A network model of global transcriptional interactions, based on clustering of transcriptionally correlated genes using publicly available floral gene expression data sets, provides a community resource for studies of reproductive gene expression.
Abstract
Floral formation, in particular anther and pollen development, is a complex biological process with critical importance for seed set and for targeted plant breeding. Many key transcription factors regulating this process have been identified; however, their direct role remains largely unknown. Using publicly available gene expression data from Arabidopsis (Arabidopsis thaliana), focusing on those studies that analyze stamen-, pollen-, or flower-specific expression, we generated a network model of the global transcriptional interactions (FlowerNet). FlowerNet highlights clusters of genes that are transcriptionally coregulated and therefore likely to have interacting roles. Focusing on four clusters, and using a number of data sets not included in the generation of FlowerNet, we show that there is a close correlation in how the genes are expressed across a variety of conditions, including male-sterile mutants. This highlights the important role that FlowerNet can play in identifying new players in anther and pollen development. However, due to the use of general floral expression data in FlowerNet, it also has broad application in the characterization of genes associated with all aspects of floral development and reproduction. To aid the dissection of genes of interest, we have made FlowerNet available as a community resource (http://www.cpib.ac.uk/anther). For this resource, we also have generated plots showing anther/flower expression from a variety of experiments: These are normalized together where possible to allow further dissection of the resource.
Anther and pollen development is a complex biological process that is indispensable for the generation of male gametophytes and the production of the next generation in flowering plants. This development includes a series of crucial events that require interactions between gametophytic and sporophytic genes in a cooperative fashion (Goldberg et al., 1993; Ma, 2005). The mature anther contains four lobes, each of which contains meiotic cells surrounded by four somatic cell layers, with the sporophytic tapetum playing an important role in pollen grain development (Goldberg et al., 1993). The tapetum serves as a nutritive tissue, providing metabolites, nutrients, and cell wall precursors for the development of pollen grains.
The use of classical genetic screens has uncovered a large number of genes involved in anther development and in the production of viable pollen. For example, in Arabidopsis (Arabidopsis thaliana), a number of genes encoding putative transcription factors involved in all aspects of anther development have been discovered. APETALA3 (AP3; Jack et al., 1994), SPOROCYTELESS/NOZZLE (SPL/NZZ; Liu et al., 2009), and EXCESS MICROSPOROCYTES1 (Zhao et al., 2002) have important roles in early anther development and differentiation, while ABORTED MICROSPORE (AMS; Sorensen et al., 2003; Xu et al., 2010), DEFECTIVE IN TAPETAL DEVELOPMENT AND FUNCTION1 (Zhu et al., 2008), DYSFUNCTIONAL TAPETUM1 (DYT1; Zhang et al., 2006), MALE STERILITY1 (MS1; Wilson et al., 2001; Alves-Ferreira et al., 2007; Ito et al., 2007; Yang et al., 2007a), MYB33/MYB65 (Millar and Gubler, 2005), and MYB80/MYB103 (Higginson et al., 2003; Zhang et al., 2007; Zhu et al., 2010) are involved in tapetum and pollen wall development (Wilson and Zhang, 2009). MYB26 (Steiner-Lange et al., 2003; Yang et al., 2007b) and RECEPTOR-LIKE PROTEIN KINASE2 (RPK2; Mizuno et al., 2007a) are involved in the later stages of anther dehiscence and pollen release. Although a number of the main regulators are known and a large number of genes directly involved in anther and pollen development have been identified, the means by which they interact and function in this process remain largely uncharacterized.
In this work, we try to address this knowledge gap by uncovering groups of genes involved in floral development, specifically focusing on anther and pollen development, through the creation of a coexpression network. This coexpression network connects genes expressed during floral development by their transcriptional regulation. Coexpressed genes have an increased likelihood of being involved in the same developmental or biochemical pathways; therefore, the correlation of gene expression is a powerful approach to analyze large data sets to identify genes involved in the same functional pathway (Peng and Weselake, 2011; Wang et al., 2012). This approach has been successfully applied to two state-dependent sets of interactions associated with seed dormancy or germination, with good correlation of transcriptional regulators of known seed dormancy and germination regulatory genes (Bassel et al., 2011).
With the recent advances in postgenomic technologies, a large number of genome-wide transcriptomic data sets have been generated, often to look at specific changes such as a transcription factor mutant compared with a wild type. Deposition of these data sets into publicly accessible online databases enables other researchers to analyze these collated data and uncover novel information, which may be tangential to the goal of the original experiment. In this study, we used publicly available gene expression data deposited in the National Center for Biotechnology Information’s Gene Expression Omnibus (Edgar et al., 2002; Barrett et al., 2013), choosing microarray data sets that included flowers, buds, anthers, or pollen materials to generate a coexpression network. Through generating the network, compact clusters of coexpressed genes were found (i.e. sets of genes having similar expression patterns across all the samples). This clustering enables the identification of novel genes that may be involved in specific biological processes based on known components within the cluster.
Additionally, we reanalyzed two previously conducted two-color microarray experiments associated with anther and pollen development (Alves-Ferreira et al., 2007; Xu et al., 2010) in order to compare gene expression across these time series as well as between the mutants. Along with the Affymetrix microarrays used to generate the correlation network, these three data sets give a clear indication of the spatial and temporal expression of anther-related genes. Plots have been created for each gene in the correlation network and are available online at http://www.cpib.ac.uk/anther.
In this work, we have generated a valuable tool for the analysis of genes involved in anther and pollen development and show how this can be used to identify new players in anther and pollen development. However, the data used for the generation of the correlation network are from all floral organs and developmental stages; therefore, FlowerNet also has broader application in the characterization of all genes associated with aspects of plant reproduction and flowering.
RESULTS
Gene Expression Plots
Arabidopsis anther, stamen, and bud microarray data sets were selected that comprised wild-type or mutant samples associated with anther and pollen development; all mutants selected displayed male-sterile phenotypes linked to a failure of pollen formation or dehiscence. The Affymetrix microarrays were renormalized together (see “Materials and Methods”) to make the data comparable between samples.
A number of additional highly relevant data sets were also available, but since they utilize different transcriptomic platforms, they cannot be integrated easily with the Affymetrix array data sets. Alves-Ferreira et al. (2007) used Agilent two-color microarrays to analyze the effect of MS1 (At5g22260) on gene expression, comparing buds between the wild type and ms1 mutants. In this experiment, the first sample consists of stage 13 flowers, with subsequent samples containing successively younger buds, until the seventh sample contains the very earliest buds and the inflorescence meristem. Xu et al. (2010) used an in-house-printed two-color microarray to compare wild-type and ams buds at pollen mother cell meiosis, pollen mitosis I, bicellular pollen, and pollen mitosis II stages. This second, developmental time course has finer temporal and spatial detail around these key stages and therefore may be used in conjunction with the one generated from the data of Alves-Ferreira et al. (2007).
Data from these two-color microarray experiments (Alves-Ferreira et al., 2007; Xu et al., 2010) were renormalized using a method similar to that of single-color arrays (see “Materials and Methods”), producing two comparative developmental time courses of gene expression in wild-type, ms1, and ams buds. This enables us to use the wild-type and mutant values as a biologically meaningful developmental time course. It is noted that this method seems more prone to noise than the use of single-color microarrays, but it is particularly useful where genes are not present on the Affymetrix microarrays, such as MS1.
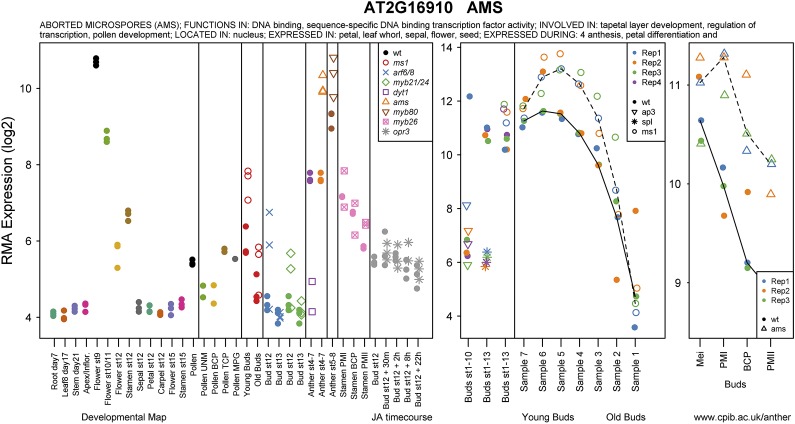
The resulting plots (e.g. for AMS; Fig. 1) from both the Affymetrix and two-color array data give immediate visual indication of how the genes are expressed across various samples. These plots show the individual replicates rather than means, allowing those genes whose behavior appears to be variable, possibly due to sensitivity to conditions or to the precise staging of the samples, to be easily visually detected. These plots are freely available online at http://www.cpib.ac.uk/anther, allowing quick analysis of anther expression data for genes of interest. Closed circles represent wild-type samples, and open symbols represent the different mutants.
Figure 1.
Plot from the Web site tool showing the expression of AMS in various anther and floral microarrays (http://www.cpib.ac.uk/anther).
