Legume root hairs remodel the interface with symbiotic rhizobia prior to initiating the tubular-growing infection thread.
Abstract
In many legumes, root entry of symbiotic nitrogen-fixing rhizobia occurs via host-constructed tubular tip-growing structures known as infection threads (ITs). Here, we have used a confocal microscopy live-tissue imaging approach to investigate early stages of IT formation in Medicago truncatula root hairs (RHs) expressing fluorescent protein fusion reporters. This has revealed that ITs only initiate 10 to 20 h after the completion of RH curling, by which time major modifications have occurred within the so-called infection chamber, the site of bacterial entrapment. These include the accumulation of exocytosis (M. truncatula Vesicle-Associated Membrane Protein721e)- and cell wall (M. truncatula EARLY NODULIN11)-associated markers, concomitant with radial expansion of the chamber. Significantly, the infection-defective M. truncatula nodule inception-1 mutant is unable to create a functional infection chamber. This underlines the importance of the NIN-dependent phase of host cell wall remodeling that accompanies bacterial proliferation and precedes IT formation, and leads us to propose a two-step model for rhizobial infection initiation in legume RHs.
Legumes possess the remarkable capacity to improve their nutrition by establishing a nitrogen-fixing root nodule symbiosis (RNS) with soil bacteria collectively called rhizobia. In many legumes such as Medicago truncatula, rhizobia penetrate across the root epidermis and outer cortex to reach the differentiating nodule tissues via sequentially constructed transcellular compartments known as infection threads (ITs; Gage, 2004). It is now well established that this mode of entry through specialized infection compartments, often referred to as accommodation, is shared with the more ancient arbuscular mycorrhizal (AM) symbiosis, from which the legume-Rhizobium RNS is thought to have evolved (Parniske, 2008; Markmann and Parniske, 2009). Furthermore, strong evidence indicates that the signaling and cellular mechanisms underlying IT formation in legumes are closely related to those used for infection compartment formation during AM infection of epidermal and outer cortical tissues (Bapaume and Reinhardt, 2012; Oldroyd, 2013).
Rhizobial infection is set in motion after an initial molecular dialogue between symbiotic partners, in which rhizobial lipo-chitooligosaccharide (LCO) Nod factors (NFs) are key signaling molecules (for review, see Oldroyd, 2013). Host responses to NF signaling include rapid and sustained nuclear-associated Ca2+ oscillations (Ca2+ spiking; Ehrhardt et al., 1996; Oldroyd and Downie, 2006; Sieberer et al., 2009; Capoen et al., 2011) and the rapid expression of early epidermal marker genes such as M. truncatula EARLY NODULIN11 (Charron et al., 2004). The activation of nuclear Ca2+ spiking is one of the most characteristic features of the so-called common symbiotic signaling pathway, common to both RNS and AM (Kistner and Parniske, 2002; Singh and Parniske, 2012). Whereas these preinfection responses to NFs are observed in the majority of elongating root hairs (RHs) early after rhizobial inoculation (Journet et al., 2001; Wais et al., 2002), ITs are only formed in a small subset of RHs, and MtENOD11 expression is strongly activated at these rhizobial infection sites (Journet et al., 2001; Boisson-Dernier et al., 2005).
ITs are tubular plant-derived structures delimited by a membrane that is contiguous with the RH plasmalemma and a layer of cell wall-like material, thus isolating the rhizobia from the host cell cytoplasm (Gage, 2004). These apoplastic infection compartments are progressively constructed along the length of the RH with their growing tip connected via a cytoplasmic bridge to the migrating RH nucleus. This broad cytoplasmic column provides the cell machinery for tip growth, which involves targeted exocytosis of membrane and extracellular materials to the growing apex of the IT (Oldroyd et al., 2011; Bapaume and Reinhardt, 2012). It is presumed that this cytoplasmic bridge shares an equivalent role to the prepenetration apparatus (PPA) formed at the onset of AM fungal infection (Genre et al., 2005, 2008). We now know that the IT tip region is formed in advance of rhizobial colonization and is progressively populated by dividing rhizobia that also physically move down the thread (Gage, 2004; Fournier et al., 2008). It has been proposed that the matrix of the growing IT tip is initially in a fluid or gel-like state compatible with bacterial growth and movement (Brewin, 2004; Fournier et al., 2008). This relative plasticity could result in part from the presence of atypical extracellular (glyco) proteins such as the repetitive Pro-rich proteins MtENOD11/MtENOD12 because their low Tyr content is presumed to limit cross linking to other wall components (Scheres et al., 1990; Pichon et al., 1992; Journet et al., 2001).
Nevertheless, the mechanism by which rhizobial IT formation is initiated in RHs is not clear. Whereas AM fungal hyphae form contact structures called hyphopodia on the exposed surface of nonhair epidermal cells prior to PPA formation and perifungal infection compartment formation (Genre et al., 2005), rhizobial entry requires that the bacteria first become entrapped between RH walls. Attachment of rhizobia close to a growing RH tip induces a continuous reorientation of tip growth, most likely the result of localized NF production (Esseling et al., 2003), eventually leading to RH curling and subsequent bacterial entrapment within a closed chamber in the center of the curl (Catoira et al., 2001; Geurts et al., 2005). Rhizobial entrapment can also occur between the cell walls of two touching RHs (Dart, 1974; Gage, 2004).
The closed chamber in curled RHs has often been termed the infection pocket (e.g. Murray, 2011; Guan et al., 2013). However, because this term is also used to designate a quite different and larger structure formed in root subepidermal tissues of legumes during intercellular infection after crack entry and involving localized cell death (Goormachtig et al., 2004), we propose to use the term infection chamber to describe the unique enclosure formed during rhizobial RH infection.
After entrapment, it has been proposed that rhizobia multiply to form a so-called microcolony (Gage et al., 1996; Limpens et al., 2003), and that IT polar growth initiates in front of this microcolony by local invagination of the RH plasmalemma combined with exocytosis of extracellular materials (Gage, 2004). Furthermore, it has been suggested that localized degradation of the chamber wall would allow the rhizobia to access the newly formed IT (Callaham and Torrey, 1981; Turgeon and Bauer, 1985). However, a detailed investigation of this particular stage of rhizobial infection is lacking, particularly concerning when and where the rhizobia/cell wall interface becomes modified. Such studies have been limited until now, notably because ITs develop only in a low proportion of curled RHs (Dart, 1974).
To attempt to answer this question, we have used a live-tissue imaging approach developed for in vivo confocal microscopy in M. truncatula (Fournier et al., 2008; Cerri et al., 2012; Sieberer et al., 2012) and particularly well adapted to time-lapse studies of the initial stages of rhizobial infection, including RH curling and IT formation. To investigate modifications occurring at the RH interface with the enclosed rhizobia during these early stages, we prepared M. truncatula plants expressing fluorescent protein fusions aimed at detecting both exocytosis activity and cell wall remodeling during the initial construction of the IT apoplastic compartment. To this end, we made use of the M. truncatula Vesicle-Associated Membrane Protein721e (MtVAMP721e; Ivanov et al., 2012), recently shown to label exocytosis sites both in growing RHs and during AM colonization (Genre et al., 2012), as well as the infection- and cell wall-associated MtENOD11 Pro-rich glycoprotein (Journet et al., 2001). Our experiments have revealed that IT development in curled RHs only initiates after a lengthy interval of 10 to 20 h, during which sustained exocytosis and MtENOD11 secretion to the infection chamber are associated with radial expansion as well as remodeling of the surrounding walls. Importantly, it was found that the infection-defective M. truncatula nodule inception-1 (Mtnin-1) mutant (Marsh et al., 2007) is impaired in chamber remodeling. Our findings led us to propose a new model for IT formation in which the infection chamber first differentiates into a globular apoplastic compartment displaying similarities to the future IT, and in which the enclosed rhizobia multiply. This is then followed by a switch from radial to tubular growth corresponding to tip-driven IT growth and associated movement of rhizobia into the extending thread. Importantly, this two-step model no longer requires that the host cell wall is degraded to allow access of the colonizing rhizobia to the newly initiated IT.
