Autophagy is responsible for the degradation of leaf chloroplasts and root plastids in rice plants and functions during energy limitation caused by interruption of photosynthesis.
Abstract
Autophagy is an intracellular process leading to vacuolar or lysosomal degradation of cytoplasmic components in eukaryotes. Establishment of proper methods to monitor autophagy was a key step in uncovering its role in organisms, such as yeast (Saccharomyces cerevisiae), mammals, and Arabidopsis (Arabidopsis thaliana), in which chloroplastic proteins were found to be recycled by autophagy. Chloroplast recycling has been predicted to function in nutrient remobilization for growing organs or grain filling in cereal crops. Here, to develop our understanding of autophagy in cereals, we established monitoring methods for chloroplast autophagy in rice (Oryza sativa). We generated transgenic rice-expressing fluorescent protein (FP) OsAuTophaGy8 (OsATG8) fusions as autophagy markers. FP-ATG8 signals were delivered into the vacuolar lumen in living cells of roots and leaves mainly as vesicles corresponding to autophagic bodies. This phenomenon was not observed upon the addition of wortmannin, an inhibitor of autophagy, or in an ATG7 knockout mutant. Markers for the chloroplast stroma, stromal FP, and FP-labeled Rubisco were delivered by a type of autophagic body called the Rubisco-containing body (RCB) in the same manner. RCB production in excised leaves was suppressed by supply of external sucrose or light. The release of free FP caused by autophagy-dependent breakdown of FP-labeled Rubisco was induced during accelerated senescence in individually darkened leaves. In roots, nongreen plastids underwent both RCB-mediated and entire organelle types of autophagy. Therefore, our newly developed methods to monitor autophagy directly showed autophagic degradation of leaf chloroplasts and root plastids in rice plants and its induction during energy limitation.
Autophagy is the primary pathway facilitating bulk degradation of intracellular components in eukaryotic cells (Ohsumi, 2001; Levine and Klionsky, 2004). Macroautophagy (hereafter referred to as autophagy) is the well-characterized autophagic process by which cytoplasm and organelles are sequestered by a double-membraned vesicle called an autophagosome and transported to the vacuole in yeast (Saccharomyces cerevisiae) and plants or the lysosome in animals (for review, see Mizushima et al., 2002; Nakatogawa et al., 2009; Li and Vierstra, 2012; Liu and Bassham, 2012; Yoshimoto, 2012). Autophagosome formation begins with the generation of a sequestering membrane called the phagophore or isolation membrane, which then elongates to engulf a portion of the cytoplasm, including organelles, and eventually, forms the autophagosome. The outer membrane of the autophagosome fuses with the vacuolar/lysosomal membrane, and the inner membrane structure called the autophagic body is degraded by the resident hydrolases within the vacuole/lysosome. The molecular events during autophagosome formation have been well characterized in the budding yeast through the identification of Autophagy (ATG) genes (Xie and Klionsky, 2007; Nakatogawa et al., 2009). Isolation and characterization of transfer-DNA insertional knockout and RNA interference knockdown mutants of Arabidopsis (Arabidopsis thaliana) ATG orthologs have revealed that molecular events for autophagosome formation in yeast are mostly conserved in plants (Doelling et al., 2002; Hanaoka et al., 2002; Yoshimoto et al., 2004; Thompson et al., 2005; Xiong et al., 2005; Fujiki et al., 2007; Phillips et al., 2008; Chung et al., 2010; Suttangkakul et al., 2011; Li et al., 2014).