FlowerNet Correlation Network
In order to investigate gene behavior across a wide range of samples, we generated a correlation network using 66 Arabidopsis Affymetrix wild-type microarrays from plant reproduction-related experiments, which included flowers, buds, anther/stamen samples, and isolated pollen samples (see “Materials and Methods”; Supplemental Files S1 and S2). These microarray data sets were mostly publicly available in the National Center for Biotechnology Information’s Gene Expression Omnibus (Honys and Twell, 2004; Schmid et al., 2005; Mandaokar et al., 2006; Yang et al., 2007a) with some previously unpublished data (Z.A. Wilson and J. Song, unpublished data). A number of other anther-related published studies were used for gene list overlay comparison; however, these could not be included in this network, or in the expression plots, since the raw data for these analyses were not publicly available at the time of construction.
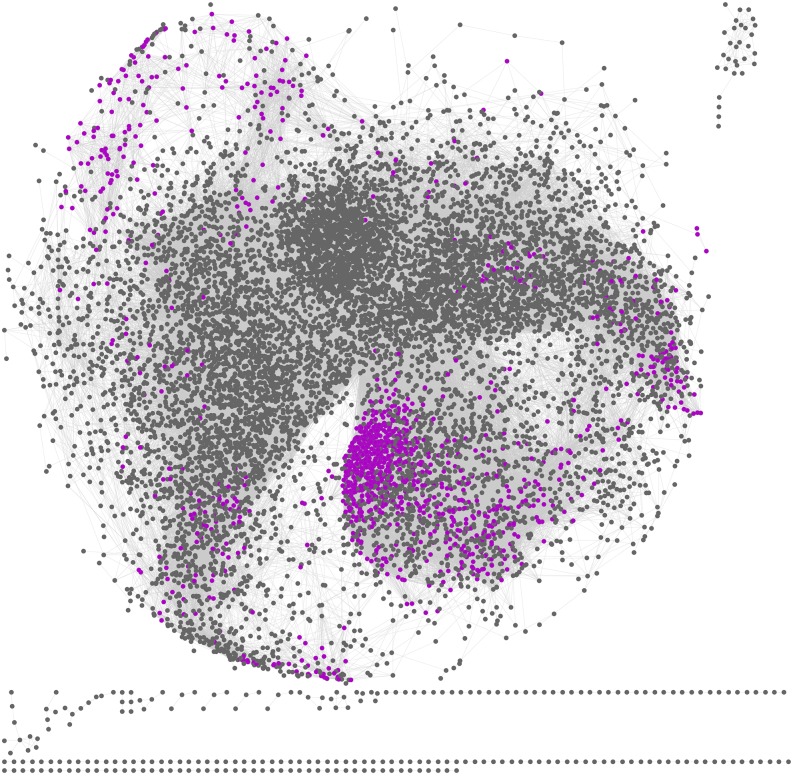
The resulting network has 605,686 edges between 10,797 genes (Fig. 2); a visualization of the network is available as a Cytoscape file (Supplemental File S3), with a navigable list of genes that are connected to each gene presented under each gene expression plot image.
Figure 2.
Cytoscape visualization of the full correlation network. Genes shown by Wellmer et al. (2004) to have higher expression in stamen are colored in purple.
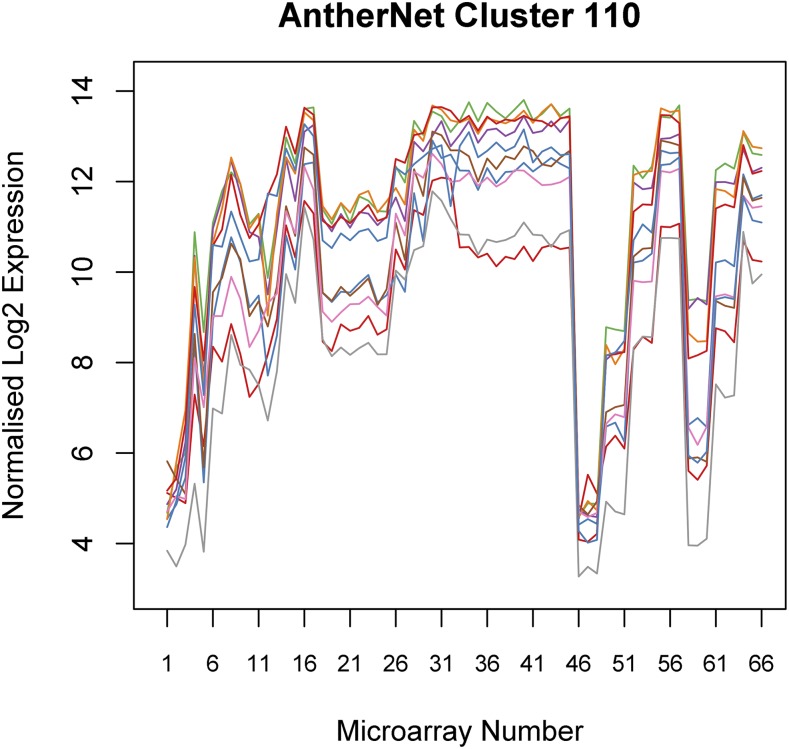
Within this network, compact highly intracorrelated clusters were identified using the TransClust algorithm (Wittkop et al., 2010) and the correlation values as an edge weight to generate small, well-connected clusters based on expression in the network (see “Materials and Methods”). The resultant clusters describe groups of genes that are highly positively correlated across all the samples included, with almost all the edges between the genes included (for cluster allocations, see Supplemental File S4). These compact clusters have extremely similar expression patterns within the samples used, avoiding the problem of chain-like clusters, where genes are several neighbors removed from those at the other end of the cluster, leading to genes in the same cluster having very diverse expression patterns. The resulting clusters contain almost all of the possible edges between the nodes involved, ensuring that all the genes considered correlate well with each other. For example, the largest cluster (cluster 1) contains 14,433 edges between 171 genes, 99.3% of the possible edges; thus, these genes have very similar expression profiles. Figure 3 shows a representative cluster (cluster 110), which demonstrates how the expression profile of each gene within the cluster has a highly correlated expression pattern in the microarrays used to generate the network.
Figure 3.
Plot of the genes in cluster 110, showing expression in the 66 microarrays that are used to generate FlowerNet. The order of the microarrays is listed in Supplemental File S1.
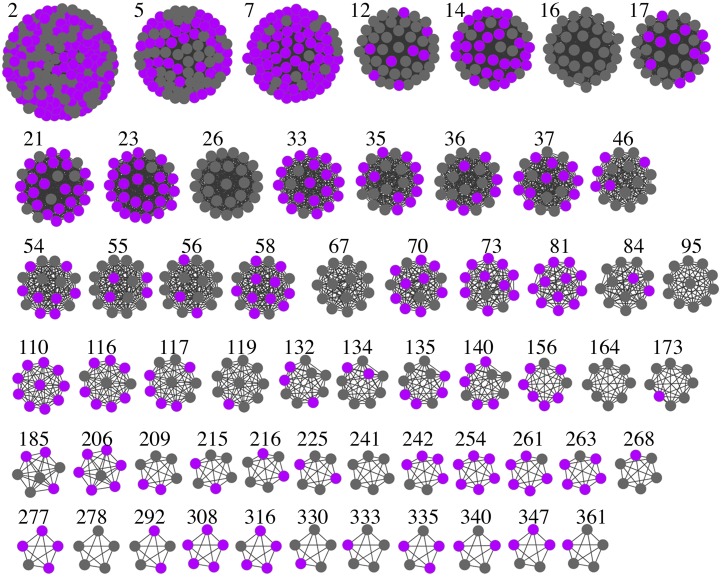
Overlaying expression data from different experiments that have not been used in the creation of the network (e.g. mutant data from the two-color microarrays detailed above) indicate that certain clusters are significantly overrepresented for these transcripts. Similarly, the majority of the clusters contain a significant overrepresentation of Gene Ontology (GO) terms, suggesting that genes within clusters are involved in the same developmental processes. A number of the clusters are enriched for general plant pathways; for example, cluster 2 is enriched in the GO term photosynthesis, cluster 11 in GA biosynthesis, and cluster 16 in water transport, although these do not show significant expression changes linked to anther/stamen-specific gene expression. Other clusters show reproduction-related enrichment; for example, cluster 67 is a set of 14 genes that all have higher expression in the carpel (and to a lesser extent the petal) samples from the AtGenExpress Developmental Map, which have not been used in the generation of the network. To focus on anther/stamen-related clusters, we used a number of data sets to select clusters of interest, such as stamen-specific transcripts (Wellmer et al., 2004), the presence of known anther-related genes within a cluster, and clusters that are highly regulated by previously described male-sterile mutants such as ams and ms1; these are shown in Figure 4.
Figure 4.
Selection of FlowerNet clusters, chosen for being bud related based on a number of criteria such as known anther-related genes and male-sterile mutant expression. Highlighted in pink are those genes found to be stamen specific (Wellmer et al., 2004).
