Disruption of four genes severely reduces the active phosphate transport but does not impact phosphate sensing capacities in Arabidopsis.
Abstract
Arabidopsis (Arabidopsis thaliana) absorbs inorganic phosphate (Pi) from the soil through an active transport process mediated by the nine members of the PHOSPHATE TRANSPORTER1 (PHT1) family. These proteins share a high level of similarity (greater than 61%), with overlapping expression patterns. The resulting genetic and functional redundancy prevents the analysis of their specific roles. To overcome this difficulty, our approach combined several mutations with gene silencing to inactivate multiple members of the PHT1 family, including a cluster of genes localized on chromosome 5 (PHT1;1, PHT1;2, and PHT1;3). Physiological analyses of these lines established that these three genes, along with PHT1;4, are the main contributors to Pi uptake. Furthermore, PHT1;1 plays an important role in translocation from roots to leaves in high phosphate conditions. These genetic tools also revealed that some PHT1 transporters likely exhibit a dual affinity for phosphate, suggesting that their activity is posttranslationally controlled. These lines display significant phosphate deficiency-related phenotypes (e.g. biomass and yield) due to a massive (80%–96%) reduction in phosphate uptake activities. These defects limited the amount of internal Pi pool, inducing compensatory mechanisms triggered by the systemic Pi starvation response. Such reactions have been uncoupled from PHT1 activity, suggesting that systemic Pi sensing is most probably acting downstream of PHT1.
Genetic redundancy is a common feature in flowering plants, resulting from the ability of angiosperms to undergo polyploidization followed by gene loss during diploidization (Van de Peer et al., 2009). This trait is responsible for the absence of a phenotype associated with a vast majority of single loss-of-function mutation compensated by homologous genes. Genome analysis makes it possible to quantify this phenomenon, revealing the presence of duplication in more than 80% of the Arabidopsis (Arabidopsis thaliana) genome (Simillion et al., 2002).
Traditionally, this phenomenon has been linked with the need of plants to adapt to a diverse range of environments. This view is illustrated by the example of ion uptake, an essential process for autotrophic organisms such as plants. In addition to the gene duplication reported within the vast majority of ion transporter families, another layer of functional redundancy is often observed. Specifically, most ions can be transported in plants by distinct nonrelated carriers (Barbier-Brygoo et al., 2011). The first physiological evidence for this phenomenon revealed the existence of a dual (high and low) affinity mechanism for potassium uptake; a transporter was found to be responsible for the high-affinity activity, whereas the low-affinity system was linked to potassium channels (Epstein et al., 1963). Progress in molecular biology since this study has confirmed these results and identified 35 potassium transporters and 15 potassium channels in the Arabidopsis genome (Mäser et al., 2001). A similar situation has been observed for nitrate absorption (Miller et al., 2007), in which 60 nitrate transporters were identified among the NITRATE TRANSPORTER1 (NRT1) and NRT2 families. Low-affinity nitrate transport activity has been linked to NRT1 proteins, henceforth called NPF (for NITRATE TRANSPORTER1/PEPTIDE TRANSPORTER family) proteins (Léran et al., 2014), whereas high-affinity transport is associated with the NRT2 family.
The third essential plant macronutrient is phosphorus, which is absorbed by plants in its mineral form as inorganic phosphate (Pi). An activity linked to the phosphate transporters named PHT1 or PT, depending on the plant species (for a recent review, see Nussaume et al., 2011). Nine PHT1 members have been identified in Arabidopsis, which are highly redundant in their expression patterns and function. Genetic approaches have been used to study PHT1;1, PHT1;4, PHT1;5, PHT1;8, and PHT1;9, which are now relatively well characterized (Misson et al., 2004; Remy et al., 2012). Nevertheless, the physiological impact from these single mutations remains limited due to the high functional redundancy between PHT1 transporters. The various PHT1 protein sequences exhibit very high homology and similarity levels (Supplemental Table S1).
PHT1;1 and PHT1;4 have the highest transcription rates in response to Pi deprivation, and consequently, the main impact on growth was observed in their double mutant. This double mutation results in a 75% to 80% reduction in Pi uptake, although the impact on plant growth is not proportional, because the reduction of biomass is only around 30% (Shin et al., 2004).
Genetic analysis of PHT1;1 reveals its substantial contribution to Pi uptake in Pi-rich medium, because the pht1;1 mutant displays a 30% to 40% decrease in Pi (Shin et al., 2004). Conversely, transcriptomic and genetic analyses of pht1;4, pht1;5, pht1;8, and pht1;9 mutants indicate a less significant role for these genes in Pi uptake in Pi-rich medium (Misson et al., 2004). PHT1;5 has a specific role that is mainly devoted to Pi translocation between roots and shoots (Nagarajan et al., 2011). The role of the other three genes appears to be restricted to Pi absorption when this element is present in limited quantities (Misson et al., 2004).
Genomic analysis has revealed that PHT1;2, PHT1;3, and PHT1;6 cluster with PHT1;1 on Arabidopsis chromosome 5 (Poirier and Bucher, 2002). PHT1;2 and PHT1;3 transcripts have mainly been identified in roots (Mudge et al., 2002), whereas PHT1;6 expression is restricted to the flowers. Creating a triple mutant to analyze the contributions of PHT1;1, PHT1;2, and PHT1;3 to Pi uptake cannot be achieved by introgression, due to the strong genetic linkage between these loci (and the absence of an identified pht1;3 mutant). To circumvent this issue, we have used RNA inactivation (RNAi) technology to simultaneously abolish the expression of these genes.
Several steps of posttranslational regulation control PHT1 targeting and accumulation at the plasma membrane, in addition to its tight transcriptional regulation (Hammond et al., 2003; Wu et al., 2003; Misson et al., 2005; Morcuende et al., 2007; Calderon-Vazquez et al., 2008; Bustos et al., 2010; Thibaud et al., 2010; Bayle et al., 2011). One of these steps involves Phosphate Transporter Traffic Facilitator1 (PHF1), a protein required to help PHT1 proteins cross the endoplasmic reticulum (González et al., 2005). The PHF1 mutation leads to a 60% to 80% decrease in Pi uptake, promoting similar physiological consequences for Pi absorption as the pht1/pht4 double mutation (Shin et al., 2004).
Here, we demonstrate the existence of a strong genetic and functional redundancy among members of the PHT1 phosphate transporter family. We combined knockout mutations and an RNAi strategy to reduce phosphate uptake to less than 5% of levels in control plants. These lines were used to investigate the impact of PHT1 activity on Pi uptake and Pi sensing in Arabidopsis.
RESULTS
Abolishing PHT1;1, PHT1;2, and PHT1;3 Activity
PHT1;1, PHT1;2, and PHT1;3 transcripts (At5g43350, At5g43370, and At5g43360) were abolished by introduction of an artificial micro-RNA (amiRNA) targeting simultaneously these three targets. This was achieved using the preestablished procedure previously described (http://wmd.weigelworld.org/cgi-bin/mirnatools.pl). Wild-type plants were transformed by this construct to generate a triple-silenced line for PHT1;1, PHT1;2, and PHT1;3 (mut3).
Homozygous lines from 10 independent transformation events were selected. All lines showed a strong reduction in internal Pi content when grown on Pi-rich medium (Supplemental Fig. S1A). The strongest reductions, ranging from 46% to 49% of the wild type, were observed in leaves, the primary location of Pi accumulation in plants. Three lines exhibiting the highest reduction of Pi content in both roots and leaves were selected for further study (mut3-1, mut3-2, and mut3-3).
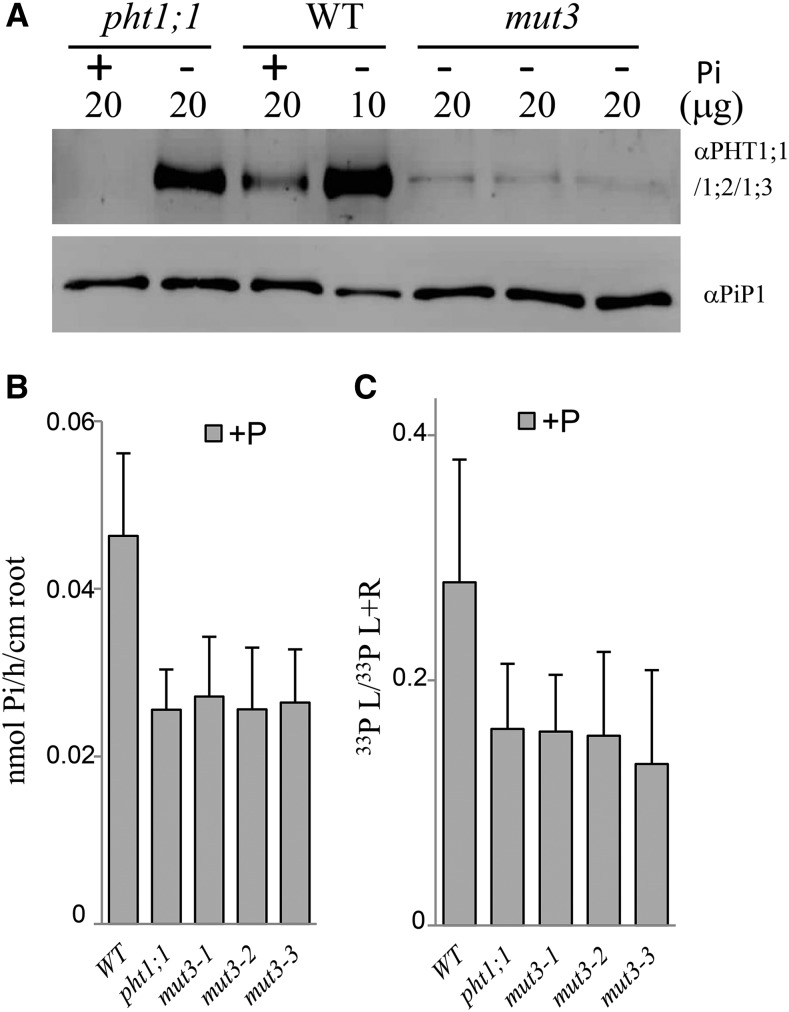
To confirm the ability of the amiRNA strategy to silence the targeted PHT1 locus, we performed SDS-PAGE with an antibody raised against two peptides specific to the PHT1;1, PHT1;2, and PHT1;3 sequences (Fig. 1A). Efficiency and specificity of the antibody were confirmed by analyzing the wild type and pht1;1 single mutant. The protein detected in the wild type 0.5 mm Pi medium (henceforth called +P) disappears in the pht1;1 mutant, illustrating that only PHT1;1 is detected by the antibody in +P. The absence of signal in pht1;1 mutant revealed the absence of PHT1;2 and PHT1;3 expression under these conditions, as these proteins can still be detected under Pi-depleted medium (approximately 0.015 mm Pi; henceforth called −P) conditions in the pht1;1 mutant background. It should be noted that the loading of the proteins in the wild-type control is reduced by one-half in –P to avoid saturation. So, the reduction of the signals observed here for mutants are more important (×2) when compared with the wild type in –P condition.
Figure 1.
Characterization of the triple mutant. A, Western-blot analysis of PHT1;1, PHT1;2, and PHT1;3 on root membrane protein extracts. No PHT1;2 or PHT1;3 proteins could be detected in pht1;1 grown in +P. Lanes were loaded with 10 or 20 μg of root proteins extracted from plants grown in –P or +P medium, as detailed in the figure. A loading control western blot, using the anti-Plasma Membrane Intrinsic Protein1 (PIP1) antibody, is shown at bottom. B, 33P uptake capacity in +P conditions. C, 33P partitioning (leaves [L] per total P) in +P conditions. For B and C, each bar corresponds to 22 to 24 individual plants. All experiments were performed at least twice. Error bars represent se. WT, Wild type; L+R, leaves + roots.