RESULTS
IT Tip Growth in M. truncatula Initiates 10 to 20 h after the Completion of RH Curling
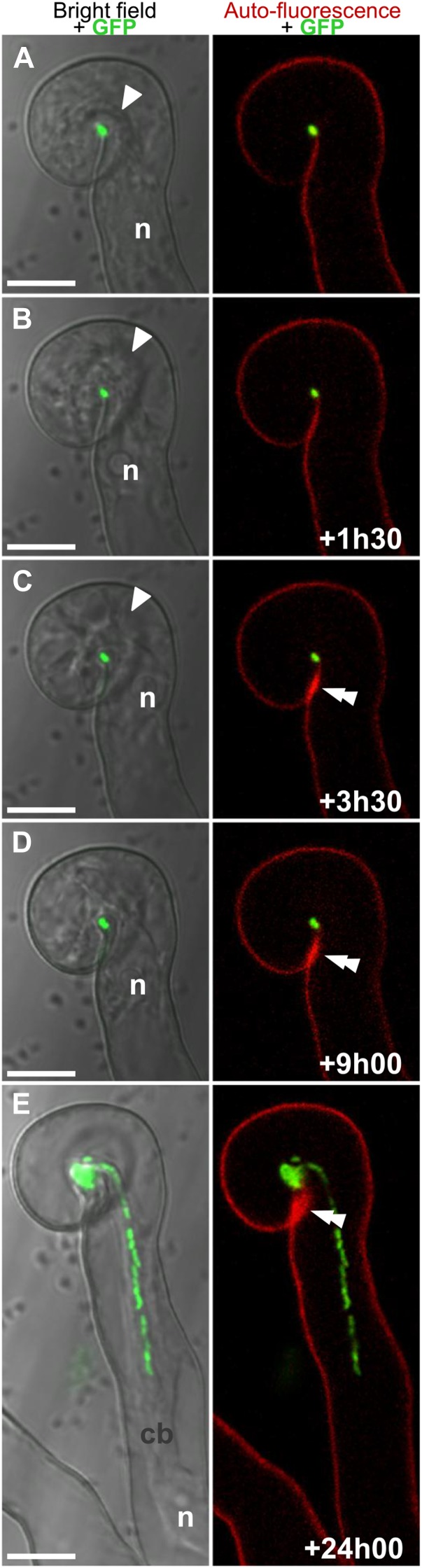
To study the cellular events associated with IT initiation, a live-tissue imaging approach was used to identify and continuously monitor RHs at different stages of curling and rhizobial entrapment (Fournier et al., 2008; Cerri et al., 2012). The observation of many such RHs surprisingly revealed that IT tip growth only initiated many hours after the completion of curling, and in no case did we observe IT formation immediately after curling. In the example shown in Figure 1, RH curling around the entrapped rhizobia is close to completion at the initial time of observation (Fig. 1A). The unchanged position of the RH tip in Figure 1, B and C (single arrowhead) indicates that curling had terminated during the first 1.5-h period. Tubular IT formation was not observed during the next 7.5 h, despite the fact that the nucleus and associated cytoplasm were stably localized in the curled tip region of the RH (Fig. 1, B–D). However, a growing IT was observed within this particular RH 15 h later (Fig. 1E) and continued its progression during the following hours (not shown). Based on the length of the IT in Figure 1E, we estimate that elongation had been underway for approximately 6–8 h (average growth rate of ITs is 4–5 µm h−1 in M. truncatula; Fournier et al., 2008). Thus, in this particular case, the delay between the completion of RH curling and IT initiation can be estimated to be around 15 to 18 h. Similar image series obtained for a number of other RHs initiating infection together revealed that the delay between the completion of bacterial entrapment and IT initiation is in the range of 10 to 20 h. During this lengthy period, the infection chamber became progressively easier to distinguish from the surrounding cytoplasm, most likely as a consequence of enlargement and surface modifications before IT initiation (Supplemental Fig. S1). To further investigate this, we exploited fluorescent cellular markers for monitoring both host exocytosis activity and possible modifications to the infection chamber extracellular matrix.
Figure 1.
IT initiation does not immediately follow rhizobial entrapment within the curled RH. Bright-field (left) and the corresponding confocal (right) images of an M. truncatula RH at different times after tip curling around GFP-labeled S. meliloti 2011. In the bright-field images, the location of the nucleus (n) is indicated, as well as the position of the RH tip (arrowhead, A–C). In the fluorescence images, the region of the cell wall adjacent to the infection chamber where autofluorescent material accumulates is indicated (double arrowhead, C–E). Note that the IT walls (E) are devoid of autofluorescent material in contrast with the rest of the RH wall. Confocal images of GFP fluorescence (single optical sections across the infection chamber) were superposed either with the laser transmission images (left) or with the cell wall autofluorescence (right). cb, Cytoplasmic bridge. Bars = 10 µm.
The GFP-MtVAMP721e Exocytosis Marker Accumulates Rapidly around the Newly Formed Infection Chamber
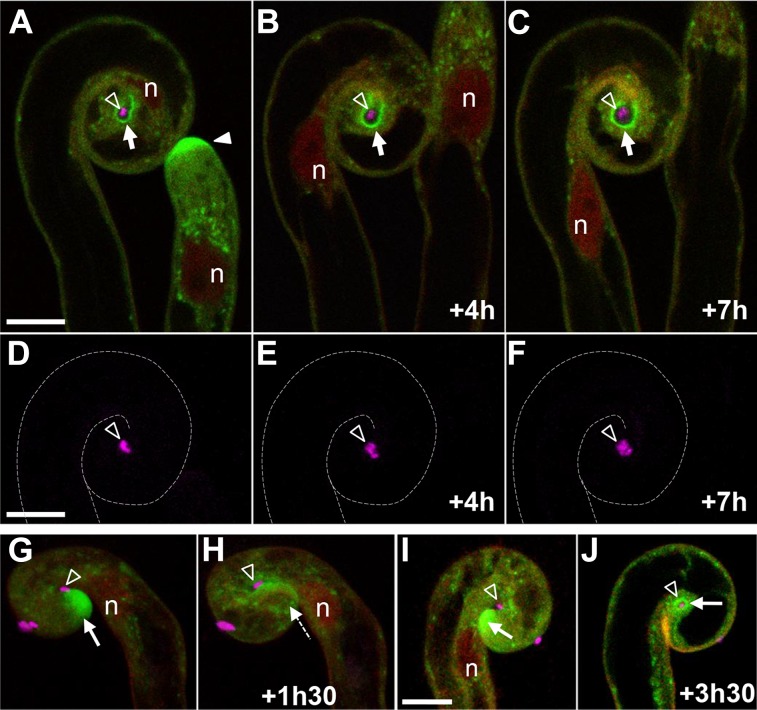
To evaluate potential exocytosis activity associated with the RH infection chamber, we made use of M. truncatula plants expressing a fluorescence-tagged MtVAMP721e (Genre et al., 2012; Ivanov et al., 2012) in their roots. In noninoculated or noncolonized RHs, the GFP-MtVAMP721e fusion protein primarily localizes to the vesicle-rich region behind the growing tip of elongating RHs and at lower levels as localized puncta elsewhere in the cytoplasm (Genre et al., 2012; Fig. 2A). Our in vivo observations have revealed that the GFP-VAMP721e fluorescent signal is no longer located on the RH tip region of fully curled RHs with entrapped rhizobia, but now surrounds the infection chamber, outlining its contours (Fig. 2A). This signal is presumably associated with the plasma membrane bordering the infection chamber, most likely corresponding to the accumulation of GFP-VAMP-labeled vesicles. By monitoring such curled RHs over time, we further discovered that the exocytosis reporter is continuously present around the infection chamber (e.g. over the entire 7-h period illustrated in Fig. 2, A–C). This suggests a lengthy period of sustained exocytosis targeted toward the infection chamber. This continuous exocytosis activity was directly associated with radial expansion of the infection chamber during the same period of time. Our results thus argue that membrane and extracellular material are actively conveyed toward the infection chamber after the completion of RH curling and before IT initiation. As illustrated in Figure 2, D–F, the continuous enlargement of the infection chamber is accompanied by progressive multiplication of the enclosed rhizobia.