Clusters Associated with Pollen Wall Formation
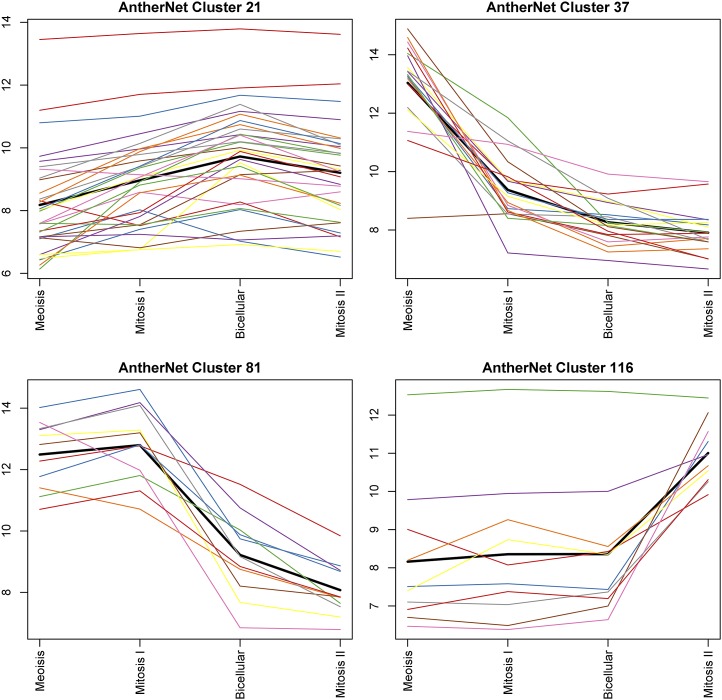
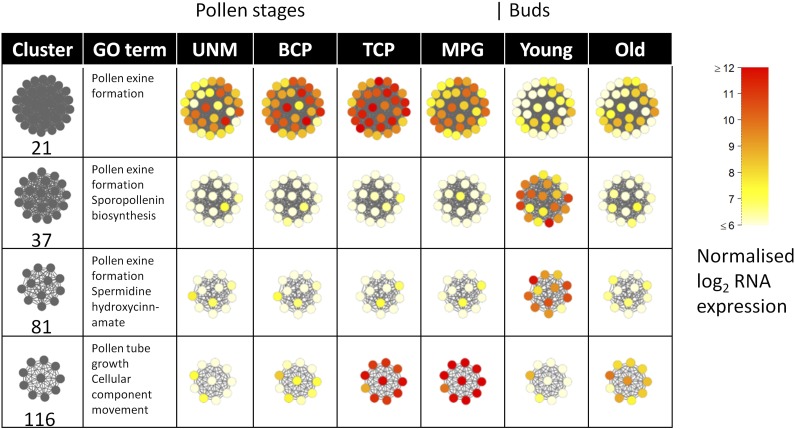
In order to validate the association between genes identified within the same cluster, four clusters were selected for detailed investigation. These were chosen based on their expression patterns, GO annotations, and behavior in other data sets. Each of these clusters has overrepresented GO terms associated with pollen or anther development. The selected clusters show enrichment of gene expression at specific time points during pollen development based upon staged bud expression data (Xu et al., 2010); cluster 37 includes genes that show up-regulation at pollen mother cell meiosis, cluster 81 at pollen mitosis I, cluster 21 at the bicellular pollen stage, and cluster 116 at pollen mitosis II (for pollen development review, see Borg et al., 2009; Table I). This allowed us to infer a good overview of the developmental expression changes during anther and pollen development through these clusters (Fig. 5).
Table I. Genes present in the four selected clusters including their predicted roles and published expression patterns.
| Cluster No. | Name; The Arabidopsis Information Resource 10 Annotation | Role | Published Expression |
|---|---|---|---|
| Cluster 21 | |||
| AT1G04290 | Thioesterase superfamily | ||
| AT1G07850 | DOMAIN OF UNKNOWN FUNCTION (DUF)064 | ||
| AT1G20080 | SYNAPTOTAGMIN2 | Protein secretion | |
| AT1G22150 | SULTR1;1; sulfate transporter | Sulfate membrane transport | |
| AT1G32250 | Calcium-binding EF-hand family | ||
| AT1G52570 | PLDALPHA2; phospholipase D α-2 | ||
| AT1G54560 | MYOSIN XI E | Golgi movement | |
| AT1G72960 | RHD3; root hair defective3 GTP-binding | ||
| AT2G25890 | Oleosin family protein | Lipid storage | |
| AT2G30290 | VSR2; vacuolar sorting receptor | Vacuolar sorting | |
| AT2G30630 | P-loop-containing nucleoside triphosphate hydrolase superfamily | Cell killing | |
| AT2G47770 | TSPO; outer membrane Trp-rich sensory protein | Abscisic acid and salt stress induced | Seed |
| AT3G03900 | APK3 | Sulfation of secondary metabolites | Pollen |
| AT3G08560 | VHA-E2 | Pollen | |
| AT3G17630 | CHX19; cation/H+ exchanger | Membrane trafficking | |
| AT3G19090 | RNA-binding protein | RNA binding | |
| AT3G25160 | Endoplasmic reticulum luman protein-retaining receptor family | Endoplasmic reticulum retention | |
| AT3G51490 | TMT3; tonoplast monosaccharide transporter | Sugar transport | Filament |
| AT3G60540 | Preprotein translocase Sec | Translocase | |
| AT4G02140 | |||
| AT4G10440 | S-Adenosyl-l-Met-dependent methyltransferases | ||
| AT4G11030 | AMP-dependent synthetase and ligase family | ||
| AT4G18920 | DUF1264 | ||
| AT4G26770 | Phosphatidate cytidyltransferase family | ||
| AT4G36600 | Late embryogenesis-abundant protein | ||
| AT4G37840 | HEXOKINASE-LIKE3 | Hexokinase | |
| AT5G10730 | NAD(P)-binding Rossmann fold superfamily | ||
| AT5G13350 | Auxin-responsive GH3 family | ||
| AT5G15490 | UDP-Glc-6-dehydrogenase family | Cell wall composition | Throughout plant |
| AT5G66020 | AtSAC1B; suppressor of actin | Pollen | |
| AT5G66150 | Glycosyl hydrolase family | ||
| Cluster 37 | |||
| AT1G01280 | CYP703A2; cytochrome P450 family 703 | Sporopollenin biosynthesis | Anther |
| AT1G02050 | LAP6; less adhesive pollen6 | Sporopollenin biosynthesis | Anther, tapetum specific |
| AT1G02813 | DUF538 | ||
| AT1G62940 | ACOS5; acyl-CoA synthetase | Sporopollenin biosynthesis | Anther, tapetum specific |
| AT1G69500 | CYP704B1; cytochrome P450 family 704 | Sporopollenin biosynthesis | Anther, tapetum specific |
| AT1G76470 | NAD(P)-binding Rossmann fold superfamily | Lignin biosynthesis | |
| AT2G42870 | PAR1; Phytochrome rapidly regulated | Cell elongation | |
| AT3G11980 | MS2; male sterility2 | Exine formation | Anther, tapetum specific |
| AT3G13220 | WHITE/BROWN COMPLEX SUBFAMILY27 (WBC27); ABC transporter-like | Exine formation | Anther, tapetum specific |
| AT3G23770 | O-Glycosyl hydrolases family17 | Exine formation | |
| AT3G42960 | ATA1; tapetum1 | Anther, tapetum specific | |
| AT4G14080 | MEE48; maternal effect embryo arrest | Exine formation | |
| AT4G20420 | TAP35/44; tapetum-specific protein | Tapetum specific | |
| AT4G34850 | LAP5; less adhesive protein | Sporopollenin biosynthesis | Anther, tapetum specific |
| AT4G35420 | DRL1/TKPR1; dihydroflavonol 4-reductase-like | Sporopollenin biosynthesis | Anther, tapetum specific |
| AT5G07230 | Bifunctional inhibitor/lipid transfer protein | Lipid transfer | |
| AT5G47600 | HSP20-like chaperone superfamily | ||
| AT5G59000 | REALLY INTERESTING NEW GENE/FYVE/PLANT HOMEODOMAIN GENE zinc finger superfamily | ||
| AT5G62080 | Bifunctional inhibitor/lipid transfer protein | Lipid transfer | |
| AT5G65410 | HB25; homeobox protein | ||
| Cluster 81 | |||
| AT1G13140 | CYP86C3; cytochrome P450, family 86 | Exine formation | |
| AT1G67990 | TSM1; tapetum-specific methyltransferase1 | Spermidine formation | Anther, tapetum specific |
| AT1G71160 | KCS7; 3-ketoacyl-CoA synthase | Lipid biosynthesis | |
| AT2G19070 | SHT; spermidine hydroxycinnamoyl transferase | Spermidine formation | Anther, tapetum specific |
| AT3G52160 | KCS15; 3-ketoacyl-CoA synthase | Lipid biosynthesis | |
| AT4G28395 | A7; anther7 | Lipid transfer | Anther |
| AT5G13380 | Auxin-responsive GH3 family | ||
| AT5G16920 | Fasciclin-like arabinogalactan family | Exine formation | |
| AT5G16960 | Zinc-binding dehydrogenase family | ||
| AT5G48210 | Exine formation | ||
| AT5G49070 | KCS21; 3-ketoacyl-CoA synthase | Lipid biosynthesis | |
| AT5G65205 | NAD(P)-binding Rossmann fold superfamily | ||
| Cluster 116 | |||
| AT1G14420 | Arabidopsis LAT59 ORTHOLOG (At59) | Cell wall modification | Pollen specific |
| AT1G23350 | Plant invertase/pectin methylesterase | Actin filament movement | |
| AT1G49290 | Actin filament movement | ||
| AT1G64740 | TUA1; α-1 tubulin | Microtubule movement | Pollen specific |
| AT2G17500 | Pectin lyase-like superfamily | Actin filament movement | |
| AT2G31500 | CPK24; calcium-dependent protein kinase24 | Regulates growth | Pollen; pollen tubes |
| AT3G17060 | Pectin lyase-like superfamily | Actin filament movement | |
| AT3G25170 | RAPID ALKALINIZATION FACTOR-LIKE26 | ||
| AT5G23270 | STP11; sugar transporter11 | Sugar transport | Pollen specific |
| AT5G39400 | PTEN1 | Phosphatase | Pollen specific |
Figure 5.