The antibody detected only a residual presence of PHT1;1, PHT1;2, and PHT1;3 proteins in the three independent mut3 lines (Fig. 1A). Quantification of the western-blot signal in the mut3 lines indicated that their levels in –P were only 4% to 8% of the wild-type control. The quantitative real-time (qRT)-PCR was able to detect transcripts for all other members of the PHT1 family, demonstrating the specificity of the silencing construct. Furthermore, all transcripts were induced in –P conditions in the roots of mut3 lines at a level similar to the wild-type control (Supplemental Fig. S1B). The PHT1;6 gene was not detected in any genotype (data not shown), as expected from its specific expression in flowers (Mudge et al., 2002).
These observations are corroborated by Pi uptake measurements under +P conditions, in which pht1;1 and the various mut3 lines are similarly affected, revealing a 42% to 45% reduction in Pi uptake (Fig. 1B). These results also indicate that PHT1;2 and PHT1;3 do not play significant roles in these conditions. Most of the PHT1 members were induced in –P conditions, which can explain the reduced impact of the pht1;1 mutation or PHT1;1/PHT1;2/PHT1;3 silencing on Pi uptake (Supplemental Fig. S1C), as well as the absence of an observable macroscopic phenotype (data not shown).
The Pi partitioning between leaves and roots was calculated to assess the impact of the silencing on root-to-shoot Pi transfer (Supplemental Fig. S1D). These values were comparable between the wild-type control and the pht1;1 and mut3 lines in –P conditions. By contrast, a 43% to 53% decrease in +P conditions (compared to the wild-type control) was observed in pht1;1 and mut3 lines (Fig. 1C). These data are in agreement with the role of PHT1;1 as the only transporter (among those modified in our experiments) that is expressed in roots in +P conditions. Therefore, our results shows an additional role for PHT1;1 in transferring Pi to the aerial parts of plants in +P conditions. Data from the single pht1;1 mutant used in this experiment (whose defects are similar to mut3 lines) confirms this assumption (Fig. 1, B and C). Analysis of transcript levels of the remaining PHT1 transporters reveals that the significant reduction of Pi content observed in mut3 lines grown in +P conditions does not trigger an induction of the PHT1;7 or PHT1;9 genes (Supplemental Fig. S1E). In addition, the slight increase of PHT1;4 and PHT1;8 transcripts observed in mut3 lines in +P remains far from the level of these transcripts accumulated in P starvation conditions (Supplemental Fig. S1E). Consequently, it can be assumed that the compensation triggered by the induction of these genes is not sufficient to offset the effects of mut3 on Pi uptake or partitioning. The classical Pi starvation markers monogalactosyldiacylglycerol synthase3 (MGD3), suppressor of yeast gpa1/yeast cyclin-dependent kinase inhibitor (PHO81)/human xenotrophic and polytronic retrovirus receptor1 (SPX3), and PHO1 Homolog1 (PHO1;H1) remained at levels similar to the wild-type control in +P conditions. Combined, these data indicate that the loss of the PHT1;1/PHT1;2/PHT1;3 cluster does not provoke a compensatory induction of remaining PHT1 transporters in +P conditions.
Investigating the Contribution of the PHT1;1, PHT1;2, and PHT1;3 Cluster to Pi Uptake
Most PHT1 transporters are induced when Pi is not present in the growth medium and can thus mask the loss of the PHT1;1/PHT1;2/PHT1;3 genes. The transporter PHT1;4 plays a major role in this phenomenon, especially when Pi is a limiting resource (Muchhal et al., 1996; Misson et al., 2004). We therefore introduced the amiRNA construct into the pht1;1/pht1;4 background (compare with Shin et al., 2004) and generated quadruple mutant lines (mut4) impaired in the expression of PHT1;1 through PHT1;4. The double mutant phf1-2/pht1;4 (Bayle et al., 2011) was also transformed with the silencing construct, generating quintuple mutant (mut5) lines. These lines were created to examine the effect of dramatically reduced PHT1 protein levels at the plasma membrane, because these lines should not express any PHT1;1/PHT1;2/PHT1;3/PHT1;4 proteins. Furthermore, they greatly reduce the proper targeting of PHT1 proteins due to the lack of PHF1.
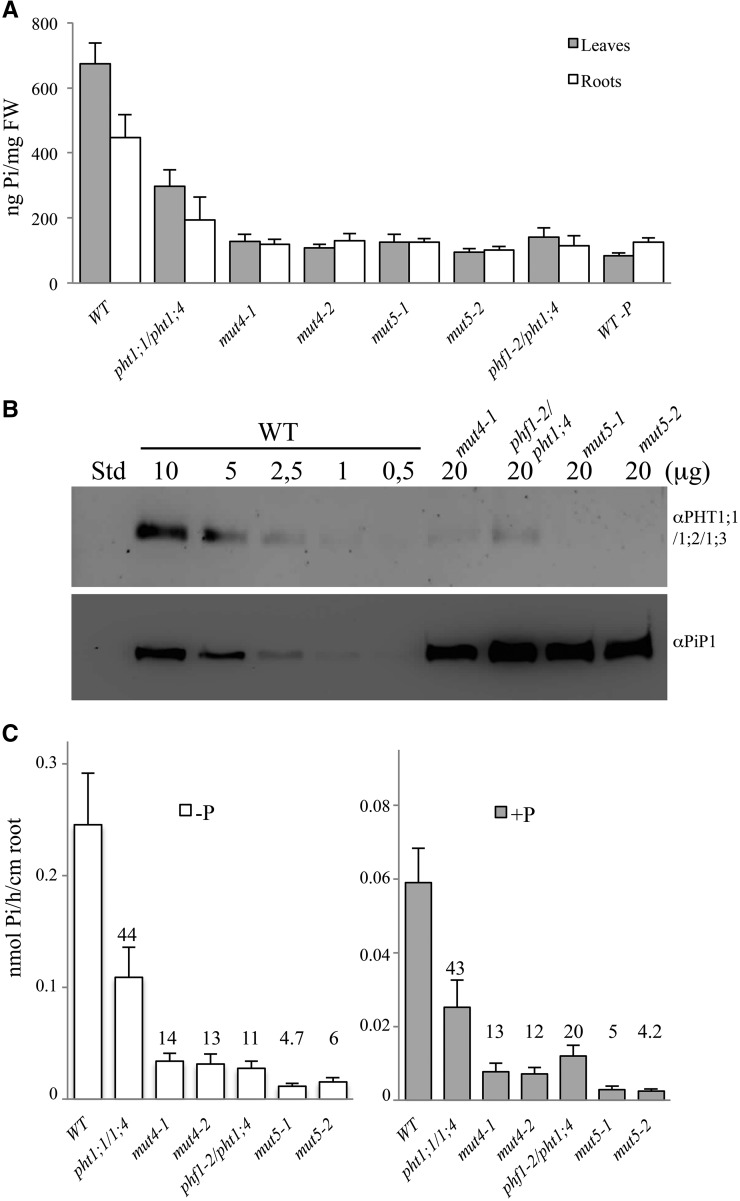
Homozygous lines of mut4 and mut5 from five independent transformation events displayed a striking reduction of their internal Pi content in roots and shoots (Supplemental Fig. S2A). We chose two lines of each type (mut4-1, mut4-2, mut5-1, and mut5-2) for further study, which present the most severe reductions in Pi leaf content (Fig. 2A). An approach combining RNAi, mutations, and altered targeting resulted in a very low amount of PHT proteins as detected by western blot (Fig. 2B). Only 5% of the wild-type intensity was detected in the mut4 line, whereas no protein could be detected in the mut5 lines, indicating an even lower quantity.
Figure 2.
Characterization of the quadruple and quintuple mutants. A, Pi content in leaves (gray bars) and roots (white bars) of plantlets grown in +P at 11 dpg, including a wild-type (WT) control grown in –P. sds are shown. B, Western-blot analysis of PHT1;1, PHT1;2, and PHT1;3 on root membrane protein extracts. No residual protein was detected in the mut5 line. The first lane was loaded with 10 μL of Bio-Rad Precision Plus Protein Standards (Std); the following lanes were loaded with 0.5 to 20 μg of root proteins extracted from plants grown in –P medium, as detailed in the figure. A loading control western blot, using the anti-PIP1 antibody, is shown at bottom. C, 33P uptake capacity is strongly affected in mut4 and mut5 lines. Pi uptake capacity was calculated as nmol Pi h–1 cm–1 root. Pi uptake was performed with 12-dpg plantlets grown in +P (gray bars) or in –P media (white bars). Each bar corresponds to 17 to 24 individual plants. Error bars represent se. Values above each bar correspond to Pi uptake percentage compared with the wild type. Data shown are from a representative experiment; experiments were performed in triplicate. FW, Fresh weight.
In the –P condition, all PHT1 transporters exhibit a significant level of induction (Misson et al., 2005) that results in a high level of Pi uptake. We measured the uptake difference between mut4 and its parental double mutant pht1;1/pht1;4 in low- and high-phosphate conditions. In –P, PHT1;2 and PHT1;3 contributed to 30% of the Pi uptake measured in the wild type (Fig. 2C). As previously reported, the phf1 mutation reduces Pi uptake by 80% (González et al., 2005). Combining the phf1 and pht1;4 mutations produced plants with 89% reduction in Pi absorption as compared with the wild type (Fig. 2C). In comparison with the wild-type control, the mut5 lines exhibited the greatest reduction in Pi uptake (approximately 95%; Fig. 2C). The reduced Pi uptake activity observed in mut5 compared with mut4 supported the view that the phf1 mutation affects membrane accumulation in all PHT1 (Bayle et al., 2011), especially for PHT1;5/PHT1;7/PHT1;8/PHT1;9.
In +P conditions, the mut4 and mut5 transformants exhibited a Pi content similar to the wild-type control grown under Pi-depleted conditions (Fig. 2A; Supplemental Fig. S2A). Next, we investigated which phosphate pool is altered in these mutants by NMR analysis, which can discriminate between free and bound phosphate. This revealed a drastic reduction in both vacuolar and (to a lesser extent) cytoplasmic free Pi (Supplemental Fig. S2B). Crucial Pi metabolites such as Glc-6-P, phospho-choline, and phospho-nucleotides also exhibited similar trends. Inductively coupled plasma (ICP) optical emission spectrometry analysis was performed in 2-week-old plantlets grown on +P to assess the global impact of the quintuple mutation on mineral content in the plants. The ICP analysis revealed that total phosphorus content of mut5-1 line was reduced by 65% and 50% in leaves and roots, respectively (Supplemental Fig. S2C). Simultaneously, Fe content in mut5-1 leaves was increased by 30% and by 53% in mut5-1 roots. Na was reduced by 30% in leaves of the mut5-1 line and by 50% in roots. All other macro- and micronutrients (Ca, K, Mg, Mn, and Zn) were unaltered when compared to the wild type (data not shown).