Figure 2.
GFP-MtVAMP721e identifies exocytotic activity surrounding the infection chamber in curled RHs. A to C, The intracellular localization of the GFP-MtVAMP721e fusion (green) in M. truncatula RHs was imaged over a 7-h period in both a curled hair and an adjacent tip-growing hair after inoculation with cCFP-labeled S. meliloti (magenta). A, The GFP-VAMP721e fusion protein fluorescence surrounds the enclosed bacteria (dark arrowhead) within the infection chamber of the curled RH (left hair, arrow), whereas GFP fluorescence localizes predominantly to the tip of the growing RH (right hair, solid arrowhead). B and C, Throughout the 7-h monitoring period, the GFP fluorescence localizes to the periphery of the infection chamber, which undergoes progressive radial expansion within the curled RH. D to F, Identical RH as in A to C, showing in more detail that the cCFP-labeled rhizobia within the infection chamber (arrowhead) have multiplied concomitantly with chamber expansion. The dashed lines indicate the RH contours. G to J, GFP-MtVAMP721e localization in RHs that are just completing curling. The completion of RH curling around an attached bacterial cell (arrowhead) occurs during the observation period (1 h 30 min [+1h30]) and is associated with the rapid loss (dashed arrow in H) of the tip-localized GFP fluorescence (arrow in G). I and J, In a second RH, two different stages are identified by GFP-MtVAMP721e localization (arrows). I, As in G, tip-localized GFP fluorescence indicates that the RH is still curling. J, After 3 h and 30 min (+3h30), curling has terminated, and GFP was found predominantly around the closed infection chamber (arrowhead), whereas the RH tip fluorescence was lost (not in focal plane). Confocal images are based on single optical sections across the infection chamber for A to C, z axis projections of seven serial optical sections encompassing the entire rhizobial microcolony for D to F, three-dimensional images reconstructed from confocal z-stacks (22 serial optical sections) for G and H, z axis projection of five serial optical sections encompassing the RH tip and attached rhizobia for I, and z axis projection of two serial optical sections across the infection chamber for J. n, Nucleus. Bars = 10 µm.
To investigate in more detail when the host membrane/cell wall interface remodeling is initiated after RH curling, we focused on the earliest stages of infection chamber formation. Tip-focused accumulation of GFP-MtVAMP721e characteristic of elongating RHs (Genre et al., 2012; Fig. 2A) persisted during RH curling around attached rhizobia (Fig. 2, G and I). However, as soon as RH tip curling was completed, fluorescence labeling at the RH tip was lost (Fig. 2H, dashed arrow). Importantly, Figure 2J shows that the accumulation of GFP-MtVAMP721e around enclosed rhizobia initiates within the first hours after the completion of RH tip curling. Taking into account the fact that rhizobial cell division within growing M. truncatula ITs takes at least 4 h (Gage, 2002), these findings imply that exocytosis of extracellular material toward the newly formed infection chamber initiates before significant rhizobial multiplication has occurred within the chamber.
The Mtnin-1 Mutant Is Impaired for Exocytosis Targeted to the Infection Chamber
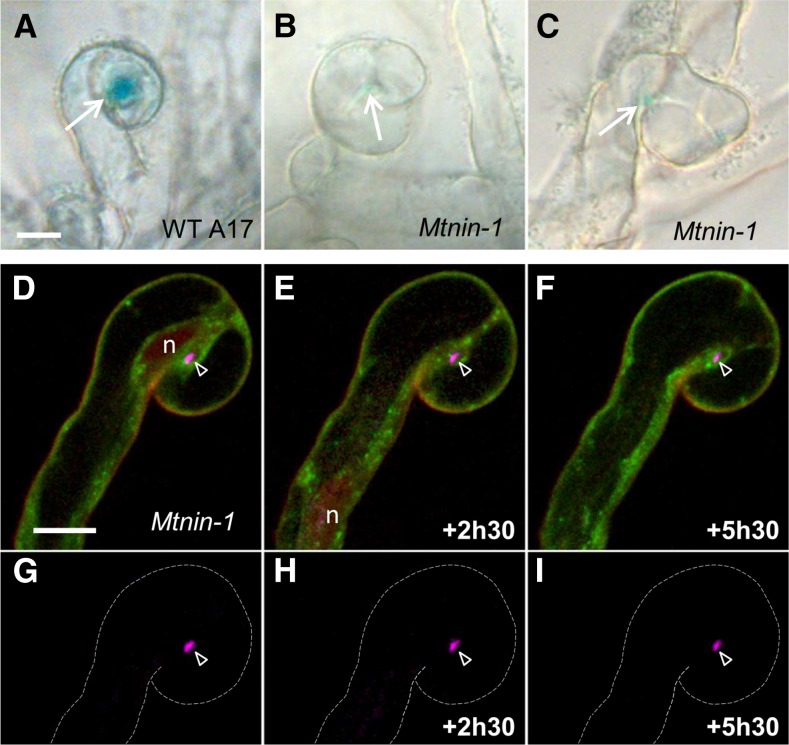
We next investigated whether exocytosis targeted to the infection chamber occurred in the case of the infection mutant Mtnin-1, which is able to enclose rhizobia by RH curling but fails to form ITs and shows impaired multiplication of the enclosed rhizobia (Marsh et al., 2007; Murray, 2011). Indeed, compared with wild-type plants where large rhizobial microcolonies were present within curled RHs (Fig. 3A), few bacteria were detectable within curled RHs in the Mtnin-1 mutant (Fig. 3, B and C and D–F). Monitoring GFP-VAMP721e fusion exocytosis-related localization in nin RH curls revealed that, in this case, the entrapment of rhizobia in curled RHs was never followed by GFP-VAMP721e accumulation around the infection chamber (Fig. 3, D–F; Supplemental Fig. S2), despite the fact that the localization of the fusion protein at the tip of growing RHs was otherwise similar to that in the wild type (Supplemental Fig. S3). Furthermore, the nucleus and associated cytoplasm moved down the RH shaft after a few hours in the nin mutant (Fig. 3, D–F), whereas they remained close to the enclosed bacteria in wild-type plants (Figs. 1, A–D and 2, A–C). The absence of exocytotic activity is consistent with the lack of radial expansion of the infection chamber in curled nin RHs (Fig. 3, G–I; Supplemental Fig. S2). Taken together, this indicates that MtNIN is required to initiate the remodeling of the infection chamber after rhizobial entrapment.
Figure 3.
In the infection-defective Mtnin mutant, entrapment of rhizobia within the infection chamber is not followed by targeted exocytosis or bacterial multiplication. A to C, Rhizobial microcolony development is strongly reduced in Mtnin-1 compared with the wild type (WT), as indicated by the level of β-galactosidase activity of S. meliloti (δ-aminolevulinate synthetase [hemA]-β-galactosidaseZ) colonies (arrows) entrapped within curled RHs of wild-type A17 (A) or Mtnin-1 (B and C) plants. D to F, The GFP-VAMP721e fusion protein does not accumulate at the periphery of the infection chamber (arrowhead) in Mtnin-1 plants expressing the exocytosis reporter, although cCFP-labeled S. meliloti (magenta) are present in the chamber. Note that, as for the wild type, background GFP-VAMP721e fluorescence was detected in both the cytoplasm and cytoplasmic bodies. G to I, Consistent with A to C, the rhizobial microcolony in the nin mutant does not visibly enlarge over the observation period compared with the wild type (see Fig. 2). The dashed lines indicate the RH contours. n, Nucleus. Bars = 10 µm.
