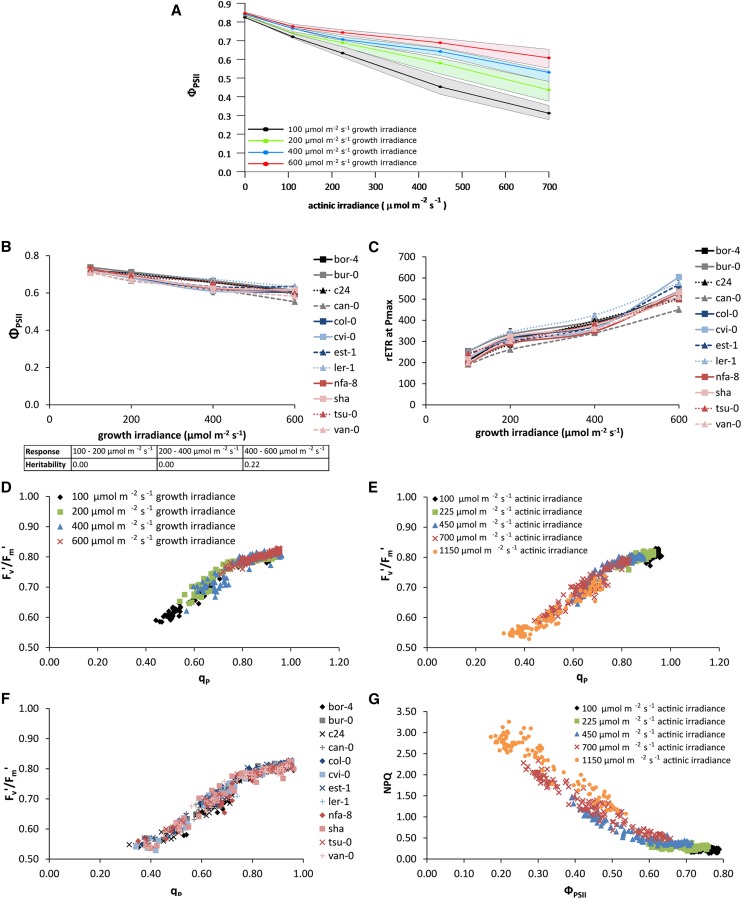
At an actinic irradiance of 600 µmol m−2 s−1, plants grown at 100 µmol m−2 s−1 have an average ΦPSII of 0.35, while for plants grown at 200 µmol m−2 s−1, the average ΦPSII is 0.48, for plants grown at 400 µmol m−2 s−1, it is 0.58, and for plants grown at 600 µmol m−2 s−1, it is 0.65 (Fig. 2A). This shows that there is long-term photosynthetic acclimation to growth irradiance for Arabidopsis. Genetic variation for this acclimation is present. For example for some accessions, the irradiance response of ΦPSII for plants grown at 600 µmol m−2 s−1 is the same as that for those grown at 400 µmol m−2 s−1, whereas for other accessions, the irradiance response of ΦPSII for plants grown at a 600 µmol m−2 s−1 differs from that of plants grown at 400 µmol m−2 s−1, while the response of plants grown at 200 and 400 µmol m−2 s−1 is similar (Supplemental Fig. S1).
Figure 2.
ΦPSII of 12 Arabidopsis accessions grown in different constant growth irradiances: 100, 200, 400, or 600 µmol m−2 s−1. A, ΦPSII light response curves measured on 24-d-old plants after reaching steady-state photosynthesis in four different actinic irradiances: 100, 225, 450, and 700 µmol m−2 s−1. The lines represent the average of all accessions, and the area around each line represents the extent of deviation among the accessions (highest value minus lowest value in the population). B, Variation in ΦPSII among 12 Arabidopsis accessions when grown at four different constant growth irradiances (100, 200, 400, and 600 µmol m−2 s−1) for 24 d and measured at actinic irradiance identical to the growth irradiance. Error bars indicate se; n = 3. The inset shows the heritabilities of the response of these values to growth irradiance level. C, Variation in maximum rETR measured at saturating actinic irradiance (Pmax) among 12 accessions when grown at four different growth irradiances (100, 200, 400, and 600 µmol m−2 s−1) for 24 d. When grown at 100 µmol m−2 s−1, the saturating actinic irradiance level used was 600 µmol m−2 s−1; at 200 µmol m−2 s−1, this was 700 µmol m−2 s−1; at 400 µmol m−2 s−1, it was 800 µmol m−2 s−1; and at 600 µmol m−2 s−1, it was 1,150 µmol m−2 s−1. Error bars indicate se; n = 3. D, Correlation of Fv′/Fm′ and qp for different growth irradiances (100, 200, 400, and 600 µmol m−2 s−1). E, Correlation of Fv′/Fm′ and qp for different actinic irradiances (100, 225, 450, 700, and 1,150 µmol m−2 s−1). F, Correlation of Fv′/Fm′ and qp for each of the 12 different accessions of Arabidopsis. G, Correlation of NPQ and ΦPSII for different actinic irradiances (100, 225, 450, 700, and 1,150 µmol m−2 s−1).
The value of ΦPSII measured at an actinic irradiance equal to growth irradiance shows, overall, an apparently linear decrease with increasing irradiance (Fig. 2B). However, at growth irradiances above 400 µmol m−2 s−1, this decline ceases for some accessions (Fig. 2B). This leads to a heritability of 0.2 for the difference in this parameter when comparing the data obtained from plants grown at an irradiance of 600 µmol m−2 s−1 with those grown at 400 µmol m−2 s−1 (Fig. 2B).
The correlation between the maximum rETR and growth irradiance shows genotypic variation (Fig. 2C). For some accessions, this relationship is triphasic (Supplemental Fig. S2), with a sharp increase over the lower irradiance range (100–200 µmol m−2 s−1), a much lower increase between 200 and 400 µmol m−2 s−1 growth irradiance, and a second sharp increase between 400 and 600 µmol m−2 s−1. For other accessions, this relationship is biphasic (Supplemental Fig. S2), while for others it is linear (Supplemental Fig. S2). Curve estimation of this relation using a cubic statistical model resulted in different fitted values for all 12 accessions (Supplemental Fig. S3). The heritability for this relation is 0.52.
The partitioning of the decrease in ΦPSII into decreases in Fv′/Fm′ and qp is shown for different growth irradiance levels (Fig. 2D), actinic irradiance levels (Fig. 2E), and genotypes (Fig. 2F). While there is some variation in the partitioning of the loss of ΦPSII between losses in Fv′/Fm′ and qp, this variation seems to be independent of genotype or growth irradiance. A negative correlation between NPQ and ΦPSII (Fig. 2E) has already been noted, but in addition, Figure 2G also shows that, considering all the data from all 12 genotypes, the correlation between ΦPSII and NPQ is curvilinear, with NPQ decreasing more strongly with ΦPSII at lower values. Overall, the correlation between ΦPSII and NPQ does not show much genotypic variation or dependency on actinic irradiance.
Long-Term Acclimation of Light Use Efficiency to a Step-Wise Increase in Growth Irradiance
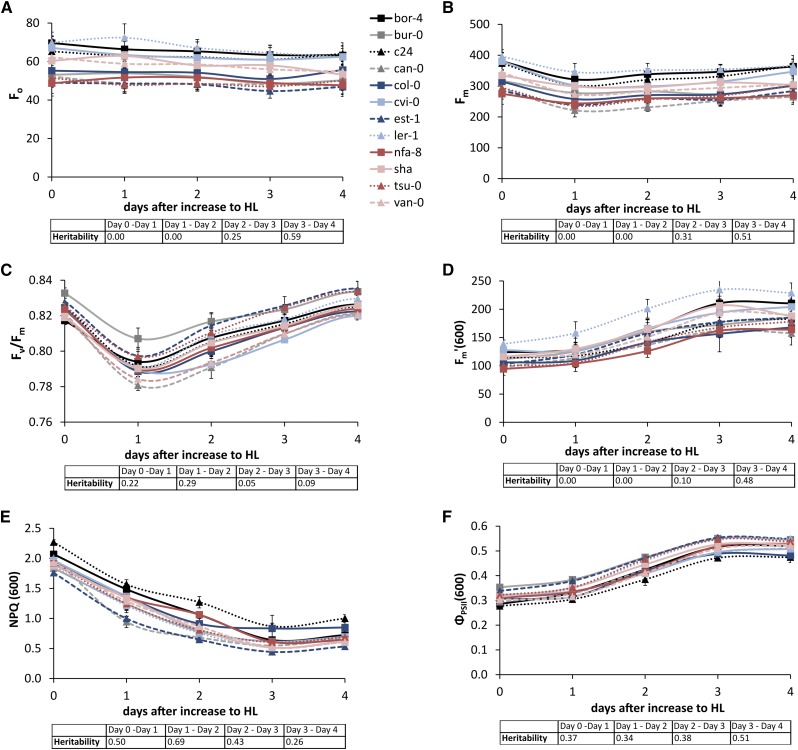
During acclimation to a step-wise increase in growth irradiance from 100 to 600 µmol m−2 s−1, Fo (initial [minimum] PSII fluorescence in the dark-adapted state) stays constant (Fig. 3A), whereas Fm decreases on the first day after the increase to high light and then recovers to its baseline value within 3 to 4 d (Fig. 3B), resulting in a decrease in Fv/Fm on day 1 after the increase to high light (Fig. 3C). This decrease in Fv/Fm is correlated significantly to the levels of ΦPSII and qp before the increase in growth irradiance (Supplemental Table S1) as well as to the levels of ΦPSII and qp on each day of the subsequent acclimation period. NPQ started to decline within the first 24 h after the irradiance increase and continued to decline until day 3 (Fig. 3E). The level of NPQ on day 1 after the increase to high light was significantly correlated to the decrease in Fv/Fm on that same day, but this correlation disappeared after the start of the acclimation of NPQ (Supplemental Table S1). The decline of NPQ on the subsequent days of the acclimation period was accompanied by an increase in ΦPSII (Fig. 3, E and F). Both NPQ and ΦPSII stabilized after 3 d of long-term acclimation.
