Medicago truncatula border cells contain elevated levels of specialized metabolites that are important in plant-microbe signaling and defense.
Abstract
Integrated metabolomics and transcriptomics of Medicago truncatula seedling border cells and root tips revealed substantial metabolic differences between these distinct and spatially segregated root regions. Large differential increases in oxylipin-pathway lipoxygenases and auxin-responsive transcript levels in border cells corresponded to differences in phytohormone and volatile levels compared with adjacent root tips. Morphological examinations of border cells revealed the presence of significant starch deposits that serve as critical energy and carbon reserves, as documented through increased β-amylase transcript levels and associated starch hydrolysis metabolites. A substantial proportion of primary metabolism transcripts were decreased in border cells, while many flavonoid- and triterpenoid-related metabolite and transcript levels were increased dramatically. The cumulative data provide compounding evidence that primary and secondary metabolism are differentially programmed in border cells relative to root tips. Metabolic resources normally destined for growth and development are redirected toward elevated accumulation of specialized metabolites in border cells, resulting in constitutively elevated defense and signaling compounds needed to protect the delicate root cap and signal motile rhizobia required for symbiotic nitrogen fixation. Elevated levels of 7,4′-dihydroxyflavone were further increased in border cells of roots exposed to cotton root rot (Phymatotrichopsis omnivora), and the value of 7,4′-dihydroxyflavone as an antimicrobial compound was demonstrated using in vitro growth inhibition assays. The cumulative and pathway-specific data provide key insights into the metabolic programming of border cells that strongly implicate a more prominent mechanistic role for border cells in plant-microbe signaling, defense, and interactions than envisioned previously.
The plant root tip includes the apical meristem and root cap initials, which are progenitor cells to all new growth in the root. Root cell division and elongation originate in the apical meristem and proceed toward the mature root. Root cap growth begins in the root cap initials and continues to the root apex. Damage or destruction of the root tip can reduce growth or kill a plant. Fortunately, the root tip in most plants is not left unguarded or defenseless. The root cap of many species produces thousands of differentiated cells that separate from the root but remain appressed to it in a water-soluble polysaccharide matrix, or mucilage, until released by exposure to water. These cells are termed border cells because they form a boundary between the root and the rhizosphere, and they are defined as the cells that disperse into suspension when root tips are placed in water (Hawes et al., 2000).
Border cells are more than by-products of root cap growth, and they provide a biotic boundary fundamental in rhizosphere modifications. For example, cocultivation of oat (Avena sativa) border cells with micromolar levels of aluminum resulted in increased border cell and mucilage production in a dose-dependent manner (Miyasaka and Hawes, 2001). In addition, border cells serve key roles in plant defense and plant-symbiont interactions. They attract and immobilize nematodes (Hawes et al., 2000), orchestrate interactions with both mutualistic fungi (Kosuta et al., 2003; Nagahashi and Douds, 2004) and pathogenic fungi (Hawes et al., 2000; Gunawardena and Hawes, 2002; Woo et al., 2004; Gunawardena et al., 2005), and bind and repel bacteria (Hawes et al., 2000).
The production of border cells appears to be tightly regulated, but little is known about this process (Brigham et al., 1998; Hawes et al., 2000). Once a full complement of border cells is produced, root cap cell division ceases and border cells remain tightly appressed to the root tip until exposed to water (Hawes et al., 2000). When border cells are removed from the root, cell division in the root cap initials resumes within 5 min, remains high for 2 h, and a complete complement of new border cells is produced within 24 h, maintaining a species-specific number of border cells (Hawes et al., 2003). Border cells are a determinate cell type that serve several functions during their journey from meristem through columella and on to peripheral cells (i.e. gravity sensing and mucilage secretion), before arriving at the outer layer of the root. Border cells undergo an increase in metabolic activity after release from the root cap, resulting in the production of specific metabolites and the secretion of mucilage and proteins into the rhizosphere (Hawes et al., 2000; Wen et al., 2007).
Two landmark publications that characterized root development in Arabidopsis (Arabidopsis thaliana) were based on comparisons of anatomically distinct cell types over a developmental time series using microarray gene expression data (Birnbaum et al., 2003; Brady et al., 2007). These studies provided a model for understanding root architecture and its relationship to root development in both space and time. A more recent study demonstrated the involvement of programmed cell death of Arabidopsis lateral root cap cells in the maintenance of root cap size (Fendrych et al., 2014). However, none of these studies addressed the biochemistry of border cells, the role of border cells in root physiology, or the signals produced by these specialized and spatially resolved cells, because Arabidopsis roots do not produce border cells but instead produce a few border-like cells in plants older than 5 d (Vicré et al., 2005; Driouich et al., 2007). In contrast, legume roots produce numerous border cells that are viable even after release from the root (Hawes et al., 1998).
Legume root biology is fundamentally important to agriculture, in part because legumes form symbiotic relationships with both mycorrhizal fungi and nodulating soil bacteria that are beneficial to plant growth and yield. Legumes also synthesize numerous natural products critical in plant defense, development, and nutrition, including flavonoids, isoflavonoids, lignin, and saponins (Dixon and Sumner, 2003). Information about the spatial localization and biosynthesis of these natural products in roots is sparse, and much of the present knowledge of root secondary metabolism comes from work done in the model legume Medicago truncatula (Achnine et al., 2005; Schliemann et al., 2008). In addition, genetic, genomic, and biochemical resources are available for M. truncatula, including a genome sequence (Young et al., 2005, 2011), high-density microarray chip sets (Stacey et al., 2006), and a gene expression atlas for many organs, including specific root tissues (Benedito et al., 2008). These resources support M. truncatula as an ideal model to investigate the basal capacity of border cells and their ability to respond metabolically to environmental stimuli.
This study integrated metabolic, transcriptional, and morphological analyses of anatomically distinct M. truncatula seedling root tissues to better characterize the spatial distribution of metabolism in legume roots. Cumulative and pathway-specific data provided compounding evidence that border cells are metabolically differentiated relative to root tips. Border cells possess a pronounced enhancement in secondary metabolism that suggests a prominent biochemical role for these unique cells in defense, plant-microbe signaling, and rhizosphere transformation. The high constitutive level of 7,4′-dihydroxyflavone (DHF) and its subsequent increase in border cells exposed to cotton root rot (Phymatotrichopsis omnivora) are reported as examples of the roles of border cells in root defense.
RESULTS
Microscopy
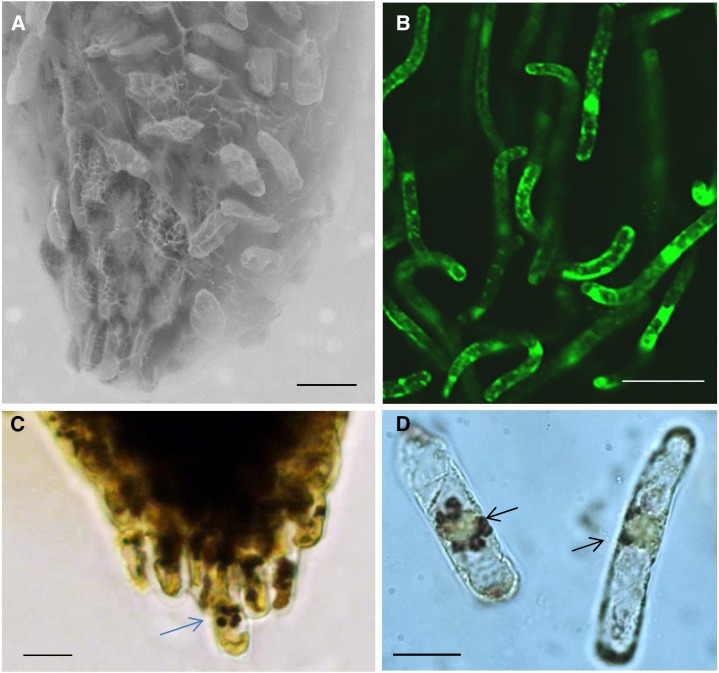
A polysaccharide matrix surrounds and adheres border cells to the tip of M. truncatula seedling roots (Fig. 1A; Supplemental Fig. S1A). Gentle agitation in or contact with water solubilizes the matrix and frees border cells from the root (Fig. 1B; Supplemental Figs. S1, B and C, and S2). M. truncatula border cells (used throughout to mean border cells with their associated mucilage) can be reproducibly harvested with over 95% viability, as determined using fluorescein diacetate viability staining (Supplemental Fig. S1, D–F) and cell counting. The number of M. truncatula seedling border cells was counted and determined to be approximately 1,700 to 2,000 per root, comparable to the numbers reported for alfalfa (Medicago sativa) seedling roots (Woo et al., 2004). Many M. truncatula border cells have an elongated appearance and thick cell walls, similar to other species (Hamamoto et al., 2006), and large iodine-stained starch bodies were clearly visible in numerous detached border cells (Fig. 1C; Supplemental Fig. S3, A and B). The relative amount of starch in border cells was lower than in most other root tip cell types, especially the columella cells (compare Supplemental Fig. S3, C and D, with Fig. 1, C and D, and Supplemental Fig. S3, A and B; Blancaflor et al., 1998; Barlow, 2003), but substantially higher than that observed in the elongation and mature root zones (Supplemental Fig. S4).
Figure 1.
Border cells and root tips of M. truncatula. A, Scanning electron microscopy of a seedling root with attached border cells and polysaccharide matrix. B, Confocal image of detached border cells stained with fluorescein diacetate. C, Border cell at the root tip containing starch granules. The blue arrow points to a border cell. D, Free border cells with starch granules. The black arrows point to the starch granules. Bars = 50 µm (A and B) and 25 µm (C and D).
Gene Expression Analysis
RNA was isolated from root tissues of 3-d-old pooled seedlings and used for the microarray gene expression comparisons of border cells with that of root tips lacking border cells and whole roots. Root tip is used in the remainder of this article to describe the terminal 2 to 4 mm of the root minus border cells, whole roots refers to unaltered roots containing border cells, and border cells were defined above. Three biological replicates were analyzed using the Affymetrix M. truncatula genome array as described by Benedito et al. (2008), a selection threshold of 2 for transcript ratios, and a Bonferroni correction P value threshold of 8.15954E-07. The raw expression data were analyzed, and each transcript was assigned an absolute expression level and a present or absent call based on the signal-to-noise ratio. Approximately 50% of the plant probe sets from the M. truncatula GeneChip array produced present calls when hybridized with biotin-labeled copy RNA from the three sample types, similar to previously reported hybridization percentages for M. truncatula (Holmes et al., 2008). Following normalization, 1,995 transcripts were identified as statistically increased and 4,519 as decreased in border cells when compared with whole seedling roots (Supplemental Table S1). Changes at the transcript level between border cells and root tip samples were more pronounced, with 5,140 transcripts higher and 7,084 transcripts lower in border cells when compared with root tips (Supplemental Table S1). The full data set has been deposited in the Array Express database and is publicly available as accession E-MEXP-2883 and in the M. truncatula Gene Expression Atlas version 3 (http://mtgea.noble.org/v3/).
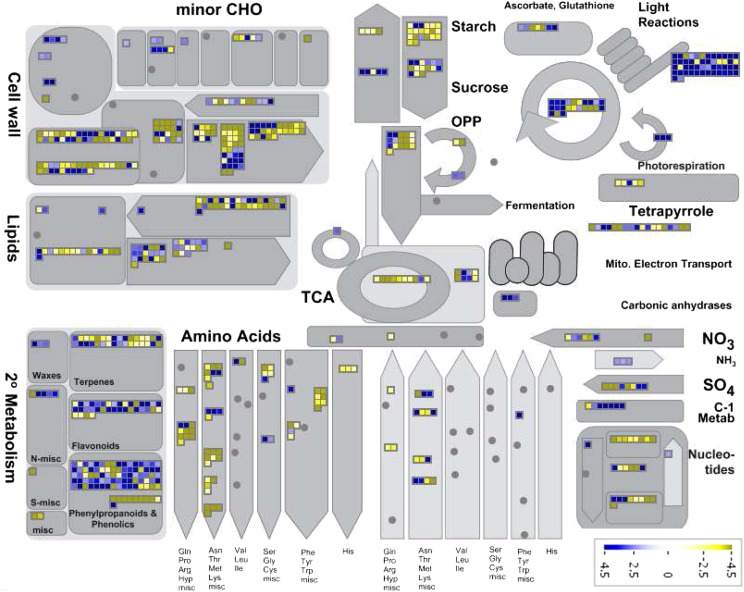
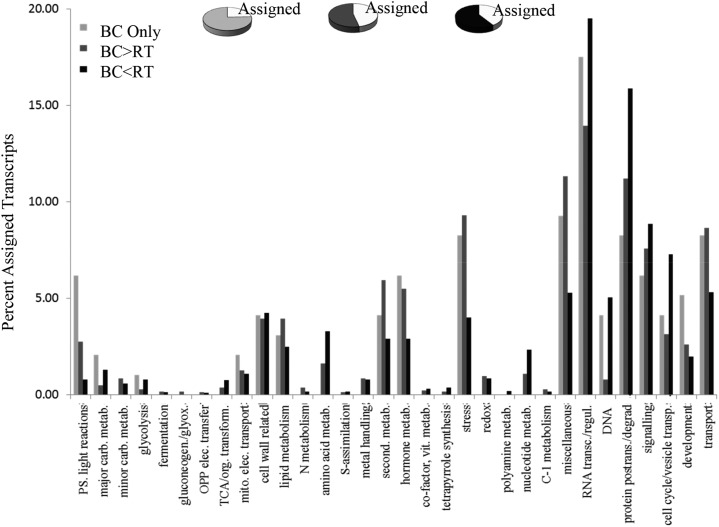
MapMan software (Thimm et al., 2004; Urbanczyk-Wochniak et al., 2006) was adopted for visualizing M. truncatula transcript data by generating species-specific mapping files for the Affymetrix Medicago spp. chip (Uppalapati et al., 2009). Differentially expressed genes from the three different sample types were functionally classified using MapMan categories (Fig. 2; Supplemental Fig. S5) and displayed on pathway diagrams. Less than 50% of differentially expressed transcripts could be assigned functional categories (Fig. 3; Supplemental Fig. S6). The assigned transcript classes most strongly differentiating border cells from whole root and root tips were associated with RNA regulation and protein posttranslational regulation (Fig. 3; Supplemental Fig. S6). In these classes, more transcripts were decreased than increased in border cells. The total number of transcripts involved in nucleotide and DNA metabolism was also lower in border cells, consistent with a determinate cell type with a slowing rate of replication and cell division. Cell wall metabolism, lipid metabolism, stress, hormone metabolism, and miscellaneous (UDP glycosyl transferases, peroxidases, oxidases, etc.) also accounted for substantial differences between border cells and whole roots and/or root tips (Fig. 3; Supplemental Fig. S6). Transcripts in these categories were higher in border cells, as were the overall number of border cell transcripts related to secondary metabolism and transport, two categories linked to defense.