The Extracellular MtENOD11 Protein Is Targeted to the Infection Chamber after Rhizobial Entrapment
To examine whether exocytotic activity in the infection chamber is associated with cell wall remodeling, we monitored the accumulation of the rhizobial infection-associated extracellular protein MtENOD11. Recently, we have found that a Yellow Fluorescent Protein-tagged MtENOD11 fusion protein, when expressed under the control of native promoter sequences (Boisson-Dernier et al., 2005), accumulates at the periphery of elongating ITs in M. truncatula RHs, and especially at the growing IT tips (J. Fournier, A. Teillet, M.-C. Auriac, D.G. Barker, and F. de Carvalho-Niebel, unpublished data). Using the same YFP-tagged protein fusion, an intense fluorescent signal could be observed within the center of the curled hair before IT formation (Fig. 4, A and B). This focalized accumulation around the entrapped bacteria is similarly observed in the less frequent situation in which the symbiotic bacteria have become entrapped between the walls of two growing RHs (Fig. 4, C and D). Once the tubular IT has initiated from the infection chamber, the fluorescent protein is also found to be associated with the growing IT tip as expected (Fig. 4C). The intense YFP-MtENOD11 fluorescence is specific to the cell wall matrix associated with rhizobial infection structures, including the infection chamber, contrasting with the very low levels of YFP fluorescence labeling the RH walls (Fig. 4, A and B; J. Fournier, A. Teillet, M.-C. Auriac, D.G. Barker, and F. de Carvalho-Niebel, unpublished data). This implies that the cell wall matrix surrounding rhizobia has a particular composition that is distinct from the normal RH extracellular matrix. In line with this, this region also differs with respect to cell wall autofluorescence. RH walls display intrinsic fluorescence that can be used to visualize the cell contours (e.g. Fig. 1, right, and Fig. 4B). Under identical conditions, the wall matrix directly surrounding the enclosed rhizobia and corresponding to the region of maximal YFP-MtENOD11 accumulation (Fig. 4A) is totally devoid of autofluorescent material (Figs. 1 and 4B). Intriguingly, a strong autofluorescent signal can be observed in a limited region of the wall adjacent to the infection chamber (Figs. 1, C–E, 4B). Additional studies will be needed to identify the chemical nature of this autofluorescent material, the accumulation of which is independent of exocytosis processes related to MtVAMP721e, and to determine whether it results from local increased secretion of phenolics or other compounds, or from enzymatic modifications to existing cell wall components. In conclusion, these observations indicate that, during the lengthy period between RH curling and IT initiation, the wall/matrix surrounding the rhizobia within the expanding infection chamber is characterized by the absence of autofluorescent material and a strong and focused accumulation of the MtENOD11 Pro-rich protein, supporting the idea that specific wall remodeling is taking place in the chamber during this period.
Figure 4.
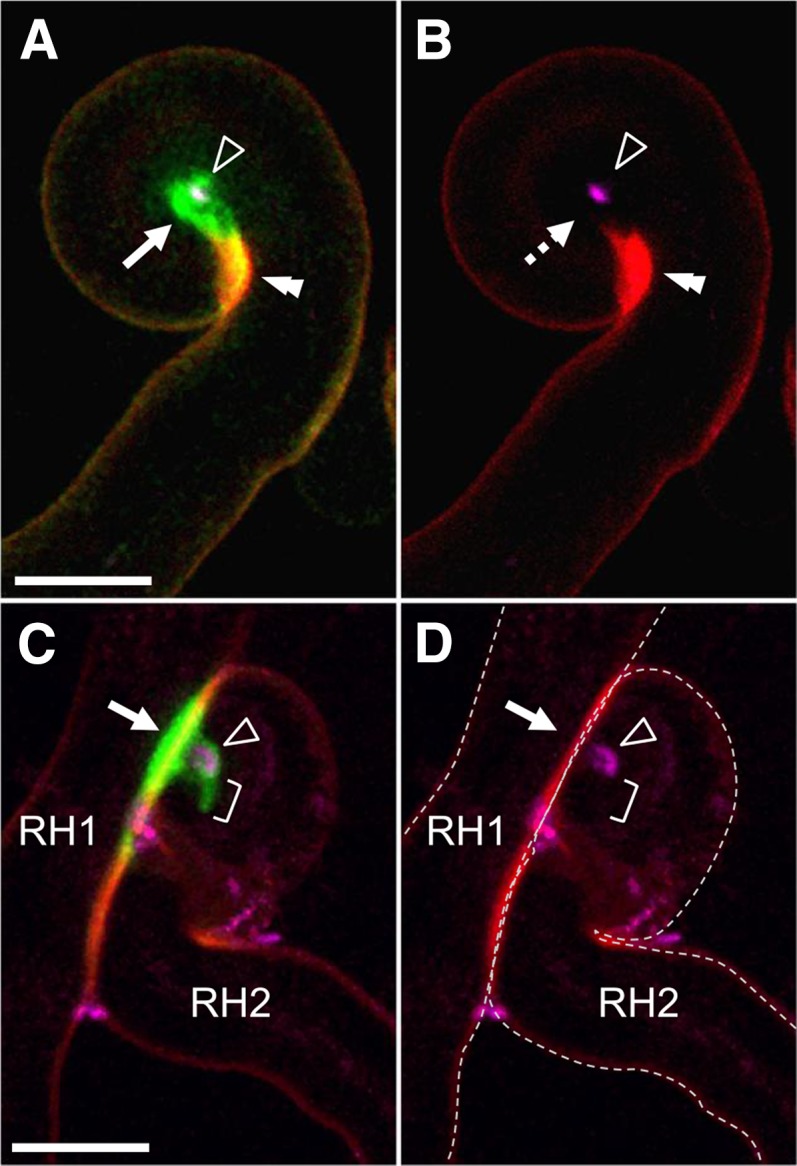
YFP-MtENOD11 accumulates within the rhizobial infection chamber preceding IT initiation. Early RH infection sites were imaged in M. truncatula roots expressing a YFP-tagged ENOD11 fusion (green) following inoculation with cCFP-expressing S. meliloti (magenta). A and B, Before IT formation, the YFP-MtENOD11 fluorescence (arrow, A) is mainly associated with the infection chamber surrounding the enclosed rhizobia (arrowhead, A and B). The double arrowhead indicates the autofluorescent wall domain (in red) adjacent to the infection chamber contrasting with the absence of autofluorescence associated with the infection chamber (dashed arrow, B). C and D, An IT is initiating from an infection chamber that has formed after rhizobia have become entrapped between two touching RHs (RH1 and RH2). YFP-MtENOD11 accumulation is associated with the site of initial rhizobial enclosure between touching walls (arrow), the protruding infection chamber formed in RH2 (arrowhead), as well as the tip of the initiating IT (bracket). Confocal images are z axis projections of 5 (A and B) or 13 (C and D) serial optical sections. A and C, Overlays of cCFP (magenta), YFP (green), and autofluorescence (red). B and D, Overlays of cCFP and autofluorescence. The dashed lines in D indicate the contours of the RH cells. Bars = 10 µm.
DISCUSSION
During the establishment of the RNS in many legumes, such as M. truncatula, root entry by rhizobia occurs via a process that initiates with the physical entrapment of the bacteria between RH cell walls followed by the formation of the tip-growing apoplastic ITs. To initiate studies on the molecular/cellular processes that accompany the transition between rhizobial entrapment and IT formation, we performed time-lapse confocal imaging on M. truncatula roots undergoing rhizobial colonization. These experiments unexpectedly revealed that there is a lengthy delay (10–20 h) before tubular IT formation is initiated from within fully curled RHs (Fig. 1). Although rarely examined in earlier studies of rhizobial infection, this finding is consistent with light microscopy experiments performed more than 30 years ago on inoculated clover roots (Callaham and Torrey, 1981). Most importantly, our live-tissue imaging studies provide strong evidence that major, NIN-dependent host cell wall remodeling occurs within the infection chamber throughout the entire period preceding IT initiation, and this discovery leads us to propose a two-stage model to explain the cellular mechanisms underlying this critical phase of rhizobial RH infection.
Remodeling of the Infection Chamber into an IT-Like Compartment
The localization of fluorescent markers labeling both the exocytosis reporter MtVAMP721e and the infection-associated secreted protein MtENOD11 has shown that the RH cell actively remodels the infection chamber during the period of 10 to 20 h preceding IT formation (Figs. 2 and 4). Indeed, this sustained exocytotic activity and concomitant deposition of extracellular material visualized with the MtENOD11 fusion protein correlates with progressive enlargement of the infection chamber (Fig. 2; Supplemental Fig. S1), a stage that has been described as the development of the refractile spot or cell wall swelling in early studies (Fåhraeus, 1957).