Figure 3.
Variation in long-term acclimation responses of 12 Arabidopsis accessions to increased growth irradiance after 24 d of growth, from 100 to 600 µmol m−2 s−1 (high light [HL]), for Fo (A), Fm (B), Fv/Fm (C), and Fm′ (D) measured with an actinic irradiance level of 600 µmol m−2 s−1, for NPQ measured with an actinic irradiance level of 600 µmol m−2 s−1 (E), and for ΦPSII measured with an actinic irradiance level of 600 µmol m−2 s−1 (F). Day 0 represents the baseline measurement taken on day 24 after sowing the plants on rockwool. All measurements were taken in the morning, 30 min after light onset. Error bars indicate se; n = 3. The insets show the heritabilities of the daily responses relative to the day before for the corresponding values [Fo, Fm, Fv/Fm, Fm′(600), NPQ(600), or ΦPSII(600)].
Genotypic variation was observed for both the decline of NPQ and the increase in ΦPSII during acclimation to increased growth irradiance (heritabilities are shown in Fig. 3, E and F). Furthermore, there was some genotypic variation for the decrease in Fv/Fm on day 1 after the increase to high light as well as its recovery on day 2 (heritabilities are shown in Fig. 3C). After 3 d, all the measured photosynthetic parameters had stabilized; some genotypic variation was also noted for the kinetics of this stabilization process, as shown by the heritabilities (Fig. 3).
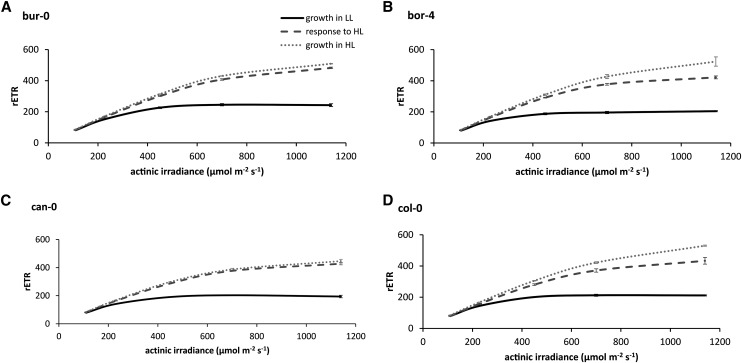
While some accessions acclimate fully (Fig. 4, A and C) to an increase in growth irradiance (i.e. after acclimation to the increased irradiance, their ΦPSII-irradiance response becomes identical to that when grown continuously at this higher irradiance), others do not (Fig. 4, B and D). Among the 12 accessions used in this study, we could distinguish only these two kinds of responses; the responses for the accessions not shown in Figure 4 are shown in Supplemental Figure S4.
Figure 4.
Variation in light response curves of rETR in four Arabidopsis accessions grown at a low growth irradiance (growth in LL), grown in LL for 24 d followed by a high growth irradiance for 4 d (response to HL), and grown in HL for 24 d (growth in HL). The low growth irradiance level is 100 µmol m−2 s−1, and the high growth irradiance level is 600 µmol m−2 s−1. Error bars indicate se; n = 3.
GWAS
ΦPSII was measured on 344 accessions of Arabidopsis 1 d before and 1 h after an increase in growth irradiance from 100 to 550 µmol m−2 s−1. ΦPSII measurements before and after the increase were correlated significantly, with a Pearson correlation coefficient of 0.436. At both time points, the measurements were normally distributed (Supplemental Fig. S5), but the phenotypic distribution, and consequently also the trait heritability, was larger 1 h after the increase in irradiance compared with before the increase in irradiance (0.26 versus 0.09). A GWAS was performed for the measurements taken 1 h after a step-wise increase in growth irradiance to identify potential candidate genes associated with the short-term high-light response of photosynthesis (Fig. 5). All of these genes are nuclear because the cytoplasmic genomes cannot be included in the analysis, because neither the mitochondrial nor the chloroplast genomes have been genotyped for the Arabidopsis population we used (nor for any other large Arabidopsis population), so there are no SNPs (or any other genetic markers) available for these genomes. Any phenotypic variation arising from the variation in the cytoplasmic genetic factors, therefore, will not be accounted for.
Figure 5.
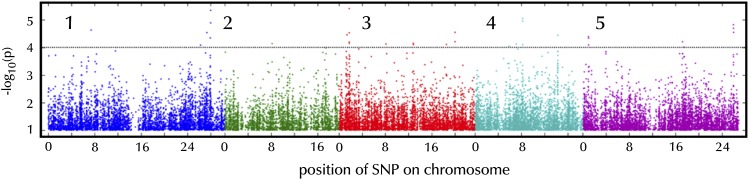
Genome-wide association analysis of ΦPSII 1 h after an increase in growth irradiance from 100 to 550 µmol m−2 s−1 based on the analysis of 344 diverse Arabidopsis accessions. Every point indicates the –log10(p) value for all SNPs that have been tested. Different colors distinguish the SNPs mapped to one of the five chromosomes of Arabidopsis. The dotted line represents the arbitrary threshold for significance of –log10(p) = 4.
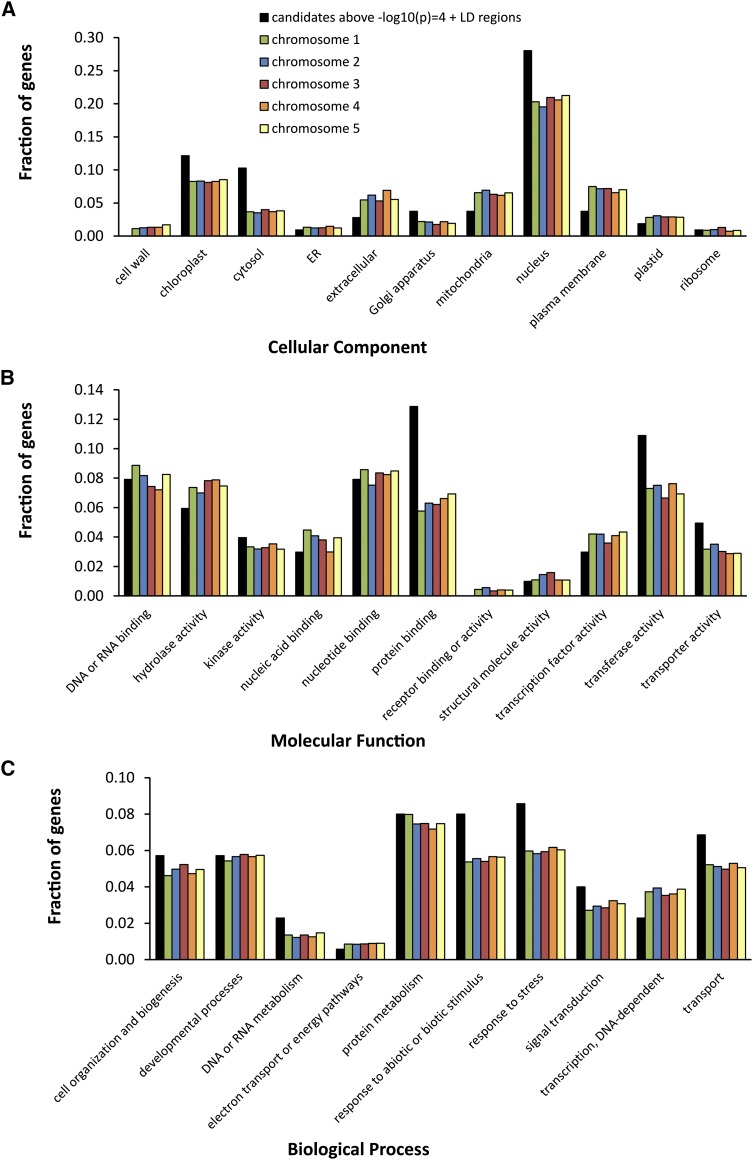
The strongest association between an SNP and the phenotype we found had a –log10(p) value of 5.31, where p is the probability of obtaining the association by chance. This strength of association was found twice, at position 27,981,096 on chromosome 1 and at position 12,784,017 on chromosome 3. Using an arbitrary threshold of significance for –log10(p) of 4, we defined the SNPs with –log10(p) values above this threshold as associated SNPs. This yielded 30 associated SNPs, corresponding to 25 candidate genes (some genes contain more than one SNP), which increases to 63 candidate genes when all genes in linkage disequilibrium (LD) with these SNPs are included (Table I). Besides information on the genes localized (either directly or in LD) to the associated SNPs, Table I shows extra information about the population genetics of the associated SNPs, such as minor allele frequency in the population, the effect size on the trait, and the percentage of variation it explains. The functions, if known, of those genes directly associated with the SNPs also are described in Table I; the functions of the genes in the LD regions with the associated SNPs are described in Supplemental Table S2. No genes reported previously to be involved in photosynthesis were found among the 63 candidate genes. The candidate gene list, however, is enriched for genes encoding proteins that are targeted to the chloroplast, cytosol, and nucleus (Fig. 6A). When examined for protein function, there is enrichment for kinase activity, protein binding, transferase activity, and transporter activity (Fig. 6B), while the processes DNA/RNA metabolism, response to abiotic or biotic stimulus, response to stress, and transport are most represented (Fig. 6C). Of the 63 candidates, 13 (nucleus-encoded) proteins are targeted to the chloroplast (Table II).