Figure 2.
Overview of border cell-to-root tip transcript ratios in major metabolic pathways visualized using MapMan. Transcripts significantly up- and down-regulated are indicated in blue and yellow, respectively. The scale bars display fold changes. CHO, Carbohydrate; OPP, oxidative pentose phosphate; TCA, tricarboxylic acid.
Figure 3.
Overview of transcript profiling results. The graph represents the percentage of transcripts assigned to each nonredundant functional category based on MapMan software. The smaller pie charts represent all transcripts, and the white sections represent the percentage of assigned transcripts. BC Only, Transcripts observed only in border cells; BC>RT, transcripts increased in border cells relative to root tips; BC<RT, transcripts decreased in border cells compared with root tips; OPP, oxidative pentose phosphate; TCA, tricarboxylic acid.
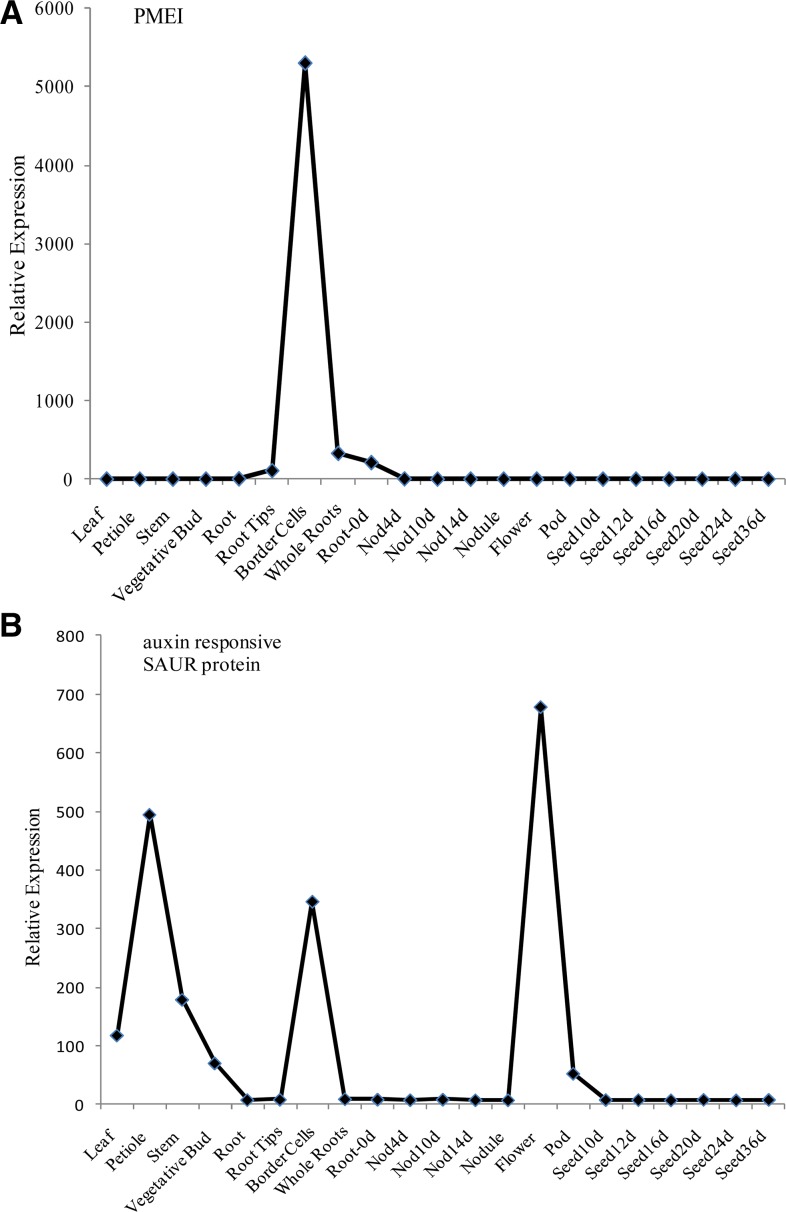
There were 396 transcripts observed only in border cells and not in other M. truncatula root tissues (Supplemental Table S1). Most of these transcripts were detected at low levels, and 75% were novel transcripts categorized as not annotated. Several other transcripts were observed at reproducible levels in only a few tissue types besides border cells. For example, a pectin methylesterase (PME) inhibitor was present in young roots prior to nodulation (equivalent to whole roots in this analysis) and in border cells but absent in all other tissues analyzed (Fig. 4A).
Figure 4.
Relative expression levels of PMEI (A) and auxin-responsive SAUR protein (B) determined using the M. truncatula Gene Expression Atlas (Benedito et al., 2008).
Quantitative real-time (qRT)-PCR was performed to provide a more rigorous quantitative measure of gene expression for select genes. Expression levels for five genes representative of central steps in primary metabolism and eight genes related to major changes in secondary metabolism were validated by qRT-PCR (Table I). Additionally, the expression levels of a PME, a known marker for root tip-border cell separation, and a pectin methylesterase inhibitor (PMEI) were also reanalyzed by qRT-PCR. Two genes important in hormone response and metabolism in different areas of the root also were analyzed. These genes, an auxin-responsive SAUR (for small auxin up RNA) protein and a lipoxygenase (LOX), showed very large expression increases in border cells compared with root tips. In total, 17 genes were analyzed by qRT-PCR, and the results from the microarray analysis were confirmed by qRT-PCR (Table I) in every case. These results are discussed in more detail in “Discussion.” A complete list of primers can be found in Supplemental Table S2.
Table I. Summary of validated genes, expression levels, and related metabolite levels.
BC/RT, Ratio of border cells compared with root tips.
| Class | Gene; Accession No. | Microarray BC/RT | qRT-PCR BC/RT | Metabolites BC/RT |
|---|---|---|---|---|
| Genes in primary metabolism | ||||
| Major carbohydrate metabolism | β-Amylase; TC94273 | 18× Increase | 15× Increase | Starch detected by microscopy |
| Glycolysis | PF6P1P; TC101885 | 12× Increase | 28× Increase | Fru derivatives 3× to 4× decrease |
| Tricarboxylic acid cycle | Citrate synthase-like; BQ153338 | 3× Increase | 2× Increase | Citrate 5× increase |
| Amino acids | Asn synthetase; TC100391 | 6× Increase | 3× Increase | Asn cumulative 3× increase in border cells |
| Not assigned (β-Ala) | β-Ureido-propionase; TC100938 | 7× Increase | 3× Increase | β-Ala 6× increase |
| Genes in secondary metabolism | ||||
| Phenylpropanoids | Phe ammonia lyase; TC101026 | 16× Increase | 30× Increase | Phe abundant in border cells |
| Lignin and lignans | CCoMT-like protein; BM814917 | 18× Increase | 24× Increase | Lignin in border cells (from literature) |
| Flavonoids | Naringenin-chalcone synthase; TC102405 | 50× Increase | 64× Increase | Apigenin 6× increase |
| Flavonoids | Flavone synthase II; BM779623 | 8× Increase | 7× Increase | DHF 13× increase |
| Isoflavonoids | Isoflavone synthase-like; TC106940 | 13× Increase | 13× Increase | Not detected |
| Terpenoids | β-Amyrin synthase; AW689929 | 3× Decrease | 1.2× Decrease | Up to 32× increase in saponins |
| Terpenoids | CYP71A8; BE943181 | 21× Increase | 91× Increase | Up to 32× increase in saponins |
| Terpenoids | (−)-Germacrene D synthase; TC94781 | 34× Increase | 167× Increase | Volatile terpenes not detected with this method |
| Additional categories | ||||
| Jasmonate | LOX; TC106479 | 224× Increase | 126× Increase | Less than in root tips |
| IAA | Auxin-responsive SAUR protein; BQ157435 | 38× Increase | 884× Increase | Not detected in border cells |
| Cell walls | PME; TC103769 | 16× Decrease | 7× Decrease | Wen et al. (1999) |
| Cell wall | PMEI; AC134522_38.4 | 45× Increase | 115× Increase | Novel |
Metabolomics
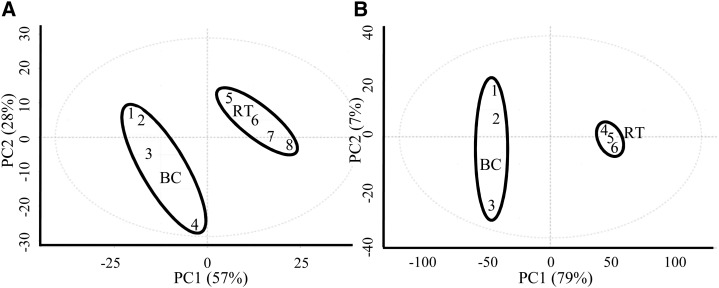
This report focuses on the metabolic comparison of anatomically distinct root tips and border cells while noting that a few prior publications have reported cumulative metabolic profiles of whole M. truncatula roots (Achnine et al., 2005; Huhman et al., 2005; Schliemann et al., 2008; Zhang et al., 2014). Metabolomics analyses were performed using a series of gas chromatography-mass spectrometry (GC-MS), liquid chromatography-mass spectrometry (LC-MS), and ultra-high-performance liquid chromatography (UPLC) experiments. The GC-MS profiling identified distinct, reproducible tissue differences (Fig. 5A) between border cells and root tips. The levels of most sugars were unchanged or lower in border cells (Table II), although Fru, Glc, Gal, Suc, and Ara were all abundant in these cells. Suc was the only sugar significantly elevated in comparison with root tips. Fru was the most abundant sugar observed in the metabolite analyses (Supplemental Table S3), and this was reflected in the high percentage of glycolytic transcripts (more than 70%) linked to Fru metabolism in border cells (Supplemental Table S1). Fru and Glc are produced from the degradation of Suc, and Glc was observed at lower levels in border cells than in root tips. However, starch, a product of Glc, was abundant in both (Fig. 1, C and D; Supplemental Fig. S3).
Figure 5.
Principal component (PC) analysis of primary (A) and secondary (B) metabolite profiling data from border cells (BC) and root tips (RT). PC1 for primary metabolites explains 57% of variance and PC2 explains 28% of variance using approximately 500 mass features. PC1 for secondary metabolites explains 79% of variance and PC2 explains 7% of variance using approximately 3,100 mass features.
Table II. Primary metabolites identified in border cells and root tips.