Gene expression levels in the four selected clusters in different pollen development stages in whole buds, based on wild-type data from Xu et al. (2010) for each gene within the cluster. The thick black line shows the average trend of all the genes in the cluster.
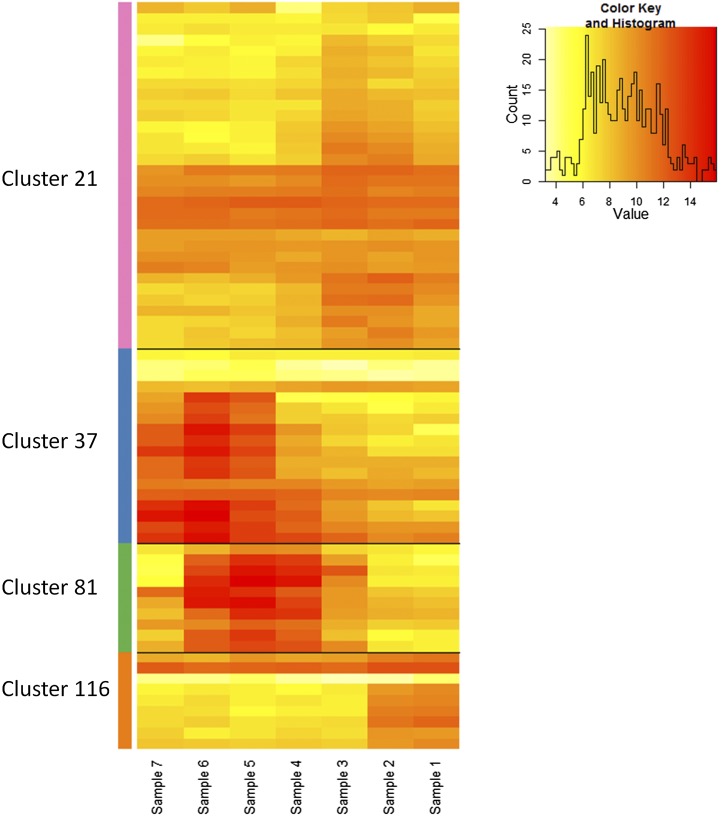
Figure 6 shows the expression patterns of individual genes within these particular clusters, looking at pollen-specific expression stages and the whole bud; both of these microarray data sets were used to create the network and illustrate the expected similar behavior of gene expression within each cluster. To interrogate these clusters further, we collected publicly available microarray data from various mutants known to have an important regulatory role during anther and pollen development. We chose eight mutants: ap3 (early stages only); spl/nzz and ems, which are involved in anther cell specification (Zhang et al., 2006; Alves-Ferreira et al., 2007; Wijeratne et al., 2007); dyt1 and ams (combination of all four stages); ms1 and myb80 (previously known as myb103), which are involved in tapetum development and mature pollen (Zhang et al., 2006; Alves-Ferreira et al., 2007; Xu et al., 2010; Phan et al., 2011); and rpk2 (flower stages 12–14), which is involved in anther dehiscence (Mizuno et al., 2007b). Wild-type data were used to initially generate the FlowerNet network, but when the corresponding mutant data were overlaid onto the network, a large proportion of genes within each cluster showed correlated expression in the specific mutants (Fig. 7). This clustering of expression was also shown as a heat map (Fig. 8), utilizing developmental staged microarray data from the ms1 mutant (Alves-Ferreira et al., 2007). This suggests not only that these genes are coexpressed but also that they may be coregulated.
Figure 6.
Gene expression levels in the four selected clusters, showing the data (Honys and Twell, 2004; Yang et al., 2007a) used to generate FlowerNet. UNM, Unicellular microspores; BCP, bicellular pollen; TCP, tricellular pollen; MPG, mature pollen grain. Young buds represent pollen mother cell mitosis I, while old buds represent mitosis II.
Figure 7.
Gene expression changes in the four selected clusters in different known anther and pollen development mutant microarray data sets; data are shown for ap3 (flower early stages only), spl/nzz (flower stages 1–13), ms1 (flower stages 1–13; Alves-Ferreira et al., 2007), ems (flower stages 8 and 9; Wijeratne et al., 2007), dyt1 (flower stages 8–10; Feng et al., 2012), ams (flower stages 9–12; Xu et al., 2010), myb80 (flower stages 9 and 10; Phan et al., 2011), and rpk2 (flower stages 12–14; Mizuno et al., 2007a). Each circle represents a gene, with red signifying up-regulation and blue for down-regulation in each mutant compared with the wild type.
Figure 8.
Gene heat map showing the levels of expression in the four selected clusters (Table I) in different development stages in whole buds, based on wild-type data from Alves-Ferreira et al. (2007). Sample 1 contains stage 13 buds, and each subsequent sample comprises the next two oldest buds until sample 7, which includes the earliest buds and the inflorescence meristem.
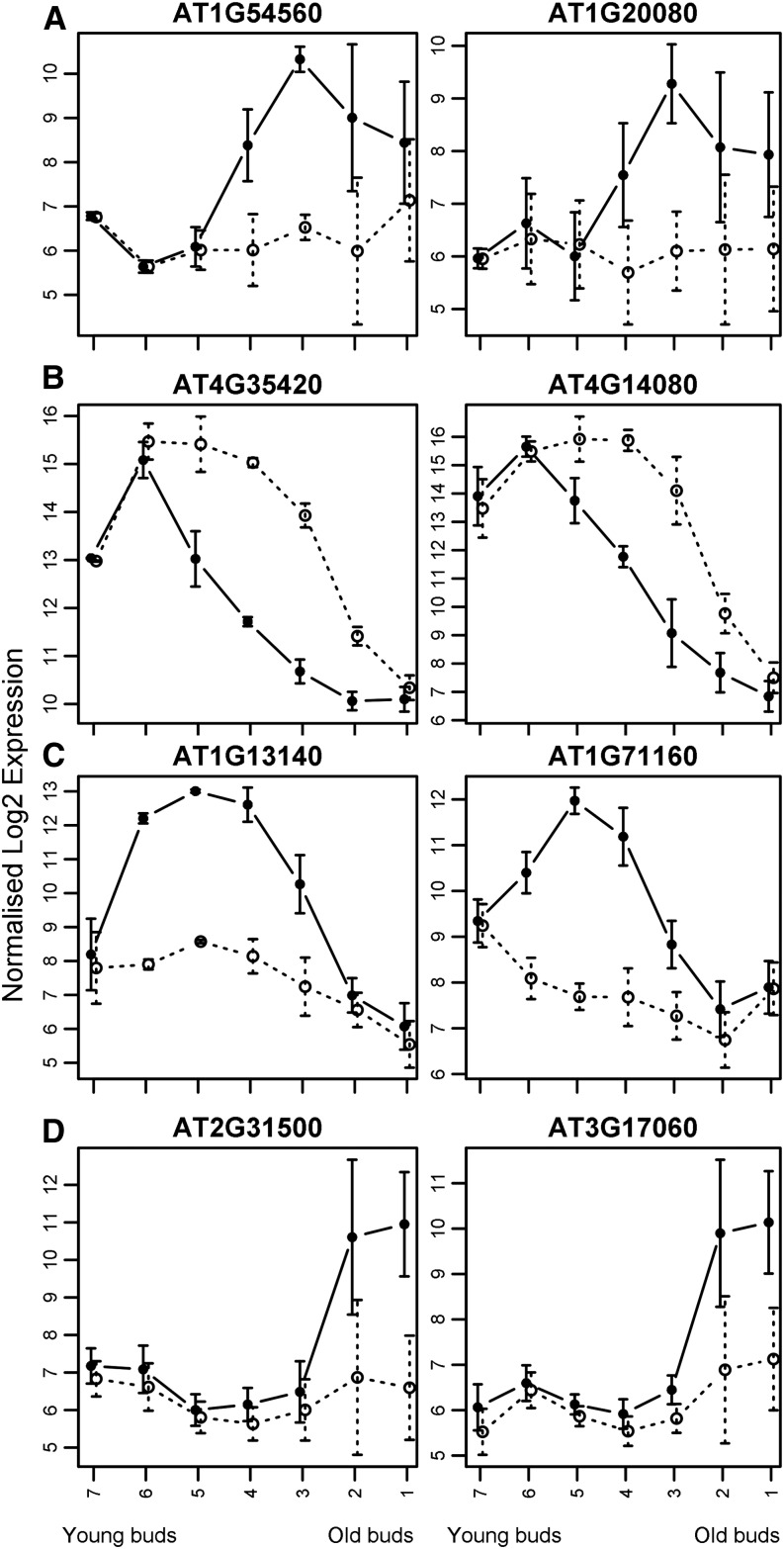
To characterize the clusters further, we compared the gene expression of these clusters in the developmental time courses generated from the data of Xu et al. (2010) and Alves-Ferreira et al. (2007). In Figure 5, each gene within the four clusters is represented by a different color line, the thick black line being the median trend line, while Figure 8 shows a heat map of expression levels of each gene within each of the four chosen clusters. From both of these sets of data, the genes in the clusters are expressed in a similar manner and in most cases at a similar level of expression. Therefore, despite only using the wild-type Affymetrix arrays to generate the network, the clusters show similar expression in the developmental time courses available in two-color Agilent arrays (Alves-Ferreira et al., 2007) as well as the various sets of corresponding mutant data. This implies that the genes within the clusters have expression patterns that are closely correlated, suggesting that they may be involved in the same process and potentially may be regulated by the same mechanism. Use of this network clustering to find clusters that change in different mutant backgrounds may allow new targets and processes to be inferred from the results. To focus on these coexpression similarities, we also looked at the individual genes in each cluster in more detail to compare expression and show that, within a cluster, the genes show very similar expression in both wild-type and mutant data (Fig. 9).