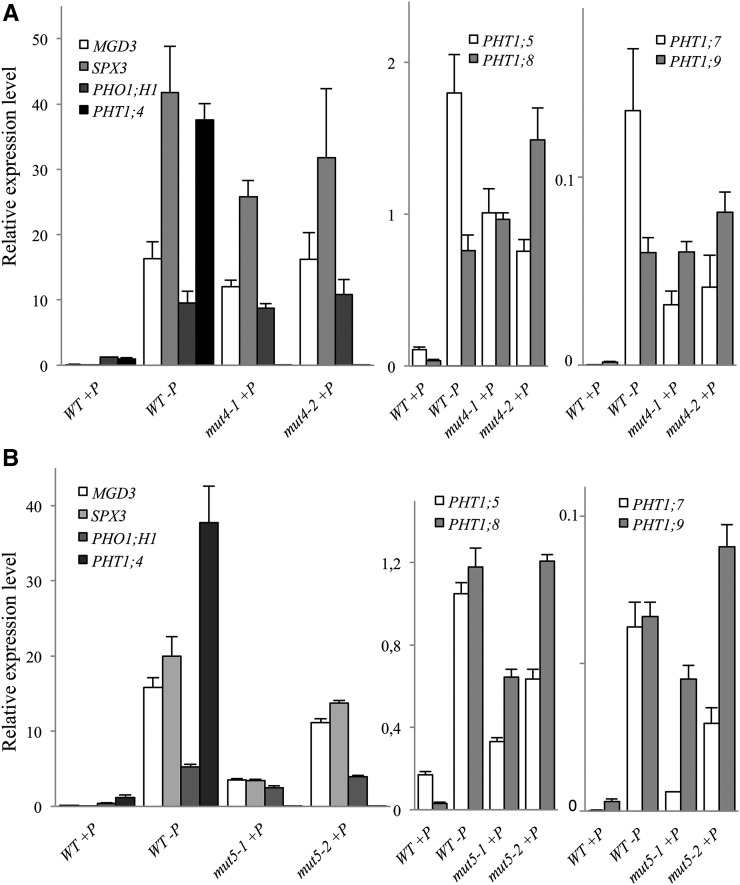
These plants also exhibited Pi starvation traits in the presence of 500 µm Pi in the growth medium. Several Phosphate Starvation Induced (PSI) markers (MGDG3, PHO1;H1, and SPX3) as well as the PHT1;5/PHT1;7/PHT1;8/PHT1;9 genes were all highly induced in mut4 and mut5 lines in +P conditions, as illustrated in our expression analysis (Fig. 3). Forty PSI genes were also tested in mut5, all of which exhibited high levels of expression in +P conditions (Supplemental Fig. S3). This observation confirms that mut4 and mut5 plants display a constitutive phosphate deficiency molecular response. Moreover, a strong induction of other PHT1 genes was observed, revealing the existence of a compensatory mechanism at the transcriptional level.
Figure 3.
Quadruple and quintuple mutant lines show transcriptional induction of PSI and PHT1 markers in Pi-sufficient growth conditions. A, qRT-PCR of several members of the PHT1 family and several PSI genes in two independent mut4 lines. B, qRT-PCR of several members of the PHT1 family and several PSI genes in two independent mut5 lines. The wild type (WT) grown on +P and on –P have been included as controls. Corresponding loci numbers and primer sequences are provided in Supplemental Table S2. Biological triplicates were performed, and all samples were analyzed with technical triplicates. sds are shown.
Reducing PHT1 Activity Affects Both High- and Low-Affinity Pi Transporters
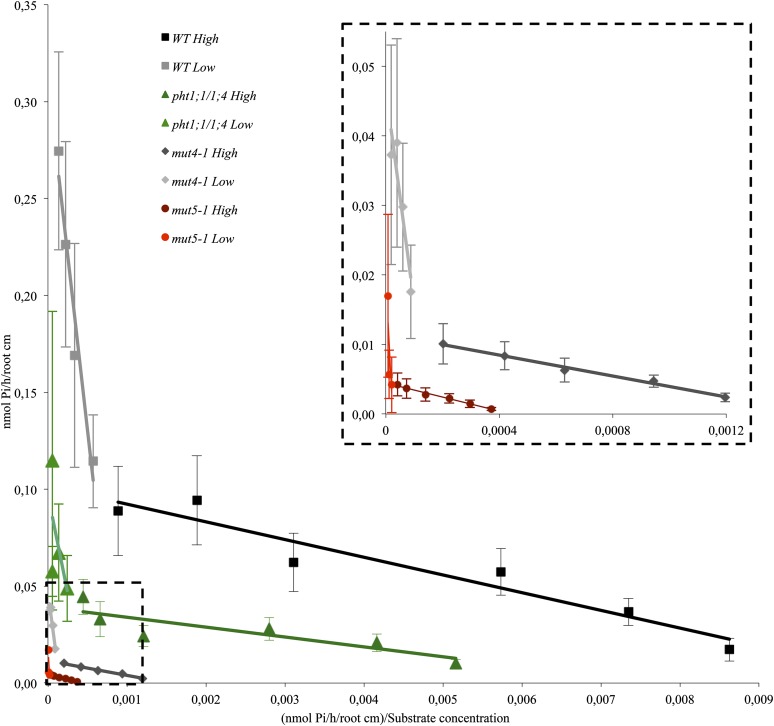
We next measured the kinetic parameters of Pi uptake in the double, quadruple, and quintuple PHT1 mutant lines, in comparison with the wild-type control. To clearly identify and illustrate the presence of high- and low-affinity Pi transporters, we used the George Eadie and Baren Hofstee representation (Fig. 4). In this representation (based on linear regression data using the means of the experimental points) the slopes correspond to the Km, and the interception point with y axis estimates the Vmax. In addition to this visual illustration, we used more accurate calculation based on nonlinear regression analysis with PRISM6 software to calculate the Km (in μm) and the Vmax (in nmol Pi h–1 cm–1 root). The Km and Vmax values (±sd) presented in Table I were obtained with the same raw data used in Figure 4. Wild-type Km values are 6.67 µm for high-affinity transport and 424 µm for low-affinity transport. Interestingly, all mutant lines displayed more or less similar Km values (as illustrated by similar slopes on the George Eadie and Baren Hofstee representation) that were quite similar to the wild-type control (Fig. 4; Table I).
Figure 4.
33P uptake kinetics demonstrates that PHT1-depleted lines are affected both in low- and high-affinity transport. Eadie and Hofstee representation of the 33P uptake kinetic parameters. The experimental points are the mean ± sd of a representative experiment with at least 11 individual plantlets for each genotype and each condition. The dotted line insert is a magnified view of the dotted line selection in the main graph. High signifies high-affinity transport (2–100 μm Pi), and Low represents low-affinity transport (0.2–2 mm Pi). WT, Wild type.
Table I. Low- and high-affinity kinetic constants ± sd of the wild type and double, quadruple, and quintuple mutant lines.
The Michaelis-Menten Km and Vmax were obtained by nonlinear regression analysis from data presented in Figure 4 using PRISM6 software.
| Genotype | High-Affinity Constants ± sd
|
Low-Affinity Constants ± sd
|
||
|---|---|---|---|---|
| Km | Vmax | Km | Vmax | |
| μm | nmol Pi h–1 cm–1 root | μm | nmol Pi h–1 cm–1 root | |
| Wild type | 6.67 ± 1,84 | 0.0920 ± 0.006 | 424 ± 209 | 0.327 ± 0.054 |
| pht1;1/pht1;4 | 6.04 ± 1.46 | 0.0409 ± 0.002 | 368 ± 263 | 0.114 ± 0.027 |
| mut4-1 | 9.07 ± 2.28 | 0.0121 ± 0.0008 | 362 ± 178 | 0.051 ± 0.008 |
| mut5-1 | 11.24 ± 3.17 | 0.0046 ± 0.0004 | 969 ± 892 | 0.029 ± 0.012 |
Only the Vmax was reduced in the mutant lines, which similarly affected high- and low-affinity transporters. Specifically, there was a 55% to 65% decrease in Pi uptake for pht1;1/pht1;4, an 84% to 86% decrease for mut4, and a 91% to 95% decrease for mut5. Because the Vmax parameter is related to the concentration of active transporters, its reduction in the various lines (when compared with the wild type) explains the reduction of Pi uptake observed in the experiments reported in Figure 2C.
The gradual reduction of PHT1 transporters described here results, therefore, in the decrease of the total uptake capacity (reduced Vmax) without affecting the affinity of total transport (unaltered Km).
Reducing Pi Uptake Strongly Affects Plant Physiology in Quadruple and Quintuple Mutants
Previous reports have demonstrated that the pht1;1/pht1;4 and phf1 mutations lead to a 40% to 70% and 80% reduction, respectively, in Pi absorption (Shin et al., 2004; González et al., 2005). Impact on plant growth appears not proportional, as these plants only have a very mild phenotype when grown in a Pi-deficient medium (below 100 µm).
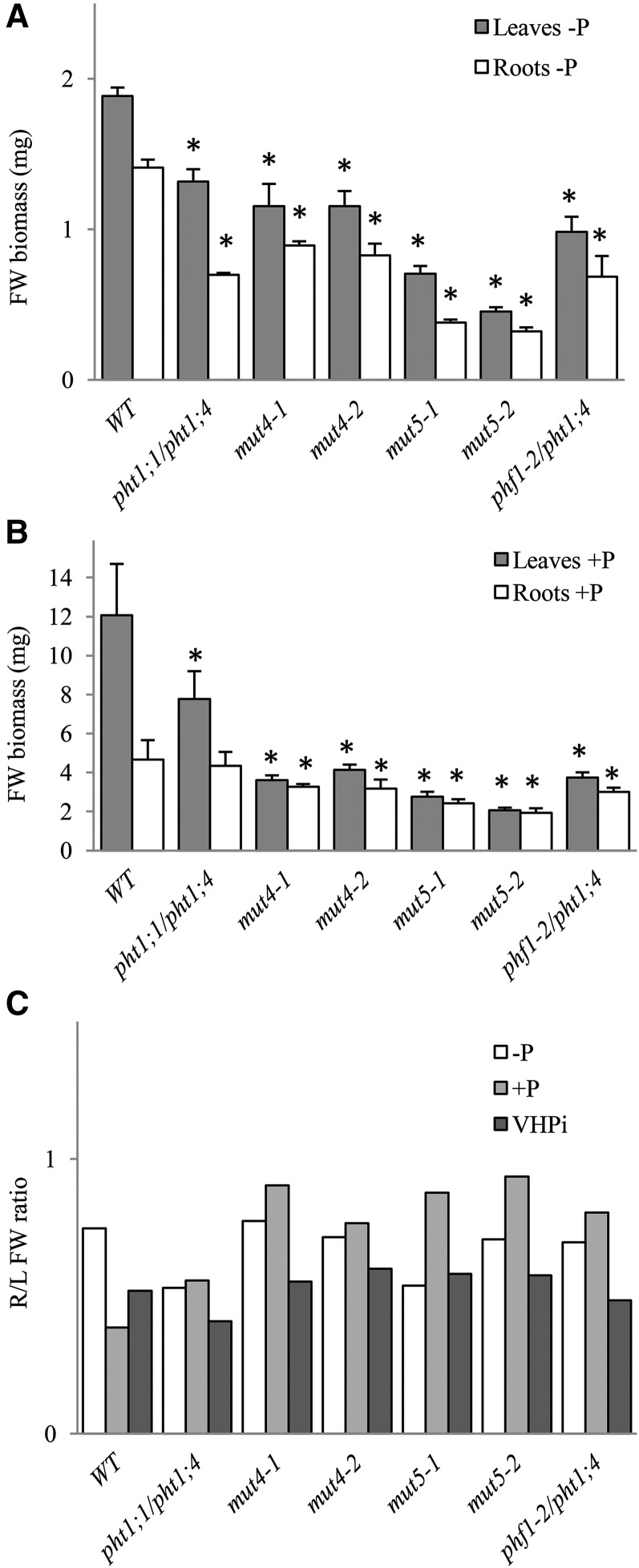
Here, we report that, even when Pi is highly limited (i.e. a –P condition in which residual Pi does not exceed 15 µm), growth of plants exhibiting less than 20% residual Pi uptake activities (phf1-2/pht1;4, mut4, or mut5; Fig. 2C) is strongly affected (Fig. 5A). Growth reduction, which can reach 78% for mut5 lines, affects both leaves and roots. The discrepancies between the wild type and mut5 lines are even more pronounced when Pi is present in a nonlimiting amount (+P). Specifically, the reduction in growth for rosettes varies from 77% to 83% and 50% to 60% for the roots (Fig. 5B). Consequently, the root-to-shoot ratio was preserved in –P and even increased under +P conditions (Fig. 5C). This is illustrated by the in vitro phenotypes of young plantlets in +P media, in which rosettes appeared strongly affected in quadruple and quintuple mutants, whereas their roots were as long as in the wild type (Fig. 6A). The control of root architecture has been related to local concentration of Pi present in the medium (for review, see Péret et al., 2011). We have investigated here the primary root (PR) length (Supplemental Fig. S4C) and the root hair response to Pi starvation (Supplemental Fig. S4B). These both well-known features of local Pi signaling were unchanged in mut4 and mut5 lines compared with the wild-type control (Supplemental Fig. S4, A–E), confirming that they conserved their ability to sense Pi deficiency at the developmental level (Reymond et al., 2006; Svistoonoff et al., 2007). External Pi strongly influenced root hair development in all lines, highlighting the importance of local signaling as revealed by differences between +P and –P (Supplemental Fig. S4, B–E). Detailed analysis of root hair number and position (measured by the length from the root tip until the first root hair) were performed as previously described (Devaiah et al., 2007; Karthikeyan et al., 2014). It revealed a statistically significant difference between the wild type and mut4 or mut5 lines. In these lines, the root hairs start to develop closer from the root tip than in the wild type (Supplemental Fig. S4E), explaining the moderate (and statistically significant) increase of root hair number measured in the first 5 mm of the root tip (Supplemental Fig. S4D). In addition, we measured the distance between two root hairs. In +P, this distance varies from 208 ± 15 to 230 ± 20 μm, depending on the genotypes. In –P, the distance was comprised between 128 ± 13 and 151 ± 18 μm. Statistical analysis of these data revealed no significant differences between the wild type and the different mutant lines. This measurement suggests that the higher root hair number in mut4 and mut5 lines compared with the wild type is due to a shorter distance between PR tip and the first root hair (Supplemental Fig. S4E) and not to a difference in cell size. Our data demonstrates that internal Pi status also influences root hair development, confirming the genetic results previously published with the analysis of Phosphate Starvation Response1 (phr1)/Phosphate Starvation Response1 Like double mutant. This mutant of the master regulatory genes controlling systemic Pi responses exhibited abnormal root hair development (Bustos et al., 2010). Nevertheless, this impact remains limited compared with impact of external Pi and underlines the complexity of Pi responses between local and systemic Pi signaling (Arnaud et al., 2014).