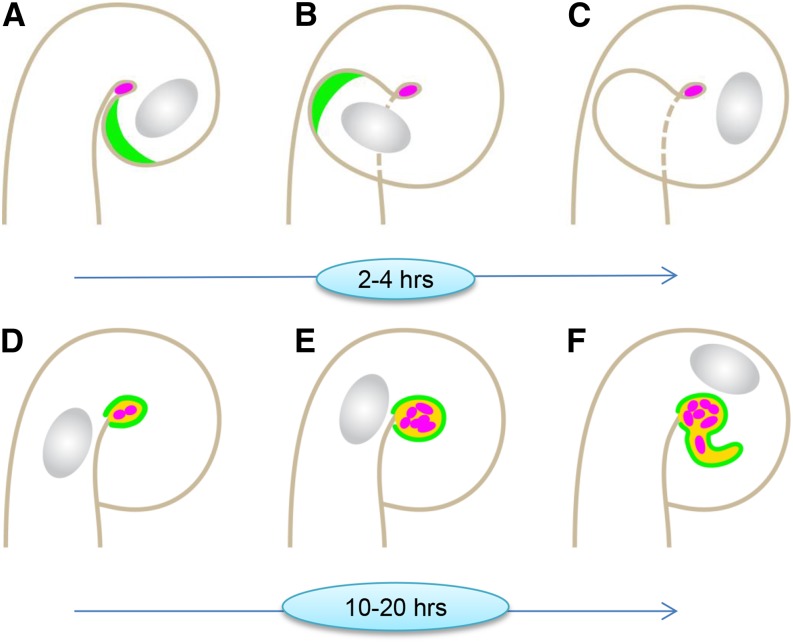
Although the early accumulation of MtENOD11 throughout the period of infection chamber remodeling is consistent with the early transcriptional activation of the gene in curled RHs before IT formation (Boisson-Dernier et al., 2005), the focalized accumulation of the MtENOD11 protein exclusively around the entrapped bacteria is an intriguing observation. The Pro-rich MtENOD11 is an atypical cell wall-associated protein with unusually low Tyr content, presumed to limit cross linking to other wall components (Journet et al., 2001). As such, the accumulation of MtENOD11 within the chamber is likely to contribute to the cell wall plasticity required for the radial expansion and the subsequent polar initiation of ITs. The occurrence of MtENOD11 within the infection chamber, therefore, leads us to propose a scenario in which the chamber progressively acquires an IT-like composition prior to tip growth initiation (Fig. 5; Supplemental Movie 1). In this scenario, the transport of exocytotic vesicles toward the membrane surrounding the infection chamber initiates within hours after the completion of RH curling (Fig. 5, A–D). Progressive deposition of new membrane and extracellular materials including MtENOD11 over the subsequent 10 to 20 h leads to radial infection chamber enlargement and conversion into a globular IT-like compartment (Fig. 5E; Supplemental Movie 1). This is accompanied by a small number of rhizobial cell divisions (described later). At the end of this first phase, a switch from radial expansion to polar tip elongation leads to the initiation of IT development (Fig. 5F; Supplemental Movie 1). In this two-step model, the Mtnin-1 mutant fails to initiate the first stage of infection chamber remodeling (corresponding to the transition between stages C and D shown in Fig. 5). In conclusion, we therefore propose that IT initiation should be viewed as a tip-growing extension emanating from the IT-like compartment already created within the infection chamber. One important consequence of this model is that there is no longer any need to hypothesize that localized host cell wall degradation is required for colonizing rhizobia to access the newly formed IT, in contrast to what was previously proposed (Callaham and Torrey, 1981; Turgeon and Bauer, 1985; Gage, 2004). Finally, it should be underlined that our findings also clearly argue against an additional suggestion that the initiation of IT development might result from a direct conversion of apical RH tip growth to inward-directed IT tip growth (e.g. Brewin, 1991; Kijne, 1992).
Figure 5.
Infection chamber remodeling paves the way for polar IT initiation in M. truncatula RHs. The localization of the exocytosis reporter and the MtENOD11 protein during RH curling, infection chamber remodeling, and IT initiation is schematically represented. A to C, During RH curling, the GFP-VAMP721e-labeled exocytosis site at the growing RH tip (A and B) is lost once RH curling is completed (C). At this stage, the infection chamber generally encloses a single rhizobial cell. D to F, Remodeling of the infection chamber starts before significant rhizobial multiplication has occurred (D) and leads to enlargement and differentiation of this new compartment accompanied by rhizobial multiplication (E) before tubular IT initiation (F). Note that the Mtnin-1 mutant fails to progress from stage C to stage D. Color code: rhizobia, pink; GFP-MtVAMP721e, green; and YFP-MtENOD11, yellow.
Plant-Microsymbiont Signal Exchange during the Two Stages of Rhizobial Entry
In light of this new two-step model for rhizobial infection, what do we know about the various factors that are involved in infection chamber remodeling and the initiation of tubular IT growth? Recent data have revealed that Ca2+ spiking that is elicited in RHs after rhizobial inoculation is strongly attenuated in fully curled hairs with entrapped rhizobia, and that sustained spiking is reactivated before and during the entry of the bacteria into the newly created IT (Sieberer et al., 2015). This suggests that there are sequential modifications in the host perception of rhizobial LCO signals during these key stages, and that the capacity to perceive these signals may be important in the triggering of IT initiation. Indeed, this is in line with the proposal that IT development within the M. truncatula RH requires that the host perceives NF-related signals via a specific entry receptor involving the LysM receptor-like kinase LYK3 (Limpens et al., 2003; Smit et al., 2007). Further evidence for distinct sequential steps prior to IT formation comes from the Sinorhizobium meliloti nodFnodL mutant, which produces abnormal NFs and cannot activate IT formation, despite the fact that rhizobial entrapment and multiplication take place (Ardourel et al., 1994) in addition to the trigger of infection-related MtENOD11 expression (Boisson-Dernier et al., 2005). Confocal imaging experiments have revealed that a single attached bacterium is often sufficient to induce initial RH-tip curling (Figs. 1 and 2, G and H), and that a small rhizobial microcolony comprising 10 to 30 bacteria is present within the infection chamber by the time the IT is initiated (Fig. 2, D–F). It is conceivable that rhizobial multiplication within the chamber may be an important parameter in generating threshold levels of bacterial signal molecules required for triggering IT initiation. Furthermore, the modified environment generated within the chamber by focused exocytosis may play a role in activating rhizobial differentiation and associated responses necessary for successful infection, including the secretion of infection-related LCOs and other important components such as acidic exopolysaccharides (for review, see Downie, 2010).
The nin mutants in Lotus japonicus, pea (Pisum sativum), or M. truncatula (Schauser et al., 1999; Borisov et al., 2003; Marsh et al., 2007) are able to respond to rhizobial inoculation by RH curling leading to rhizobial entrapment (a stage depicted in Fig. 5C) but do not progress to tubular IT initiation. We now show that this is most likely the result of the failure of infection chamber remodeling and associated microcolony development. In L. japonicus, the recently characterized NODULATION PECTATE LYASE (Xie et al., 2012) that is required for proper infection is regulated by NIN. It is therefore possible that the infection-defective phenotype of Mtnin-1 results at least in part from the lack of expression of an orthologous M. truncatula pectate lyase. Identifying and characterizing NIN targets in M. truncatula will be important for understanding the infection chamber remodeling process. The two-step model for infection is also consistent with the phenotypes of other legume mutants defective for rhizobial infection. For example, Ljcyclops (Yano et al., 2008) or M. truncatula lumpy infection (Kuppusamy et al., 2004; Guan et al., 2013) mutants appear to be impaired at the intermediate stage (depicted in Fig. 5E) after rhizobial microcolony development but preceding tubular IT initiation. It will now be important to identify and study additional host and bacterial cell wall-associated components involved in the development of the rhizobial infection chamber compartment, including cell wall-modifying enzymes proposed to be required for IT formation such as L. japonicus NODULATION PECTATE LYASE (Xie et al., 2012) or the Rhizobium leguminosarum bv trifolii cell-bound CellulaseC2 (Robledo et al., 2008).