Italicized amino acids are nonstandard. BC/RT, Ratio of border cells compared with root tips; MEOX, methyloxime; TMS, trimethylsilyl.
| Metabolites | BC/RT | P | Ion | Retention Time |
|---|---|---|---|---|
| fold change | min | |||
| Sugars | ||||
| Suc TMS | 3.68 | 0.0092 | 361.2 | 45.1907 |
| Fru 5-TMS MEOX2 | 0.29 | 0.0003 | 217.2 | 32.0651 |
| Fru 5-TMS MEOX1 | 0.27 | 0.0006 | 217.2 | 31.8745 |
| Xyl 4-TMS MEOX2 | 0.65 | 0.0219 | 217.1 | 27.1635 |
| Ara 4-TMS MEOX1 | 0.62 | 0.0015 | 217.1 | 27.6668 |
| Rib 4-TMS MEOX | 0.55 | 0.0089 | 308.2 | 27.3277 |
| Fru-6-P 6-TMS MEOX | 0.38 | 0.0149 | 315.1 | 39.9454 |
| Glc-6-P TMS MEOX1 | 0.21 | 0.0020 | 387.1 | 40.1773 |
| Gal 5-TMS MEOX1 | 0.20 | 0.0001 | 319.2 | 32.2821 |
| Glc 5-TMS MEOX2 | 0.20 | 0.0001 | 319.2 | 32.4089 |
| Organic acids | ||||
| Succinic acid 2-TMS | 5.78 | 0.0060 | 247.1 | 18.8065 |
| Citric acid TMS | 5.39 | 0.0109 | 273.1 | 30.8361 |
| Shikimic acid 4-TMS | 4.17 | 0.0014 | 204.1 | 30.5913 |
| BA TMS | 3.96 | 0.0010 | 194.1 | 17.1936 |
| Hexanoic acid TMS | 3.92 | 0.0002 | 173.1 | 12.1131 |
| Nicotinic acid TMS | 3.55 | 0.0032 | 180.1 | 18.487 |
| Propionic acid 3-TMS | 3.53 | 0.0029 | 189.1 | 19.1171 |
| Fumaric acid 2-TMS | 3.08 | 0.0056 | 245.1 | 19.7831 |
| Hexadecanoic acid TMS | 2.81 | 0.0045 | 313.3 | 35.5837 |
| Malic acid 3-TMS | 2.53 | 0.0188 | 233.1 | 23.2597 |
| Butanoic acid 4-TMS | 2.13 | 0.0362 | 174.1 | 28.4915 |
| Nonanoic acid | 1.96 | 0.0251 | 215.1 | 20.1453 |
| Pyruvic acid TMS MEOX1 | 1.66 | 0.0113 | 174.1 | 11.3521 |
| Amino acids | ||||
| Gln 3-TMS | 97.50 | 0.0226 | 246.1 | 29.9694 |
| Ile TMS | 23.76 | 0.0018 | 188.2 | 15.1115 |
| Pro +CO2 2-TMS | 9.66 | 0.0013 | 186.1 | 25.7291 |
| Pro 2-TMS | 7.48 | 0.0046 | 216.1 | 18.4395 |
| Pro TMS | 6.10 | 0.0002 | 172.1 | 15.1135 |
| Gly 3-TMS | 7.49 | 0.0017 | 174.1 | 18.6267 |
| Gly 2-TMS | 6.35 | 0.0008 | 204.1 | 13.399 |
| Homo-Ser 3-TMS | 7.47 | 0.0018 | 218.2 | 22.317 |
| Ser 4-TMS | 6.65 | 0.0011 | 290.1 | 25.3873 |
| Ser 2-TMS | 4.36 | 0.0008 | 219.1 | 17.2432 |
| Ser 3-TMS | 4.06 | 0.0017 | 204.1 | 19.9493 |
| Thr 3-TMS | 4.92 | 0.0044 | 218.1 | 20.6207 |
| Thr 2-TMS | 3.01 | 0.0032 | 219.1 | 18.2625 |
| Asn +CO2 4-TMS | 4.58 | 0.0003 | 232.1 | 33.5789 |
| Asn 4-TMS | 3.05 | 0.0161 | 188.1 | 32.0088 |
| Asn 2-TMS | 2.84 | 0.0010 | 159.1 | 26.077 |
| Asn 3-TMS | 2.60 | 0.0118 | 231.2 | 27.6817 |
| Ala +CO2 2-TMS | 3.26 | 0.0020 | 160.1 | 21.0853 |
| Ala 3-TMS | 3.21 | 0.0007 | 188.2 | 20.0924 |
| Val TMS ester | 2.72 | 0.0001 | 156.1 | 12.5797 |
| Val 2-TMS | 2.36 | 0.0169 | 218.1 | 16.0488 |
| Tyr 2-TMS | 1.70 | 0.0416 | 219.1 | 33.4378 |
| Lys 3-TMS | 0.51 | 0.0014 | 174.1 | 31.7517 |
| Glu 3-TMS | 0.72 | 0.0062 | 246.1 | 26.4894 |
| Phe 2-TMS | 0.67 | 0.0101 | 218.1 | 26.8129 |
| CoA synthesis | ||||
| β-Ala 3-TMS | 5.53 | 0.0007 | 174.1 | 21.8571 |
| Miscellaneous | ||||
| Urea 2-TMS | 5.19 | 0.0022 | 171.1 | 16.8071 |
Several organic acids were found at higher levels in border cells, including four (Table II) intermediates in the tricarboxylic acid cycle. The level of citrate, an early tricarboxylic acid intermediary, was 5-fold higher in border cells than in root tips. Fumarate, a precursor for the amino acids Asp and Asn, also was more abundant in border cells compared with root tips (Table II). The level of malic acid was approximately 2.5-fold higher when compared with root tips. Malate is an important precursor in the formation of pyruvate and thus of the branched-chain amino acids and CoA (Fig. 6).
Figure 6.
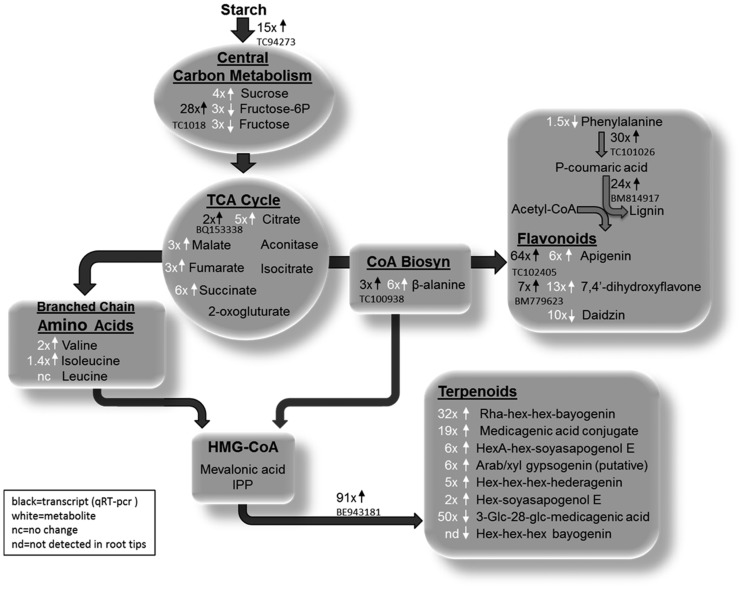
Systems model of major metabolic and transcriptional differences in M. truncatula border cells. Cumulative constitutive data provide evidence that border cells have enhanced metabolic capacity and content relative to root tips. Starch reserves in border cells are directed toward increased secondary metabolism as opposed to fueling continued cell growth and division. The enhanced secondary metabolism of border cells fortifies them as front-line defenders in plant-pathogen interactions and important ambassadors in mutualistic signaling. TCA, Tricarboxylic acid; HMG-CoA, hydroxy-methyl glutaryl coenzyme A; IPP, isopentenyl pyrophosphate; Hex, hexose; HexA, hexose acid.
Nineteen standard and two nonstandard amino acids were identified in border cells (Table II; Supplemental Table S3). Of these, 10 were significantly higher in border cells, while the levels of 11 amino acids were statistically unchanged (±2-fold; P < 0.05) compared with root tips. The most abundant amino acids in border cells were among those most elevated in comparison with root tips. These included Asn, one of the most abundant metabolites in border cells, and Ser, homo-Ser, and Gly (Table I; Supplemental Table S3). Pro and Thr were also highly abundant in border cells, with levels that were elevated in comparison with root tips. The branched-chain amino acids Ile, Val, and Leu, precursors for CoA biosynthesis, also were observed at high levels in border cells. Phe, the precursor for phenylpropanoids, was abundant in both root tips and border cells (Table I; Supplemental Table S3).
An ultra-high-pressure liquid chromatography (UHPLC) coupled to hybrid quadrupole time-of-flight mass spectrometry method was used to profile a range of saponins and flavonoids because of their importance to defense and signaling in legumes. Consistent with the Metabolomics Standard Initiative (Sumner et al., 2007), this profiling method provided a significant number of confident identifications for a number of differentially accumulated secondary metabolites by comparison with authentic standards and a number of tentative identifications through accurate mass matches to metabolites within public databases (Table III). Distinct and reproducible tissue differences in secondary metabolite levels were observed (Fig. 5B). Almost 20 saponins and sapogenin aglycones were observed at increased levels in border cells. Five of these were observed at a 10-fold or greater excess in comparison with root tips. Root tips also contained many saponins; some of these were in excess of the levels observed in border cells or not detected in border cells (Table III).
Table III. Secondary metabolites identified in border cells and root tips.
BC/RT, Ratio of border cells compared with root tips; m/z, mass-to-charge ratio; nd in bc, not detected in border cells; nd in rt, not detected in root tips. The minimum area of the border cell peak is 1-100th of the internal standard.
| Metabolites | Identification Sourcea | BC/RT | P | Retention Time | m/z |
|---|---|---|---|---|---|
| fold change | min | ||||
| Phenolics | |||||
| 4-Hydroxy-7-methoxy flavone | 1 | nd in rt | 0.0042 | 12.30 | 267.0685 |
| Epicatechin pentose | 2 | 21.45 | 0.0005 | 8.88 | 421.2085 |
| Epicatechin pentose | 2 | 15.30 | 0.0000 | 9.42 | 421.2073 |
| DHF | 1 | 12.51 | 0.0004 | 7.42 | 253.0492 |
| Apigenin | 1 | 5.92 | 0.0002 | 9.96 | 269.0445 |
| Luteolin 7-glucoside | 2 | 1.96 | 0.0040 | 3.91 | 447.097 |
| Naringenin chalcone 4-O-glucoside | 2 | 1.92 | 0.0034 | 3.92 | 433.115 |
| 4-Methylumbelliferone | 1 | 1.68 | 0.0012 | 5.98 | 175.0391 |
| Kaempferol-3-O-rutinoside | 1 | 0.33 | 0.0031 | 6.05 | 593.1519 |
| Unknown phenolic | 2 | 0.30 | 0.0005 | 5.27 | 577.156 |
| Daidzin | 1 | 0.10 | 0.0028 | 4.56 | 415.1044 |
| Saponins or sapogenins | |||||
| Hex-Hex-Hex-bayogenin | 2 | nd in rt | 0.0002 | 13.88 | 973.5013 |
| Rha-Hex-Hex-bayogenin | 2 | 31.90 | 0.0016 | 13.97 | 957.5092 |
| Rha-Hex-Hex-Hex-bayogenin | 2 | 22.38 | 0.0072 | 8.46 | 1,119.5665 |
| 3-Glc-Glc-28-Ara-Rha-Xyl medicagenic acid | 1 | 19.25 | 0.0017 | 13.12 | 1,087.4988 |
| Rha-Hex-Hex-bayogenin | 2 | 18.63 | 0.0001 | 13.38 | 957.5135 |
| 3-Glc-malonyl-medicagenic acid | 2 | 12.50 | 0.0266 | 16.76 | 705.3861 |
| HexA-Hex-soyasapogenol E | 2 | 6.32 | 0.0000 | 18.74 | 793.4370 |
| 3-Glc-malonyl-medicagenic acid | 2 | 6.30 | 0.0006 | 17.92 | 705.3877 |
| 3-Rha-Gal-GlcA-soyasapogenol B | 2 | 5.92 | 0.0004 | 17.02 | 941.5163 |
| Ara/Xyl-gypsogenin | 3 | 5.81 | 0.0046 | 11.63 | 1,221.5609 |
| 3-Glc-malonyl-medicagenic acid | 2 | 5.31 | 0.0392 | 14.79 | 705.3874 |
| Hex-Hex-Hex-hederagenin | 2 | 4.90 | 0.0014 | 14.74 | 957.5079 |
| Hex-Hex-Hex-bayogenin | 2 | 4.67 | 0.0003 | 11.11 | 973.5040 |
| Hex-gypsogenic acid | 2 | 4.56 | 0.0009 | 16.29 | 647.3811 |
| 3-Glc-28-Ara-Rha-Xyl medicagenic acid | 2 | 4.50 | 0.0291 | 13.68 | 1,073.5138 |
| 3-Rha-Xyl-GlcA-gypsogenic acid | 2 | 4.33 | 0.0053 | 18.53 | 939.4972 |
| 3-Glc-malonyl-medicagenic acid | 2 | 2.89 | 0.0287 | 17.41 | 705.3878 |
| 3-Glc-medicagenic acid | 1 | 2.10 | 0.0029 | 17.02 | 663.3768 |
| Echinocystic acid | 2 | 1.66 | 0.0277 | 26.10 | 471.3476 |
| Rha-Hex-Hex-bayogenin | 2 | 1.59 | 0.0142 | 15.54 | 957.5125 |
| Hex-gypsogenic acid | 2 | 1.33 | 0.0178 | 18.22 | 647.3803 |
| 3-Rha-Gal-GlcA-soyasapogenol B | 2 | 1.18 | 0.0431 | 16.47 | 941.5138 |
| Hex-soyasapogenol E | 2 | 0.45 | 0.0056 | 21.44 | 617.4059 |
| 3-Rha-Gal-GlcA-soyasapogenol B | 2 | 0.13 | 0.0021 | 18.89 | 941.5139 |
| Hex-Hex-Rha-bayogenin | 2 | 0.12 | 0.0032 | 12.00 | 957.5054 |
| Rha-Hex-Hex-Hex-bayogenin | 2 | 0.07 | 0.0001 | 11.17 | 1,119.5549 |
| 3-Glc-28-Glc-medicagenic acid | 1 | 0.02 | 0.0000 | 13.03 | 825.4293 |
| Hex-Hex-Hex-bayogenin | 2 | 0.01 | 0.0000 | 12.41 | 973.5075 |
| Hex-Rha-Hex-Hex-hederagenin | 2 | nd in bc | 0.0000 | 12.85 | 1,103.5693 |
| Rha-Hex-Hex-Hex-bayogenin | 2 | nd in bc | 0.0004 | 12.00 | 1,119.5616 |
| Hex-Hex-Hex-bayogenin | 2 | nd in bc | 0.0004 | 11.64 | 973.5012 |
| Hex-Hex-Hex-medicagenic acid | 2 | nd in bc | 0.0000 | 12.70 | 987.4860 |
| Hex-Hex-Rha-bayogenin | 2 | nd in bc | 0.0000 | 11.18 | 957.5043 |
Identification codes are as follows: 1, identification using authentic standards; 2, putative identification using accurate mass (database search ± 6 ppm; echinocystic acid has been identified by Tava et al. [2011]: [M-H]− 471); and 3, putative identification using tandem mass spectrometry. The aglycone has the same m/z as aglycone B reported by Pollier et al. (2011).