Figure 9.
Two exemplar genes from the four clusters, cluster 21 (A), cluster 37 (B), cluster 81 (C), and cluster 116 (D), showing expression patterns using the wild type (black circles) and ms1 (white circles). Data are from Alves-Ferreira et al. (2007). Error bars show sd.
Cluster 21: Pollen Exine Formation
Cluster 21 contains 31 genes with 464 edges between them (99.78% of the possible edges), which are involved in pollen exine formation, and contains three characterized genes that are known to be pollen specific, Arabidopsis SUPPRESSOR OF ACTIN (AtSAC1b), ADENOSINE-5′-PHOSPHOSULFATE KINASE (APK3), and VACUOLAR H+-ATPASE SUBUNIT E (VHA-E2; Despres et al., 2003; Dettmer et al., 2010; Mugford et al., 2010); 23 of these genes were found to be specifically expressed in stamens (Wellmer et al., 2004). The majority of the genes within this cluster have an unknown role, with the GO term pollen exine formation overrepresented; however, their specific function is currently undetermined. These genes are up-regulated in the stamen of stage 12 flowers (using the data from Schmid et al. [2005]) and in pollen; the majority are down-regulated in old ms1 buds, particularly in the second and third samples of the time course from Alves-Ferreira et al. (2007; sample 1 equates to mature flowers and sample 7 to immature buds/inflorescence meristem). This corresponds with a peak in the wild-type bicellular pollen development stage of buds in the data from Xu et al. (2010), with the majority of the genes within this cluster being down-regulated in the ams mutant from the same data set. The majority of these genes have higher expression in the tapetum compared with other anther tissues at pollen mitosis II (Z.A. Wilson and G. Vizcay-Barrena, unpublished data). Twenty-one of these genes were found to be down-regulated in stage 5 to 8 myb80 anthers (Phan et al., 2011), suggesting that this cluster acts downstream of AMS and MYB80.
Of the three pollen-specific genes, AtSAC1b shows specific expression in pollen, with higher levels at the uninucleate stage (Despres et al., 2003), VHA-E2 is specific to the vegetative cell in the pollen (Strompen et al., 2005), while APK3 has expression throughout development, with strongest expression in pollen grains (Mugford et al., 2009). The single mutants in all of these three genes have no phenotype, most likely because of functional redundancy of other isoforms expressed within the pollen: only in the triple mutant of APK1 isoforms (apk1 apk3 apk4) is there a pollen lethality phenotype (Mugford et al., 2010). APK3 is an ADENOSINE-5′-PHOSPHOSULFATE kinase involved in sulfur metabolism, while AtSAC1b has been suggested to regulate the PHOSPHOINOSITOL-4-phosphate pool and could control ATP transport (Despres et al., 2003). This links with VHA-E2, which is a vacuolar H+-ATPase that plays an important role in maintaining the pH of endomembrane compartments in eukaryotic cells (Dettmer et al., 2010). The double and triple knockouts of AtSAC1b and VHA-E2 would be interesting to study to see if they have a similar phenotype to APK3 and discover what role these families of genes have in pollen development. Another two published genes from this cluster, AT5G15490 (UDP-GLUCOSE DEHYDROGENASE; Reboul et al., 2011) and AT3G51490 (TONOPLAST MONOSACCHARIDE TRANSPORTER1; Wormit et al., 2006), are involved in cell wall composition and sugar transport, which also may be important for pollen wall formation.
By focusing research on the other cluster members within the group, we may be able to infer further roles of this cluster, as many of the genes have predicted roles in protein movement and in secretion. It will be interesting to discover the roles they play in anther and pollen development.
Cluster 37: Sporopollenin Biosynthesis
Cluster 37 is a 20-node clique, with all edges between them present, and contains a number of genes known to be tapetum specific and linked to sporopollenin biosynthesis. Of these 20 genes, 11 were found to be stamen specific (Wellmer et al., 2004); however, two of these 20 genes are not included in the array analysis. The genes in this cluster are up-regulated in flowers at stage 9 to 11 (Schmid et al., 2005) and are mostly up-regulated in the ms1 mutant, particularly in the third to fifth samples of the time course generated from the data of Alves-Ferreira et al. (2007), while the majority of these genes are down-regulated in ams mutants, particularly at the meiosis and pollen mitosis I stages (Xu et al., 2010). This corresponds to a peak in expression in wild-type data from the same data set at the meiosis stage, with the majority of the genes more highly expressed in the tapetum than in the other anther tissues at the tetrad stage (Z.A. Wilson and G. Vizcay-Barrena, unpublished data). Collectively, this suggests that these genes are downstream of AMS and are up-regulated by AMS, while they are negatively regulated by MS1 to give a specific expression pattern.
A large proportion of these genes are involved in the biosynthesis of sporopollenin, which is a major constituent of exine in the outer pollen wall. Recently in Arabidopsis, a number of genes have been demonstrated to be involved in sporopollenin biosynthesis: ACYL-COENZYME A SYNTHETASE (ACOS5), CYTOCHROME P450 (CYP703A2), CYP704B1, LESS ADHESIVE POLLEN3 (LAP3), LAP5 (POLYKETIDE SYNTHASE B), LAP6 (POLYKETIDE SYNTHASE A), MS2, FACELESS POLLEN1 (FLP1), and TETRAKETIDE α-PYRONE REDUCTASE1 (TKPR1 [DIHYDROFLAVONOL 4-REDUCTASE-LIKE1]; Ariizumi and Toriyama, 2011); all of these except LAP3 and FLP1 are found within this cluster. Lallemand et al. (2013) demonstrated that ACOS5, LAP5, LAP6, TKPR1, CYP703A2, and CYP704B1 all interact together on the endoplasmic reticulum of plant cells, and all of these interacting genes are found in this cluster. Of the studied genes within this cluster, the phenotypes range from an absence of production of pollen due to compromised pollen walls (ACOS5 and TKPR1; de Azevedo Souza et al., 2009; Tang et al., 2009; Grienenberger et al., 2010) to having irregular exine layers (CYP703A2, CYP704B1, ATP-BINDING CASSETTE G26 [ABCG26], LAP5, LAP6, and MS2; Morant et al., 2007; Dobritsa et al., 2009, 2010; Kim et al., 2010; Choi et al., 2011), with varying phenotypes from loss of fertility to reduced pollen viability to no effect on pollen viability. The sporopollenin precursor biosynthesis gene WBC27 (ABCG26) is also within this cluster and is involved in the transport of these sporopollenin precursors from the tapetum, facilitating exine formation on the pollen surface (Choi et al., 2011). Two other genes of unknown function are believed to play a role in pollen exine formation, At3g23770 and At4g14080 (MATERNAL EFFECT EMBRYO ARREST48), and two further unknown genes are tapetum specific, At3g4290 (TAPETUM1) and At4g20420, based on their description in AtEnsembl (Flicek et al., 2013; http://atensembl.arabidopsis.info/index.html). Therefore, 12 out of 20 genes in this cluster have a direct role in the tapetum, with one-half of the genes in this cluster involved in exine/sporopollenin formation. The rest of the genes in this cluster have an unknown function, including two putative lipid transfer proteins, At5g07230 and At5g62080, which also may be involved in sporopollenin biosynthesis. This strongly suggests that the remaining six unknown genes within this cluster could play a role in sporopollenin synthesis in some manner, and studying the knockouts of these genes could further our understanding of the sporopollenin biosynthesis pathway and transport to the pollen coat wall.
Cluster 81: Pollen Spermidine Formation
Cluster 81 is a 12-gene clique, with all 12 of these genes found to be stamen specific (Wellmer et al., 2004) and up-regulated in flower stages 9 to 11 (Schmid et al., 2005). These genes are differentially expressed (mostly down-regulated) in both ms1 and ams mutants throughout pollen developmental stages (Alves-Ferreira et al., 2007; Xu et al., 2010) as well as tapetum specific (Z.A. Wilson and G. Vizcay-Barrena, unpublished data) and particularly expressed in the pollen mitosis I stage (Xu et al., 2010). Two genes (At5g49070 and At3g52160) were also down-regulated in myb80 anthers.