Is Infection Chamber Remodeling a Specificity of the Rhizobium-Legume Symbiosis?
As underlined in the introduction, there are a number of striking similarities between the early infection stages of the rhizobial and AM associations, in terms of both host/microbe signaling pathways and the mechanisms involved in the respective host-regulated apoplastic infection processes (Parniske, 2008; Bapaume and Reinhardt, 2012). AM fungi also penetrate plant roots via a host-constructed transcellular compartment equivalent to the rhizobial IT. The development of this specialized perifungal compartment is prefigured by the formation of the transient cytoplasmic PPA, which links the migrating cell nucleus to the site of AM fungal attachment via the hyphopodium (Genre et al., 2005, 2008). In addition, Rich et al. (2014) have recently proposed that host cell-driven modifications to the cell wall at the site of hyphopodium contact would precede Rhizophagus irregularis entry into the Petunia hybrida root. Thus, by analogy with the creation of the IT precursor within the enclosed space formed by RH curling, it is possible that hyphopodium attachment also creates an enclosed environment within which host secretion and associated wall remodeling generates a specialized compartment, thus allowing the AM hyphae to cross the host cell wall. Future studies focused on this key step preceding AM fungal cell penetration will now be needed to address this intriguing question.
MATERIALS AND METHODS
Biological Materials
The Medicago truncatula super numeric nodules-2 mutant was used for most of the experiments described in this article because its enhanced infection phenotype (Schnabel et al., 2005) greatly facilitates the identification of RH infection sites, while at the same time it possesses a normal wild-type infection process (Fournier et al., 2008; Cerri et al., 2012; Sieberer et al., 2012). The M. truncatula nin-1 mutant, provided by Giles Oldroyd (John Innes Center) and Tatiana Vernié (Laboratoire des Interactions Plantes Micro-organismes), and M. truncatula ‘Jemalong A17’ were also used in this study, and all plants were grown as described previously (Fournier et al., 2008). Strains of Sinorhizobium meliloti 2011 constitutively expressing either GFP (Sm 2011-GFP) or the Cerulean version of Cyan Fluorescent Protein (CFP; Sm 2011-cCFP; both provided by Patrick Smit, University of Wageningen) or carrying the pXLGD4 plasmid containing a hemA-β-galactosidaseZ fusion (Sm 2011-lacZ; Ardourel et al., 1994) were propagated as described (Cerri et al., 2012).
Expression of Fluorescent Protein Fusions in M. truncatula Roots
The GFP-MtVAMP721e fusion (Ivanov et al., 2012) was expressed under the control of the Arabidopsis (Arabidopsis thaliana) UBIQUITIN3 (UBQ3) promoter in a pK7WGF2-R-derived binary vector carrying the Red Root selection marker, which comprises the Discosoma sp. red fluorescent protein (DsRed) coding sequence driven by the Arabidopsis UBQ10 promoter (Smit et al., 2005; Limpens et al., 2009). The construction of the chimeric gene expressing YFP-labeled MtENOD11 is described in detail in J. Fournier, A. Teillet, M.-C. Auriac, D.G. Barker, and F. de Carvalho-Niebel (unpublished data). In this construct, YFP has been inserted between the N-terminal ENOD11 signal peptide (75 bp) and the remaining repetitive Pro-rich domain (450 bp), and expression is driven by a 1-kb fragment of the endogenous ENOD11 promoter (pE11) that is sufficient for both preinfection and infection-related expression (Boisson-Dernier et al., 2005). The pUBQ3-GFP-MtVAMP721e and pE11-YFP-MtENOD11 constructs were introduced into Agrobacterium rhizogenes ARquaI (Quandt et al., 1993), and composite sunn-2 or nin-1 plants were produced via A. rhizogenes-mediated transformation as described in Boisson-Dernier et al. (2001). Composite plants with roots constitutively expressing the GFP-MtVAMP721e fusion were selected under the stereomicroscope for moderate and uniform fluorescence levels. Those with roots expressing pE11-YFP-MtENOD11 were selected on Fåhraeus medium supplemented with 25 µg mL−1 kanamycin.
In Vivo Microscopy of Rhizobial Infection Sites in RHs
Rhizobial inoculation for in vivo microscopic observation was performed essentially as described (Fournier et al., 2008; Sieberer et al., 2012). In brief, plants were placed in 12- × 12-cm petri dishes containing a modified Fåhraeus medium (0.5% [w/v] Phytagel; Sigma-Aldrich) supplemented with 50 nM 2-amino ethoxyvinyl Gly. Roots were covered with a gas-permeable plastic film (Lumox Film, Starsted), and plants were grown with the dishes slightly tilted to encourage the growth of the roots along the plastic film. Inoculation with Sm 2011-GFP or Sm 2011-cCFP strains was performed by introducing 0.5 to 1 mL of an aqueous suspension of exponentially growing bacteria (optical density at 600 nm = 0.001; approximately 106 bacteria mL−1) under the plastic film. To investigate the early stages of rhizobial infection, roots of inoculated plants were observed 1 to 4 d postinoculation. Curled RHs with enclosed fluorescent bacteria as well as bacterial entrapment between cell walls of adjacent RHs were identified as potential sites for imaging. In addition, preference was given to curled RHs where the nucleus was located close to the enclosed rhizobia and associated with significant quantities of cytoplasm since experience had shown that these were more likely to initiate ITs. Plants were returned to the culture room between observations. Data were obtained from a total of 18 experiments in sunn-2 and two experiments in cv Jemalong A17 and Mtnin-1. The results presented are representative of observations recorded on 27 (Fig. 1), 24 (Fig. 2), 10 (Fig. 3, D–I), and 15 (Fig. 4) rhizobial infection sites, monitored using 20, 15, two, and 12 independent plants, respectively. The duration of the interval between curl closure and IT initiation was evaluated for seven sites (five independent experiments) and corroborated by data from an additional 20 sites monitored over longer intervals.
Confocal Microscopy
Selected infection sites were imaged with a Leica TCS SP2 AOBS confocal laser scanning microscope equipped with a long-distance 40× HCX Apo L (numerical aperture, 0.80) water immersion objective. The argon laser bands of 458, 488, and 514 nm were used to excite CFP, GFP, and YFP, respectively, and a 561-nm diode to excite the DsRed and observe cell wall autofluorescence. Specific emission windows used for CFP, GFP, YFP, DsRed, and autofluorescence signals were 465 to 485 nm, 500 to 530 nm, 525 to 550 nm, 600 to 630 nm, and 620 to 720 nm, respectively, and emitted fluorescence was false colored in magenta (CFP), green (GFP or YFP), and red (DsRed and/or wall autofluorescence). The images shown are single confocal sections, maximal projections of selected planes of a z-stack, or three-dimensional reconstructions of confocal image stacks. Images were acquired and projected using Leica confocal software and processed using Leica confocal software, ImageJ (http://imagej.nih.gov/ij/) or Volocity version 6.0.1 (PerkinElmer) software.
β-Galactosidase Assay
Histochemical staining for β-galactosidase activity after inoculation with S. meliloti strain Sm 2011-lacZ was performed 3 d postinoculation using 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside as substrate (Boivin et al., 1990). Data in Figure 3 (A–C) are representative of results obtained in 10 cv Jemalong A17 and 10 Mtnin-1 plants.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Radial expansion of the infection chamber before thread initiation.
Supplemental Figure S2. Infection chamber development and associated rhizobial multiplication are blocked in curled RHs of the Mtnin mutant.
Supplemental Figure S3. Before infection chamber closure, the localization of GFP-VAMP721e in Mtnin-1 RHs is similar to that in a wild-type plant.
Supplemental Movie S1. Animation illustrating the two-step model for rhizobial RH infection initiation.