Flavonoids are important in plant defense, symbiosis, development, and pollination (Modolo et al., 2007). Two flavonoids observed at higher levels in border cells were apigenin and DHF (Table III). Naringenin chalcone 4′-O-glucoside, a glycosylated form of an apigenin precursor, also was identified in border cells at elevated levels. Supplemental Table S3 contains a comprehensive list of primary and secondary metabolites observed in this study with the supporting chromatographic retention times and mass spectral m/z values.
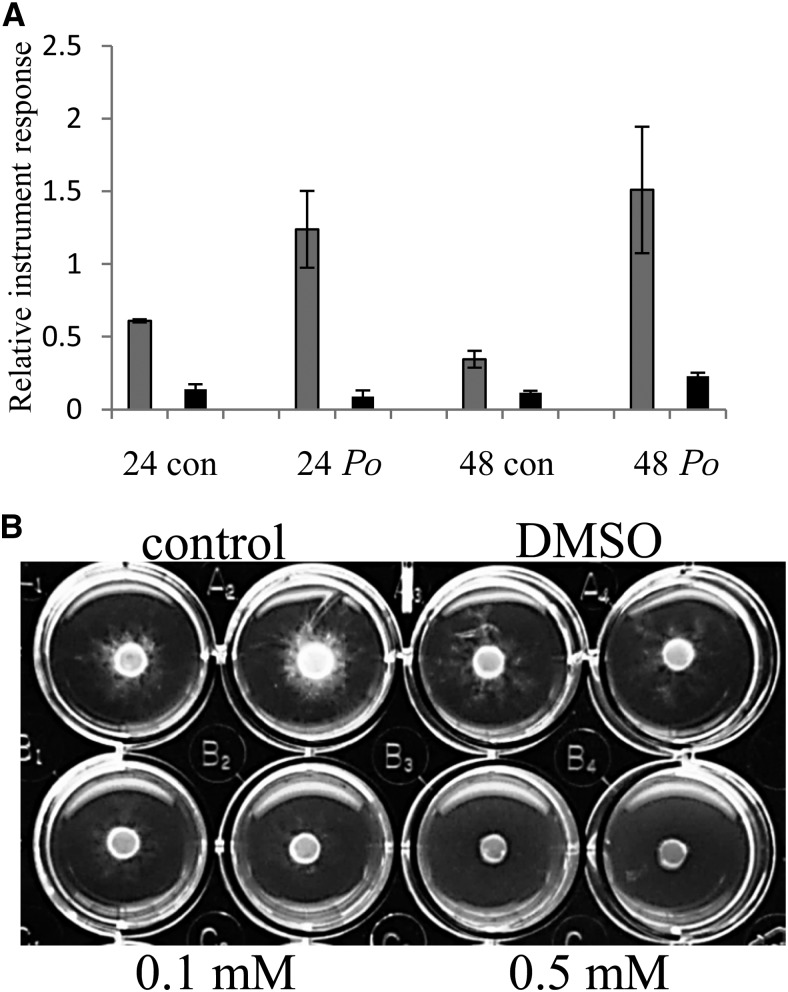
M. truncatula seedlings were treated with water or a suspension of mycelia from the nonsporulating fungus cotton root rot, a devastating root pathogen that attacks many plants, including legumes (Marek et al., 2009; Uppalapati et al., 2009, 2010). Border cells and root tips without border cells were collected 24 and 48 h after inoculation and analyzed by UHPLC-quadrupole time-of-flight mass spectrometry. The flavone DHF was found constitutively at higher levels in border cells and was further increased approximately 2-fold in border cells after a 24-h exposure to cotton root rot, while there was no change in root tips (Fig. 7A). After a 48-h exposure to cotton root rot, the level of DHF had increased further in border cells but was unchanged in root tips. The level of DHF in control border cells dropped at the 48-h time point (Fig. 7A). DHF was tested for antifungal properties against cotton root rot and was effective at concentrations as low as 100 µm (Fig. 7B). DHF showed greater growth inhibition than catechol, a known potent inhibitor of cotton root rot growth (Greathouse and Rigler, 1940), and similar to or slightly better than that of the phytoalexin medicarpin (Naoumkina et al., 2007; Table IV).
Figure 7.
A, Relative abundance of DHF in water control (con) and cotton root rot (Po)-treated border cells (gray bars) and root tips (black bars) after 24 h (24) and 48 h (48) of exposure. n = 3 or 4, and error bars represent se. B, In vitro assay of antifungal activity after 5 d of no-treatment control, dimethyl sulfoxide (DMSO) only, and two concentrations of DHF against cotton root rot. Fungal growth was quantified on a scale of 0 to 5, with 0 meaning no growth and 5 meaning no inhibition: control and DMSO = 5, 0.1 mm = 1, and 0.5 mm = 0.
Table IV. In vitro antifungal activity of catechol and flavonoids against cotton root rot.
Qualitative measurement of fungal infection is shown. Inhibition effect is scored by numbers, with 5 representing no inhibition and 0 representing complete inhibition. Dashes indicate no data.
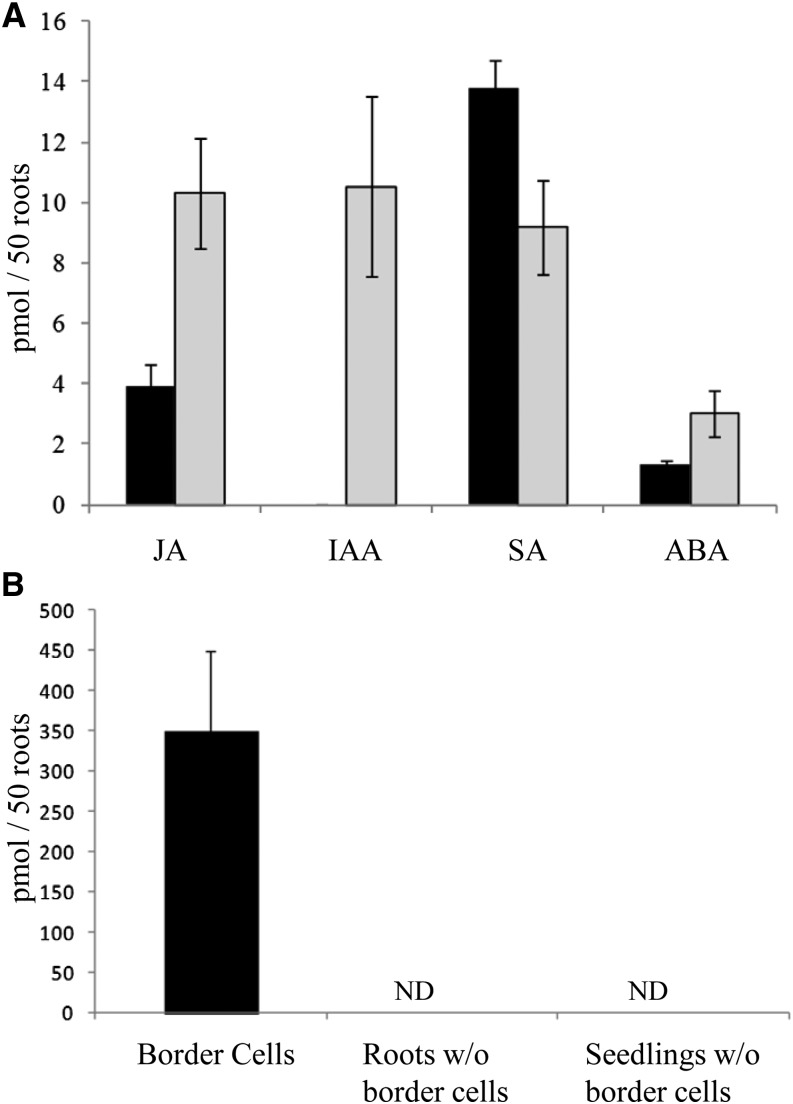
Phytohormones were analyzed in root tips without border cells and border cells using UHPLC coupled to triple quadrupole tandem mass spectrometry. Phytohormone analyses in root tips revealed high concentrations of jasmonic acid (JA), indole acetic acid (IAA), and salicylic acid (SA) along with a lesser concentration of abscisic acid (ABA; Fig. 8A). SA was also abundant in border cells, while JA and ABA were less abundant, and IAA was not detected in border cells (Fig. 8A).
Figure 8.
A, Phytohormone content of border cells (black bars) and root tips (gray bars). B, Headspace hexanal concentration of border cells, roots without (w/o) border cells, and seedlings without border cells. Volatiles were analyzed from border cells collected from 100 roots and compared on the basis of 50 roots. Error bars represent se. ND, Not detected.
The volatile compound hexanal was observed in border cells (350 ± 100 pmol per 50 roots) using solid-phase microextraction (SPME) GC-MS analysis but not in roots without border cells or whole seedlings without border cells (Fig. 8B). Volatiles from root tips were not measured because excising the tip would cause a wounding response.
DISCUSSION
Border cells are determinate cells capable of responding to many different stimuli encountered in the rhizosphere (Hawes et al., 2000) and are differentiated from other root cells. The differentiation of border cells begins with important developmental cues and heightened hormonal activity, resulting in substantial differences in primary and secondary metabolism fueled through intracellular starch-based energy production (Fig. 6). Discussions follow that provide detailed gene expression and metabolomics data supporting our conclusion that border cells are differentially programmed with enhanced secondary metabolism. We further demonstrate that the differential metabolic programming and composition of border cells facilitate a unique role in root growth, development, defense, and plant-microbe signaling.
Border Cell Production Includes PME, PMEI, and Localized Hormone Activity
Border cells are produced within 2 to 3 d after germination and originate from the root cap meristem initials (Brigham et al., 1998; Hawes et al., 2000). They transition through columella cells, peripheral cells, and, ultimately, into border cells (Brigham et al., 1998; Hawes et al., 2000). Border cells are released from the root, in part by PME activity. This enzyme demethylates pectin and allows cell wall degradation, which leads to the separation of border cells from roots. Inhibition of this gene blocks the normal detachment of border cells (Wen et al., 1999). Expression levels for PME were low in border cells, as determined by both microarray and qRT-PCR data (Table I; Supplemental Table S1), and substantially higher in root tips (7- to 16-fold higher). The action of PME in roots has been well documented (Wen et al., 1999), and confirmation of PME expression in M. truncatula root tips and border cells provided reassurance of our experimental approaches.
Interestingly, a novel transcript for a gene annotated as a PMEI was greatly enhanced in border cells (115-fold higher; Fig. 4A; Table I). PMEI activity has been characterized in kiwifruit (Actinidia chinensis) and Arabidopsis (Balestrieri et al., 1990; Raiola et al., 2004) and in pepper (Capsicum annuum) leaves (An et al., 2008), where it also exhibited antifungal properties. However, to our knowledge, this is the first time a putative PMEI has been identified in border cells. PMEI and PME form a complex in a 1:1 stoichiometric ratio (Di Matteo et al., 2005). Therefore, PMEI expression in border cells appears to be a negative regulator of PME activity and associated with border cell detachment. PMEI levels increase once PME activity is no longer needed.