Figure 5.
Biomass and roots-to-leaves ratio of mut4 and mut5 lines. A, Average biomass of leaves (gray bars) and roots (white bars) expressed as mg of fresh weight (FW) in 12-dpg plantlets grown in –P conditions (n = 30–80 plantlets). B, Average biomass of leaves (gray bars) and roots (white bars) expressed as mg of fresh weight in 12-dpg plantlets grown in +P conditions (n = 15–30 plantlets). C, Ratio of roots (R) to leaves (L; fresh weight biomass) in –P (white), +P (pale gray), and VHPi (10 mm ; dark gray) conditions. All experiments were reproduced at least twice. For A and B, bilateral Student’s t tests were conducted to assess statistical differences between the wild type (WT) and the other genotypes, comparing leaf biomass to wild-type leaf biomass and root biomass to wild-type root biomass. In A, the single asterisk indicates P < 0.001. In B, the single asterisk indicates P < 0.05.
Figure 6.
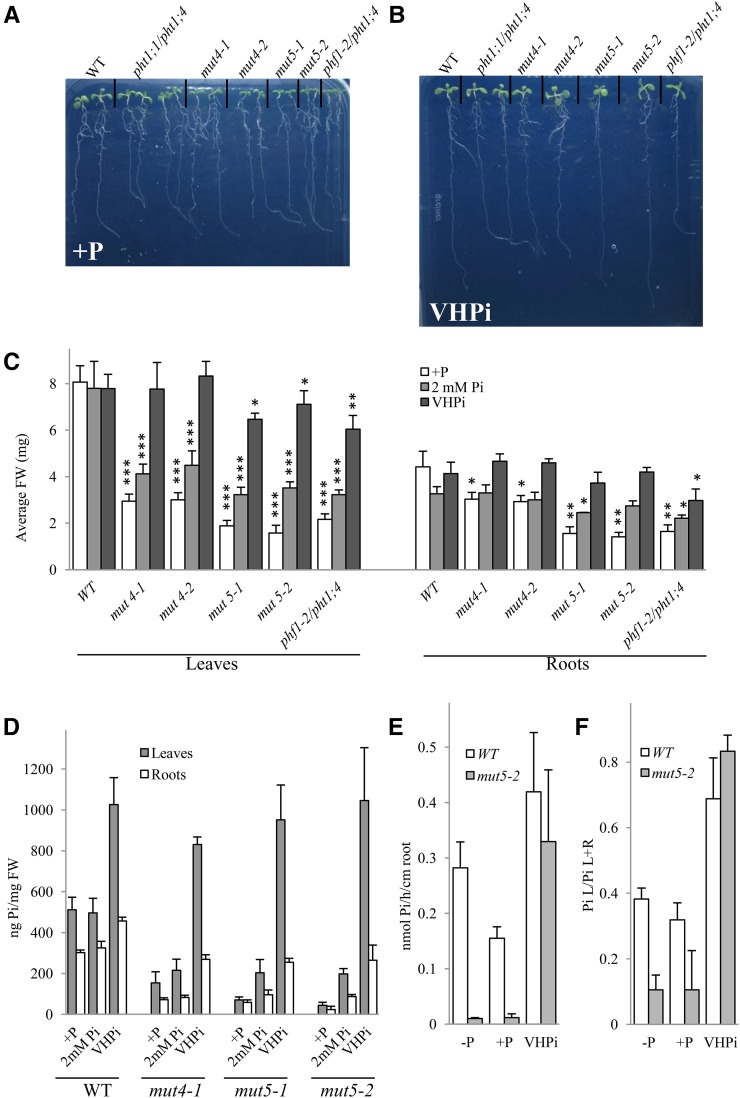
Physiological characterization of mut4 and mut5 lines on VHPi. In vitro phenotyping of the lines used in this study in +P (A) and VHPi (10 mm Pi; B) at 8 dpg. C, Average biomass of leaves (left) and roots (right) in +P (white bars), 2 mm Pi (light-gray bars), and 10 mm Pi (dark-gray bars) at 12 dpg. Bilateral Student’s t tests were conducted to assess statistical differences between the wild type (WT) and the other genotypes for every growing condition and comparing leaves to wild-type leaves biomass and roots to wild-type root biomass. The single asterisk indicates P < 0.5; the double asterisk indicates P < 0.001, and the triple asterisk indicates P < 0.0001. D, Pi content in leaves (gray bars) and roots (white bars) of wild-type, mut4, and mut5 lines in +P, 2 mm Pi, and VHPi at 12 dpg. E, 33P uptake capacity of the wild type and mut5-2 in –P, +P, and VHPi growth/incubation conditions. F, 33P partitioning of the plants from E. For E and F, 20 to 24 plants were used for each genotype/condition, and plantlets were analyzed at 9 dpg. All experiments were repeated at least twice. FW, Fresh weight.
As expected, there was significant anthocyanin accumulation in mut4 and mut5 rosettes in +P conditions, which increased when plants were grown under Pi-deficient conditions (Supplemental Fig. S4F).
When plantlets were grown in the greenhouse under short-day conditions (which favor rosette development), only the mut5 lines were significantly affected. These lines exhibited a delay in flowering time (Supplemental Fig. S5A) and showed a significant reduction (approximately 30%) in seed yield (Supplemental Fig. S5B). The size and weight of seeds in mut5 lines were respectively reduced by 15% and 30% (Supplemental Fig. S5C), whereas the other mutant lines were not affected (data not shown). Germination and viability of mut5 seeds was not affected by these modifications. By contrast, a more dramatic phenotype was observed under long-day conditions. Here, rapid flower induction did not allow enough time for mut5 plants to develop a substantial aerial biomass, resulting in dwarf plants that barely produced any seeds (data not shown).
Pi Accumulation Is Uncoupled from PHT1 Activity in mut5 Lines
We next examined whether mut4 and mut5 plants are only affected in Pi transport or if they display any other defects. The reduced Pi uptake capacity was therefore compensated by providing massive amounts of Pi (2 and 10 mm) to the culture medium. These Pi doses did not modify wild-type growth, confirming that the standard +P medium used (500 µm) was optimal for plant growth in our conditions. Interestingly, they improved plant growth for all mutants tested. The 10 mm Pi condition, henceforth referred to as very high Pi (VHPi), restored growth close to wild-type levels for all mutants (Fig. 6, A–C). VHPi conditions promoted a significant accumulation of Pi in wild-type leaves (+100% compared with +P) although effects were milder in roots (+50%), as determined by Pi content measurements (Fig. 6D). Pi levels in the leaves of mut4 and mut5 mutants were similar to the wild-type control, whereas Pi accumulation in their roots was less pronounced. Interestingly, feeding plants with 2 mm Pi only impacted Pi accumulation in the roots and leaves of mut5 mutants (Fig. 6D). VHPi growth conditions also allowed the restoration of anthocyanin to wild-type levels in all mutant and transgenic lines (Supplemental Fig. S4B). Eventual modifications of mineral content due to the very high Pi conditions were investigated by ICP. In addition to 30% reduction of total P content in mut5-1 compared to the wild type, no modification of other cations was observed (Supplemental Fig. S2C).
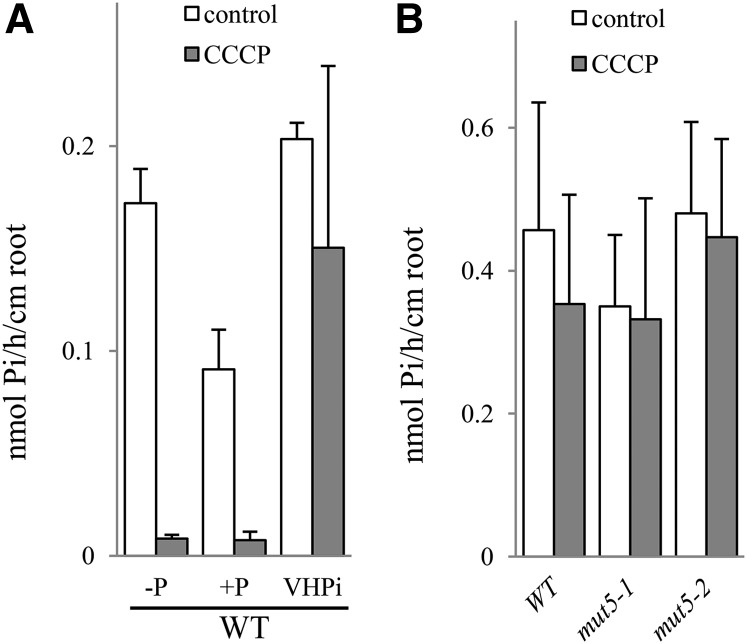
To determine whether the 5% Pi uptake detected in the mut5 line is active (due to residual PHT1 activity) or passive, 33P uptake experiments were performed in plants grown under VHPi conditions. Both the wild type and mut5 lines showed comparable Pi uptake as well as distribution between leaves and roots (Fig. 6, E and F), suggesting that Pi entry into the plant does not require PHT1;1/PHT1;2/PHT1;3/PHT1;4 transport under VHPi conditions. Applying the protonophore carbonyl cyanide 3-chloro phenylhydrazone (CCCP) dissipated the membrane potential, which drastically reduced Pi uptake (by 94%) in low Pi (Supplemental Fig. S6A). Moreover, the addition of CCCP to the incubation media also provoked a nearly complete inhibition of root-to-leaf transfer in our experimental conditions (Supplemental Fig. S6B). This demonstrates that Pi uptake and translocation require active phosphate transport coupled to a proton gradient in low-Pi conditions.
The CCCP effect was then assayed in wild-type plantlets grown and incubated in the 3 Pi supply experimental conditions. As expected, the addition of CCCP to –P and +P incubation solutions resulted in drastic reductions (93% to 95%) in Pi transport (Fig. 7A). Strikingly, CCCP had no effect on Pi uptake when applied to the VHPi incubation solution, revealing that Pi absorption is mostly a passive process under these conditions (Fig. 7A). Additional experiments using two independent mut5 lines grown in VHPi conditions revealed that 33P uptake was not modified by the addition of CCCP (Fig. 7B), revealing the passive nature of Pi uptake in VHPi conditions (both for the wild type and mut5 lines). VHPi (10 mm external Pi) is not a physiological concentration, and this created a very high ionic gradient across the plasma membrane. In addition, CCCP abolished membrane potential, but Pi uptake remained unchanged. It suggests the existence of proteins such as channels or unspecific transporters involved in Pi uptake, which are not activated or regulated by membrane potential. Additional elements including the increase of apoplastic pathway and/or abolition of Pi export/secretion are also likely favored in VHPi conditions.