Supplementary Material
Acknowledgments
We thank Patrick Smit (Wageningen University) for providing the S. meliloti 2011 strains expressing cCFP or GFP, Tatiana Vernié (Laboratoire des Interactions Plantes-Micro-organismes) and Giles Oldroyd (John Innes Center) for providing the Mtnin-1 seeds, and Alain Jauneau and colleagues (Fédération de Recherche 3450 imagery facility) for assistance with confocal microscopy.
Glossary
- RNS
root nodule symbiosis
- IT
infection thread
- AM
arbuscular mycorrhizal
- NF
Nod factor
- RH
root hair
- PPA
prepenetration apparatus
- pE11
ENOD11 promoter
Footnotes
This work was supported in part by the French National Research Agency (grant no. ANR–08–BLAN–0029–01 to D.G.B. and fellowship to A.T.), the Partenariat Hubert Curien Galilée (grant no. 30111WJ to A.G. and D.G.B.), and the French National Laboratoire d’Excellence TULIP initiative (grant no. ANR–10–LABX–41).
References
- Ardourel M, Demont N, Debellé F, Maillet F, de Billy F, Promé JC, Dénarié J, Truchet G (1994) Rhizobium meliloti lipooligosaccharide nodulation factors: different structural requirements for bacterial entry into target root hair cells and induction of plant symbiotic developmental responses. Plant Cell 6: 1357–1374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bapaume L, Reinhardt D (2012) How membranes shape plant symbioses: signaling and transport in nodulation and arbuscular mycorrhiza. Front Plant Sci 3: 223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisson-Dernier A, Andriankaja A, Chabaud M, Niebel A, Journet EP, Barker DG, de Carvalho-Niebel F (2005) MtENOD11 gene activation during rhizobial infection and mycorrhizal arbuscule development requires a common AT-rich-containing regulatory sequence. Mol Plant Microbe Interact 18: 1269–1276 [DOI] [PubMed] [Google Scholar]
- Boisson-Dernier A, Chabaud M, Garcia F, Bécard G, Rosenberg C, Barker DG (2001) Agrobacterium rhizogenes-transformed roots of Medicago truncatula for the study of nitrogen-fixing and endomycorrhizal symbiotic associations. Mol Plant Microbe Interact 14: 695–700 [DOI] [PubMed] [Google Scholar]
- Boivin C, Camut S, Malpica CA, Truchet G, Rosenberg C (1990) Rhizobium meliloti genes encoding catabolism of trigonelline are induced under symbiotic conditions. Plant Cell 2: 1157–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borisov AY, Madsen LH, Tsyganov VE, Umehara Y, Voroshilova VA, Batagov AO, Sandal N, Mortensen A, Schauser L, Ellis N. , et al (2003) The Sym35 gene required for root nodule development in pea is an ortholog of Nin from Lotus japonicus. Plant Physiol 131: 1009–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewin NJ. (1991) Development of the legume root nodule. Annu Rev Cell Biol 7: 191–226 [DOI] [PubMed] [Google Scholar]
- Brewin NJ. (2004) Plant cell wall remodelling in the rhizobium-legume symbiosis. Crit Rev Plant Sci 23: 293–316 [Google Scholar]
- Callaham DA, Torrey JG (1981) The structural basis for infection of root hairs of Trifolium repens by Rhizobium. Can J Bot 59: 1647–1664 [Google Scholar]
- Capoen W, Sun J, Wysham D, Otegui MS, Venkateshwaran M, Hirsch S, Miwa H, Downie JA, Morris RJ, Ané JM. , et al (2011) Nuclear membranes control symbiotic calcium signaling of legumes. Proc Natl Acad Sci USA 108: 14348–14353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catoira R, Timmers ACJ, Maillet F, Galera C, Penmetsa RV, Cook D, Dénarié J, Gough C (2001) The HCL gene of Medicago truncatula controls Rhizobium-induced root hair curling. Development 128: 1507–1518 [DOI] [PubMed] [Google Scholar]
- Cerri MR, Frances L, Laloum T, Auriac MC, Niebel A, Oldroyd GED, Barker DG, Fournier J, de Carvalho-Niebel F (2012) Medicago truncatula ERN transcription factors: regulatory interplay with NSP1/NSP2 GRAS factors and expression dynamics throughout rhizobial infection. Plant Physiol 160: 2155–2172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charron D, Pingret JL, Chabaud M, Journet EP, Barker DG (2004) Pharmacological evidence that multiple phospholipid signaling pathways link Rhizobium nodulation factor perception in Medicago truncatula root hairs to intracellular responses, including Ca2+ spiking and specific ENOD gene expression. Plant Physiol 136: 3582–3593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dart PJ. (1974) The infection process. InQuispel A, ed, The biology of nitrogen fixation. North-Holland Publishing Co., Amsterdam, pp 381-429 [Google Scholar]
- Downie JA. (2010) The roles of extracellular proteins, polysaccharides and signals in the interactions of rhizobia with legume roots. FEMS Microbiol Rev 34: 150–170 [DOI] [PubMed] [Google Scholar]
- Ehrhardt DW, Wais R, Long SR (1996) Calcium spiking in plant root hairs responding to Rhizobium nodulation signals. Cell 85: 673–681 [DOI] [PubMed] [Google Scholar]
- Esseling JJ, Lhuissier FG, Emons AMC (2003) Nod factor-induced root hair curling: continuous polar growth towards the point of nod factor application. Plant Physiol 132: 1982–1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fåhraeus G. (1957) The infection of clover root hairs by nodule bacteria studied by a simple glass slide technique. J Gen Microbiol 16: 374–381 [DOI] [PubMed] [Google Scholar]
- Fournier J, Timmers ACJ, Sieberer BJ, Jauneau A, Chabaud M, Barker DG (2008) Mechanism of infection thread elongation in root hairs of Medicago truncatula and dynamic interplay with associated rhizobial colonization. Plant Physiol 148: 1985–1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gage DJ. (2002) Analysis of infection thread development using Gfp- and DsRed-expressing Sinorhizobium meliloti. J Bacteriol 184: 7042–7046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gage DJ. (2004) Infection and invasion of roots by symbiotic, nitrogen-fixing rhizobia during nodulation of temperate legumes. Microbiol Mol Biol Rev 68: 280–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gage DJ, Bobo T, Long SR (1996) Use of green fluorescent protein to visualize the early events of symbiosis between Rhizobium meliloti and alfalfa (Medicago sativa). J Bacteriol 178: 7159–7166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genre A, Chabaud M, Faccio A, Barker DG, Bonfante P (2008) Prepenetration apparatus assembly precedes and predicts the colonization patterns of arbuscular mycorrhizal fungi within the root cortex of both Medicago truncatula and Daucus carota. Plant Cell 20: 1407–1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genre A, Chabaud M, Timmers T, Bonfante P, Barker DG (2005) Arbuscular mycorrhizal fungi elicit a novel intracellular apparatus in Medicago truncatula root epidermal cells before infection. Plant Cell 17: 3489–3499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genre A, Ivanov S, Fendrych M, Faccio A, Zársky V, Bisseling T, Bonfante P (2012) Multiple exocytotic markers accumulate at the sites of perifungal membrane biogenesis in arbuscular mycorrhizas. Plant Cell Physiol 53: 244–255 [DOI] [PubMed] [Google Scholar]
- Geurts R, Fedorova E, Bisseling T (2005) Nod factor signaling genes and their function in the early stages of Rhizobium infection. Curr Opin Plant Biol 8: 346–352 [DOI] [PubMed] [Google Scholar]
- Goormachtig S, Capoen W, Holsters M (2004) Rhizobium infection: lessons from the versatile nodulation behaviour of water-tolerant legumes. Trends Plant Sci 9: 518–522 [DOI] [PubMed] [Google Scholar]