Similar to cluster 37, the majority of these genes have suggestive roles in pollen exine formation (five), lipid biosynthesis (three), and lipid transfer (one), with the two studied genes in this cluster being involved in spermidine (exine) formation. Both SPERMIDINE HYDROXYCINNAMOYL TRANSFERASE (SHT) and TAPETUM-SPECIFIC METHYLTRANSFERASE1 (TMS1) are specifically expressed in tapetum cells (Fellenberg et al., 2008; Grienenberger et al., 2009). SHT mutants display irregularities in the pollen wall, with reduced autofluorescence of the pollen wall suggesting a reduction in the exine coat components. They also have a reduction of two spermidine derivatives, while in the overexpressing plants there was an increased peak for these derivatives and increased autofluorescence (Grienenberger et al., 2009). Grienenberger et al. (2009) demonstrated that SHT can catalyze the last methylation step in the biosynthesis of acylated spermidine conjugates. The TMS1 mutant showed impaired silique development (Fellenberg et al., 2008) and was shown to be involved in the biosynthesis of spermidine conjugates at a later step than SHT (Grienenberger et al., 2009).
A number of the unknown genes in this cluster also have potential roles in pollen exine and lipid biosynthesis, based on their description in AtEnsembl (Flicek et al., 2013). Therefore, it would be interesting to study the rest of the genes within this cluster to further the understanding of exine biosynthesis.
Cluster 116: Pollen Tube Formation/Growth
Cluster 116 is a 10-gene clique, with seven of these genes found to be stamen specific (Wellmer et al., 2004), with the genes showing increased expression in pollen and in stamen at stages 12 and 15 (Schmid et al., 2005). These genes are down-regulated in both ms1 (samples 2 and 1) and ams (pollen mitosis II) mutants (Alves-Ferreira et al., 2007; Xu et al., 2010). The genes have highest expression in pollen mitosis II (Xu et al., 2010). One-half of the genes in this cluster are known to be pollen specific, such as At59 (Kulikauskas and McCormick, 1997), α-1 TUBULIN (Carpenter et al., 1992), and Arabidopsis PHOSPHATASE AND TENSIN HOMOLOG1 (Gupta et al., 2002), and have roles such as microtubule/filament movement, cell wall modification, and sugar transport (Schneidereit et al., 2005), so they could play roles in pollen tube growth and formation, as does CALCIUM-DEPENDENT PROTEIN KINASE24 (Zhao et al., 2013).
DISCUSSION
FlowerNet highlights clusters of genes that are similarly transcriptionally regulated and, therefore, may play closely related functional roles. Focusing on four specific clusters, we show that there is a close relationship between the genes in how they are expressed from a number of different data sets from wild-type tissues or staged samples and also in male-sterile mutant expression data sets, which were not used to make FlowerNet. This provides evidence for the robustness of this method and the network generated. Therefore, FlowerNet could play a valuable role in identifying new players in anther and pollen development, as, while the main transcription factors are known, their exact interaction and primary targets are still uncharacterized. Cluster 37 is a strong indication of how this tool can be used successfully, as 12 out of 20 genes in this cluster have a direct role in the tapetum, with one-half of the genes in this cluster known to be involved in exine/sporopollenin formation. The rest of the genes in this cluster have an unknown function, including two putative lipid transfer proteins, At5g07230 and At5g62080, which also may be involved in sporopollenin biosynthesis. This strongly suggests that the remaining unknown genes within this cluster also could play a role in sporopollenin synthesis; studying the knockouts of these genes could further our understanding of the sporopollenin biosynthesis pathway and transport to the pollen coat wall. In addition to the characterization of genes associated with anther and pollen development, the FlowerNet correlation network is also a valuable tool for the analysis of genes generally involved in flowering and plant reproduction, since the data used to generate the network have come from microarray experiments from various organs and developmental stages throughout floral development.
To aid the global dissection of anther and pollen development, we have made the gene expression plots available as a community resource (http://www.cpib.ac.uk/anther), allowing visualization of the expression across different microarray experiments that have been normalized together. Included in this resource is a list of highly correlated genes based on the FlowerNet clusters to extract further information from these plots. Additionally, the FlowerNet network is available as a Cytoscape file for specific analysis of the network and resulting clusters. These tools will be extremely helpful in the flower development community and will aid the identification of novel genes on which to focus and also in the selection of putatively redundant family members for the generation of mutants carrying multiple mutations. For example, the majority of cluster 21 has not been studied, despite the fact that many of these genes have been linked to pollen development. The mutant phenotypes that have been shown in this cluster are only from triple mutants of families, indicating the high level of redundancy of the genes within this cluster. This shows how important it is to understand how similar genes from the same family behave transcriptionally in relation to each other to further our understanding of pollen development. This clustering tool allows selective focusing on specific targets to develop our understanding of anther and pollen development.
MATERIALS AND METHODS
Gene Expression Plots and Microarray Normalization
The gene expression plots consist of three separate panels of data relating to different microarray platforms. The first panel consists of Affymetrix microarrays, where the raw .cel files were background corrected and normalized using the robust microarray averaging procedure (Irizarry et al., 2003), with a custom chip definition file from the CustomCDF project (Ath1121501_At_TAIRG.cdf v14.0.0; Dai et al., 2005), using the Bioconductor affy package in the programming language R. This chip definition file maps the individual probes on the Affymetrix chip, using recent sequencing information contained in The Arabidopsis Information Resource (Lamesch et al., 2012), to their corresponding genes and results in 21,313 unique genes being considered. This mapping eliminates the many-many relationship that exists between the Affymetrix probe sets and gene targets as traditionally used. In particular, this bijective mapping ensures that gene Arabidopsis Genome Initiative codes may be used as the primary identifiers in a correlation network with no question of how to deal with multiple probe sets, with sometimes markedly differing behaviors, corresponding to the same gene. The resulting probe sets have varying numbers of probes with a minimum of three, although the majority of probe sets have the 11 probes from an original Affymetrix probe set.
The second panel includes the two-color microarrays described by Alves-Ferreira et al. (2007), where bud samples were collected from stage 13 through the earliest buds and the inflorescence meristem. In this data set, sample 1 consists of the pooled mature stage 13 flowers from a number of plants, with subsequent samples comprising successively younger buds per sample, until the final sample (sample 7), which consists of the remaining early-stage buds and the inflorescence meristem. These two-color microarrays consist of one channel for wild-type buds and the second channel for mutant buds (ap3, spl/nzz, and ms1). The typical analysis for such two-color microarrays is to find lists of genes that are up- or down-regulated at each sample but not to compare across the different samples. Using the Bioconductor (Gentleman et al., 2004) package limma (Smyth and Speed, 2003; Smyth, 2005), the 33 .gpr files were background corrected (using the normexp method; Ritchie et al., 2007), then the two channels within each array (wild type and mutant) were normalized to have similar distributions (using the median method) and then quantile normalized across the 33 arrays. This last step of quantile normalization is nonstandard for two-color arrays, discussed by Yang and Thorne (2003), but is a routine part of single-color microarray normalization and allows the comparison of fluorescence intensity between the different arrays and enabled comparisons of wild-type and mutant values as a biologically meaningful developmental time course.
The third panel includes data from an in-house-printed two-color microarray generated by Xu et al. (2010), which was used to compare between wild-type and ams buds at pollen mother cell meiosis, pollen mitosis I, bicellular pollen, and pollen mitosis II stages. These 12 .gpr files also have been normalized in the same way as the data from Alves-Ferreira et al. (2007), allowing the comparison of genes across this time course.
Each of these three microarray platforms contains probe sets corresponding to a different set of genes, so some genes are not present in all three panels, such as the key gene MS1, which is not present on the Affymetrix chip.
Correlation Network Generation
Sixty-six wild-type Arabidopsis (Arabidopsis thaliana) Affymetrix .cel files from both public (Honys and Twell, 2004; Schmid et al., 2005; Mandaokar et al., 2006; Yang et al., 2007) and unpublished experiments on functionally wild-type buds and anthers were used to generate a correlation network, which we term FlowerNet. These chips include 10 pollen samples, 28 whole buds, and 28 anther/stamen samples; for full details, see Supplemental File S1. A number of chips from other published experiments were unable to be included, as the raw data were not available at the time of construction.
To generate the network, the 66 raw .cel files were background corrected and normalized using robust microarray averaging and the CustomCDF as discussed above. The genes were then filtered by keeping those probe sets that have at least one sample with expression greater than or equal to 6 (on the log2 scale). To identify interactions between the expressed genes, the Pearson correlation coefficient between all pairs of genes was calculated. A cutoff was then applied to the resulting correlation matrix to produce a set of edges. Previous studies (Bassel et al., 2011; Dekkers et al., 2013) show that having approximately one-half million edges gives a good number from which to generate meaningful clusters. To this end, we choose a correlation of 0.88 as the cutoff, giving 605,686 edges between 10,797 genes, which is the 0.38% most significant positive correlations (Supplemental File S5); this is chosen semiarbitrarily to balance sensitivity and specificity. Each correlation above this cutoff is considered to be an edge, and a table of edges between nodes is exported into Cytoscape version 2.8.1 (Shannon et al., 2003; Smoot et al., 2011), along with the correlation value between each edge. The yGraph Organic layout was used to display the resulting networks, although such layouts should be viewed as arbitrary.
From these correlation networks, the Cytoscape plug-in ClusterMaker version 1.11 (Morris et al., 2011) was used to partition the overall network into distinct clusters. In particular, the transitivity clustering method (Wittkop et al., 2010) was used, with parameters Max Subcluster Size = 400 and Max Time = 10, using the correlation values as an edge weight to generate small, well-connected clusters in the network (Supplemental File S3). To verify that these clusters are relatively insensitive to this choice of cutoff, we generated an additional set of clusters with cutoff of 0.9; this keeps 324,977 edges. Comparing these clusters with the FlowerNet clusters, of the 92 new clusters with 10 or more genes, on average, 65.2% of the genes are in a single FlowerNet cluster.