Figure 7.
33P uptake capacity in VHPi is mainly a passive process. A, Wild-type (WT) uptake capacity is reduced by CCCP addition, except in VHPi conditions. Wild-type plants were grown for 10 d in –P, +P, and VHPi before incubation in the corresponding media in control (white bars) or +10 μm CCCP (gray bars) conditions. B, In VHPi conditions, mut5 lines show the same uptake capacity as the wild type, both in the control (white bars) and with +CCCP addition (gray bars). Pi uptake capacity was calculated as nmol Pi h–1 cm–1 root. Data shown are from a representative experiment; experiments were performed in either duplicate or triplicate. Each bar corresponds to 20 to 24 plants. Error bars represent se.
Experiments performed with plants grown on –P media indicated an effect of CCCP treatment even on mut5 lines (Supplemental Fig. S6A), suggesting that the very weak activities observed in mut5 lines grown on –P medium are related to an active process, likely associated with residual PHT1 activity.
In conclusion, VHPi abolished active Pi uptake in the wild type and mut5 lines, probably by activation of several down-regulation mechanisms that act at the transcriptional and posttranscriptional levels, as previously reported for PHT1 transporters (Bayle et al., 2011). Nevertheless, if inactive PHT1 proteins may be present in the wild type, this is not the case in mut5 lines where the pool of PHT1 is decreased at least by 95%. This experiment offers, therefore, the opportunity to uncouple PHT1 presence from Pi accumulation inside the plants.
Induction of Pi Starvation Markers Does Not Require PHT1 Presence or Pi Uptake Activity
The dual role of yeast (Saccharomyces cerevisiae) phosphate transporters as Pi sensors has led to their description as transceptors (Ozcan et al., 1996; Lorenz and Heitman, 1998). In plants, a similar duality was reported for the Arabidopsis transporter Chlorina1 (CHL1), which also acts as a nitrate receptor (Ho et al., 2009). Here, we have investigated whether the PHT1 family similarly performs a double role regarding phosphate transport and perception. Our approach, to circumvent genetic redundancy, resulted with mut5 line in a drastic reduction (approximately 20-fold) of the PHT1 pool of transporters, with a subsequent diminution in Pi uptake activity. These plants therefore exhibit a strong reduction in Pi content when grown in either poor or rich Pi media. Consequently, these plants are constitutively phosphate deficient and induced in both conditions markers typically associated with Pi deficiency (Fig. 3; Supplemental Fig. S3), suggesting that phosphate sensing is unaltered. It may be noticed that compensatory overexpression of other PHT1 members (e.g. PHT1;5, PHT1;7, PHT1;8, and PHT1;9) in mut5 lines remains ineffective, due to the targeting defect caused by the phf1-2 mutation on PHT1 proteins (Fig. 2C).
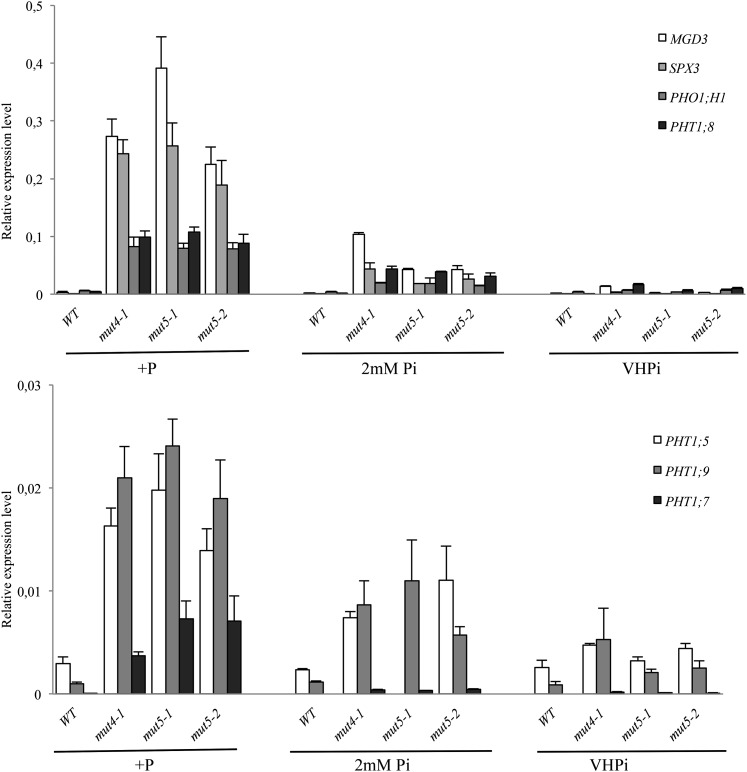
Moreover, several markers typically used to report Pi deficiency (e.g. MGD3, SPX3, PHO1;H1, PHT1;5, PHT1;7, PHT1;8, and PHT1;9) have revealed constitutive phosphate deficiency responses in mut4 and mut5 lines grown under +P conditions. This trait can be repressed by adding 2 or 10 mm Pi in a dose response manner (Fig. 8). The 10 mm Pi (i.e. the VHPi condition) restores the repression to levels observed in wild-type plants grown in +P. This strongly suggests that transcriptional repression of the Pi deficiency response can be associated with internal Pi concentration (or Pi-derived metabolites), and that it does not require the presence of PHT1 transporters involved in Pi uptake (Fig. 8).
Figure 8.
VHPi conditions restore transcriptional repression of PHT1 and PSI markers in mut5 lines. qRT-PCR of several members of the PHT1 family and several PSI genes in the wild type (WT), a mut4 line, and two independent mut5 lines. Plantlets were grown on +P as well as 2 and 10 mm Pi media (VHPi) for 11 d before RNA extraction from roots. Corresponding loci numbers and primer sequences are provided in Supplemental Table S2. Biological triplicates were performed, and all samples were analyzed with technical triplicates. sds are shown.
DISCUSSION
Pi Uptake Relies Greatly upon the PHT1;1, PHT1;2, and PHT1;3 Cluster
Deciphering the contribution of PHT1;1, PHT1;2, and PHT1;3 to Pi uptake remains challenging due to their close proximity on chromosome 5, which is associated with a high sequence identity at the nucleic acid level. A fourth gene, PHT1;6, is also located in this cluster but was omitted from this study because its expression is not detected in the roots (Mudge et al., 2002; Misson et al., 2005), signifying that it does not contribute to Pi uptake. On the other hand, PHT1;1 has a known role in Pi uptake when this nutrient is not limited (Muchhal et al., 1996). This contribution, confirmed in our study, provides one-half of the Pi uptake under this condition. One crucial explanation has been provided by the study of transcriptional regulation of PHT1 genes, indicating that all PHT1 family members are induced by Pi starvation (Mudge et al., 2002; Misson et al., 2005). Whereas most members exhibited only a very low transcript level in Pi-rich medium and were highly induced in Pi-limiting conditions, PHT1;1 presented clearly distinct features. Specifically, PHT1;1 exhibited a high basal level in +P medium but only a 3- to 4-fold increase in –P. The induction varies from 25-fold to more than 70-fold for other PHT1 members (Misson et al., 2005). Nevertheless, the very high homologies between PHT1;1 and PHT1;2 (e.g. 99% identity and 99.8% similarity) impair the distinction between these two genes (Mudge et al., 2002). Although the position of PHT1;3 is slightly more distant from PHT1;1 (94% identity and 96% similarity; Supplemental Table S1), it is often difficult to distinguish it from PHT1;1 and PHT1;2 using molecular tools. As with most PHT1 members, PHT1;3 is reported to be highly induced by Pi starvation (Mudge et al., 2002; Misson et al., 2005).
The genetic approaches used here to compare pht1;1 and mut3 allowed us to establish that PHT1;2 and PHT1;3 do not contribute significantly to Pi uptake when plants are grown in Pi-rich medium. They are nevertheless detected in pht1;1 mutant grown in –P, suggesting that the activities of PHT1;2 and PHT1;3 are restricted to a Pi-deficient environment, corroborating previous results (Mudge et al., 2002; Misson et al., 2005). In these conditions, both transporters participate in approximately one-third of Pi uptake, whereas 15% to 20% of the uptake results from PHT1;1 activity, as shown both here and previously (Shin et al., 2004). Combined, the three genes thus account for nearly one-half of the Pi uptake in Pi-limiting conditions. This impact is not so clear in the mut3 line, probably due to incomplete silencing of the transporters. However, we cannot exclude if other compensatory mechanisms (such as modification of the activity of the other PHT1 transporters) could make up for a partial reduction in PHT1 level under these conditions. Strikingly, phosphate transporters are able to compensate for the loss of activity of specific PHT1 members by inducing other family members. This mechanism is triggered by an internal Pi deficiency (Thibaud et al., 2010), and it is likely that the required threshold to trigger it is not reached in mut3 lines. This was well established in a previous study (Shin et al., 2004), where the double mutant pht1;1/pht1;4 exhibited a stronger reduction in Pi uptake (70%) in +P than in any of the single mutants (pht1;1 only reduced Pi uptake by approximately 30% in this condition). The crucial contribution of PHT1;4 is very clear in this study, as illustrated by highly significant differences of Pi uptake between mut3 and mut4.
The role of these transporters is not limited to Pi uptake, as they also appear to be involved in Pi translocation between roots and leaves. In the event that compensatory mechanisms occurring within –P complicate the analysis of mutants, pht1;1 plants grown under +P conditions can be used to examine the root-to-leave Pi translocation. This approach clearly highlights the role of PHT1;1 in this process. Analysis of the expression patterns of other PHT1 members (such as PHT1;3 or PHT1;4) revealed their involvement in the vascular tissues (Mudge et al., 2002; Misson et al., 2005). Specifically, this suggests that PHT1;8 and PHT1;9 (Remy et al., 2012) and other PHT1 members such as PHT1;5 (Nagarajan et al., 2011) play a crucial role in Pi translocation between organs. A result also observed in other species such as rice (Oryza sativa) for OsPT8 (Jia et al., 2011) or OsPT2 (Ai et al., 2009).
PHF1 Activity Impacts Several PHT1 Members
Previous studies based on cell biology experiments have demonstrated that PHT1;1, PHT1;2, and PHT1;4 require PHF1 presence to ensure proper targeting to the plasma membrane (González et al., 2005; Bayle et al., 2011). In rice, OsPHF1 is also necessary for the proper targeting of the phosphate transporters OsPT2 and OsPT8 (Chen et al., 2011). An 80% decrease in Pi uptake observed in the phf1 mutant, both here and previously (González et al., 2005), provides genetic proof that this protein can impact several PHT1 members. This previous study also demonstrated that a portion of PHT1 can still reach the plasma membrane despite the absence of PHF1. This is confirmed in this study: introduction of the phf1-2 mutation reduced Pi absorption by 60% in mut5 compared with mut4. This clearly illustrates that at least some (if not all) of the remaining PHT1 transporters (i.e. PHT1;5/PHT1;7/PHT1;8/PHT1;9) are also dependent on PHF1 activity. Furthermore, due to the very strong effect of the phf1 mutation on PHT1 activity (González et al., 2005; Bayle et al., 2011), mutant plants exhibit a very low Pi content, even if this nutrient is abundant in the growth medium. No compensatory effect can take place in this background because all PHT1 genes are already highly induced, suggesting that no Pi uptake mechanisms exist independently of this transporter family. This explains the additive impact of the pht1;4 or pht1;1/pht1;2/pht1;3/pht1;4 mutations introduced into the phf1 background.