Table I. List of candidate genes localized to SNPs.
Candidate genes are those genes containing the SNP and those genes in LD with the SNP associated with a –log10(p) > 4 for ΦPSII measured 1 h after a step-wise increase in irradiance from 100 to 550 µmol m−2 s−1, selected upon a GWAS on 344 Arabidopsis accessions. Chr., Chromosome; Gene, the gene to which the associated SNP localizes (if two genes are indicated, the SNP is mapped between the two genes); SNP Pos., the chromosome position(s) of the SNP(s); MAF, minor allele frequency (the letters in parentheses indicate the Columbia-0 allele [C] or the non-Columbia-0 allele [NC]); −log10(p), the significance level of the associated SNP expressed as –log10(p); Effect Size, the contribution of the Columbia-0 allele of the SNP on ФPSII; Percentage Genetic Variation, the percentage of genetic variation explained by the SNP; Description, the annotation of the gene function as indicated in The Arabidopsis Information Resource (TAIR; www.arabidopsis.org); LD, the genes found to be in LD (r > 0.45) with the indicated SNP.
| Chr. | Gene | SNP Pos. | MAF | −log10(p) | Effect Size | Percentage Genetic Variation | Description | LD |
|---|---|---|---|---|---|---|---|---|
| 1 | AT1G21080/AT1G21090 | 7,384,441 | 0.146 (C) | 4.6 | 0.018 | 9 | DNAJ heat-shock N-terminal domain-containing protein/cupredoxin superfamily protein | AT1G21060 to AT1G21140 |
| 1 | AT1G69770 | 26,249,116 | 0.254 (NC) | 4.3 | −0.014 | 8.1 | Chromomethylase3, involved in methylating cytosine residues at non-CG sites | – |
| 1 | AT1G72560 | 27,329,236 | 0.246 (NC) | 4.6 | 0.014 | 8.1 | PAUSED, a karyopherin | – |
| 1 | AT1G74180 | 27,899,243 | 0.351 (C) | 4.4 | 0.013 | 8.6 | Receptor-like protein14, located in chloroplast | AT1G74190 |
| 1 | AT1G74440 | 27,979,318; 27,981,096 | 0.447 (C); 0.365 (NC) | 5.0; 5.3 | 0.013; 0.013 | 9.0; 9.8 | Unknown protein | – |
| 2 | AT2G26280 | 11,189,311 | 0.196 (NC) | 4.3 | −0.018 | 8.3 | CID7, CTC-interacting domain7, functions in DNA binding and mismatch repair, located in chloroplast | AT2G26290 |
| 3 | AT3G04910 | 1,353,894 | 0.155 (NC) | 4.3 | −0.016 | 8.3 | WNK1, With No Lys Kinase1, Ser/Thr protein kinase, whose transcription is regulated by circadian rhythm | AT3G04880 to AT3G04910 |
| 3 | AT3G05860 | 1,750,265; 1,750,573; 1,750,946; 1,751,042 | 0.175; 0.167; 0.167; 0.211 (C) | 4.2; 4.5; 4.2; 5.4 | 0.016; 0.017; 0.016; 0.016 | 8.3; 9.1; 8.3; 9.8 | MADS box transcription factor family protein | AT3G05790 to AT3G05890 |
| 3 | AT3G22910 | 8,120,853 | 0.307 (NC) | 4.1 | 0.015 | 7.9 | ATPase E1-E2 type family protein/haloacid dehalogenase-like hydrolase family protein | – |
| 3 | AT3G31410 | 12,784,017 | 0.377 (C) | 5.3 | −0.015 | 8.7 | Transposable element | – |
| 3 | AT3G44230 | 15,932,189 | 0.184 (NC) | 4.1 | −0.016 | 6.6 | Unknown protein | – |
| 3 | AT3G54010 | 20,000,766 | 0.289 (NC) | 4.7 | −0.014 | 9.6 | Immunophilin-like protein | AT3G54000 |
| 4 | AT4G11300 | 6,872,903 | 0.480 (C) | 4.2 | 0.014 | 7.6 | Unknown protein | – |
| 4 | AT4G14250 | 8,209,018; 8,209,226 | 0164; 0.187 (C) | 4.9; 5.1 | 0.017; 0.016 | 9.3; 9.1 | Pseudogene | – |
| 4 | AT4G21760 | 11,561,583 | 0.053 (NC) | 4.2 | 0.023 | 6.3 | β-Glucosidase47, involved in carbohydrate metabolic process | AT4G21750 to AT4G21770 |
| 5 | AT5G03760 | 987,180; 987,216; 988,003 | 0.216; 0.272; 0.240 (C) | 4.9; 4.3; 4.4 | 0.014; 0.014; 0.014 | 7.7; 8.5; 8.3 | Cellulose synthase-like A9 | AT5G03750 |
| 5 | AT5G42870 | 17,186,178 | 0.345 (C) | 4.3 | −0.012 | 8 | PAH2, a phosphatidate phosphohydrolase | – |
| 5 | AT5G43390 | 17,424,158 | 0.450 (C) | 4.0 | −0.012 | 8.5 | Uncharacterized conserved protein, located in chloroplast | – |
| 5 | AT5G64960 | 25,956,134 | 0.465 (C) | 4.3 | −0.013 | 9.9 | CDKC2, expression modifies the location of spliceosomal components | AT5G64910 to AT5G65030 |
| 5 | AT5G64980 | 25,963,073 | 0.465 (C) | 4.5 | −0.013 | 9.9 | Unknown protein | |
| 5 | AT5G65000/AT5G65005 | 25,967,700 | 0.354 (NC) | 4.9 | 0.013 | 9.4 | Nucleotide-sugar transporter family protein/polynucleotidyl transferase | |
| 5 | AT5G65010 | 25,968,943 | 0.284 (NC) | 4.2 | 0.014 | 10.1 | ASN2, Asn synthetase2 | |
| 5 | AT5G65030 | 25,975,808 | 0.480 (C) | 4.4 | −0.013 | 10.4 | Unknown protein |
Figure 6.
Gene Ontology enrichment analysis for cellular component (A), molecular function (B), and biological process (C). The graphs depict the fractions of genes from a list [gene candidate list directly localized to, and in LD regions of, the SNPs associated in GWAS above –log10(p) = 4 and lists of all genes on chromosomes 1–5] annotated to different Gene Ontology categories.
Table II. List of candidate genes that encode proteins predicted to localize at the chloroplast, selected from 63 genes found to be directly localized to, or be in LD with, the associated SNPs identified in the GWAS.