Supplemental Data
The following supplemental materials are available.
Supplemental File S1. Description of microarray chips used to generate FlowerNet and expression plots.
Supplemental File S2. Normalized data used in FlowerNet and the expression plots.
Supplemental File S3. FlowerNet Cytoscape file.
Supplemental File S4. Cluster allocations.
Supplemental File S5. Histogram of edge correlations.
Supplementary Material
Acknowledgments
We thank Dr. Will Townsend for help with the Web interface.
Glossary
- GO
Gene Ontology
Footnotes
This work was supported by the Biotechnology and Biological Sciences Research Council (grant no. BB/J001295/1), the Royal Society, and the Wolfson Foundation.
Articles can be viewed without a subscription.
References
- Alves-Ferreira M, Wellmer F, Banhara A, Kumar V, Riechmann JL, Meyerowitz EM (2007) Global expression profiling applied to the analysis of Arabidopsis stamen development. Plant Physiol 145: 747–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariizumi T, Toriyama K (2011) Genetic regulation of sporopollenin synthesis and pollen exine development. Annu Rev Plant Biol 62: 437–460 [DOI] [PubMed] [Google Scholar]
- Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Holko M, et al. (2013) NCBI GEO: archive for functional genomics data sets: update. Nucleic Acids Res 41: D991–D995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassel GW, Lan H, Glaab E, Gibbs DJ, Gerjets T, Krasnogor N, Bonner AJ, Holdsworth MJ, Provart NJ (2011) Genome-wide network model capturing seed germination reveals coordinated regulation of plant cellular phase transitions. Proc Natl Acad Sci USA 108: 9709–9714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borg M, Brownfield L, Twell D (2009) Male gametophyte development: a molecular perspective. J Exp Bot 60: 1465–1478 [DOI] [PubMed] [Google Scholar]
- Carpenter JL, Ploense SE, Snustad DP, Silflow CD (1992) Preferential expression of an alpha-tubulin gene of Arabidopsis in pollen. Plant Cell 4: 557–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H, Jin JY, Choi S, Hwang JU, Kim YY, Suh MC, Lee Y (2011) An ABCG/WBC-type ABC transporter is essential for transport of sporopollenin precursors for exine formation in developing pollen. Plant J 65: 181–193 [DOI] [PubMed] [Google Scholar]
- Dai M, Wang P, Boyd AD, Kostov G, Athey B, Jones EG, Bunney WE, Myers RM, Speed TP, Akil H, et al. (2005) Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res 33: e175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Azevedo Souza C, Kim SS, Koch S, Kienow L, Schneider K, McKim SM, Haughn GW, Kombrink E, Douglas CJ (2009) A novel fatty acyl-CoA synthetase is required for pollen development and sporopollenin biosynthesis in Arabidopsis. Plant Cell 21: 507–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekkers BJW, Pearce S, van Bolderen-Veldkamp RP, Marshall A, Widera P, Gilbert J, Drost HG, Bassel GW, Müller K, King JR, et al. (2013) Transcriptional dynamics of two seed compartments with opposing roles in Arabidopsis seed germination. Plant Physiol 163: 205–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Despres B, Bouissonnié F, Wu HJ, Gomord V, Guilleminot J, Grellet F, Berger F, Delseny M, Devic M (2003) Three SAC1-like genes show overlapping patterns of expression in Arabidopsis but are remarkably silent during embryo development. Plant J 34: 293–306 [DOI] [PubMed] [Google Scholar]
- Dettmer J, Liu TY, Schumacher K (2010) Functional analysis of Arabidopsis V-ATPase subunit VHA-E isoforms. Eur J Cell Biol 89: 152–156 [DOI] [PubMed] [Google Scholar]
- Dobritsa AA, Lei Z, Nishikawa S, Urbanczyk-Wochniak E, Huhman DV, Preuss D, Sumner LW (2010) LAP5 and LAP6 encode anther-specific proteins with similarity to chalcone synthase essential for pollen exine development in Arabidopsis. Plant Physiol 153: 937–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobritsa AA, Shrestha J, Morant M, Pinot F, Matsuno M, Swanson R, Møller BL, Preuss D (2009) CYP704B1 is a long-chain fatty acid omega-hydroxylase essential for sporopollenin synthesis in pollen of Arabidopsis. Plant Physiol 151: 574–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE (2002) Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30: 207–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fellenberg C, Milkowski C, Hause B, Lange PR, Böttcher C, Schmidt J, Vogt T (2008) Tapetum-specific location of a cation-dependent O-methyltransferase in Arabidopsis thaliana. Plant J 56: 132–145 [DOI] [PubMed] [Google Scholar]
- Feng B, Lu D, Ma X, Peng Y, Sun Y, Ning G, Ma H (2012) Regulation of the Arabidopsis anther transcriptome by DYT1 for pollen development. Plant J 72: 612–624 [DOI] [PubMed] [Google Scholar]
- Flicek P, Ahmed I, Amode MR, Barrell D, Beal K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fairley S, et al. (2013) Ensembl 2013. Nucleic Acids Res 41: D48–D55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. (2004) Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5: R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg RB, Beals TP, Sanders PM (1993) Anther development: basic principles and practical applications. Plant Cell 5: 1217–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grienenberger E, Besseau S, Geoffroy P, Debayle D, Heintz D, Lapierre C, Pollet B, Heitz T, Legrand M (2009) A BAHD acyltransferase is expressed in the tapetum of Arabidopsis anthers and is involved in the synthesis of hydroxycinnamoyl spermidines. Plant J 58: 246–259 [DOI] [PubMed] [Google Scholar]
- Grienenberger E, Kim SS, Lallemand B, Geoffroy P, Heintz D, Souza CdeA, Heitz T, Douglas CJ, Legrand M (2010) Analysis of TETRAKETIDE α-PYRONE REDUCTASE function in Arabidopsis thaliana reveals a previously unknown, but conserved, biochemical pathway in sporopollenin monomer biosynthesis. Plant Cell 22: 4067–4083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R, Ting JT, Sokolov LN, Johnson SA, Luan S (2002) A tumor suppressor homolog, AtPTEN1, is essential for pollen development in Arabidopsis. Plant Cell 14: 2495–2507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higginson T, Li SF, Parish RW (2003) AtMYB103 regulates tapetum and trichome development in Arabidopsis thaliana. Plant J 35: 177–192 [DOI] [PubMed] [Google Scholar]
- Honys D, Twell D (2004) Transcriptome analysis of haploid male gametophyte development in Arabidopsis. Genome Biol 5: R85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP (2003) Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4: 249–264 [DOI] [PubMed] [Google Scholar]
- Ito T, Nagata N, Yoshiba Y, Ohme-Takagi M, Ma H, Shinozaki K (2007) Arabidopsis MALE STERILITY1 encodes a PHD-type transcription factor and regulates pollen and tapetum development. Plant Cell 19: 3549–3562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack T, Fox GL, Meyerowitz EM (1994) Arabidopsis homeotic gene APETALA3 ectopic expression: transcriptional and posttranscriptional regulation determine floral organ identity. Cell 76: 703–716 [DOI] [PubMed] [Google Scholar]
- Kim SS, Grienenberger E, Lallemand B, Colpitts CC, Kim SY, Souza CdeA, Geoffroy P, Heintz D, Krahn D, Kaiser M, et al. (2010) LAP6/POLYKETIDE SYNTHASE A and LAP5/POLYKETIDE SYNTHASE B encode hydroxyalkyl α-pyrone synthases required for pollen development and sporopollenin biosynthesis in Arabidopsis thaliana. Plant Cell 22: 4045–4066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulikauskas R, McCormick S (1997) Identification of the tobacco and Arabidopsis homologues of the pollen-expressed LAT59 gene of tomato. Plant Mol Biol 34: 809–814 [DOI] [PubMed] [Google Scholar]
- Lallemand B, Erhardt M, Heitz T, Legrand M (2013) Sporopollenin biosynthetic enzymes interact and constitute a metabolon localized to the endoplasmic reticulum of tapetum cells. Plant Physiol 162: 616–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamesch P, Berardini TZ, Li D, Swarbreck D, Wilks C, Sasidharan R, Muller R, Dreher K, Alexander DL, Garcia-Hernandez M, et al. (2012) The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res 40: D1202–D1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Huang J, Parameswaran S, Ito T, Seubert B, Auer M, Rymaszewski A, Jia G, Owen HA, Zhao D (2009) The SPOROCYTELESS/NOZZLE gene is involved in controlling stamen identity in Arabidopsis. Plant Physiol 151: 1401–1411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H. (2005) Molecular genetic analyses of microsporogenesis and microgametogenesis in flowering plants. Annu Rev Plant Biol 56: 393–434 [DOI] [PubMed] [Google Scholar]
- Mandaokar A, Thines B, Shin B, Lange BM, Choi G, Koo YJ, Yoo YJ, Choi YD, Choi G, Browse J (2006) Transcriptional regulators of stamen development in Arabidopsis identified by transcriptional profiling. Plant J 46: 984–1008 [DOI] [PubMed] [Google Scholar]