All cell types in the root tip of Arabidopsis seedlings synthesize auxin (Ljung et al., 2005). The auxin is transported basipetally by polar transport from most root tip cells, and IAA levels in columella cells are lower than in the rest of the root tip. In contrast, the quiescent center maintains a high concentration of IAA (Ljung et al., 2005), and high auxin levels contribute to the mitotic activity in meristematic cells, while the auxin gradient in other root tip cells is tightly connected to differentiation and development (Ljung et al., 2005). Border cells in Medicago spp. progress from columella cells and are the most mature cells in the root tip; thus, the lack of detectable auxin in border cells and high levels of auxin in the root tip (Fig. 8A) correlate with auxin-related root development. In border cells, elevated auxin-related transcripts are the predominant class of hormone transcripts (40% of border cells compared with root tips); many of these are negative regulators of auxin. For example, multiple auxin-responsive SAUR transcripts are elevated in border cells. These proteins are short-lived nuclear proteins hypothesized to be negative regulators of auxin synthesis and transport (Kant et al., 2009). Overall, 19 of 21 SAUR transcripts were higher in border cells, and the transcript level for one SAUR verified by qRT-PCR was especially elevated (more than 900-fold; Table I). Expression of this SAUR was not observed in other root tissues contained within the Medicago spp. gene atlas (Fig. 4B). SAURs are involved in auxin signal transduction (Davies, 2004) and in auxin-induced cell elongation (Knauss et al., 2003) and likely play a role in the elongation of border cells. Transcript levels of several auxin response factors, transcription factors that regulate auxin-mediated responses, were lower in border cells (Supplemental Table S1), further confirming a lack of auxin in these cells.
JA expression is high in root tips (Birnbaum et al., 2003), and levels of JA in this study also were higher in root tips than in border cells (Fig. 8A). LOX genes function early in the JA pathway, but the transcript levels for three LOX transcripts were 84- to 225-fold higher (microarray data) in border cells. The level of one LOX was further validated by qRT-PCR and confirmed as 126-fold higher in border cells (Table I). This LOX catalyzes an early step in the oxylipin pathway, and its product is the branch-point compound for either JA synthesis or the synthesis of hexanal (Supplemental Fig. S7B), a stress volatile produced in response to biotic and abiotic stimuli (Chehab et al., 2006). Hexanal was the only volatile detected by SPME analysis of border cells, and there was no discernible peak for hexanal in roots or seedlings without border cells (Fig. 8B). Transcript levels for hydroperoxide lyase, the next enzyme in the biosynthetic pathway for hexanal, were 4-fold higher in border cells than in root tips (Supplemental Table S1). Levels of allene oxide synthase and allene oxide cyclase transcripts, enzymes that catalyze subsequent reactions in the JA pathway, were either unchanged or higher in root tips (Supplemental Table S1). Elevated levels of JA, hexanal (Fig. 8), and transcripts for enzymes in the biosynthetic pathway of these compounds support an important role for border cells in stress metabolism and in protection against pathogen and insect attack, processes in which oxylipins have an established role (Reymond and Farmer, 1998; Uppalapati et al., 2009).
SA is a key signaling molecule synthesized in response to both biotic and abiotic stress (Horvath et al., 2007). It is abundant in root tips and even more abundant in border cells (Fig. 8A), yet there are few transcripts annotated as SA related in either tissue (Supplemental Table S1). Two pathways for SA synthesis are proposed in plants, one of which is through benzoic acid (BA; Chen et al., 2009). Border cells and root tips contain BA (Table II; Supplemental Table S3), and only one additional hydroxyl group differentiates SA from BA. Benzoic acid 2-hydroxylase (BA2H) activity has been detected in tobacco (Nicotiana tabacum) and rice (Oryza sativa), and the tobacco protein has been partially purified, although a gene has not yet been isolated (Chen et al., 2009). Removal of border cells from root tips could be enough stress to cause an as yet unannotated Medicago spp. BA2H to synthesize SA from BA to aid in root tip defense (Naoumkina et al., 2010).
Border Cells Utilize Starch for Energy and Carbon
Border cells are detached root cells and unable to benefit directly from energy sources transported through the vascular system. However, border cells contain starch-filled plastids (Fig. 1, C and D), which are a common source of stored energy and carbon (Blancaflor et al., 1998; Barlow, 2003). Starch is synthesized from Glc made available when Suc is degraded, and over 70% of Suc transcripts in border cells and root tips are annotated as degradation related (Supplemental Table S1). Fifty-six percent of the starch-annotated transcripts in root tips are associated with synthesis, and root cap columella cells are packed with starch bodies, as determined through iodine staining (Supplemental Fig. S3C), supporting the importance of the root tip as a site for the conversion of Glc to starch. Starch levels are lower in border cells than in their progenitor root cap cells, and 80% of observed border cell transcripts involved in starch metabolism were annotated as degradation genes (Supplemental Table S1). In addition, the transcript level of the starch-degrading enzyme β-amylase was validated by qRT-PCR as approximately 15-fold higher in border cells (Table I). Cumulative transcriptomic and microscopic data indicate that starch reserves accumulated during border cell production are utilized as an energy and carbon source.
Primary Metabolism Is Reduced in Border Cells and Redirected toward Secondary Metabolism
Seventy percent of transcripts involved in primary metabolism were lower in border cells compared with root tips (Table V). Exceptions to this trend are discussed in more detail below and include compounds that serve important roles in supplying primary metabolic precursors for the synthesis of important secondary metabolites.
Table V. Comparison of border cell and root tip transcripts increased/decreased in primary and secondary metabolism.
BC/RT, Ratio of border cells compared with root tips.
| Category | No. of Transcripts Increased in BC/RT | No. of Transcripts Decreased in BC/RT |
|---|---|---|
| Carbohydrate metabolism (major and minor) | 32 | 55 |
| Glycolysis | 7 | 23 |
| Tricarboxylic acid/organic acid transformation | 9 | 22 |
| Amino acid metabolism | 39 | 96 |
| Secondary metabolism | 142 | 82 |
| Flavonoids | 56 | 20 |
| Terpenoids | 24 | 19 |
Levels of many amino acids were much higher in border cells than in root tips (Table II), while overall transcript levels for amino acid synthesis in border cells was decreased (Fig. 3; Supplemental Table S1), suggesting that most amino acids were synthesized during early border cell development. Alternatively, border cell amino acids may have originated from protein degradation; this is less likely, because the percentage of protein degradation transcripts elevated in border cells and root tips was equivalent, with fewer total degradation transcripts in border cells. In addition, border cells are reported to actively synthesize proteins even after release from the root (Brigham et al., 1995; Wen et al., 2007). Thus, the origin of increased amino acids in border cells is unknown, but evidence supports their utilization as precursors for the synthesis of proteins and metabolites.
Asn was the most abundant amino acid in border cells and was increased compared with root tips. Transcript levels for Asn synthetase also were elevated 3- to 6-fold (Table I), although the transcript levels for most other amino acid syntheses were decreased in border cells. Asn is an end-point amino acid that serves as a major nitrogen transport and storage compound in plant cells (Ta et al., 1984). Amino acids also serve as precursors for the rapid production of defense compounds. For example, Phe is the entry point for lignin, flavonoid, and salicylate biosynthesis. Phe was present in border cells and root tips, and lignin transcripts plus multiple flavonoids and SA (compounds and/or transcripts) important in plant defense were identified in both (Tables I and III; Fig. 6).
Transcript levels for the enzyme that reversibly converts Fru-6-P to Fru-1,6-P were 28-fold higher in border cells as determined by qRT-PCR (Table I). Fru is a product of Suc degradation and the most abundant sugar measured. Fru is a precursor metabolite in glycolysis, and the glycolysis of Fru yields ATP and NADH with an end product of pyruvate. Pyruvate can be metabolized in the tricarboxylic acid cycle to form acetyl-CoA, central to the process of shuttling carbon from primary to secondary metabolism.
The level of citrate was up 5-fold (Table II) in border cells, and the correlated transcript level of citrate synthase, as measured by qRT-PCR, was slightly higher (Table I). Citrate is an important intermediary in the tricarboxylic acid cycle (Fig. 6) and also serves as a substrate for the cytosolic production of acetyl-CoA, an essential precursor in the synthesis of secondary metabolites. As a side note, citrate is secreted in response to aluminum (Li et al., 2000), and the importance of border cells in protecting the root tip from aluminum has been documented (Miyasaka and Hawes, 2001). Prior literature and increased levels of citrate in border cells support their role as a major quantitative source for the secretion of citrate.
CoA is important in numerous metabolic processes, especially in providing carbon substrates for secondary metabolism. Precursors for CoA biosynthesis, including β-Ala and the branched-chain amino acids Ile and Val, were found at increased levels in border cells (Table II). β-Ala is a precursor of pantothenate and CoA, and the transcript for β-ureidopropionase, an enzyme involved in β-Ala synthesis, was determined to be 3-fold higher in border cells by qRT-PCR (Table I). CoA is necessary for the production of acetyl-CoA, a central metabolite in the shuttling of carbon from primary to secondary metabolism, and the synthesis of many natural products, including flavonoids and terpenoids.
Although flux was not measured, the integrated metabolite and transcript data strongly support that the carbon and energy necessary for growth and development are redirected toward secondary metabolism in border cells.
Secondary Metabolism Is Enhanced in Border Cells
Border cells are rich in secondary metabolites and contain numerous elevated transcripts for secondary metabolism (Figs. 2 and 3; Table V). This is highly evident in the differential MapMan visualizations, where the majority of transcripts for waxes, phenylpropanoids, phenolics, and flavonoids were distinctly higher in border cells (Fig. 2). The elevation in secondary metabolism transcripts is in sharp contrast to the decrease of many primary metabolite transcripts, indicating that energy and carbon from primary metabolism are channeled into border cell secondary metabolism.
Border cells are mature lignified cells (Hamamoto et al., 2006), and the transcript level for caffeoyl coenzyme A O-methyltransferase (CCoMT), an important enzyme in phenylpropanoid-based monolignol biosynthesis, was determined by qRT-PCR and found to be 24-fold higher in border cells (Table I). The relative increase in CCoMT supports continued lignification and secondary cell wall reinforcement in border cells after detachment from the root. Lignin provides strengthened cell walls for enhanced protection against mechanical damage and during encounters with plant pathogens (Vance et al., 1980).
Many triterpene saponins and flavonoids were identified in border cells, and the levels of many of these compounds were dramatically higher than in root tips. Saponins have reported allelopathic, antimicrobial, and insecticidal properties important in plant protection (Shao et al., 2005) and are generally toxic to cold-blooded animals and insects (Tava and Odoardi, 1996; Waterman, 1996). Legumes have a rich variety of saponins (Huhman and Sumner, 2002; Dixon and Sumner, 2003; Huhman et al., 2005; Pollier et al., 2011), many of which are present in M. truncatula root tissue. Triterpene saponins were quantitatively the most abundant class of secondary metabolites identified in border cells, and the largest fold increases in border cell metabolites were observed for saponins.
Much of the biosynthetic pathway for triterpenoid saponins is unknown, but the first committed step is the cyclization of 2,3-oxidosqualene by β-amyrin synthase to form β-amyrin (Hayashi et al., 2001; Suzuki et al., 2002). This is the starting point for the synthesis of at least seven different sapogenins (the aglycone form of saponins). The transcript levels for β-amyrin synthase were similar in border cells and root tips, as measured by qRT-PCR (Table I), suggesting that early steps in saponin biosynthesis occur at approximately equivalent rates in root tissues. The transcript level of CYP93E, a cytochrome P450 isoform of the enzyme reported to catalyze the hydroxylation of β-amyrin and sophoradiol in soybean (Glycine max) and licorice (Glycyrrhiza glabra) to form the first soyasapogenin (Shibuya et al., 2006; Seki et al., 2008), was 7-fold higher in border cells, while the cytochrome P450 that functions in the first committed step of the oleanate sapogenol pathway, CYP716A12 (Carelli et al., 2011), is 3-fold higher in root tips (Supplemental Table S1). Subsequent oxidation of the triterpene aglycone skeletons is believed to involve several currently unknown cytochrome P450s. The transcript level of a proposed cytochrome P450 family member associated with terpenoid biosynthesis, CYP71A8 (Naoumkina et al., 2010), was 91-fold higher in border cells, implying a tissue/cell specificity for certain steps in terpenoid biosynthesis. The transcript for another gene involved in sesquiterpenoid biosynthesis, (−)-germacrene synthase, was validated by qRT-PCR and found in border cells at levels 167-fold higher than in root tips. Unfortunately, the product of this enzyme, (−)-germacrene D, was not observed in the GC-MS analysis due to its volatility. However, these data cooperatively support the elevated biosynthesis of terpenes in border cells and a role for border cell terpenoids in defense and rhizosphere modification.
Glycosylation typically influences the bioactivity of secondary metabolites as well as their cellular localization, stability, and metabolism (Modolo et al., 2007). More specifically, glycosylation can increase the biological activity of triterpenoid saponins in comparison with the aglycone, presumably due to the increased ability of the molecules to complex in fungal membranes, and the activity is dependent on the number of sugar molecules in the attached chain (Haridas et al., 2001; Osbourn, 2003). In border cells, multiple sapogenin aglycones and various conjugated forms of each sapogenin were identified (e.g. bayogenin, Rha-hex-hex-bayogenin, and hex-hex-hex-bayogenin), thereby increasing the saponin diversity (Table III) and potential defense compounds useful in responses to rhizosphere microbes and environmental stresses.