| Gene | SNP | Description |
|---|---|---|
| AT1G21060 | In LD with 7384441 | Unknown protein |
| AT1G21065 | In LD with 7384441 | Unknown protein |
| AT1G74180 | Directly localized to 27899243 | Receptor-like protein14, located in chloroplast |
| AT1G74440 | 27979318; 27981096 | Unknown protein |
| AT2G26280 | Directly localized to 11189311 | CID7, CTC-interacting domain7, functions in DNA binding and mismatch repair |
| AT3G05790 | In LD with 1750265; 1750573; 1750946; 1751042 | Lon-protease4, for degradation of abnormal, damaged, and unstable protein |
| AT3G05810 | In LD with 1750265; 1750573; 1750946; 1751042 | Chromatin assembly/disassembly protein |
| AT4G21770 | In LD with 11561583 | Pseudouridine synthase, involved in RNA modification |
| AT5G43390 | Directly localized to 17424158 | Uncharacterized conserved protein |
| AT5G64930 | In LD with 25956134; 25963073; 25967700; 25968943; 25975808 | Regulator of expression of pathogenesis-related genes, participates in signal transduction pathways involved in plant defense |
| AT5G64940 | In LD with 25956134; 25963073; 25967700; 25968943; 25975808 | Oxidative stress-related ABC1-like (for ATP-binding cassette) protein |
| AT5G65000 | Directly localized to 25967700 | Nucleotide-sugar transporter family protein/polynucleotidyl transferase |
| AT5G65020 | In LD with 25956134; 25963073; 25967700; 25968943; 25975808 | Annexin2, calcium-binding protein |
DISCUSSION
Short-Term Response of Plants to Increased Irradiances
In 12 genotypically diverse Arabidopsis accessions grown at 100 µmol m−2 s−1, there is variability in the responses of ΦPSII to short-term (minutes time scale) changes in irradiance (Fig. 1, A and B). There is more phenotypic variance for electron transport rate the closer the actinic irradiance level is to light saturation (Fig. 1B), which would be expected given that increasing irradiances move the leaf from light limitation to light saturation (Björkman, 1981; Evans and Poorter, 2001). In a light-limited leaf, ΦPSII is close to a maximum value (approximately 0.80–0.83) that is similar across many groups of plants (Björkman and Demmig, 1987). The value for ΦPSII at steady state is due to the balance between supply-side processes, which give rise to the formation of excited states of chlorophyll a in PSII following light absorption by PSII, and demand-side processes, which dissipate these excited states of chlorophyll a photochemically (Genty et al., 1989). If supply exceeds demand, then the light use efficiency for photosynthesis must decrease. For plants grown at 100 µmol m−2 s−1, such a decrease has already occurred at an actinic irradiance equal to the growth irradiance (Fig. 1A), which implies that, even at low growth irradiances, acclimation did not maximize light use efficiency. In a light-saturated leaf, ΦPSII is limited by electron transport or metabolic factors that generally can differ greatly between species (Seemann, 1989; Murchie and Horton, 1997; Valladares et al., 1997) and within species (Balaguer et al., 2001; Walters et al., 2003; Ptushenko et al., 2013).
Values for heritability generally range from 0 to 1 (Visscher et al., 2008), where a value of 1 means that all of the observed phenotypic variance is due solely to genetic variation. The heritability for ΦPSII ranges around 0.5, independent of the actinic irradiance at which it is measured (Fig. 1B), from which we conclude that the amount of genetic variation for short-term responses of ΦPSII to increased irradiance is independent of the level of the irradiance. To further dissect the variation for photosynthetic light use efficiency, ΦPSII can be broken down into its Fv′/Fm′ and qp components (Genty et al., 1989). Overall, Fv′/Fm′ and qp decrease as ΦPSII decreases, but not in parallel (Harbinson et al., 1989). In the absence of photodamage or slowly reversible down-regulation of PSII (Demmig-Adams and Adams, 2006), decreases in Fv′/Fm′ are due to the activation of a nonphotochemical dissipation mechanism that gives rise to the qE component of NPQ (Demmig et al., 1987). Decreases in qp are due to the reduction of QA (the primary electron-accepting plastoquinone of PSII), although the relationship between QA redox state and qp is nonlinear (Kramer et al., 2004). In response to a moderate increase in irradiance (from 100 to 200 µmol m−2 s−1), the decrease in ΦPSII is due only to decreases in qp (Fig. 1, C and D). The loss of qp at low irradiances, which is due to an overexcitation of PSII compared with PSI and not to a limitation of electron transport (Genty and Harbinson, 1996), is still correlated with a loss of light use efficiency for carbon dioxide fixation (Hogewoning et al., 2012). The lack of any decrease in Fv′/Fm′ at the lowest measurement irradiances is paralleled by the pattern of heritability for the irradiance responses of qp and Fv′/Fm′ (Fig. 1, C and D).
The parameters Fv′/Fm′ and NPQ quantify the effect of inducible (i.e. not present in the dark-adapted state) nonphotochemical dissipation in PSII on the efficiency of open PSII traps (Fv′/Fm′) and the quenching of Fm (NPQ). Even if Fm′ changes, the Fv′/Fm′ calculated using a measured value of Fo′ (the yield of chlorophyll fluorescence from PSII in the light-adapted state when all QA is oxidized) should be unaffected by state transitions, in contrast to the NPQ parameter. In our case, Fo′ was calculated from Fv (variable PSII fluorescence in the dark-adapted state), Fm, and Fm′ (Oxborough and Baker, 1997), and while this approach allows a good estimate of Fo′, it cannot estimate the impact of fluorescence quenching due to state-transitions (qT quenching [Horton and Hague, 1988; Quick and Stitt, 1989]) on Fo′ and is limited to low irradiances (Walters and Horton, 1991; Rintamäki et al., 1997), so the greatest impact of using a calculated Fo′ in place of measured Fo′ will be at low irradiances. Therefore, an Fv′/Fm′ based on a calculated Fo′ is likely to be better correlated with NPQ than would be an Fv′/Fm′ based on a measured Fo′, especially at low irradiances. Figure 1, E to G, show a high correlation of NPQ, qp, and Fv′/Fm′ with ΦPSII measured at light saturation (actinic light of 700 µmol m−2 s−1). Fv′/Fm′ and qp are more strongly correlated with ΦPSII than is NPQ (Fig. 1, E–G), which is to be expected, as ΦPSII is the product of qp and Fv′/Fm′. The correlation between NPQ and Fv′/Fm′ (Fig. 1H) also is expected, given that under the experimental conditions used, both these parameters will be predominantly affected by the energy-dependent quenching mechanism that gives rise to qE. These results suggest that there is little variability in the extent to which the loss of ΦPSII can be absorbed via the thermal dissipation processes that give rise to NPQ and produce the decrease in Fv′/Fm′.
ΦPSII in Plants Grown at Different Constant Growth Irradiances
In response to increasing growth irradiances, all accessions of Arabidopsis showed a smaller loss of ΦPSII with increasing actinic irradiance (Fig. 2A). As a result, ΦPSII measured at an actinic irradiance identical to the growth irradiance decreases only slightly with growth irradiance (maximally, 25%; Fig. 2B). This response shows that Arabidopsis has considerable flexibility in its photosynthetic apparatus and responds strongly to high irradiances, a trait not found in all species (Murchie and Horton, 1997). Genotypic variation for this trait is minor among the 12 genotypes used in this study (Fig. 2B). More genotypic variation can be found in the relationship between the rETR at light saturation and growth irradiance (Fig. 2C). For some accessions this relation is linear, for some it is biphasic, and for some it is triphasic, confirming the presence of separate low and high light responses in Arabidopsis (Bailey et al., 2001).
The pattern of partitioning of losses in ΦPSII between Fv′/Fm′ and qp is independent of growth irradiance (Fig. 2D), actinic irradiance (Fig. 2E), and genotype (Fig. 2F). These results imply that nonphotochemical dissipation in PSII is highly and consistently regulated across diverse genotypes grown under a range of growth irradiances. Another implication is that when evaluating and comparing the development and extent of the inducible nonphotochemical dissipation processes that give rise to decreases in Fv′/Fm′ and to increases in NPQ, the underlying change in PSII efficiency should be taken into account (Fig. 2G).
Long-Term Responses to a Step-Wise Increase in Growth Irradiance
The decrease in Fv/Fm (0.02–0.04) 1 d after a step increase in growth irradiance from 100 to 600 µmol m−2 s−1 is due to a decrease in Fm, which is followed by a recovery in Fv/Fm, an increase in ФPSII, and a decrease in NPQ (Fig. 3, A–C). The decrease in Fv/Fm was correlated to the values for ФPSII and qp before the increase in irradiance as well as to the values for ФPSII, qp, and NPQ on day 1 after the increase in irradiance (Supplemental Table S1). Slowly reversible decreases in Fv/Fm are an indicator of photodamage or of slowly reversible down-regulation of PSII (Walters and Horton, 1991; Demmig-Adams and Adams, 1992, 2006; Niyogi, 1999). We were not able to distinguish between these two mechanisms in our experiments, but overall, the phenomenon of slowly reversible loss of Fv/Fm seems to occur if the short-term protection mechanisms that give rise to NPQ are insufficient to protect PSII from damage.
The kinetics of NPQ decay and ΦPSII recovery after a step increase in irradiance show genotypic variation (Fig. 3, E and F), implying that there is variation in the regulation of photosynthetic recovery after an irradiance increase, whereas the ultimate extent of acclimation after 4 d shows no genotypic variation. The extents of the changes in ΦPSII in response to a step increase in irradiance found by use are similar to those found by Yin et al. (2012), but their ΦPSII values decreased for 2 d following the increase in irradiance before recovering on day 3, whereas our data showed a recovery of ΦPSII beginning on day 1 after the increase in irradiance. This difference could be caused by the relatively greater irradiance increase they used (from 120–950 µmol m−2 s−1 versus from 100–600 µmol m−2 s−1 in our case), causing more extensive slowly reversible loss of Fv/Fm. Yin et al. (2012) identified two potential regulatory mechanisms that are variable among three Arabidopsis accessions in response to increased irradiance: One is the abundance of kinases that facilitate state transitions, and the other is a mechanism to facilitate lateral protein traffic in the membrane by diluting chlorophyll-protein complexes with additional lipids and carotenoids. The variation we found for the kinetics of NPQ decay and ΦPSII recovery is likely to reflect these different mechanisms found by Yin et al. (2012) and, in addition, other possible regulatory mechanisms yet undefined.