- Guan D, Stacey N, Liu C, Wen J, Mysore KS, Torres-Jerez I, Vernié T, Tadege M, Zhou C, Wang ZY, et al. (2013) Rhizobial infection is associated with the development of peripheral vasculature in nodules of Medicago truncatula. Plant Physiol 162: 107–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov S, Fedorova EE, Limpens E, De Mita S, Genre A, Bonfante P, Bisseling T (2012) Rhizobium-legume symbiosis shares an exocytotic pathway required for arbuscule formation. Proc Natl Acad Sci USA 109: 8316–8321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Journet EP, El-Gachtouli N, Vernoud V, de Billy F, Pichon M, Dedieu A, Arnould C, Morandi D, Barker DG, Gianinazzi-Pearson V (2001) Medicago truncatula ENOD11: a novel RPRP-encoding early nodulin gene expressed during mycorrhization in arbuscule-containing cells. Mol Plant Microbe Interact 14: 737–748 [DOI] [PubMed] [Google Scholar]
- Kijne JW. (1992) The Rhizobium infection process. InStacey G, Burris RH, Evans HJ, eds, Biological Nitrogen Fixation. Chapman and Hall, New York, pp 349-398 [Google Scholar]
- Kistner C, Parniske M (2002) Evolution of signal transduction in intracellular symbiosis. Trends Plant Sci 7: 511–518 [DOI] [PubMed] [Google Scholar]
- Kuppusamy KT, Endre G, Prabhu R, Penmetsa RV, Veereshlingam H, Cook DR, Dickstein R, Vandenbosch KA (2004) LIN, a Medicago truncatula gene required for nodule differentiation and persistence of rhizobial infections. Plant Physiol 136: 3682–3691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limpens E, Franken C, Smit P, Willemse J, Bisseling T, Geurts R (2003) LysM domain receptor kinases regulating rhizobial Nod factor-induced infection. Science 302: 630–633 [DOI] [PubMed] [Google Scholar]
- Limpens E, Ivanov S, van Esse W, Voets G, Fedorova E, Bisseling T (2009) Medicago N2-fixing symbiosomes acquire the endocytic identity marker Rab7 but delay the acquisition of vacuolar identity. Plant Cell 21: 2811–2828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markmann K, Parniske M (2009) Evolution of root endosymbiosis with bacteria: How novel are nodules? Trends Plant Sci 14: 77–86 [DOI] [PubMed] [Google Scholar]
- Marsh JF, Rakocevic A, Mitra RM, Brocard L, Sun J, Eschstruth A, Long SR, Schultze M, Ratet P, Oldroyd GE (2007) Medicago truncatula NIN is essential for rhizobial-independent nodule organogenesis induced by autoactive calcium/calmodulin-dependent protein kinase. Plant Physiol 144: 324–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray JD. (2011) Invasion by invitation: rhizobial infection in legumes. Mol Plant Microbe Interact 24: 631–639 [DOI] [PubMed] [Google Scholar]
- Oldroyd GED. (2013) Speak, friend, and enter: signalling systems that promote beneficial symbiotic associations in plants. Nat Rev Microbiol 11: 252–263 [DOI] [PubMed] [Google Scholar]
- Oldroyd GED, Downie JA (2006) Nuclear calcium changes at the core of symbiosis signalling. Curr Opin Plant Biol 9: 351–357 [DOI] [PubMed] [Google Scholar]
- Oldroyd GED, Murray JD, Poole PS, Downie JA (2011) The rules of engagement in the legume-rhizobial symbiosis. Annu Rev Genet 45: 119–144 [DOI] [PubMed] [Google Scholar]
- Parniske M. (2008) Arbuscular mycorrhiza: the mother of plant root endosymbioses. Nat Rev Microbiol 6: 763–775 [DOI] [PubMed] [Google Scholar]
- Pichon M, Journet EP, Dedieu A, de Billy F, Truchet G, Barker DG (1992) Rhizobium meliloti elicits transient expression of the early nodulin gene ENOD12 in the differentiating root epidermis of transgenic alfalfa. Plant Cell 4: 1199–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quandt HJ, Puhler A, Broer I (1993) Transgenic root nodules of Vicia hirsuta: a fast and efficient system for the study of gene expression in indeterminate-type nodules. Mol Plant Microbe Interact 6: 699–706 [Google Scholar]
- Rich MK, Schorderet M, Reinhardt D (2014) The role of the cell wall compartment in mutualistic symbioses of plants. Front Plant Sci 5: 238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robledo M, Jiménez-Zurdo JI, Velázquez E, Trujillo ME, Zurdo-Piñeiro JL, Ramírez-Bahena MH, Ramos B, Díaz-Mínguez JM, Dazzo F, Martínez-Molina E. , et al (2008) Rhizobium cellulase CelC2 is essential for primary symbiotic infection of legume host roots. Proc Natl Acad Sci USA 105: 7064–7069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauser L, Roussis A, Stiller J, Stougaard J (1999) A plant regulator controlling development of symbiotic root nodules. Nature 402: 191–195 [DOI] [PubMed] [Google Scholar]
- Scheres B, Van De Wiel C, Zalensky A, Horvath B, Spaink H, Van Eck H, Zwartkruis F, Wolters AM, Gloudemans T, Van Kammen A. , et al (1990) The ENOD12 gene product is involved in the infection process during the pea-Rhizobium interaction. Cell 60: 281–294 [DOI] [PubMed] [Google Scholar]
- Schnabel E, Journet EP, de Carvalho-Niebel F, Duc G, Frugoli J (2005) The Medicago truncatula SUNN gene encodes a CLV1-like leucine-rich repeat receptor kinase that regulates nodule number and root length. Plant Mol Biol 58: 809–822 [DOI] [PubMed] [Google Scholar]
- Sieberer BJ, Chabaud M, Fournier J, Timmers AC, Barker DG (2012) A switch in Ca2+ spiking signature is concomitant with endosymbiotic microbe entry into cortical root cells of Medicago truncatula. Plant J 69: 822–830 [DOI] [PubMed] [Google Scholar]
- Sieberer BJ, Chabaud M, Timmers AC, Monin A, Fournier J, Barker DG (2009) A nuclear-targeted cameleon demonstrates intranuclear Ca2+ spiking in Medicago truncatula root hairs in response to rhizobial nodulation factors. Plant Physiol 151: 1197–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieberer BJ, Fournier J, Timmers ACJ, Chabaud M, Barker DG (2015) Nuclear Ca2+ signaling reveals active bacterial-host signaling throughout rhizobial infection in root hairs of Medicago truncatula. Inde Bruijn FJ, ed, Biologial Nitrogen Fixation. Wiley/Blackwell, Hoboken, NJ, USA: in press [Google Scholar]
- Singh S, Parniske M (2012) Activation of calcium- and calmodulin-dependent protein kinase (CCaMK), the central regulator of plant root endosymbiosis. Curr Opin Plant Biol 15: 444–453 [DOI] [PubMed] [Google Scholar]
- Smit P, Limpens E, Geurts R, Fedorova E, Dolgikh E, Gough C, Bisseling T (2007) Medicago LYK3, an entry receptor in rhizobial nodulation factor signaling. Plant Physiol 145: 183–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit P, Raedts J, Portyanko V, Debellé F, Gough C, Bisseling T, Geurts R (2005) NSP1 of the GRAS protein family is essential for rhizobial Nod factor-induced transcription. Science 308: 1789–1791 [DOI] [PubMed] [Google Scholar]
- Turgeon BG, Bauer WD (1985) Ultrastructure of infection-thread development during the infection of soybean by Rhizobium japonicum. Planta 163: 328–349 [DOI] [PubMed] [Google Scholar]
- Wais RJ, Wells DH, Long SR (2002) Analysis of differences between Sinorhizobium meliloti 1021 and 2011 strains using the host calcium spiking response. Mol Plant Microbe Interact 15: 1245–1252 [DOI] [PubMed] [Google Scholar]
- Xie F, Murray JD, Kim J, Heckmann AB, Edwards A, Oldroyd GED, Downie JA (2012) Legume pectate lyase required for root infection by rhizobia. Proc Natl Acad Sci USA 109: 633–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano K, Yoshida S, Müller J, Singh S, Banba M, Vickers K, Markmann K, White C, Schuller B, Sato S, et al. (2008) CYCLOPS, a mediator of symbiotic intracellular accommodation. Proc Natl Acad Sci USA 105: 20540–20545 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.