Flavonoids are associated with symbiosis, signaling, plant development, and plant defense (Kape et al., 1991; Stafford, 1997; Shirley, 1998; Aoki et al., 2000; Forkmann and Martens, 2001), and there is a net increase in flavonoids in border cells (Table III). Phenylpropanoids are the precursors of flavonoids, isoflavonoids, anthocyanins, and lignin and are synthesized from the primary amino acid Phe. An early step in the biosynthesis of these compounds is the conversion of Phe to cinnamic acid by Phe ammonia lyase. Phe was abundant in border cells, and three isoforms of the Phe ammonia lyase transcript were identified at increased levels (Supplemental Table S1). This increase was confirmed for one isoform by qRT-PCR (Table I) as 30-fold higher in border cells. Isoflavone synthase is the branch point of phenylpropanoids into isoflavone synthesis, and the transcript for an isoflavone synthase-like protein was increased 13-fold in border cells. These data demonstrate elevated isoflavone synthesis in border cells.
Secreted flavonoids are important in the signaling processes between plants and other organisms in the rhizosphere. One example is nodulation gene induction in rhizobacteria (Peck et al., 2006), where apigenin and DHF are among the most potent inducers of nod genes in Sinorhizobium meliloti during presymbiotic interactions with Medicago spp. (Zhang et al., 2007). Multiple flavonoids involved in rhizobial signaling and symbiosis (Modolo et al., 2007) were substantially elevated in border cells. Specifically, apigenin was 6-fold higher, DHF was increased by more than 12-fold, and naringenin-chalcone glucoside was 2-fold higher in border cells than in root tips (Table III). Transcripts associated with the production of apigenin, naringenin-chalcone synthase, and flavone synthase II also were measured by qRT-PCR at substantially increased levels in border cells (64-fold and approximately 7-fold, respectively; Table I). These data illustrate that border cells contain substantially greater quantitative amounts of important flavonoid signaling molecules than do other root tissues. Thus, border cells are equipped to recruit motile rhizobia to inoculate root hairs and initiate nodule development as the root continues its developmental processes.
Flavonoids also serve as defense compounds (Ferreyra et al., 2012), and the differentially accumulated specialized metabolites in border cells likely serve important defense roles as well as signaling roles. Hence, the metabolic responses of root tips and border cells were measured in response to exposure to cotton root rot, a devastating fungal root rot pathogen with limited treatment options and no known resistance in Medicago spp. or any crop species (Uppalapati et al., 2010). The flavone DHF was strongly increased in border cells after exposure to cotton root rot. The constitutively high levels of DHF in border cells doubled after 24 h of exposure to the fungus and increased even further after 48 h (Fig. 7A). Yet the levels of DHF remained low at all tested time points in the root tip. To further demonstrate the roles of DHF and border cells in plant defense, DHF was tested for growth inhibition against cotton root rot. DHF showed strong fungal growth inhibition and was as potent an antifungal agent as medicarpin and 20-fold better than catechol, one of the few reported chemical treatments for cotton root rot (Table IV). This novel bioprotection against cotton root rot, in conjunction with high levels of DHF in nonelicited border cells and a robust increase in DHF after exposure to cotton root rot, led us to conclude that border cells produce DHF and other secondary metabolites as phytoanticipins and/or phytoalexins to protect the critically important meristematic root tip.
A Systems Model for Metabolic Programming and Enhanced Metabolic Capacity of M. truncatula Border Cells
Integrated transcriptome and metabolome data detailing differences observed between M. truncatula border cells and root tips are provided in Figure 6. This figure includes a model describing the enhanced secondary metabolic capacity of border cells. Border cells begin their life cycle as root cap initial cells, develop as columella and peripheral root cells, and then transition into border cells. Differentiated border cells are characterized by large expression differences when compared with root tips, and the most dramatic differences are in hormone-associated transcripts. Border cells contain SA, JA, and ABA, but IAA is not detected in border cells. This contrasts with the root tip, which contains the highest concentration of IAA in roots. The volatile compound hexanal is produced by border cells and is absent in roots without border cells. Iodine staining revealed starch deposition in the root cap and border cells, and comparative microarray data revealed increases in border cell β-amylase. These data support starch reserves as a critical energy source and carbon reserve for detached border cells. Transcript data document an overall general decrease in primary metabolism, with exceptions associated with branched-chain amino acid and β-Ala biosynthesis, which are associated with CoA biosynthesis and carbon shuttling into secondary metabolic pathways (i.e. flavonoid and triterpene biosynthesis). Flavonoid transcripts and related metabolites are substantially increased in border cells, and many triterpenoid transcripts and metabolites also are observed at elevated levels.
The cumulative pathway-specific data provide compounding evidence that primary and secondary metabolism are differentially regulated in border cells relative to root tips. Although flux was not measured, the integrated metabolite and transcript data strongly support that carbon and energy are reallocated from biosynthesis for growth and development toward enhanced secondary metabolism in border cells. Quantitative increases in specific secondary metabolites indicate an important role for border cells in defense and plant-microbe interactions, a hypothesis validated by the antifungal effect of DHF against cotton root rot. Future work will concentrate on expanding our understanding of the molecular and metabolic basis for border cells in plant-microbe signaling and defense and the specificity of plant-microbe interactions.
MATERIALS AND METHODS
Sample Growth Conditions and Collection
Medicago truncatula ‘A17’ seeds were scarified by soaking in concentrated sulfuric acid for 5 min and then rinsed three times with chilled, distilled water. Scarified seeds were sterilized in bleach for 10 min, rinsed three times with distilled water, and placed on sterile filter paper atop 1% (w/v) agar water plates to germinate at 24°C in a dark growth chamber for 3 d. Twenty microliters of a mycelial suspension of cotton root rot (Phymatotrichopsis omnivora) was pipetted along the germinated root 2 d after plating, and the plates were returned to the dark. Border cells and root tips were collected 24 and 48 h later. For the metabolite analyses, border cells were collected from 40 replicate seedlings, and the resulting root tips without border cells were excised and collected. Five whole roots consisting of the complete radicle were collected separately. All samples were frozen immediately in liquid N2.
Histology
Border cells from 10 seedlings were collected in water and incubated for 5 min with fluorescein diacetate (50 ng µL−1) to detect live cells and propidium iodide (500 ng µL−1) to stain dead cells. Dead and live cells were counted using a hemocytometer. The count was repeated at least three times. A Nikon Microphot-FX microscope was used for cell counts and starch body visualization.
Starch-stained plastids in border cells were confirmed by examining five or more seedlings on three separate occasions. Dilute iodine stain (one-fourth strength) was added to visualize starch in seedling roots and detached border cells. Seedling roots were sectioned to a thickness of 70 µm and iodine stained to image starch bodies in columella cells. Differential interference contrast microscopy was utilized to visualize starch bodies in detached border cells.
Roots and border cells stained with fluorescein diacetate or double stained with fluorescein diacetate and propidium iodide were imaged with a Leica TCS SP2 AOBS confocal laser scanning microscope (Leica Microsystems) using a 63× HCX Plan-Apo water-immersion objective with a numerical aperture of 1.2. Fluorescein diacetate was detected by illuminating with the 488-nm line of the argon laser, and emission was detected at 510 nm. Propidium iodide was detected after illumination with the 543-nm line of the argon laser, and emission was detected at 617 nm.
Roots with appressed border cells and matrix were attached to a specimen holder frozen in liquid nitrogen and imaged on a Hitachi TM3000 tabletop scanning electron microscope. Environmental scanning electron microscopy of border cells floating off seedling roots was performed using an FEI Quanta 600F environmental scanning electron microscope at 6.5 Torr and 5°C. The root was placed on a thin strip of agar with water droplets initially surrounding the root tip.
Ruthenium Red at a concentration of 0.02% (w/v) in distilled water was used to stain acidic pectins in mucilage and border cells released from seedling roots. The roots were placed in a drop of stain on a microscope slide and monitored for 20 to 30 min. Images were made using a Nikon Microphot-FX once sufficient color developed.
Fungal Growth and Inhibition Assays
Cotton root rot cultures were grown at 28°C on sterile plates of 18 g L−1 potato dextrose agar (PDA; 1 g L−1 malt extract, 1 g L−1 yeast extract, and 1 g L−1 peptone). Fungal inhibition was assayed on PDA plates for 5 d. Flavonoid molecules were redissolved in DMSO to make 25 and 5 mm stock solutions, which were diluted to 1 mL with PDA medium to final concentrations of 0.5 and 0.1 mm. A 2-mm fungal plug of cotton root rot was incubated on the assay plates, and fungal growth was recorded every 12 h starting at 48 h. The fungal growth for each tested molecule was scored on a qualitative scale from 0 to 5, with 0 as no growth and 5 as the most growth, by comparing with PDA-only medium and the DMSO control (20 µL in 1 mL of PDA). Catechol at concentrations of 5 and 10 mm was used as a positive control. Formononetin, which bears no antifungal activity, was used as the negative control. All experiments were replicated four times.
Gene Expression Analysis
Border cells for each replicate were collected by gently agitating roots directly in Qiagen buffer RLT from approximately 150 to 200 seedlings. Root tips were collected by agitating roots in water (detailed below in “Mass Spectrometry Analysis”), and the root tips (2–4 mm) minus border cells from 10 roots were excised and frozen immediately in liquid N2. Five whole roots were excised from seedlings and frozen immediately in liquid N2. Three biological replicates were performed for each tissue sample. Total RNA was isolated using the Qiagen RNeasy Plant Mini Kit. RNA was quantified and evaluated for purity using the Nanodrop Spectrophotometer ND-100 (NanoDrop Technologies) and Bioanalyzer 2100 (Agilent). Four micrograms of total RNA was used for the expression analysis of each sample using the Affymetrix GeneChip Medicago Genome Array (Affymetrix). Probe labeling, chip hybridization, and scanning were performed according to the manufacturer’s instructions for one-cycle labeling (Affymetrix). Data normalization between chips was conducted using robust multichip average (Irizarry et al., 2003). Presence/absence calls for each probe set were obtained using dCHIP (Forkmann and Martens, 2001). Gene selections based on associative Student’s t test (Dozmorov and Centola, 2003) were made using Matlab (MathWorks). A selection threshold of 2 for transcript ratios and a Bonferroni-corrected P value threshold of 8.15954E-07 were used (where the threshold was derived from 0.05/n, and n is the number of probe sets on the chip). The false discovery rate of all significant genes was monitored with Q values obtained by EDGE software (Storey and Tibshirani, 2003; Leek et al., 2006). Transcriptome data were preprocessed independently and integrated using MapMan software tools customized for Medicago spp. (Urbanczyk-Wochniak and Sumner, 2007).
Genes of interest were selected for further confirmation by qRT-PCR following the manufacturer’s protocols (Power SYBR Green; Life Technologies). Total RNA was isolated as above, and primer pairs were designed using Primer3 software and located in the same region of the gene as the microarray probes whenever possible. LinRegPCR was used to assess amplification efficiency, and the expression data were analyzed according to Czechowski et al. (2004) for samples without a control. Ubiquitin and helicase genes were used for normalization, as these were stably expressed in the microarray experiments. The mean ratio of the two normalization genes was used to present the data in Table I. A list of the primer pairs used for qRT-PCR is included in Supplemental Table S2. The melting curves of all primer pairs except those for β-amylase and citrate synthase showed amplification of a single product. The curve for β-amylase showed a minor secondary product, and the curve for citrate synthase also showed multiple products, but both were of satisfactory quality for this purpose.
Mass Spectrometry Analysis
Border cells were collected by gently agitating roots in water for 30 to 60 s, and an equal volume of methanol was added to stop enzyme activity during drying. For the cotton root rot experiment, border cells were collected directly in 80% methanol and 20% (v/v) water. The border cells were frozen in liquid N2, dried, ground in an Eppendorf tube, and extracted for 2 h in 80% methanol and 20% (v/v) water containing 20 μg mL−1 umbelliferone as an internal standard. After border cells were removed, roots were rinsed in water and root tips were excised and frozen in liquid N2. Whole roots containing border cells and root tips were separated from the seedling hypocotyl and frozen in liquid nitrogen. The secondary metabolite analyses consisted of three replicates for each tissue. Samples were lyophilized, ground, and extracted with 80% methanol and 20% (v/v) water as above. Samples were centrifuged, and the supernatant was analyzed by UPLC coupled to a Waters Premier hybrid quadrupole time-of-flight mass spectrometer. Separations were achieved using a Waters Acquity UPLC, 2.1- × 150-mm BEH C18 column, mobile phases of 0.1% (v/v) aqueous acetic acid (A) and acetonitrile (B), and a linear gradient of 95%:5% to 30%:70% eluents A:B in 30 min. The mass spectrometer was operated in negative electrospray ionization mode. Raffinose was used as the reference compound. Peak picking, alignment, and quantification were performed using Waters MarkerLynx software. The cotton root rot samples were examined using a Waters Acquity UPLC coupled with LECO’s fast-acquisition-speed Unique HT time-of-flight mass spectrometer operated in negative electrospray ionization mode, followed by ChromaTOF software deconvolution. All conditions for UPLC were as listed above, except that eluent A was 0.1% (v/v) formic acid in water. Compounds were normalized relative to the internal standard, then to the total ion abundance. Metabolite identifications were achieved via comparison of retention time and accurate mass with those of authentic standards. Tentative identifications were performed by matching experimental accurate mass data to those in plant metabolite databases and public literature within a 5-ppm mass accuracy tolerance.