After full acclimation to a step-wise increase in irradiance from 100 to 600 µmol m−2 s−1, all accessions had a photosynthetic capacity higher than that achieved under a constant growth irradiance of 100 µmol m−2 s−1 (Fig. 4; Supplemental Fig. S4). In contrast to our results, a similar study performed by Athanasiou et al. (2010) revealed significant variation in the ability of different accessions to acclimate to an increased irradiance. While Athanasiou et al. (2010) increased irradiance after 8 weeks of growth at 100 µmol m−2 s−1, we increased it after 3.5 weeks of growth. Plant age or size, therefore, might have an effect on the capacity of photosynthesis to respond to the increase in irradiance. We also grew the plants hydroponically, while Athanasiou et al. (2010) used soil-based cultivation, and differences in plant nutritional state might have contributed to the different responses of photosynthesis. Whatever the explanation, the fact of these differences implies that there are extra dimensions to the irradiance responses of photosynthesis in Arabidopsis that need to be understood. There is genotypic variation for the ability of leaves grown at 100 µmol m−2 s−1 before a step increase in irradiance to 600 µmol m−2 s−1 to acclimate their photosynthetic capacity to that found in leaves grown at a constant irradiance of 600 µmol m−2 s−1 (Fig. 4). A possible role for leaf anatomy in limiting the response of photosynthetic capacity in leaves subjected to an increase in irradiance has been reported (Oguchi et al., 2003), and there is variation in leaf architecture in Arabidopsis (Pérez-Pérez et al., 2002).
Natural Genetic Variation and GWAS
The fact that there is genetic variation for photosynthesis could mean that natural selection has favored different optima depending on the local environment, especially since it is hard to imagine that genetic drift is the sole cause of variation for such an important trait (Alonso-Blanco et al., 2009; Trontin et al., 2011). To investigate this, it is crucial to identify the genes involved and the effect of alleles of those genes on the photosynthesis phenotype. Identifying causal allelic variation is easier when there is substantial genetic variation for a trait, as quantified by the heritability values (Barton and Keightley, 2002) and the effect size of the quantitative trait locus (Falke and Frisch, 2011). It is hard to predict how much heritability is required before a trait will be amenable to genetic analysis, as this depends on the number of loci that contribute to the heritability (i.e. the genetic complexity of the trait) and on the population used in the study (Visscher et al., 2008; Brachi et al., 2011). In this study of 12 Arabidopsis accessions, it is clear that there are some photosynthetic traits for which heritability values look more promising for further genetic analysis than others
The heritability calculated for ΦPSII in the 344 accessions used for the GWAS (0.09 before and 0.26 1 h after the increase in growth irradiance; Supplemental Fig. S5) is different from the heritability calculated for the population of 12 accessions (0.59 before and 0.53 after the increase in growth irradiance; Fig. 1B). Different heritabilities for the same trait in different populations can be explained by different allele frequencies (Visscher et al., 2008); in this case, it looks like the small set of 12 diverse accessions already captured most of the genetic variation for ΦPSII also found in the larger set of 344 accessions. Even though alleles get more heterogeneous when studying a bigger natural population (Brachi et al., 2011; Gibson, 2012), if the larger heterogeneity found in the set of 344 accessions does not contribute to larger phenotypic variation than in the set of 12 accessions but merely a dilution of the phenotypic effect of extreme genotypes, the result will be a reduction in heritability.
Successful GWAS have identified an overrepresentation in a priori candidate genes (Atwell et al., 2010) or only a small number of genes that were associated above the Bonferroni threshold of –log10(p) = 6.50 (Chao et al., 2012; Meijón et al., 2014). The Bonferroni threshold is a very stringent statistical test correcting for multiple testing (Holm, 1979). In our study, no genetic associations are detected with a –log10(p) above the Bonferroni threshold (Fig. 5). As the change in photosynthesis efficiency in response to an increase in irradiance is a highly polygenic trait, we expect that the effects of the individual underlying genes will be small. Such genes are likely to remain hidden in associations that do not exceed the Bonferroni threshold because of epistatic and interactive effects (Gibson, 2010; Korte and Farlow, 2013). By lowering the –log10(p) threshold to 4, some of these hidden associations were revealed (Fig. 5), allowing us to select 63 candidate genes that either contained an associated SNP or were in LD with the SNP (Table I). No genes reported previously to be involved in photosynthesis were found among these candidate genes. To provide validation for our approach, a Gene Ontology enrichment analysis was performed from which we could extract the ontology classes that are relevant for the natural variation of our trait (Fig. 6; Huang et al., 2009). There was enrichment for the regulation of protein abundance (enriched ontology classes: Golgi apparatus, kinase activity, transferase activity, protein binding, protein metabolism, and response to [a]biotic stimulus or to stress) as well as to responses limited to the nucleus (enriched ontology class: nucleus) and the chloroplast (enriched ontology classes: chloroplast, cytosol, and transporter activity). Out of the 63 candidate genes, 13 of the (nucleus-) encoded proteins localized to the chloroplast (Table II). Close analysis of the functions of these 13 genes validates the enrichment for abiotic stress responses, as some genes are involved with the sensing of a stress (receptor, oxidative stress protein, and calcium-binding protein), others with regulating a stress response (chromatin assembly/disassembly, RNA modification, and regulator of the expression of defense genes), and others are involved with the act of responding to (photosynthetic) stress (DNA mismatch repair, protease for degrading abnormal and damaged proteins, and carbohydrate transport; Table II). Only four genes have no known function.
CONCLUSION
In conclusion, we have demonstrated that, for Arabidopsis accessions, there is genotypic variation for the short-term response of photosynthetic light use efficiency to a step-wise increase in growth irradiance as well as its long-term acclimation. The data also show that, over the range of growth irradiances employed, the light use efficiency of photosynthesis (measured by ΦPSII) acclimates strongly to the level of growth irradiance, so that the ΦPSII, measured at an actinic irradiance identical to the growth irradiance, only decreases slightly with higher levels of growth irradiance. A broader phenotypic distribution is found by measuring ΦPSII at light saturation (Figs. 1B, 2A, and 4; Supplemental Fig. S5). In relation to productivity and yield, the ability of photosynthesis to acclimate has been shown to increase the plant fitness of Arabidopsis in a greenhouse environment where natural fluctuations in irradiance and other environmental factors would have occurred (Athanasiou et al., 2010). If there was a desire to breed Arabidopsis for improved photosynthetic properties leading to increased yield, more might be gained by focusing on the dynamic regulatory acclimation response of photosynthesis to different light environments instead of investigating acclimation to stable environments (Leister, 2012). The response described here will be useful when selecting light environments for optimal, uniform growth of Arabidopsis and also serves as a reminder that, while often grown at low irradiances (100–200 µmol m−2 s−1), Arabidopsis has photosynthetic responses that are more typical of a high-light-adapted plant, in accordance with its natural habitat, which is disturbed sites, possibly with light shade (Pigliucci, 1998; Mitchell-Olds and Schmitt, 2006). In addition, we have shown that for those traits for which there is considerable heritability, it is possible to use GWAS to identify novel candidates for genetic components of the plant photosynthetic response to light.
MATERIALS AND METHODS
Plant Material and Growth Conditions
The measurements described here can be divided into GWAS and non-GWAS measurements. As the non-GWAS measurements were used as a pilot for the GWAS, many of the methods and genotypes used are common to both the non-GWAS and GWAS measurements.
The non-GWAS measurements were made on 12 genotypically diverse accessions of Arabidopsis (Arabidopsis thaliana), which were used throughout the experiments in this study (Table III), grown in three replications. All these accessions are part of a core set of 360 natural accessions, which represents the global genetic diversity in Arabidopsis (http://www.naturalvariation.org/hapmap; Li et al., 2010). For the GWAS, we used 344 accessions of the set of 360 accessions; included in the set of 344 accessions were all of the accessions used in the non-GWAS. The 16 accessions that were not used were lines CS28051, CS28108, CS28808, CS28631, CS76086, CS76104, CS76110, CS76112, CS76118, CS76121, CS76138, CS76196, CS76212, CS76254, CS76257, and CS76302.