Pi Uptake Activity Relies on PHT1 Transporters
Other Pi transporters in addition to the PHT1 family have been identified in plants, including plastidic PHT2 carriers (Daram et al., 1998), mitochondrial PHT3 transporters (Zhu et al., 2012), and PHT4 proteins. Among the six members of the PHT4 family, one is located in the Golgi membrane and five are found in the plast envelope (Guo et al., 2008). Another key component, PHO1, was identified in a genetic screen searching for low shoot Pi content, as pho1 mutation impaired xylem loading of Pi (Poirier et al., 1991) and it appears to be involved in Pi efflux (Stefanovic et al., 2011). Additional phosphate carriers have also been reported in the plastidic membrane, where they couple the exchange between phosphorylated metabolites and Pi (Rausch and Bucher, 2002). Although the vast majority of these proteins contribute to the distribution of Pi into various cell compartments, they have no role in Pi uptake; so far, this function has only been attributed to PHT1 proteins. This feature is peculiar to Pi, as several distinct transporter families have been identified for several other ions (e.g. nitrate, K, Fe, etc.). Nevertheless, the severity of the phenotypes observed in mut4 and mut5 lines (which respectively impair 86% and 95% of Pi uptake) confirms that PHT1 proteins are the primary Pi uptake carriers. In addition, because the residual Pi uptake activity is likely related to the other PHT1 proteins present in these lines (PHT1;5/PHT1;7/PHT1;8/PHT1;9), it can be assumed that PHT1 proteins are probably the only plant carriers devoted to Pi uptake. This situation is clearly distinct from yeast, where different high (PHO84 and PHO89)- and low (PHO87 and PHO90)-affinity transporters have been reported (Wykoff and O’Shea, 2001).
The exact nature of the PHT1 transporters involved in Pi uptake in mut5 plants remains elusive. However, our transcriptomic data suggest that PHT1;5, PHT1;7, PHT1;8, and/or PHT1;9 could be involved in Pi uptake, as these transporters are highly expressed under +P conditions in mut5 lines (at a level similar to wild-type plants grown in –P). Possibly, translocation of these proteins to the epidermal plasma membrane could account in part for Pi uptake. Specific antibodies and reporter lines will be necessary to further investigate this point.
Dual Affinity for Pi Uptake Appears Conserved in the Different PHT1 Mutant Lines
Several previous in planta uptake kinetics studies using radioactive Pi tracers identified two distinct phases, suggesting the existence of low- and high-affinity systems (Cogliati and Clarkson, 1983; Drew et al., 1984). Pi uptake activity has so far only been attributed to a single class of PHT1 carriers (Nussaume et al., 2011). This view is reinforced by this study, in which PHT1 proteins appear to be the main (and probably the only) participants in this process. Previous studies (reviewed by Nussaume et al., 2011) obtained Km measurements from several PHT1 transporters, through their heterologous expression in yeast or Xenopus spp. oocytes. These results reveal a wide range of values (from 3 µm to more than 850 µm), suggesting that distinct PHT1 members from several species might encode high- or low-affinity phosphate transporters. See, for example, studies performed in rice (Ai et al., 2009), Medicago truncatula (Liu et al., 2008), or barley (Hordeum vulgare; Rae et al., 2003). One caveat regarding these results is that experiments in heterologous species could lack essential regulatory proteins found in planta. In addition, several posttranslational regulatory steps affecting PHT1 have been identified (González et al., 2005; Bayle et al., 2011; Chen et al., 2011). This could explain the wide range of Km measurements, as the values from yeast often greatly exceed those found in plants. Another complication is that plant protoplasts are not suitable for examining the activity of a particular Pi transporter, as the measured signal is often diluted by the presence of endogenous PHT1 members. It is also possible that endogenous regulatory proteins do not recognize the assayed transporters, due to their heterologous nature.
One previous study endeavored to identify the amino acid modifications responsible for these variations in affinity observed in the M. truncatula Pi transporters (Liu et al., 2008). Analysis of the transporter sequences revealed restricted nonconservative amino acid changes between the low-affinity transporters MtPT1, MtPT2, and MtPT3 and the high-affinity transporter MtPT5. Nevertheless, sequence comparisons with other PHT1 transporters that exhibit distinct affinities for Pi do not reveal any correlation with these results (Nussaume et al., 2011). This suggests either independent evolution in separate species or that the amino acids identified from the M. truncatula data cannot account for affinity changes in other species. Alternatively, some PHT1 members could present a dual affinity, a view put forth in this study for Arabidopsis. A characteristic already identified for other transporters such as the K Cup1/Arabidopsis K+ Transporter1 (Fu and Luan, 1998) and the nitrate NRT1;1 (Parker and Newstead, 2014; Sun et al., 2014). The posttranslational modifications for PHT1 involved in such mechanism remain to be identified.
Physiological Impact of Genetic Modifications to Pi Uptake
A well-known feature of plant mineral nutrition is the occurrence of high genetic redundancy, which is probably related to vital functions that provide autotrophic abilities to plants. This affects the uptake of ions as well as steps involved in their fixation into organic matter. Here, we have examined a distinct situation for Pi uptake, as a single PHT1 mutation reducing up to one-third of Pi uptake activity (Shin et al., 2004) barely has any effect on plant physiology. Reducing Pi uptake by 60% to 80% through multiple PHT1 knockout (pht1;1/pht1;4; Shin et al., 2004) or through alteration of regulatory proteins (phf1; González et al., 2005) requires prolonged growth under low-Pi conditions to affect plant growth. Only the extreme reduction (95%) of Pi uptake, mainly within mut5 lines (and mut4 to a lesser extent), could produce a strong plant growth defect in our high- and low-Pi supply conditions. This clearly illustrates the absence of a linear relationship between Pi uptake and accumulation and plant growth. Nevertheless, plants can use various adaptations to cope with reduced Pi uptake. These include well-known modifications of the root-to-shoot ratio to favor soil exploration during adverse conditions. Our mutant lines are also capable of reducing seed size to maintain significant seed productivity. We did not observe any obvious physiological impact on the seeds produced regarding longevity or germination rate.
Pi Deficiency Sensing
Previous studies have established the existence of local and systemic Pi signaling (Thibaud et al., 2010; Péret et al., 2011). The first one affect mostly root architecture parameters and was linked to the Pi content in the growth culture medium. The second one has been connected to the control of phosphate homeostasis and appears to be driven by internal Pi pool. In agreement with such view, only systemic Pi responses were found affected in the different mutant lines produced for this study.
The restoration of Pi sensing in mut5 lines by massive amount of external Pi offers opportunity to disconnect internal Pi content from PHT1 activity. This suggested that systemic Pi perception is most probably triggered by mechanisms acting downstream of PHT1 activity. In such case, phosphate transporters would be distinct from nitrate transporter CHL1, which acts as a transceptor (Ho et al., 2009). This is reinforced by a very recent report on OsSPX4 regulatory proteins in rice (Lv et al., 2014). These results demonstrate that an internal high-Pi concentration promotes interactions between SPX4 and PHR2 (the PHR1 rice homolog). This interaction prevents its ability to bind DNA and activate the Pi starvation response. This is also in agreement with the capability of phosphite, a nonmetabolizable form of Pi, to prevent activation of the Pi starvation response (Varadarajan et al., 2002). Altogether, these data suggest that plants are able to sense internal Pi and/or its derivatives, a view reinforcing previous publication (Liu et al., 1998). PHT1 transporters nevertheless play a crucial role in the control of the Pi starvation response, as they are the primary contributors of Pi delivery into cells. This might explain the importance of the regulatory steps that fine tune the level of PHT1 activity according to the external medium Pi concentration, of which at least four have been identified (Bayle et al., 2011; Huang et al., 2013; Lin et al., 2013; Park et al., 2014).
CONCLUSION
Genetic approaches and their tools are extremely useful for investigating the physiological roles of proteins, including structure/function analyses. Specifically, they can abolish endogenous activity, which can otherwise mask the phenotype of interest. However, examining mineral nutrition in plants is complicated with these approaches, due to the significant gene duplication underlying these processes. In this study, we produced mut4 and mut5 lines to diminish this complexity to focus our study on the specific modification of PHT1 transporters. The mut4 line is favored over mut5 for this purpose, as the presence of the phf1 defect could reduce the production of modified PHT1 introduced in this background. The low endogenous Pi uptake activity in mut4 should provide very good genetic background for such purpose. Thanks to genetic code degeneration, mutations of PHT1;1, PHT1;2, or PHT1;3 offer the opportunity to produce a gene allele untargeted by the silencing without alteration of amino acid sequence. It thus appears that all PHT1 members can be analyzed in this background. Therefore, this study provides both physiological data and unique genetic tools for future investigations of Pi uptake.
MATERIALS AND METHODS
Plant Material and Growth Conditions
The wild-type Arabidopsis (Arabidopsis thaliana) ecotypes Wassilewskija and Colombia were used in this study. The double mutant pht1;1/pht1;4 (Shin et al., 2004) was obtained from Dr. Maria Harrison’s laboratory (Boyce Thompson Institute for Plant Research), and the single pht1;1 mutant line was provided by Dr. Kashchandra G. Raghothama (Purdue University).
For in vitro analyses, seeds were surface sterilized and sown in vitro on square petri plates with solid modified Murashige and Skoog medium (Arnaud et al., 2014) containing 0.15 mm MgSO4, 2.1 mm NH4NO3, 1.9 mm KNO3, 0.34 mm CaCl2, 0.5 µm KI, 10 µm H3BO3, 10 µm MnSO4, 3 µm ZnSO4, 0.1 µm CuSO4, 0.1 µm CoCl2, 1 µm Na2MoO4, 3.4 mm MES (pH 5.8), 0.5% (w/v) Suc, and 0.8% (w/v) agar. Media was supplemented with 2 μm FeCl2, unless otherwise stated. Then, Pi was added at 0.5 mm NaH2PO4 (complete media or +P) or 10 mm NaH2PO4 (VHPi). In the Pi-deficient medium (–P), approximately 0.015 mm residual Pi was introduced exclusively by the Sigma agar stock, and 0.5 mm NaCl was added to replace the equivalent amount of sodium provided by NaH2PO4. For physiological analyses and RNA extractions, seeds from the different wild-type and mutant lines were cultivated as previously described (Misson et al., 2004). For Southern analysis, plants were grown in individual pots on soil. Four to six weeks in long-daylight conditions were necessary to induce flowering, after which DNA extraction was performed (Sarrobert et al., 2000).
Multiple Mutant Generations by RNAi
A 21-bp sequence common to PHT1;1, PHT1;2, and PHT1;3 was used to synthesize the following set of primers: micro-RNA (miRNA)-s, gaTTGGCAAACATAAACACGGTAtctctcttttgtattcc; miRNA-a, gaTACCGTGTTTATGTTTGCCAAtcaaagagaatcaatga; miRNA*s, gaTAACGTGTTTATGATTGCCATtcacaggtcgtgatatg; and miRNA*a, gaATGGCAATCATAAACACGTTAtctacatatatattcct.
For PCR template, we used the pRS300 vector containing Arabidopsis MIR319a in a pBSK backbone (Schwab et al., 2010). Two oligonucleotides based on the template plasmid sequence and located outside of the pBSK multiple cloning site were necessary to generate bigger PCR products. These oligonucleotides (A and B) were designed with the Web MicroRNA Designer (http://wmd3.weigelworld.org/cgi-bin/webapp.cgi), yielding the following sequences: A, CACCTGCAAGGCGATTAAGTTGGGTAAC; and B, GCGGATAACAATTTCACACAGGAAACAG.
The PCR fragment was first cloned in pGEMT-easy for sequencing. Next, the amiRNA precursor was released by enzymatic restriction and cloned into pENTR before subcloning into the binary vector pGWB2 (Nakagawa et al., 2007).
Subsequently, the wild type, pht1;1/pht1;4 (Shin et al., 2004), and phf1-2/pht1;4 (Bayle et al., 2011) were transformed by a simplified floral dip method (Logemann et al., 2006).
At least 15 independent transgenic lines were generated for every transformation event, and progeny were sown in hygromycin-containing petri dishes to select lines with a single genetic transfer DNA insertion. Southern blot (with the hygromycin phosphotransferase gene probe) was then used to select lines with simple transfer DNA insertions. Finally, their Pi content was analyzed at the T3 homozygous state. Two to three lines with reduced Pi content were retained for physiological characterization.