For GC-MS analyses, dried polar extracts were derivatized with methoxyamine hydrochloride in pyridine followed by trimethylsilyl derivatization using N-methyl-N-trimethylsilyltrifluoroacetamide and analyzed as reported previously (Broeckling et al., 2005). Four replicates were performed. Mass spectra deconvolution and metabolite identification were performed using AMDIS software (http://chemdata.nist.gov/dokuwiki/doku.php?id=chemdata:amdis) and a custom, in-house electron ionization-mass spectrometry metabolite library. Peak picking, alignment, and quantification were achieved using MET-IDEA software (Broeckling et al., 2006; http://bioinfo.noble.org/download). Normalization was performed as described above.
Phytohormone analyses were based upon Pan et al. (2010), with some modifications. Briefly, border cells were collected from 50 seedlings and frozen in liquid nitrogen. Root tips without border cells were collected from the same seedlings, frozen in liquid nitrogen, and ground to a fine powder. Ten milligrams of root tip tissue and all border cell tissue was extracted in 1 mL of isopropanol:water:HCl (2:1:0.002) for 1 h at 4°C with 50 pmol of the deuterated standards 2H5-IAA (C/D/N Isotopes), 2H6-SA (Sigma), 2H-JA (TCI), and 2H6-ABA (Icon). Dichloromethane at 0.5 mL was added to each sample, and samples were shaken for another 30 min at 4°C. Samples were centrifuged, and two phases formed. One milliliter of the bottom layer was transferred to a 2-mL glass vial, and the solvent was dried under nitrogen. The residue was redissolved in 0.1 mL of methanol and diluted to 1 mL with 1% (v/v) acetic acid. The solution was applied to a conditioned Waters Hydrophilic/Lipophilic Balanced HLB column, and the column was rinsed with 1 mL of 1% (v/v) acetic acid. The rinse was discarded, and phytohormones were eluted from the column using 1.8 mL of 80% methanol containing 1% (v/v) acetic acid and collected in a 2-mL autosampler glass vial. Solvents were dried under nitrogen and redissolved in 50 μL of 50% methanol in 1% (v/v) acetic acid. Ten microliters was injected onto an Agilent 1290/6430 UHPLC/MS TripleQuad system. Separations were achieved using a Waters Acquity UPLC, 2.1- × 150-mm BEH C18 column with a mobile phase of 0.1% (v/v) aqueous formic acid (A) and acetonitrile (B) and a linear gradient of 5% to 46% (v/v) acetonitrile in 25 min. Phytohormones were detected and quantified using multiple reaction monitoring as described (Pan et al., 2010). At least four replicates were performed for each tissue.
Volatiles emitted from border cells, root tissues without border cells, and seedlings without border cells were extracted using an SPME fiber and analyzed by GC-MS. Prior to the analysis, each sample was prepared in a 10-mL glass vial. Border cells were collected from 100 seedlings into approximately 0.5 mL of water. The collected border cells were transferred into a 10-mL glass vial, and the vial was tightly capped. For the analysis of root volatiles, the root portion of 50 seedlings without border cells was placed inside a 10-mL glass vial. The top part of the seedlings was covered with aluminum foil. The analysis of volatiles from whole seedling minus border cells was conducted on 50 seedlings in a tightly capped 10-mL glass vial. A divinylbenzene/carboxen/polydimethylsiloxane (50/30 μm, 2 cm) SPME fiber (Supelco) was used to extract the headspace volatiles for 60 min at 30°C. SPME-absorbed volatiles were desorbed at 250°C for 90 s in a splitless gas chromatography injector. Separation of volatiles was achieved with an Agilent 6890/5973 GC-MS device equipped with a 60-m-length, 0.25-mm-i.d., 0.25-μm-film thickness fused silica capillary column (DB-5; Agilent). Helium was used as the carrier gas with a flow rate of 1 mL min−1. The column temperature was held at 40°C for 2 min and then programmed at 5°C min−1 to 250°C and held for 3 min. Mass spectrometry conditions were as follows: ion source, 200°C; electron energy, 70 eV; quadrupole temperature, 150°C; GC-MS interface zone, 280°C; and scan range, 35 to 350 mass units. The SPME fiber was heated to 250°C for 20 min to remove carryover between extractions, and three replicates of each tissue were analyzed. Hexanal was identified by its Kovats retention index and by comparison with the mass spectra of an authentic standard. The amount of hexanal was quantified using a standard curve of hexanal as the external standard and calculated on a per-plant basis.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Microscopy of border cell and root tips.
Supplemental Figure S2. Ruthenium red staining of border cells and mucilage.
Supplemental Figure S3. Starch granules in different root cell types.
Supplemental Figure S4. Iodine-stained seedling root.
Supplemental Figure S5. Border cell/whole root transcript ratios visualized within MapMan.
Supplemental Figure S6. Overview of border cell and whole root transcript profiling results.
Supplemental Figure S7. Phytohormone analyses of roots.
Supplemental Table S1. Transcript data for border cells, root tips, and whole roots.
Supplemental Table S2. Primer pairs for qRT-PCR.
Supplemental Table S3. Metabolomics data compliant with the Metabolomics Standards Initiative.
Supplementary Material
Acknowledgments
We thank Vagner Benedito for assistance in analyzing qRT-PCR data; Jin Nakashima, Terry Colberg, and Elison Blancaflor for help with microscopy; and Zhentian Lei, Elison Blancaflor, and Michael Udvardi for a careful reading of the article.
Glossary
- qRT
quantitative real-time
- PME
pectin methylesterase
- GC-MS
gas chromatography-mass spectrometry
- UHPLC
ultra-high-pressure liquid chromatography
- m/z
mass-to-charge ratio
- DHF
7,4′-dihydroxyflavone
- JA
jasmonic acid
- IAA
indole acetic acid
- SA
salicylic acid
- ABA
abscisic acid
- SPME
solid-phase microextraction
- PMEI
pectin methylesterase inhibitor
- BA
benzoic acid
- PDA
potato dextrose agar
- DMSO
dimethyl sulfoxide
- UPLC
ultra-high-performance liquid chromatography
Footnotes
This work was supported by the Samuel Roberts Noble Foundation, the National Science Foundation (grant nos. DBI 0400580, 1139489, and 1126719), and the LECO Corporation.
Articles can be viewed without a subscription.
References
- Achnine L, Huhman DV, Farag MA, Sumner LW, Blount JW, Dixon RA (2005) Genomics-based selection and functional characterization of triterpene glycosyltransferases from the model legume Medicago truncatula. Plant J 41: 875–887 [DOI] [PubMed] [Google Scholar]
- An SH, Sohn KH, Choi HW, Hwang IS, Lee SC, Hwang BK (2008) Pepper pectin methylesterase inhibitor protein CaPMEI1 is required for antifungal activity, basal disease resistance and abiotic stress tolerance. Planta 228: 61–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki T, Akashi T, Ayabe S (2000) Flavonoids of leguminous plants: structure, biological activity, and biosynthesis. J Plant Res 113: 475–488 [Google Scholar]
- Balestrieri C, Castaldo D, Giovane A, Quagliuolo L, Servillo L (1990) A glycoprotein inhibitor of pectin methylesterase in kiwi fruit (Actinidia chinensis). Eur J Biochem 193: 183–187 [DOI] [PubMed] [Google Scholar]
- Barlow PW. (2003) The root cap: cell dynamics, cell differentiation and cap function. J Plant Growth Regul 21: 261–286 [Google Scholar]
- Benedito VA, Torres-Jerez I, Murray JD, Andriankaja A, Allen S, Kakar K, Wandrey M, Verdier J, Zuber H, Ott T, et al. (2008) A gene expression atlas of the model legume Medicago truncatula. Plant J 55: 504–513 [DOI] [PubMed] [Google Scholar]
- Birnbaum K, Shasha DE, Wang JY, Jung JW, Lambert GM, Galbraith DW, Benfey PN (2003) A gene expression map of the Arabidopsis root. Science 302: 1956–1960 [DOI] [PubMed] [Google Scholar]
- Blancaflor EB, Fasano JM, Gilroy S (1998) Mapping the functional roles of cap cells in the response of Arabidopsis primary roots to gravity. Plant Physiol 116: 213–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady SM, Orlando DA, Lee JY, Wang JY, Koch J, Dinneny JR, Mace D, Ohler U, Benfey PN (2007) A high-resolution root spatiotemporal map reveals dominant expression patterns. Science 318: 801–806 [DOI] [PubMed] [Google Scholar]
- Brigham LA, Woo HH, Nicoll SM, Hawes MC (1995) Differential expression of proteins and mRNAs from border cells and root tips of pea. Plant Physiol 109: 457–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brigham LA, Woo HH, Wen F, Hawes MC (1998) Meristem-specific suppression of mitosis and a global switch in gene expression in the root cap of pea by endogenous signals. Plant Physiol 118: 1223–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broeckling CD, Huhman DV, Farag MA, Smith JT, May GD, Mendes P, Dixon RA, Sumner LW (2005) Metabolic profiling of Medicago truncatula cell cultures reveals the effects of biotic and abiotic elicitors on metabolism. J Exp Bot 56: 323–336 [DOI] [PubMed] [Google Scholar]
- Broeckling CD, Reddy IR, Duran AL, Zhao X, Sumner LW (2006) MET-IDEA: data extraction tool for mass spectrometry-based metabolomics. Anal Chem 78: 4334–4341 [DOI] [PubMed] [Google Scholar]
- Carelli M, Biazzi E, Panara F, Tava A, Scaramelli L, Porceddu A, Graham N, Odoardi M, Piano E, Arcioni S, et al. (2011) Medicago truncatula CYP716A12 is a multifunctional oxidase involved in the biosynthesis of hemolytic saponins. Plant Cell 23: 3070–3081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chehab EW, Raman G, Walley JW, Perea JV, Banu G, Theg S, Dehesh K (2006) Rice HYDROPEROXIDE LYASES with unique expression patterns generate distinct aldehyde signatures in Arabidopsis. Plant Physiol 141: 121–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Zheng Z, Huang J, Lai Z, Fan B (2009) Biosynthesis of salicylic acid in plants. Plant Signal Behav 4: 493–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski T, Bari RP, Stitt M, Scheible WR, Udvardi MK (2004) Real-time RT-PCR profiling of over 1400 Arabidopsis transcription factors: unprecedented sensitivity reveals novel root- and shoot-specific genes. Plant J 38: 366–379 [DOI] [PubMed] [Google Scholar]
- Davies PJ. (2004) Plant hormones. InDavies PJ, ed, Biosynthesis, Signal Transduction, Action, Ed 3 Kluwer Academic, Boston: pp 282–302 [Google Scholar]
- Di Matteo A, Giovane A, Raiola A, Camardella L, Bonivento D, De Lorenzo G, Cervone F, Bellincampi D, Tsernoglou D (2005) Structural basis for the interaction between pectin methylesterase and a specific inhibitor protein. Plant Cell 17: 849–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon RA, Sumner LW (2003) Legume natural products: understanding and manipulating complex pathways for human and animal health. Plant Physiol 131: 878–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dozmorov I, Centola M (2003) An associative analysis of gene expression array data. Bioinformatics 19: 204–211 [DOI] [PubMed] [Google Scholar]
- Driouich A, Durand C, Vicré-Gibouin M (2007) Formation and separation of root border cells. Trends Plant Sci 12: 14–19 [DOI] [PubMed] [Google Scholar]
- Fendrych M, Van Hautegem T, Van Durme M, Olvera-Carrillo Y, Huysmans M, Karimi M, Lippens S, Guérin CJ, Krebs M, Schumacher K, et al. (2014) Programmed cell death controlled by ANAC033/SOMBRERO determines root cap organ size in Arabidopsis. Curr Biol 24: 931–940 [DOI] [PubMed] [Google Scholar]
- Ferreyra MLF, Rius SP, Casati P (2012) Flavonoids: biosynthesis, biological functions, and biotechnological applications. Front Plant Sci 3: 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forkmann G, Martens S (2001) Metabolic engineering and applications of flavonoids. Curr Opin Biotechnol 12: 155–160 [DOI] [PubMed] [Google Scholar]
- Greathouse GA, Rigler N (1940) The chemistry of resistance of plants to Phymatotrichum root rot. V. Influence of alkaloids on growth of fungi. Phytopathology 30: 475–485 [Google Scholar]
- Gunawardena U, Hawes MC (2002) Tissue specific localization of root infection by fungal pathogens: role of root border cells. Mol Plant Microbe Interact 15: 1128–1136 [DOI] [PubMed] [Google Scholar]