Table III. Origins of the 12 accessions of Arabidopsis used in this study.
| Accession | Full Name | Latitude | Longitude | Origin |
|---|---|---|---|---|
| Bor-4 | Borky | 49.4 | 16.2 | Czech Republic |
| Bur-0 | Burren | 53 | −9 | Ireland |
| C24 | C24 | 41.2 | −8.4 | Portugal |
| Can-0 | Canary Islands | 29.2 | −13.5 | Spain |
| Col-0 | Columbia | – | – | Unknown |
| Cvi-0 | Cape Verde Islands | 15.1 | −23.6 | Cape Verde |
| Est-1 | Estonia | 58.3 | 25.3 | Estonia |
| Ler-1 | Landsberg erecta | 52.7 | 15.2 | Poland |
| NFA-8 | NFA | 51.4 | −0.6 | United Kingdom |
| Sha | Shahdara | 38.3 | 68.5 | Tajikistan |
| Tsu-0 | Tsushima | 34.4 | 136.3 | Japan |
| Van-0 | Vancouver | 49.3 | −123 | Canada |
For both GWAS and non-GWAS experiments, seeds were presown on filter paper in petri dishes wetted with 0.5 mL of demineralized water and placed in the dark at 4°C for 4 d to stratify. Once stratified, the seeds were planted in a hydroponic cultivation system based on rockwool blocks (Grodan Rockwool Group; 40 × 40 × 40 mm in size). The blocks were positioned and secured using a frame (Fig. 7) consisting of a base plate made from a sheet of perforated stainless steel, a second polyvinyl chloride (PVC) frame that was held 15 mm above the stainless steel base and into which the blocks were placed, and a black nonreflective foamed PVC coversheet drilled with countersunk holes 60 mm apart and 3 mm diameter that were positioned over the centers of the blocks. The seeds were placed on the rockwool surface exposed in these holes, and the plants then grew up through the holes with the leaves spreading across the surface of the upper, black PVC sheet. The three layers of the growing system were secured by stainless steel screws and spacers and placed in a basin to which nutrient solution could be added. The base plate was supported 5 mm above the floor of the basin, allowing nutrient solution to pass freely and uniformly under the growing frame and circulate through the frame via the holes in the perforated metal base plate and the 10-mm spaces between the blocks. The black plastic upper plate prevented the growth of algae on the rockwool blocks and offered a good background for imaging of the plants.
Figure 7.

Growing system for Arabidopsis used in this study. A, Top view showing the nonreflective cover with small holes covering rock wool blocks on which Arabidopsis is germinated and grown. B, When the nonreflective cover is removed, the rock wool block can be placed in 4- × 4-cm square holes, on a stainless steel grid, which is supported by short pins allowing nutrient solution to spread evenly underneath the rock wool blocks
For the smaller scale non-GWAS experiments, the growing frame was 180 × 180 mm in size (Fig. 7) and could hold nine individual rockwool blocks in a 3 × 3 array. Three seeds were sown per accession, and the total of 36 seeds were randomized over four growing systems, with the limitation that two seeds of the same accession were never planted in the same growing system. For the GWAS experiment, the growing frame was 390 × 85 cm and could hold 720 rockwool blocks in a 60 × 12 array. Two of these 60 × 12 growing systems, each in a separate basin, were combined to potentially grow a total of 1,440 plants (allowing for four replicates of 360 genotypes) for the GWAS. Each growing frame was notionally subdivided into two blocks, giving a total of four growing blocks. Four seeds were sown per accession in the GWAS experiment, and each set of 344 seeds was randomized over each of the four blocks.
For both GWAS and non-GWAS experiments, plants were grown hydroponically using a nutrient solution developed for Arabidopsis (pH 7; electrical conductivity 1.4 mS cm−1) consisting of 1.7 mmol of NH4+, 4.5 mmol of K+, 0.4 mmol of Na+, 2.3 mmol of Ca2+, 1.5 mmol of Mg2+, 4.4 mmol of NO3−, 0.2 mmol of Cl−, 3.5 mmol of SO42−, 0.6 mmol of HCO3−, 1.12 mmol of PO43−, 0.23 mmol of SiO32−, 21 µmol of Fe2+ (chelated using 3% [w/v] diethylene triaminopentaacetic acid), 3.4 µmol of Mn2+, 4.7 µmol of Zn2+, 14 µmol of BO33−, 6.9 µmol of Cu2+, and less than 0.1 µmol MoO44−, which was added to the basins containing the growing frames for 5 min three times per week.
For the non-GWAS experiments, plants were grown at either constant irradiances of 100, 200, 400, or 600 µmol m−2 s−1 (Philips MASTER TL5 HO fluorescent tubes, 80 W) or the irradiance was increased from 100 to 600 µmol m−2 s−1 at noon, 24 d after sowing on rockwool. In all cases, the photoperiod was a 10-h-d/14-h-night cycle, temperature was 20°C/18°C (day/night), relative humidity was 70%, and CO2 levels were ambient. Following the step increase in irradiance, the photosynthetic acclimation response was measured over 5 d by measuring light response curves once per day in the morning 30 min after light onset. Other conditions were kept similar, although an increase of leaf temperature due to energy absorbed by the black cover needed for imaging could not be prevented. The highest irradiance we used in this study was well within the adaptive range of the accessions used, so even the highest irradiance used was nonstressful insofar that it provoked no significant sign of light stress (e.g. a decrease in the dark-adapted Fv/Fm or anthocyanin formation). At the end of the acclimation period to increased growth irradiance in this second experiment, when ΦPSII had stabilized, light response curves were compared with the curves of plants grown at either a constant low (100 µmol m−2 s−1) or high (600 µmol m−2 s−1) growth irradiance. Plants were measured using chlorophyll fluorescence imaging for the first time at 30 min after light onset on day 24 after sowing on rockwool. Depending on the experiment, this imaging was continued daily at the same time until plants began to overlap.
For the GWAS experiments, plants were grown at a constant irradiance of 100 µmol m−2 s−1 (Philips MASTER TL5 HO fluorescent tubes, 80 W). The irradiance was increased to 550 µmol m−2 s−1 on day 25 after sowing, at the onset of irradiance. In all cases, the photoperiod was a 10-h-d/14-h-night cycle, temperature was 20°C/18°C (day/night), relative humidity was 70%, and CO2 levels were ambient. The plants were imaged 1 h after light onset on day 24 (measurement before the increase in irradiance) as well as 1 h after light onset on day 25 (measurement after the increase in irradiance).
Chlorophyll a Fluorescence Imaging and Analysis
For the non-GWAS experiments, chlorophyll a fluorescence was measured using an imaging fluorimeter (Open FluorCam; P.S.I.; http://www.psi.cz) driven by the Fluorcam software package (FluorCam7). Fluorescence was detected by a camera of which the electronic shutter time and sensitivity were adapted to the irradiance being used. Measurements of the dark-adapted Fo and Fm were made after 20 min of dark adaptation. Images of the dark-adapted Fo were measured using nonactinic measuring flashes provided by light-emitting diodes. Next, a 1-s duration pulse of saturating light (6,500 µmol m−2 s−1) generated by the same and other light-emitting diode panels was given to produce the Fm. An image of Fv/Fm was then calculated. To measure the irradiance responses of parameters describing the operation and regulation of PSII, the plants were illuminated with a series of increasing actinic irradiances (100, 225, 450, 700, and 1,150 µmol m−2 s−1). Each irradiance was applied for 15 min, after which the Ft (steady-state fluorescence yield) and Fm′ yield were measured. Pilot experiments showed that using these irradiances for 15 min was sufficient to allow Ft and Fm′ to stabilize after each irradiance increase. The Fm′ fluorescence yield was measured during a 1-s duration pulse of saturating light (6,500 µmol m−2 s−1). Values for Fo, Fm, Ft, and Fm′ in the images were averaged over all pixels per plant; derived values for ΦPSII, Fv/Fm, NPQ, qp, rETR, Fo′, and Fv′/Fm′ were calculated using these averages of Fo, Fm, and Fm′ (Oxborough and Baker, 1997; Baker, 2008).
For imaging of the 344 accessions used for GWAS, we used a laboratory-built high-throughput chlorophyll fluorescence imager. This system imaged plants in groups of 12 (a 3 × 4 array). Chlorophyll fluorescence was measured at 730 nm and excited using radiation from Phlatlight light-emitting diodes (Luminus; peak emission wavelength of 624 nm). ΦPSII was imaged at growth room irradiance, and the irradiances supplied by the growth room (produced by fluorescent tubes) and the imager (produced by light-emitting diodes) were matched by comparing ΦPSII (measured using a chlorophyll fluorimeter [MiniPam; Walz]) in leaves under the growth room irradiance and the imager irradiance. The matching of the irradiances provided by the growth room lights and the actinic irradiance of the imager meant that there was only a minor disturbance of photosynthesis as a result of positioning the camera over the plants; a 30-s recovery time was found to be enough to allow the disappearance of any disturbance before the imaging procedure for ΦPSII was begun.
Genetic Variation
To estimate the genetic variation for a parameter, we calculated its heritability. Heritability, in this case broad-sense heritability, is a term used in quantitative genetics that describes the portion of the total phenotypic variance in a population that is contributed by genetic variance (Visscher et al., 2008).
Genetic variance and the total phenotypic variance within an experiment were calculated with an ANOVA using type III sums of squares in a general linear model in the IBM statistical software program SPSS. The genetic variance was estimated as the proportion of variance explained by differences between genotypes based on measurement of three plants per genotype. se values for heritability were calculated using the heritabilities of three repeated experiments. Heritability for a response was calculated by first calculating the response values for a trait between two time points or two light steps. These response values were always calculated relative to the initial value of the first measurement of the two. When a relation between different parameters in different environments needed to be parameterized, we estimated the curve of the relation using regression statistics and parameterized it using the statistical model that fitted closest to the curve with the IBM statistical software program SPSS. We then used the parameterized value for each individual to calculate the variances in the population.