- Millar AA, Gubler F (2005) The Arabidopsis GAMYB-like genes, MYB33 and MYB65, are microRNA-regulated genes that redundantly facilitate anther development. Plant Cell 17: 705–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno S, Osakabe Y, Maruyama K, Ito T, Osakabe K, Sato T, Shinozaki K, Yamaguchi-Shinozaki K (2007a) Receptor-like protein kinase 2 (RPK 2) is a novel factor controlling anther development in Arabidopsis thaliana. Plant J 50: 751–766 [DOI] [PubMed] [Google Scholar]
- Mizuno S, Osakabe Y, Maruyama K, Ito T, Sato T, Shinozaki K, Yamaguchi-Shinozaki K (2007b) RPK2 is a novel factor controlling anther development in Arabidopsis thaliana. Plant Cell Physiol 48: S229. [DOI] [PubMed] [Google Scholar]
- Morant M, Jørgensen K, Schaller H, Pinot F, Møller BL, Werck-Reichhart D, Bak S (2007) CYP703 is an ancient cytochrome P450 in land plants catalyzing in-chain hydroxylation of lauric acid to provide building blocks for sporopollenin synthesis in pollen. Plant Cell 19: 1473–1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris JH, Apeltsin L, Newman AM, Baumbach J, Wittkop T, Su G, Bader GD, Ferrin TE (2011) clusterMaker: a multi-algorithm clustering plugin for Cytoscape. BMC Bioinformatics 12: 436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mugford SG, Matthewman CA, Hill L, Kopriva S (2010) Adenosine-5′-phosphosulfate kinase is essential for Arabidopsis viability. FEBS Lett 584: 119–123 [DOI] [PubMed] [Google Scholar]
- Mugford SG, Yoshimoto N, Reichelt M, Wirtz M, Hill L, Mugford ST, Nakazato Y, Noji M, Takahashi H, Kramell R, et al. (2009) Disruption of adenosine-5′-phosphosulfate kinase in Arabidopsis reduces levels of sulfated secondary metabolites. Plant Cell 21: 910–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng FY, Weselake RJ (2011) Gene coexpression clusters and putative regulatory elements underlying seed storage reserve accumulation in Arabidopsis. BMC Genomics 12: 286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan HA, Iacuone S, Li SF, Parish RW (2011) The MYB80 transcription factor is required for pollen development and the regulation of tapetal programmed cell death in Arabidopsis thaliana. Plant Cell 23: 2209–2224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reboul R, Geserick C, Pabst M, Frey B, Wittmann D, Lütz-Meindl U, Léonard R, Tenhaken R (2011) Down-regulation of UDP-glucuronic acid biosynthesis leads to swollen plant cell walls and severe developmental defects associated with changes in pectic polysaccharides. J Biol Chem 286: 39982–39992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie ME, Silver J, Oshlack A, Holmes M, Diyagama D, Holloway A, Smyth GK (2007) A comparison of background correction methods for two-colour microarrays. Bioinformatics 23: 2700–2707 [DOI] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Schölkopf B, Weigel D, Lohmann JU (2005) A gene expression map of Arabidopsis thaliana development. Nat Genet 37: 501–506 [DOI] [PubMed] [Google Scholar]
- Schneidereit A, Scholz-Starke J, Sauer N, Büttner M (2005) AtSTP11, a pollen tube-specific monosaccharide transporter in Arabidopsis. Planta 221: 48–55 [DOI] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13: 2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T (2011) Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics 27: 431–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK. (2005) Limma: linear models for microarray data. InBioinformatics and Computational Biology Solutions Using R and Bioconductor. Springer-Verlag, New York, pp 397–420 [Google Scholar]
- Smyth GK, Speed T (2003) Normalization of cDNA microarray data. Methods 31: 265–273 [DOI] [PubMed] [Google Scholar]
- Sorensen AM, Kröber S, Unte US, Huijser P, Dekker K, Saedler H (2003) The Arabidopsis ABORTED MICROSPORES (AMS) gene encodes a MYC class transcription factor. Plant J 33: 413–423 [DOI] [PubMed] [Google Scholar]
- Steiner-Lange S, Unte US, Eckstein L, Yang C, Wilson ZA, Schmelzer E, Dekker K, Saedler H (2003) Disruption of Arabidopsis thaliana MYB26 results in male sterility due to non-dehiscent anthers. Plant J 34: 519–528 [DOI] [PubMed] [Google Scholar]
- Strompen G, Dettmer J, Stierhof YD, Schumacher K, Jürgens G, Mayer U (2005) Arabidopsis vacuolar H-ATPase subunit E isoform 1 is required for Golgi organization and vacuole function in embryogenesis. Plant J 41: 125–132 [DOI] [PubMed] [Google Scholar]
- Tang LK, Chu H, Yip WK, Yeung EC, Lo C (2009) An anther-specific dihydroflavonol 4-reductase-like gene (DRL1) is essential for male fertility in Arabidopsis. New Phytol 181: 576–587 [DOI] [PubMed] [Google Scholar]
- Wang S, Yin Y, Ma Q, Tang X, Hao D, Xu Y (2012) Genome-scale identification of cell-wall related genes in Arabidopsis based on co-expression network analysis. BMC Plant Biol 12: 138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellmer F, Riechmann JL, Alves-Ferreira M, Meyerowitz EM (2004) Genome-wide analysis of spatial gene expression in Arabidopsis flowers. Plant Cell 16: 1314–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijeratne AJ, Zhang W, Sun Y, Liu W, Albert R, Zheng Z, Oppenheimer DG, Zhao D, Ma H (2007) Differential gene expression in Arabidopsis wild-type and mutant anthers: insights into anther cell differentiation and regulatory networks. Plant J 52: 14–29 [DOI] [PubMed] [Google Scholar]
- Wilson ZA, Morroll SM, Dawson J, Swarup R, Tighe PJ (2001) The Arabidopsis MALE STERILITY1 (MS1) gene is a transcriptional regulator of male gametogenesis, with homology to the PHD-finger family of transcription factors. Plant J 28: 27–39 [DOI] [PubMed] [Google Scholar]
- Wilson ZA, Zhang DB (2009) From Arabidopsis to rice: pathways in pollen development. J Exp Bot 60: 1479–1492 [DOI] [PubMed] [Google Scholar]
- Wittkop T, Emig D, Lange S, Rahmann S, Albrecht M, Morris JH, Böcker S, Stoye J, Baumbach J (2010) Partitioning biological data with transitivity clustering. Nat Methods 7: 419–420 [DOI] [PubMed] [Google Scholar]
- Wormit A, Trentmann O, Feifer I, Lohr C, Tjaden J, Meyer S, Schmidt U, Martinoia E, Neuhaus HE (2006) Molecular identification and physiological characterization of a novel monosaccharide transporter from Arabidopsis involved in vacuolar sugar transport. Plant Cell 18: 3476–3490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Yang C, Yuan Z, Zhang D, Gondwe MY, Ding Z, Liang W, Zhang D, Wilson ZA (2010) The ABORTED MICROSPORES regulatory network is required for postmeiotic male reproductive development in Arabidopsis thaliana. Plant Cell 22: 91–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Vizcay-Barrena G, Conner K, Wilson ZA (2007a) MALE STERILITY1 is required for tapetal development and pollen wall biosynthesis. Plant Cell 19: 3530–3548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Xu Z, Song J, Conner K, Vizcay Barrena G, Wilson ZA (2007b) Arabidopsis MYB26/MALE STERILE35 regulates secondary thickening in the endothecium and is essential for anther dehiscence. Plant Cell 19: 534–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YH, Thorne NP (2003) Normalization for two-color cDNA microarray data. Lecture Notes-Monograph Series 40: 403–418 [Google Scholar]
- Zhang W, Sun Y, Timofejeva L, Chen C, Grossniklaus U, Ma H (2006) Regulation of Arabidopsis tapetum development and function by DYSFUNCTIONAL TAPETUM1 (DYT1) encoding a putative bHLH transcription factor. Development 133: 3085–3095 [DOI] [PubMed] [Google Scholar]
- Zhang ZB, Zhu J, Gao JF, Wang C, Li H, Li H, Zhang HQ, Zhang S, Wang DM, Wang QX, et al. (2007) Transcription factor AtMYB103 is required for anther development by regulating tapetum development, callose dissolution and exine formation in Arabidopsis. Plant J 52: 528–538 [DOI] [PubMed] [Google Scholar]
- Zhao DZ, Wang GF, Speal B, Ma H (2002) The excess microsporocytes1 gene encodes a putative leucine-rich repeat receptor protein kinase that controls somatic and reproductive cell fates in the Arabidopsis anther. Genes Dev 16: 2021–2031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao LN, Shen LK, Zhang WZ, Zhang W, Wang Y, Wu WH (2013) Ca2+-dependent protein kinase11 and 24 modulate the activity of the inward rectifying K+ channels in Arabidopsis pollen tubes. Plant Cell 25: 649–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Chen H, Li H, Gao JF, Jiang H, Wang C, Guan YF, Yang ZN (2008) Defective in Tapetal development and function 1 is essential for anther development and tapetal function for microspore maturation in Arabidopsis. Plant J 55: 266–277 [DOI] [PubMed] [Google Scholar]
- Zhu J, Zhang G, Chang Y, Li X, Yang J, Huang X, Yu Q, Chen H, Wu T, Yang Z (2010) AtMYB103 is a crucial regulator of several pathways affecting Arabidopsis anther development. Sci China Life Sci 53: 1112–1122 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.