RNA Extraction and qRT-PCR
Total RNA was extracted from rosettes and roots of 10- to 14-d-old plantlets, using the RNeasy plant mini kit (Qiagen) with DNase treatment. qRT-PCR analyses were performed following reverse transcription (Invitrogen VILO Kit) and amplification (Light Cycler 480, Roche). The primer efficiency factor was measured for each gene, using Glyceraldehyde 3-phosphate dehydrogenase C subunit1, Rotamase Cyp3, and AT1G32050 as reference genes. Analyzed genes were selected as systemically or locally Pi starvation induced, as previously detailed (Thibaud et al., 2010). Primer sequences are provided in Supplemental Table S2.
Western-Blot Analysis
Membrane protein extraction was performed (according to Witte et al., 2004) from leaves and roots of in vitro plantlets grown in 15 or 500 μm Pi. To optimize protein yield from root samples, the procedure was slightly modified by extending the sample grinding. Following SDS-PAGE (10% [w/v] acrylamide gel separation) and transfer onto nitrocellulose membranes, PHT1;1, PHT1;2, and PHT1;3 were immunodetected with a primary rabbit polyclonal antibody against the common peptide CEERVEDDUKDPQK. Goat anti-rabbit IgG Horse Radish Peroxidase (Sigma, A6154) was used as the secondary antibody. Signal was acquired and quantified in a G-Box (Chemin XL Syngene). Anti-PIP1 antibody was used as a loading control (Boursiac et al., 2005).
Inorganic Pi, Total P, Anthocyanin Content, and NMR Analysis
Pi content was calculated as previously described (Misson et al., 2004) from rosettes and roots of in vitro grown plantlets. Total P content was analyzed by ICP optical emission spectrometry as previously described (Hirsch et al., 2006) in leaves and roots of 2-week-old plantlets.
Anthocyanin content was measured at 12 d post germination (dpg) in leaves of in vitro grown plantlets on MS/10 medium containing different quantities of Pi, as previously described (Ticconi et al., 2001).
In vivo 31P-NMR spectra were recorded on an AMX 400 Bruker spectrometer (http://www.bruker.com), as previously described (Aubert et al., 1998) on roots of hydroponically grown plants in short-day conditions.
33P Uptake Measurements
Phosphate uptake experiments were performed as previously described (Narang et al., 2000) either with plantlets grown in –P or in +P conditions. Twenty to 24 seedlings per condition were incubated separately in a phosphate incubation solution for 2 h. The incubation solution contains 0.15 µCi mL–1 33PO4, 5 mm MES, 0.1 mm CaCl2, and 50 or 500 µm KH2PO4. When measuring high-affinity uptake, the plantlets were grown in –P (approximately 15 µm residual P in the agar), but incubation was performed in 50 µm KH2PO4 instead of 15 µm to avoid a putative limitation of the uptake during the 2-h experiment. When measuring low-affinity uptake, plantlets were grown in +P (500 µm) and incubated in a solution with 500 µm KH2PO4. Our experimental conditions reflect uptake rates under steady-state growth conditions. In addition, we checked that uptake rates were identical after 1 h and after 2 h of incubation.
For Vmax and Km determination, plantlets grown in –P were incubated in solutions ranging from 2 to 100 μm P (2, 5, 10, 20, 50, and 100 μm KH2PO4), and another pool of plants was grown in +P and then incubated in solutions ranging from 0.2 to 2 mm P (0.2, 0.5, 1, and 2 mm KH2PO4). This protocol allowed a better estimation of the kinetic constants for discriminating high- and low-affinity transport. Consequently, the low-affinity Km value reflects only low-affinity transport, as the high-affinity transporters are not induced in the plantlets grown in +P. The Km and Vmax values ± sd were obtained by nonlinear regression using the PRISM 6.0 software (GraphPad).
After incubation in the uptake solution, plantlets were transferred to a chilled desorption medium (5 mm MES, 0.1 mm CaCl2, and 1 mm KH2PO4) for 2 h at 4°C. The seedlings (or the split rosettes and roots) were then dried in scintillation vials at 60°C overnight, after which 2 mL of scintillation cocktail (InstaGel, PerkinElmer) was added, and the radioactivity was measured with a scintillation analyzer (Tri-Carb, Packard Instrument Company). The amount of phosphate absorbed during the experiment was calculated and normalized per cm of root.
When using the protonophore CCCP, this was added in the incubation solution at 10 μm final concentration. CCCP was prepared as a 100 mm stock solution in pure dimethyl sulfoxide (DMSO). Consequently, an additional control experiment with the equivalent amount of DMSO was conducted in parallel to every CCCP experiment. In our experimental conditions, DMSO addition was without effect in 33P uptake or partitioning.
Analysis of Root Architecture
Seedlings were photographed at different times after germination, and PR length was measured with the ImageJ software (http://rsb.info.nih.gov/ij/). The lateral root number was calculated after stereomicroscopy observation of 7- or 8-dpg plantlets to include the earlier stages of lateral root formation. All experiments were performed at least three times.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Pi content, qRT-PCR, 33P uptake, and partitioning in triple mutant lines.
Supplemental Figure S2. Pi content, 31P-NMR spectra, and mineral content in mut4 and mut5 lines.
Supplemental Figure S3. qRT-PCR of mut5 lines reveals a general induction of PSI genes.
Supplemental Figure S4. In vitro phenotype of mut4 and mut5 lines on −P medium containing 0.010 mm Fe reveals full perception of P deficiency.
Supplemental Figure S5. Flowering time, grain yield, and seed size are affected in quintuple mutant lines.
Supplemental Figure S6. CCCP addition inhibits both 33P uptake and roots-to-leaves translocation.
Supplemental Table S1. PHT1 transporter family comparison at the amino acid level.
Supplemental Table S2. List of the primers used in qRT-PCR experiments.
Supplementary Material
Acknowledgments
We thank Dr. Richard Bligny (Laboratoire de Physiologie Cellulaire Végétale, Commissariat à l’Energie Atomique et aux Energies Alternatives-Grenoble) for NMR analysis, Paul Soreau (Groupe de Recherches Appliquées en Phytotechnologie, Commissariat à l'Energie Atomique et aux Energies Alternatives, Cadarache) for ICP analysis, Dr. Maria Harrison (Boyce Thompson Institute for Plant Research) for providing seeds of pht1;1/pht1;4 double mutant, Dr. Kashchandra G. Raghothama (Purdue University) for providing seeds of the pht1;1 mutant, The Arabidopsis Information Resource (http://www.arabidopsis.org) as a source of data, Dr. Benjamin Péret (Laboratoire de Biologie du Développement des Plantes, Commissariat à l'Energie Atomique et aux Energies Alternatives, Cadarache) for suggestions while preparing the article, and Dr. Brandon Loveall (Improvence) for English proofreading.
Glossary
- Pi
inorganic phosphate
- qRT
quantitative real-time
- PSI
Phosphate Starvation Induced
- PR
primary root
- VHPi
very high inorganic phosphate
- CCCP
carbonyl cyanide 3-chloro phenylhydrazone
- dpg
days post germination
- DMSO
dimethyl sulfoxide
Footnotes
This work was supported by the Commissariat à l’Energie Atomique et aux Energies Alternatives, France.
Articles can be viewed without a subscription.
References
- Ai P, Sun S, Zhao J, Fan X, Xin W, Guo Q, Yu L, Shen Q, Wu P, Miller AJ, et al. (2009) Two rice phosphate transporters, OsPht1;2 and OsPht1;6, have different functions and kinetic properties in uptake and translocation. Plant J 57: 798–809 [DOI] [PubMed] [Google Scholar]
- Arnaud C, Clément M, Thibaud MC, Javot H, Chiarenza S, Delannoy E, Revol J, Soreau P, Balzergue S, Block MA, et al. (2014) Identification of phosphatin, a drug alleviating phosphate starvation responses in Arabidopsis. Plant Physiol 166: 1479–1491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubert S, Curien G, Bligny R, Gout E, Douce R (1998) Transport, compartmentation, and metabolism of homoserine in higher plant cells: carbon-13- and phosphorus-31-nuclear magnetic resonance studies Plant Physiol 116: 547–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbier-Brygoo H, De Angeli A, Filleur S, Frachisse JM, Gambale F, Thomine S, Wege S (2011) Anion channels/transporters in plants: from molecular bases to regulatory networks. Annu Rev Plant Biol 62: 25–51 [DOI] [PubMed] [Google Scholar]
- Bayle V, Arrighi JF, Creff A, Nespoulous C, Vialaret J, Rossignol M, Gonzalez E, Paz-Ares J, Nussaume L (2011) Arabidopsis thaliana high-affinity phosphate transporters exhibit multiple levels of posttranslational regulation. Plant Cell 23: 1523–1535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boursiac Y, Chen S, Luu DT, Sorieul M, van den Dries N, Maurel C (2005) Early effects of salinity on water transport in Arabidopsis roots: molecular and cellular features of aquaporin expression. Plant Physiol 139: 790–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustos R, Castrillo G, Linhares F, Puga MI, Rubio V, Pérez-Pérez J, Solano R, Leyva A, Paz-Ares J (2010) A central regulatory system largely controls transcriptional activation and repression responses to phosphate starvation in Arabidopsis. PLoS Genet 6: e1001102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderon-Vazquez C, Ibarra-Laclette E, Caballero-Perez J, Herrera-Estrella L (2008) Transcript profiling of Zea mays roots reveals gene responses to phosphate deficiency at the plant- and species-specific levels. J Exp Bot 59: 2479–2497 [DOI] [PubMed] [Google Scholar]
- Chen J, Liu Y, Ni J, Wang Y, Bai Y, Shi J, Gan J, Wu Z, Wu P (2011) OsPHF1 regulates the plasma membrane localization of low- and high-affinity inorganic phosphate transporters and determines inorganic phosphate uptake and translocation in rice. Plant Physiol 157: 269–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogliati DH, Clarkson DT (1983) Physiological changes in, and phosphate uptake by potato plants during development of and recovery from phosphate deficiency. Physiol Plant 58: 287–294 [Google Scholar]
- Daram P, Brunner S, Persson BL, Amrhein N, Bucher M (1998) Functional analysis and cell-specific expression of a phosphate transporter from tomato. Planta 206: 225–233 [DOI] [PubMed] [Google Scholar]
- Devaiah BN, Karthikeyan AS, Raghothama KG (2007) WRKY75 transcription factor is a modulator of phosphate acquisition and root development in Arabidopsis. Plant Physiol 143: 1789–1801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drew MC, Saker LR, Barber SA, Jenkins W (1984) Changes in the kinetics of phosphate and potassium absorption in nutrient-deficient barley roots measured by a solution-depletion technique. Planta 160: 490–499 [DOI] [PubMed] [Google Scholar]
- Epstein E, Rains DW, Elzam OE (1963) Resolution of dual mechanisms of potassium absorption by barley roots. Proc Natl Acad Sci USA 49: 684–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu HH, Luan S (1998) AtKuP1: a dual-affinity K+ transporter from Arabidopsis. Plant Cell 10: 63–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González E, Solano R, Rubio V, Leyva A, Paz-Ares J (2005) PHOSPHATE TRANSPORTER TRAFFIC FACILITATOR1 is a plant-specific SEC12-related protein that enables the endoplasmic reticulum exit of a high-affinity phosphate transporter in Arabidopsis. Plant Cell 17: 3500–3512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo B, Jin Y, Wussler C, Blancaflor EB, Motes CM, Versaw WK (2008) Functional analysis of the Arabidopsis PHT4 family of intracellular phosphate transporters. New Phytol 177: 889–898 [DOI] [PubMed] [Google Scholar]