Genome-Wide Association Analysis
The analysis was performed using the publicly available Web application for genome-wide association mapping in Arabidopsis (gwas.gmi.oeaw.ac.at) using the accelerated mixed-model option (Seren et al., 2012). GWAS was performed for ΦPSII measured 1 h after a step-wise increase in irradiance from 100 to 550 µmol m−2 s−1 by providing a list with average values of three biological replicates per accession. The Web site already knows the 215,000 SNPs used for mapping and will output a list with association scores for all these SNPs. For our analysis, we classified the SNPs with an association score greater than 4 as associated SNPs. A core set of candidate genes was selected by cataloging the genes that contained the associated SNPs in their coding region. The same Web application was used to calculate the level of LD for an SNP and to catalog the genes in these LD regions (Seren et al., 2012); these genes were added to the core set of genes to form the complete list of candidate genes. A description for all candidate genes was obtained from TAIR (www.arabidopsis.org).
Gene Ontology Enrichment Analysis
All Gene Ontology annotations were downloaded from TAIR (www.arabidopsis.org). The Gene Ontology enrichment analysis was performed for three categories: cellular component, molecular function, and biological process (Ashburner et al., 2000). Per category, the fraction of genes annotated to a certain ontology class for the list of candidate genes from GWAS was compared with the fraction of genes annotated to the same ontology class in a control group consisting of all the genes in the genome of Arabidopsis.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Light response curves per accession in different constant growth irradiances.
Supplemental Figure S2. rETR measured at saturating actinic irradiance per accession in different growth irradiances.
Supplemental Figure S3. Variation in parameterized value of the relation of maximum rETR with growth irradiance.
Supplemental Figure S4. Light response curves of rETR of all 12 accessions grown in LL, response to HL, or grown in HL.
Supplemental Figure S5. Histograms of ΦPSII before and after increased irradiance for 344 accessions used for the GWAS.
Supplemental Table S1. Correlation photosynthetic parameters during acclimation.
Supplemental Table S2. Annotations of all genes found in LD with the SNPS associated with a −log10(p) > 4 in GWAS.
Supplementary Material
Acknowledgments
We thank Pádraic Flood for designing the trays used to grow Arabidopsis uniformly and image them for chlorophyll fluorescence without background noise, Rob van der Schoor for help in obtaining the chlorophyll fluorescence imaging data, and the Unifarm section of Wageningen University for providing the nutrient solutions and taking care of the Arabidopsis plants.
Glossary
- NPQ
nonphotochemical quenching
- LHCII
light-harvesting complex II
- Fv/Fm
maximum photochemical efficiency of PSII in the dark-adapted state
- GWAS
genome-wide association study
- SNP
single-nucleotide polymorphism
- ΦPSII
light use efficiency of photosystem II
- rETR
relative linear electron transport rate
- Fv′/Fm′
quantum efficiency of the open PSII reaction centers
- qp
PSII efficiency factor
- Fo
initial (minimum) PSII fluorescence in the dark-adapted state
- Fm
maximum PSII fluorescence in the dark-adapted state
- LD
linkage disequilibrium
- Fm′
maximum PSII fluorescence in the light-adapted state
- PVC
polyvinyl chloride
- Ft
steady-state fluorescence yield
- TAIR
The Arabidopsis Information Resource
Footnotes
This work was supported by the Dutch Ministry of Economic Affairs.
Articles can be viewed without a subscription.
References
- Allen JF. (1992) How does protein phosphorylation regulate photosynthesis? Trends Biochem Sci 17: 12–17 [DOI] [PubMed] [Google Scholar]
- Alonso-Blanco C, Aarts MGM, Bentsink L, Keurentjes JJB, Reymond M, Vreugdenhil D, Koornneef M (2009) What has natural variation taught us about plant development, physiology, and adaptation? Plant Cell 21: 1877–1896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alter P, Dreissen A, Luo FL, Matsubara S (2012) Acclimatory responses of Arabidopsis to fluctuating light environment: comparison of different sunfleck regimes and accessions. Photosynth Res 113: 221–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asada K. (2006) Production and scavenging of reactive oxygen species in chloroplasts and their functions. Plant Physiol 141: 391–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. (2000) Gene Ontology: tool for the unification of biology. Nat Genet 25: 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athanasiou K, Dyson BC, Webster RE, Johnson GN (2010) Dynamic acclimation of photosynthesis increases plant fitness in changing environments. Plant Physiol 152: 366–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atwell S, Huang YS, Vilhjálmsson BJ, Willems G, Horton M, Li Y, Meng D, Platt A, Tarone AM, Hu TT, et al. (2010) Genome-wide association study of 107 phenotypes in Arabidopsis thaliana inbred lines. Nature 465: 627–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey S, Walters RG, Jansson S, Horton P (2001) Acclimation of Arabidopsis thaliana to the light environment: the existence of separate low light and high light responses. Planta 213: 794–801 [DOI] [PubMed] [Google Scholar]
- Baker NR. (2008) Chlorophyll fluorescence: a probe of photosynthesis in vivo. Annu Rev Plant Biol 59: 89–113 [DOI] [PubMed] [Google Scholar]
- Balaguer L, Martinez-Ferri E, Valladares F, Perez-Corona ME, Baquedano FJ, Castillo FJ, Manrique E (2001) Population divergence in the plasticity of the response of Quercus coccifera to the light environment. Funct Ecol 15: 124–135 [Google Scholar]
- Ballottari M, Dall’Osto L, Morosinotto T, Bassi R (2007) Contrasting behavior of higher plant photosystem I and II antenna systems during acclimation. J Biol Chem 282: 8947–8958 [DOI] [PubMed] [Google Scholar]
- Barton NH, Keightley PD (2002) Understanding quantitative genetic variation. Nat Rev Genet 3: 11–21 [DOI] [PubMed] [Google Scholar]
- Björkman O. (1981) Responses to different quantum flux densities. In OL Lange, PS Nobel, CB Osmond, H Ziegler, eds, Physiological Plant Ecology. I. Responses to the Physical Environment. Springer-Verlag, New York, pp 57–107 [Google Scholar]
- Björkman O, Demmig B (1987) Photon yield of O2 evolution and chlorophyll fluorescence characteristics at 77 K among vascular plants of diverse origins. Planta 170: 489–504 [DOI] [PubMed] [Google Scholar]
- Brachi B, Morris GP, Borevitz JO (2011) Genome-wide association studies in plants: the missing heritability is in the field. Genome Biol 12: 232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao DY, Silva A, Baxter I, Huang YS, Nordborg M, Danku J, Lahner B, Yakubova E, Salt DE (2012) Genome-wide association studies identify heavy metal ATPase3 as the primary determinant of natural variation in leaf cadmium in Arabidopsis thaliana. PLoS Genet 8: e1002923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker JP, Boekema EJ (2005) Supramolecular organization of thylakoid membrane proteins in green plants. Biochim Biophys Acta 1706: 12–39 [DOI] [PubMed] [Google Scholar]
- Demmig B, Winter K, Krüger A, Czygan FC (1987) Photoinhibition and zeaxanthin formation in intact leaves: a possible role of the xanthophyll cycle in the dissipation of excess light energy. Plant Physiol 84: 218–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demmig-Adams B, Adams WW (1992) Photoprotection and other responses of plants to high light stress. Annu Rev Plant Physiol 43: 599–626 [Google Scholar]
- Demmig-Adams B, Adams WW III (2006) Photoprotection in an ecological context: the remarkable complexity of thermal energy dissipation. New Phytol 172: 11–21 [DOI] [PubMed] [Google Scholar]
- El-Lithy ME, Rodrigues GC, van Rensen JJS, Snel JFH, Dassen HJHA, Koornneef M, Jansen MAK, Aarts MGM, Vreugdenhil D (2005) Altered photosynthetic performance of a natural Arabidopsis accession is associated with atrazine resistance. J Exp Bot 56: 1625–1634 [DOI] [PubMed] [Google Scholar]
- Evans JR, Poorter H (2001) Photosynthetic acclimation of plants to growth irradiance: the relative importance of specific leaf area and nitrogen partitioning in maximizing carbon gain. Plant Cell Environ 24: 755–767 [Google Scholar]
- Falke KC, Frisch M (2011) Power and false-positive rate in QTL detection with near-isogenic line libraries. Heredity (Edinb) 106: 576–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flood PJ, Harbinson J, Aarts MGM (2011) Natural genetic variation in plant photosynthesis. Trends Plant Sci 16: 327–335 [DOI] [PubMed] [Google Scholar]
- Foyer CH, Neukermans J, Queval G, Noctor G, Harbinson J (2012) Photosynthetic control of electron transport and the regulation of gene expression. J Exp Bot 63: 1637–1661 [DOI] [PubMed] [Google Scholar]
- Genty B, Briantais JM, Baker NR (1989) The relationship between the quantum yield of photosynthetic electron transport and quenching of chlorophyll fluorescence. Biochim Biophys Acta 990: 87–92 [Google Scholar]