- Gunawardena U, Rodriguez M, Straney D, Romeo JT, VanEtten HD, Hawes MC (2005) Tissue-specific localization of pea root infection by Nectria haematococca: mechanisms and consequences. Plant Physiol 137: 1363–1374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamamoto L, Hawes MC, Rost TL (2006) The production and release of living root cap border cells is a function of root apical meristem type in dicotyledonous angiosperm plants. Ann Bot (Lond) 97: 917–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haridas V, Higuchi M, Jayatilake GS, Bailey D, Mujoo K, Blake ME, Arntzen CJ, Gutterman JU (2001) Avicins: triterpenoid saponins from Acacia victoriae (Bentham) induce apoptosis by mitochondrial perturbation. Proc Natl Acad Sci USA 98: 5821–5826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawes MC, Bengough G, Cassab G, Ponce G (2003) Root caps and rhizosphere. J Plant Growth Regul 21: 352–367 [Google Scholar]
- Hawes MC, Brigham LA, Wen F, Woo HH, Zhu Y (1998) Function of root border cells in plant health: pioneers in the rhizosphere. Annu Rev Phytopathol 36: 311–327 [DOI] [PubMed] [Google Scholar]
- Hawes MC, Gunawardena U, Miyasaka S, Zhao X (2000) The role of root border cells in plant defense. Trends Plant Sci 5: 128–133 [DOI] [PubMed] [Google Scholar]
- Hayashi H, Huang P, Kirakosyan A, Inoue K, Hiraoka N, Ikeshiro Y, Kushiro T, Shibuya M, Ebizuka Y (2001) Cloning and characterization of a cDNA encoding β-amyrin synthase involved in glycyrrhizin and soyasaponin biosyntheses in licorice. Biol Pharm Bull 24: 912–916 [DOI] [PubMed] [Google Scholar]
- Holmes P, Goffard N, Weiller GF, Rolfe BG, Imin N (2008) Transcriptional profiling of Medicago truncatula meristematic root cells. BMC Plant Biol 8: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath E, Szalai G, Janda T (2007) Induction of abiotic stress tolerance by salicylic acid signaling. J Plant Growth Regul 26: 290–300 [Google Scholar]
- Huhman DV, Berhow MA, Sumner LW (2005) Quantification of saponins in aerial and subterranean tissues of Medicago truncatula. J Agric Food Chem 53: 1914–1920 [DOI] [PubMed] [Google Scholar]
- Huhman DV, Sumner LW (2002) Metabolic profiling of saponins in Medicago sativa and Medicago truncatula using HPLC coupled to an electrospray ion-trap mass spectrometer. Phytochemistry 59: 347–360 [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP (2003) Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res 31: e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kant S, Bi YM, Zhu T, Rothstein SJ (2009) SAUR39, a small auxin-up RNA gene, acts as a negative regulator of auxin synthesis and transport in rice. Plant Physiol 151: 691–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kape R, Parniske M, Werner D (1991) Chemotaxis and nod gene activity of Bradyrhizobium japonicum in response to hydroxycinnamic acids and isoflavonoids. Appl Environ Microbiol 57: 316–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knauss S, Rohrmeier T, Lehle L (2003) The auxin-induced maize gene ZmSAUR2 encodes a short-lived nuclear protein expressed in elongating tissues. J Biol Chem 278: 23936–23943 [DOI] [PubMed] [Google Scholar]
- Kosuta S, Chabaud M, Lougnon G, Gough C, Dénarié J, Barker DG, Bécard G (2003) A diffusible factor from arbuscular mycorrhizal fungi induces symbiosis-specific MtENOD11 expression in roots of Medicago truncatula. Plant Physiol 131: 952–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leek JT, Monsen E, Dabney AR, Storey JD (2006) EDGE: extraction and analysis of differential gene expression. Bioinformatics 22: 507–508 [DOI] [PubMed] [Google Scholar]
- Li XF, Ma JF, Matsumoto H (2000) Pattern of aluminum-induced secretion of organic acids differs between rye and wheat. Plant Physiol 123: 1537–1544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ljung K, Hull AK, Celenza J, Yamada M, Estelle M, Normanly J, Sandberg G (2005) Sites and regulation of auxin biosynthesis in Arabidopsis roots. Plant Cell 17: 1090–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marek SM, Hansen K, Romanish M, Thorn RG (2009) Molecular systematics of the cotton root rot pathogen, Phymatotrichopsis omnivora. Persoonia 22: 63–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyasaka SC, Hawes MC (2001) Possible role of root border cells in detection and avoidance of aluminum toxicity. Plant Physiol 125: 1978–1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modolo LV, Blount JW, Achnine L, Naoumkina MA, Wang X, Dixon RA (2007) A functional genomics approach to (iso)flavonoid glycosylation in the model legume Medicago truncatula. Plant Mol Biol 64: 499–518 [DOI] [PubMed] [Google Scholar]
- Nagahashi G, Douds DD Jr (2004) Isolated root caps, border cells, and mucilage from host roots stimulate hyphal branching of the arbuscular mycorrhizal fungus, Gigaspora gigantea. Mycol Res 108: 1079–1088 [DOI] [PubMed] [Google Scholar]
- Naoumkina M, Farag MA, Sumner LW, Tang Y, Liu CJ, Dixon RA (2007) Different mechanisms for phytoalexin induction by pathogen and wound signals in Medicago truncatula. Proc Natl Acad Sci USA 104: 17909–17915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naoumkina MA, Modolo LV, Huhman DV, Urbanczyk-Wochniak E, Tang Y, Sumner LW, Dixon RA (2010) Genomic and coexpression analyses predict multiple genes involved in triterpene saponin biosynthesis in Medicago truncatula. Plant Cell 22: 850–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osbourn AE. (2003) Saponins in cereals. Phytochemistry 62: 1–4 [DOI] [PubMed] [Google Scholar]
- Pan X, Welti R, Wang X (2010) Quantitative analysis of major plant hormones in crude plant extracts by high-performance liquid chromatography-mass spectrometry. Nat Protoc 5: 986–992 [DOI] [PubMed] [Google Scholar]
- Peck MC, Fisher RF, Long SR (2006) Diverse flavonoids stimulate NodD1 binding to nod gene promoters in Sinorhizobium meliloti. J Bacteriol 188: 5417–5427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollier J, Morreel K, Geelen D, Goossens A (2011) Metabolite profiling of triterpene saponins in Medicago truncatula hairy roots by liquid chromatography Fourier transform ion cyclotron resonance mass spectrometry. J Nat Prod 74: 1462–1476 [DOI] [PubMed] [Google Scholar]
- Raiola A, Camardella L, Giovane A, Mattei B, De Lorenzo G, Cervone F, Bellincampi D (2004) Two Arabidopsis thaliana genes encode functional pectin methylesterase inhibitors. FEBS Lett 557: 199–203 [DOI] [PubMed] [Google Scholar]
- Reymond P, Farmer EE (1998) Jasmonate and salicylate as global signals for defense gene expression. Curr Opin Plant Biol 1: 404–411 [DOI] [PubMed] [Google Scholar]
- Schliemann W, Ammer C, Strack D (2008) Metabolite profiling of mycorrhizal roots of Medicago truncatula. Phytochemistry 69: 112–146 [DOI] [PubMed] [Google Scholar]
- Seki H, Ohyama K, Sawai S, Mizutani M, Ohnishi T, Sudo H, Akashi T, Aoki T, Saito K, Muranaka T (2008) Licorice beta-amyrin 11-oxidase, a cytochrome P450 with a key role in the biosynthesis of the triterpene sweetener glycyrrhizin. Proc Natl Acad Sci USA 105: 14204–14209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao H, He X, Achnine L, Blount JW, Dixon RA, Wang X (2005) Crystal structures of a multifunctional triterpene/flavonoid glycosyltransferase from Medicago truncatula. Plant Cell 17: 3141–3154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibuya M, Hoshino M, Katsube Y, Hayashi H, Kushiro T, Ebizuka Y (2006) Identification of beta-amyrin and sophoradiol 24-hydroxylase by expressed sequence tag mining and functional expression assay. FEBS J 273: 948–959 [DOI] [PubMed] [Google Scholar]
- Shirley BW. (1998) Flavonoids in seeds and grains: physiological function, agronomic importance and the genetics of biosynthesis. Seed Sci Res 8: 415–422 [Google Scholar]
- Stacey G, Libault M, Brechenmacher L, Wan J, May GD (2006) Genetics and functional genomics of legume nodulation. Curr Opin Plant Biol 9: 110–121 [DOI] [PubMed] [Google Scholar]
- Stafford HA. (1997) Roles of flavonoids in symbiotic and defense functions in legume roots. Bot Rev 63: 27–39 [Google Scholar]
- Storey JD, Tibshirani R (2003) Statistical significance for genomewide studies. Proc Natl Acad Sci USA 100: 9440–9445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumner LW, Amberg A, Barrett D, Beale MH, Beger R, Daykin CA, Fan TWM, Fiehn O, Goodacre R, Griffin JL, et al. (2007) Proposed minimum reporting standards for chemical analysis. Metabolomics 3: 211–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H, Achnine L, Xu R, Matsuda SP, Dixon RA (2002) A genomics approach to the early stages of triterpene saponin biosynthesis in Medicago truncatula. Plant J 32: 1033–1048 [DOI] [PubMed] [Google Scholar]
- Ta TC, Joy KW, Ireland RJ (1984) Amino acid metabolism in pea leaves: utilization of nitrogen from amide and amino groups of [15N]asparagine. Plant Physiol 74: 822–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tava A, Odoardi M (1996) Saponins from Medicago ssp.: chemical characterization and biological activity against insects. InWaller GR, Yamasaki K, eds, Advances in Experimental Medicine and Biology, Vol 405 Plenum Press, New York, pp 97–109 [DOI] [PubMed] [Google Scholar]
- Tava A, Scotti C, Avato P (2011) Biosynthesis of saponins in the genus Medicago. Phytochem Rev 10: 459–469 [Google Scholar]
- Thimm O, Bläsing O, Gibon Y, Nagel A, Meyer S, Krüger P, Selbig J, Müller LA, Rhee SY, Stitt M (2004) MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J 37: 914–939 [DOI] [PubMed] [Google Scholar]
- Uppalapati SR, Marek SM, Lee HK, Nakashima J, Tang Y, Sledge MK, Dixon RA, Mysore KS (2009) Global gene expression profiling during Medicago truncatula-Phymatotrichopsis omnivora interaction reveals a role for jasmonic acid, ethylene, and the flavonoid pathway in disease development. Mol Plant Microbe Interact 22: 7–17 [DOI] [PubMed] [Google Scholar]
- Uppalapati SR, Young CA, Marek SM, Mysore KS (2010) Phymatotrichum (cotton) root rot caused by Phymatotrichopsis omnivora: retrospects and prospects. Mol Plant Pathol 11: 325–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbanczyk-Wochniak E, Sumner LW (2007) MedicCyc: a biochemical pathway database for Medicago truncatula. Bioinformatics 23: 1418–1423 [DOI] [PubMed] [Google Scholar]
- Urbanczyk-Wochniak E, Usadel B, Thimm O, Nunes-Nesi A, Carrari F, Davy M, Bläsing O, Kowalczyk M, Weicht D, Polinceusz A, et al. (2006) Conversion of MapMan to allow the analysis of transcript data from solanaceous species: effects of genetic and environmental alterations in energy metabolism in the leaf. Plant Mol Biol 60: 773–792 [DOI] [PubMed] [Google Scholar]
- Vance CP, Kirk TK, Sherwood RT (1980) Lignification as a mechanism of disease resistance. Annu Rev Phytopathol 18: 259–288 [Google Scholar]
- Vicré M, Santaella C, Blanchet S, Gateau A, Driouich A (2005) Root border-like cells of Arabidopsis: microscopical characterization and role in the interaction with rhizobacteria. Plant Physiol 138: 998–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterman PG. (1996) Saponins used in food and agriculture. InWaller GR, Yamasaki K, eds, Advances in Experimental Medicine and Biology, Vol 405 Plenum Press, New York, p 569 [Google Scholar]
- Wen F, VanEtten HD, Tsaprailis G, Hawes MC (2007) Extracellular proteins in pea root tip and border cell exudates. Plant Physiol 143: 773–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen F, Zhu Y, Hawes MC (1999) Effect of pectin methylesterase gene expression on pea root development. Plant Cell 11: 1129–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo HH, Hirsch AM, Hawes MC (2004) Altered susceptibility to infection by Sinorhizobium meliloti and Nectria haematococca in alfalfa roots with altered cell cycle. Plant Cell Rep 22: 967–973 [DOI] [PubMed] [Google Scholar]
- Young ND, Cannon SB, Sato S, Kim D, Cook DR, Town CD, Roe BA, Tabata S (2005) Sequencing the genespaces of Medicago truncatula and Lotus japonicus. Plant Physiol 137: 1174–1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young ND, Debellé F, Oldroyd GE, Geurts R, Cannon SB, Udvardi MK, Benedito VA, Mayer KF, Gouzy J, Schoof H, et al. (2011) The Medicago genome provides insight into the evolution of rhizobial symbioses. Nature 480: 520–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Subramanian S, Zhang Y, Yu O (2007) Flavone synthases from Medicago truncatula are flavanone-2-hydroxylases and are important for nodulation. Plant Physiol 144: 741–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JY, Cruz DE, Carvalho MH, Torres-Jerez I, Kang Y, Allen SN, Huhman DV, Tang Y, Murray J, Sumner LW, et al. (2014) Global reprogramming of transcription and metabolism in Medicago truncatula during progressive drought and after rewatering. Plant Cell Environ 37: 2553–2576 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.