- Hammond JP, Bennett MJ, Bowen HC, Broadley MR, Eastwood DC, May ST, Rahn C, Swarup R, Woolaway KE, White PJ (2003) Changes in gene expression in Arabidopsis shoots during phosphate starvation and the potential for developing smart plants. Plant Physiol 132: 578–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch J, Marin E, Floriani M, Chiarenza S, Richaud P, Nussaume L, Thibaud MC (2006) Phosphate deficiency promotes modification of iron distribution in Arabidopsis plants. Biochimie 88: 1767–1771 [DOI] [PubMed] [Google Scholar]
- Ho CH, Lin SH, Hu HC, Tsay YF (2009) CHL1 functions as a nitrate sensor in plants. Cell 138: 1184–1194 [DOI] [PubMed] [Google Scholar]
- Huang TK, Han CL, Lin SI, Chen YJ, Tsai YC, Chen YR, Chen JW, Lin WY, Chen PM, Liu TY, et al. (2013) Identification of downstream components of ubiquitin-conjugating enzyme PHOSPHATE2 by quantitative membrane proteomics in Arabidopsis roots. Plant Cell 25: 4044–4060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia H, Ren H, Gu M, Zhao J, Sun S, Zhang X, Chen J, Wu P, Xu G (2011) The phosphate transporter gene OsPht1;8 is involved in phosphate homeostasis in rice. Plant Physiol 156: 1164–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karthikeyan AS, Jain A, Nagarajan VK, Sinilal B, Sahi SV, Raghothama KG (2014) Arabidopsis thaliana mutant lpsi reveals impairment in the root responses to local phosphate availability. Plant Physiol Biochem 77: 60–72 [DOI] [PubMed] [Google Scholar]
- Léran S, Varala K, Boyer JC, Chiurazzi M, Crawford N, Daniel-Vedele F, David L, Dickstein R, Fernandez E, Forde B, et al. (2014) A unified nomenclature of NITRATE TRANSPORTER 1/PEPTIDE TRANSPORTER family members in plants. Trends Plant Sci 19: 5–9 [DOI] [PubMed] [Google Scholar]
- Lin WY, Huang TK, Chiou TJ (2013) Nitrogen limitation adaptation, a target of microRNA827, mediates degradation of plasma membrane-localized phosphate transporters to maintain phosphate homeostasis in Arabidopsis. Plant Cell 25: 4061–4074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Muchhal US, Uthappa M, Kononowicz AK, Raghothama KG (1998) Tomato phosphate transporter genes are differentially regulated in plant tissues by phosphorus. Plant Physiol 116: 91–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Versaw WK, Pumplin N, Gomez SK, Blaylock LA, Harrison MJ (2008) Closely related members of the Medicago truncatula PHT1 phosphate transporter gene family encode phosphate transporters with distinct biochemical activities. J Biol Chem 283: 24673–24681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logemann E, Birkenbihl RP, Ulker B, Somssich IE (2006) An improved method for preparing Agrobacterium cells that simplifies the Arabidopsis transformation protocol. Plant Methods 2: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz MC, Heitman J (1998) The MEP2 ammonium permease regulates pseudohyphal differentiation in Saccharomyces cerevisiae. EMBO J 17: 1236–1247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv Q, Zhong Y, Wang Y, Wang Z, Zhang L, Shi J, Wu Z, Liu Y, Mao C, Yi K, et al. (2014) SPX4 negatively regulates phosphate signaling and homeostasis through its interaction with PHR2 in rice. Plant Cell 26: 1586–1597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mäser P, Thomine S, Schroeder JI, Ward JM, Hirschi K, Sze H, Talke IN, Amtmann A, Maathuis FJ, Sanders D, et al. (2001) Phylogenetic relationships within cation transporter families of Arabidopsis. Plant Physiol 126: 1646–1667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller AJ, Fan X, Orsel M, Smith SJ, Wells DM (2007) Nitrate transport and signalling. J Exp Bot 58: 2297–2306 [DOI] [PubMed] [Google Scholar]
- Misson J, Raghothama KG, Jain A, Jouhet J, Block MA, Bligny R, Ortet P, Creff A, Somerville S, Rolland N, et al. (2005) A genome-wide transcriptional analysis using Arabidopsis thaliana Affymetrix gene chips determined plant responses to phosphate deprivation. Proc Natl Acad Sci USA 102: 11934–11939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misson J, Thibaud MC, Bechtold N, Raghothama K, Nussaume L (2004) Transcriptional regulation and functional properties of Arabidopsis Pht1;4, a high affinity transporter contributing greatly to phosphate uptake in phosphate deprived plants. Plant Mol Biol 55: 727–741 [DOI] [PubMed] [Google Scholar]
- Morcuende R, Bari R, Gibon Y, Zheng W, Pant BD, Bläsing O, Usadel B, Czechowski T, Udvardi MK, Stitt M, et al. (2007) Genome-wide reprogramming of metabolism and regulatory networks of Arabidopsis in response to phosphorus. Plant Cell Environ 30: 85–112 [DOI] [PubMed] [Google Scholar]
- Muchhal US, Pardo JM, Raghothama KG (1996) Phosphate transporters from the higher plant Arabidopsis thaliana. Proc Natl Acad Sci USA 93: 10519–10523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudge SR, Rae AL, Diatloff E, Smith FW (2002) Expression analysis suggests novel roles for members of the Pht1 family of phosphate transporters in Arabidopsis. Plant J 31: 341–353 [DOI] [PubMed] [Google Scholar]
- Nagarajan VK, Jain A, Poling MD, Lewis AJ, Raghothama KG, Smith AP (2011) Arabidopsis Pht1;5 mobilizes phosphate between source and sink organs and influences the interaction between phosphate homeostasis and ethylene signaling. Plant Physiol 156: 1149–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Kurose T, Hino T, Tanaka K, Kawamukai M, Niwa Y, Toyooka K, Matsuoka K, Jinbo T, Kimura T (2007) Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J Biosci Bioeng 104: 34–41 [DOI] [PubMed] [Google Scholar]
- Narang RA, Bruene A, Altmann T (2000) Analysis of phosphate acquisition efficiency in different Arabidopsis accessions. Plant Physiol 124: 1786–1799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussaume L, Kanno S, Javot H, Marin E, Pochon N, Ayadi A, Nakanishi TM, Thibaud MC (2011) Phosphate import in plants: focus on the PHT1 transporters. Front Plant Sci 2: 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozcan S, Dover J, Rosenwald AG, Wölfl S, Johnston M (1996) Two glucose transporters in Saccharomyces cerevisiae are glucose sensors that generate a signal for induction of gene expression. Proc Natl Acad Sci USA 93: 12428–12432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park BS, Seo JS, Chua NH (2014) NITROGEN LIMITATION ADAPTATION recruits PHOSPHATE2 to target the phosphate transporter PT2 for degradation during the regulation of Arabidopsis phosphate homeostasis. Plant Cell 26: 454–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker JL, Newstead S (2014) Molecular basis of nitrate uptake by the plant nitrate transporter NRT1.1. Nature 507: 68–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Péret B, Clément M, Nussaume L, Desnos T (2011) Root developmental adaptation to phosphate starvation: better safe than sorry. Trends Plant Sci 16: 442–450 [DOI] [PubMed] [Google Scholar]
- Poirier Y, Bucher M (2002) Phosphate transport and homeostasis in Arabidopsis. In CR Somerville, ed, The Arabidopsis Book. American Society of Plant Biologists, Rockville, MD, pp 1–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirier Y, Thoma S, Somerville C, Schiefelbein J (1991) Mutant of Arabidopsis deficient in xylem loading of phosphate. Plant Physiol 97: 1087–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rae AL, Cybinski DH, Jarmey JM, Smith FW (2003) Characterization of two phosphate transporters from barley; evidence for diverse function and kinetic properties among members of the Pht1 family. Plant Mol Biol 53: 27–36 [DOI] [PubMed] [Google Scholar]
- Rausch C, Bucher M (2002) Molecular mechanisms of phosphate transport in plants. Planta 216: 23–37 [DOI] [PubMed] [Google Scholar]
- Remy E, Cabrito TR, Batista RA, Teixeira MC, Sá-Correia I, Duque P (2012) The Pht1;9 and Pht1;8 transporters mediate inorganic phosphate acquisition by the Arabidopsis thaliana root during phosphorus starvation. New Phytol 195: 356–371 [DOI] [PubMed] [Google Scholar]
- Reymond M, Svistoonoff S, Loudet O, Nussaume L, Desnos T (2006) Identification of QTL controlling root growth response to phosphate starvation in Arabidopsis thaliana. Plant Cell Environ 29: 115–125 [DOI] [PubMed] [Google Scholar]
- Sarrobert C, Thibaud MC, Contard-David P, Gineste S, Bechtold N, Robaglia C, Nussaume L (2000) Identification of an Arabidopsis thaliana mutant accumulating threonine resulting from mutation in a new dihydrodipicolinate synthase gene. Plant J 24: 357–367 [DOI] [PubMed] [Google Scholar]
- Schwab R, Ossowski S, Warthmann N, Weigel D (2010) Directed gene silencing with artificial microRNAs. Methods Mol Biol 592: 71–88 [DOI] [PubMed] [Google Scholar]
- Shin H, Shin HS, Dewbre GR, Harrison MJ (2004) Phosphate transport in Arabidopsis: Pht1;1 and Pht1;4 play a major role in phosphate acquisition from both low- and high-phosphate environments. Plant J 39: 629–642 [DOI] [PubMed] [Google Scholar]
- Simillion C, Vandepoele K, Van Montagu MC, Zabeau M, Van de Peer Y (2002) The hidden duplication past of Arabidopsis thaliana. Proc Natl Acad Sci USA 99: 13627–13632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanovic A, Arpat AB, Bligny R, Gout E, Vidoudez C, Bensimon M, Poirier Y (2011) Over-expression of PHO1 in Arabidopsis leaves reveals its role in mediating phosphate efflux. Plant J 66: 689–699 [DOI] [PubMed] [Google Scholar]
- Sun J, Bankston JR, Payandeh J, Hinds TR, Zagotta WN, Zheng N (2014) Crystal structure of the plant dual-affinity nitrate transporter NRT1.1. Nature 507: 73–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svistoonoff S, Creff A, Reymond M, Sigoillot-Claude C, Ricaud L, Blanchet A, Nussaume L, Desnos T (2007) Root tip contact with low-phosphate media reprograms plant root architecture. Nat Genet 39: 792–796 [DOI] [PubMed] [Google Scholar]
- Thibaud MC, Arrighi JF, Bayle V, Chiarenza S, Creff A, Bustos R, Paz-Ares J, Poirier Y, Nussaume L (2010) Dissection of local and systemic transcriptional responses to phosphate starvation in Arabidopsis. Plant J 64: 775–789 [DOI] [PubMed] [Google Scholar]
- Ticconi CA, Delatorre CA, Abel S (2001) Attenuation of phosphate starvation responses by phosphite in Arabidopsis. Plant Physiol 127: 963–972 [PMC free article] [PubMed] [Google Scholar]
- Van de Peer Y, Fawcett JA, Proost S, Sterck L, Vandepoele K (2009) The flowering world: a tale of duplications. Trends Plant Sci 14: 680–688 [DOI] [PubMed] [Google Scholar]
- Varadarajan DK, Karthikeyan AS, Matilda PD, Raghothama KG (2002) Phosphite, an analog of phosphate, suppresses the coordinated expression of genes under phosphate starvation. Plant Physiol 129: 1232–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witte CP, Noël LD, Gielbert J, Parker JE, Romeis T (2004) Rapid one-step protein purification from plant material using the eight-amino acid StrepII epitope. Plant Mol Biol 55: 135–147 [DOI] [PubMed] [Google Scholar]
- Wu P, Ma L, Hou X, Wang M, Wu Y, Liu F, Deng XW (2003) Phosphate starvation triggers distinct alterations of genome expression in Arabidopsis roots and leaves. Plant Physiol 132: 1260–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wykoff DD, O’Shea EK (2001) Phosphate transport and sensing in Saccharomyces cerevisiae. Genetics 159: 1491–1499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W, Miao Q, Sun D, Yang G, Wu C, Huang J, Zheng C (2012) The mitochondrial phosphate transporters modulate plant responses to salt stress via affecting ATP and gibberellin metabolism in Arabidopsis thaliana. PLoS ONE 7: e43530. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.