- Genty B, Harbinson J (1996) Regulation of light utilization for photosynthetic electron transport. In NR Baker, ed, Photosynthesis and the Environment. Springer, Dordrecht, The Netherlands, pp 67–99 [Google Scholar]
- Gibson G. (2010) Hints of hidden heritability in GWAS. Nat Genet 42: 558–560 [DOI] [PubMed] [Google Scholar]
- Gibson G. (2012) Rare and common variants: twenty arguments. Nat Rev Genet 13: 135–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbinson J, Genty B, Baker NR (1989) Relationship between the quantum efficiencies of photosystems I and II in pea leaves. Plant Physiol 90: 1029–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogewoning SW, Wientjes E, Douwstra P, Trouwborst G, van Ieperen W, Croce R, Harbinson J (2012) Photosynthetic quantum yield dynamics: from photosystems to leaves. Plant Cell 24: 1921–1935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm S. (1979) A simple sequentially rejective multiple test procedure. Scand J Stat 6: 65–70 [Google Scholar]
- Horton P, Hague A (1988) Studies on the induction of chlorophyll fluorescence in isolated barley protoplasts. IV. Resolution of non-photochemical quenching. Biochim Biophys Acta 932: 107–115 [Google Scholar]
- Huang W, Sherman BT, Lempicki RA (2009) Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 37: 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung HS, Niyogi KK (2009) Quantitative genetic analysis of thermal dissipation in Arabidopsis. Plant Physiol 150: 977–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Plagnol V, Hu TT, Toomajian C, Clark RM, Ossowski S, Ecker JR, Weigel D, Nordborg M (2007) Recombination and linkage disequilibrium in Arabidopsis thaliana. Nat Genet 39: 1151–1155 [DOI] [PubMed] [Google Scholar]
- Korte A, Farlow A (2013) The advantages and limitations of trait analysis with GWAS: a review. Plant Methods 9: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouřil R, Dekker JP, Boekema EJ (2012) Supramolecular organization of photosystem II in green plants. Biochim Biophys Acta 1817: 2–12 [DOI] [PubMed] [Google Scholar]
- Kouřil R, Wientjes E, Bultema JB, Croce R, Boekema EJ (2013) High-light vs. low-light: effect of light acclimation on photosystem II composition and organization in Arabidopsis thaliana. Biochim Biophys Acta 1827: 411–419 [DOI] [PubMed] [Google Scholar]
- Kramer DM, Johnson G, Kiirats O, Edwards GE (2004) New fluorescence parameters for the determination of QA redox state and excitation energy fluxes. Photosynth Res 79: 209–218 [DOI] [PubMed] [Google Scholar]
- Leakey ADB, Press MC, Scholes JD (2003) Patterns of dynamic irradiance affect the photosynthetic capacity and growth of dipterocarp tree seedlings. Oecologia 135: 184–193 [DOI] [PubMed] [Google Scholar]
- Leister D. (2012) How can the light reactions of photosynthesis be improved in plants? Front Plant Sci 3: 199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Huang Y, Bergelson J, Nordborg M, Borevitz JO (2010) Association mapping of local climate-sensitive quantitative trait loci in Arabidopsis thaliana. Proc Natl Acad Sci USA 107: 21199–21204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell K, Johnson GN (2000) Chlorophyll fluorescence: a practical guide. J Exp Bot 51: 659–668 [DOI] [PubMed] [Google Scholar]
- Meijón M, Satbhai SB, Tsuchimatsu T, Busch W (2014) Genome-wide association study using cellular traits identifies a new regulator of root development in Arabidopsis. Nat Genet 46: 77–81 [DOI] [PubMed] [Google Scholar]
- Mitchell-Olds T, Schmitt J (2006) Genetic mechanisms and evolutionary significance of natural variation in Arabidopsis. Nature 441: 947–952 [DOI] [PubMed] [Google Scholar]
- Müller P, Li XP, Niyogi KK (2001) Non-photochemical quenching: a response to excess light energy. Plant Physiol 125: 1558–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murchie EH, Horton P (1997) Acclimation of photosynthesis to irradiance and spectral quality in British plant species: chlorophyll content, photosynthetic capacity and habitat preference. Plant Cell Environ 20: 438–448 [Google Scholar]
- Murchie EH, Niyogi KK (2011) Manipulation of photoprotection to improve plant photosynthesis. Plant Physiol 155: 86–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niyogi KK. (1999) Photoprotection revisited: genetic and molecular approaches. Annu Rev Plant Physiol Plant Mol Biol 50: 333–359 [DOI] [PubMed] [Google Scholar]
- Oguchi R, Hikosaka K, Hirose T (2003) Does the photosynthetic light-acclimation need change in leaf anatomy? Plant Cell Environ 26: 505–512 [Google Scholar]
- Oxborough K, Baker NR (1997) Resolving chlorophyll a fluorescence images of photosynthetic efficiency into photochemical and non-photochemical components: calculation of qP and Fv′/Fm′ without measuring Fo′. Photosynth Res 54: 135–142 [Google Scholar]
- Pérez-Pérez JM, Serrano-Cartagena J, Micol JL (2002) Genetic analysis of natural variations in the architecture of Arabidopsis thaliana vegetative leaves. Genetics 162: 893–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pigliucci M. (1998) Ecological and evolutionary genetics of Arabidopsis. Trends Plant Sci 3: 485–489 [Google Scholar]
- Portes MT, Alves TH, Souza GM (2008) Time-course of photosynthetic induction in four tropical woody species grown in contrasting irradiance habitats. Photosynthetica 46: 431–440 [Google Scholar]
- Powles SB. (1984) Photoinhibition of photosynthesis induced by visible-light. Annu Rev Plant Physiol 35: 15–44 [Google Scholar]
- Ptushenko VV, Ptushenko EA, Samoilova OP, Tikhonov AN (2013) Chlorophyll fluorescence in the leaves of Tradescantia species of different ecological groups: induction events at different intensities of actinic light. Biosystems 114: 85–97 [DOI] [PubMed] [Google Scholar]
- Quick WP, Stitt M (1989) An examination of factors contributing to non-photochemical quenching of chlorophyll fluorescence in barley leaves. Biochim Biophys Acta 977: 287–296 [Google Scholar]
- Rabinowitch E. (1951) Photosynthesis. Interscience Publishers, New York [Google Scholar]
- Rintamäki E, Salonen M, Suoranta UM, Carlberg I, Andersson B, Aro EM (1997) Phosphorylation of light-harvesting complex II and photosystem II core proteins shows different irradiance-dependent regulation in vivo: application of phosphothreonine antibodies to analysis of thylakoid phosphoproteins. J Biol Chem 272: 30476–30482 [DOI] [PubMed] [Google Scholar]
- Seemann JR. (1989) Light adaptation/acclimation of photosynthesis and the regulation of ribulose-1,5-bisphosphate carboxylase activity in sun and shade plants. Plant Physiol 91: 379–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seren Ü, Vilhjálmsson BJ, Horton MW, Meng D, Forai P, Huang YS, Long Q, Segura V, Nordborg M (2012) GWAPP: a web application for genome-wide association mapping in Arabidopsis. Plant Cell 24: 4793–4805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims DA, Pearcy RW (1993) Sunfleck frequency and duration affects growth-rate of understorey plant, Alocasia macrorrhiza. Funct Ecol 7: 683–689 [Google Scholar]
- Tikkanen M, Grieco M, Kangasjärvi S, Aro EM (2010) Thylakoid protein phosphorylation in higher plant chloroplasts optimizes electron transfer under fluctuating light. Plant Physiol 152: 723–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikkanen M, Suorsa M, Gollan PJ, Aro EM (2012) Post-genomic insight into thylakoid membrane lateral heterogeneity and redox balance. FEBS Lett 586: 2911–2916 [DOI] [PubMed] [Google Scholar]
- Trontin C, Tisné S, Bach L, Loudet O (2011) What does Arabidopsis natural variation teach us (and does not teach us) about adaptation in plants? Curr Opin Plant Biol 14: 225–231 [DOI] [PubMed] [Google Scholar]
- Valladares F, Allen MT, Pearcy RW (1997) Photosynthetic responses to dynamic light under field conditions in six tropical rainforest shrubs occuring along a light gradient. Oecologia 111: 505–514 [DOI] [PubMed] [Google Scholar]
- Visscher PM, Hill WG, Wray NR (2008) Heritability in the genomics era: concepts and misconceptions. Nat Rev Genet 9: 255–266 [DOI] [PubMed] [Google Scholar]
- Walters RG. (2005) Towards an understanding of photosynthetic acclimation. J Exp Bot 56: 435–447 [DOI] [PubMed] [Google Scholar]
- Walters RG, Horton P (1991) Resolution of components of non-photochemical chlorophyll fluorescence quenching in barley leaves. Photosynth Res 27: 121–133 [DOI] [PubMed] [Google Scholar]
- Walters RG, Shephard F, Rogers JJM, Rolfe SA, Horton P (2003) Identification of mutants of Arabidopsis defective in acclimation of photosynthesis to the light environment. Plant Physiol 131: 472–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wientjes E, van Amerongen H, Croce R (2013) LHCII is an antenna of both photosystems after long-term acclimation. Biochim Biophys Acta 1827: 420–426 [DOI] [PubMed] [Google Scholar]
- Yin L, Fristedt R, Herdean A, Solymosi K, Bertrand M, Andersson MX, Mamedov F, Vener AV, Schoefs B, Spetea C (2012) Photosystem II function and dynamics in three widely used Arabidopsis thaliana accessions. PLoS ONE 7: e46206. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.