Analyses of numerous autophagy-deficient (atg) mutants of Arabidopsis have shown the importance of autophagy for nutrient recycling in plants, similar to its role in yeast and animals. Almost all atg mutants exhibit sensitivity to nitrogen- or carbon-limited conditions: their survival in nitrogen-free medium or darkness is significantly reduced compared with wild-type plants (Doelling et al., 2002; Hanaoka et al., 2002; Yoshimoto et al., 2004; Thompson et al., 2005; Xiong et al., 2005; Chung et al., 2010; Suttangkakul et al., 2011; Li et al., 2014). Chloroplasts are the main source of carbon or nitrogen recycling in plants, because the majority of plant nutrients are distributed to chloroplasts, such that chloroplastic proteins account for 75% to 80% of the total leaf nitrogen in C3 plants (Makino et al., 2003). Rubisco, which is the CO2-fixation enzyme in photosynthesis, is particularly predominant and accounts for around 50% of the total soluble protein in leaves. We have previously reported that there are autophagic pathways for chloroplastic proteins, including Rubisco (Ishida et al., 2014). During the early stage of natural leaf senescence or energy limitation, a portion of the stromal proteins is partially transported to the vacuole by a type of autophagic bodies called Rubisco-containing bodies (RCBs; Ishida et al., 2008; Izumi et al., 2010). At the late stage of sugar starvation-induced senescence, entire chloroplasts are transported into the vacuole by an autophagic process termed chlorophagy (Wada et al., 2009). These chloroplast autophagy pathways likely contribute to carbon and nitrogen utilization throughout the entire plant in Arabidopsis (Guiboileau et al., 2012; Izumi et al., 2013; Ono et al., 2013).
Nutrient remobilization initiating with chloroplastic protein degradation is an important factor determining the productivity of crop plants. In important cereals, such as rice (Oryza sativa) and barley (Hordeum vulgare), chloroplastic proteins are degraded during leaf senescence, and the released nitrogen is remobilized to growing organs and finally, stored within seeds (Friedrich and Huffaker, 1980; Mae et al., 1983). Remobilized nitrogen accounts for 45% of the total nitrogen of newly expanding leaves in rice (Mae and Ohira, 1981). The contribution of remobilized nitrogen to the grain-filling nitrogen has been estimated to be 50% to 90% in rice, wheat (Triticum aestivum), and maize (Zea mays), although it is variable depending on cultivar and growth conditions, particularly nitrogen fertilization (Masclaux et al., 2001). Despite the importance of degradation in this remobilization, to date, there is no direct evidence for the involvement of autophagy in chloroplastic protein degradation in cereals. Furthermore, unlike AtATG7 mutants, the retrotransposon Tos17-insertional knockout rice of OsATG7 (Osatg7) exhibits a male sterility phenotype (Kurusu et al., 2014), indicating that there are some differences in the roles of autophagy depending on plant species. Most studies to date of autophagy in plant species other than Arabidopsis have focused on transcriptional analysis or morphological observation (Ghiglione et al., 2008; Chung et al., 2009; Xia et al., 2011). To show the relevance of autophagic recycling in cereals and investigate any potential roles that differ from those in Arabidopsis, direct monitoring of the progression and activity of autophagy in living cells or intact tissues is essential.
A vital step in the study of autophagy in yeast, mammals, and Arabidopsis was the establishment of fluorescent protein (FP)-based methods for monitoring autophagy, in which fusion proteins consisting of FPs and ATG proteins or specific organelle-localized proteins were not only visual markers for autophagy under fluorescence microscopy but also, biochemical autophagy markers, such as for immunoblotting (Yoshimoto et al., 2004; Thompson et al., 2005; Cheong and Klionsky, 2008; Mizushima et al., 2010). There is experimental evidence indicating that acidotropic fluorescent dyes, such as monodansylcadaverine or Lyso-tracker, which have been used as autophagy markers, do not specifically label autophagic structures (Bampton et al., 2005; Merkulova et al., 2014). By contrast, FP-ATG8 fusions are currently considered to be the most reliable autophagy markers in mammals and Arabidopsis (Mizushima et al., 2010; Merkulova et al., 2014). For chloroplast autophagy in Arabidopsis, chloroplast stroma-localized FPs or FP-labeled Rubisco are not only visual markers for RCBs but also, good indicators for autophagy progression during leaf senescence (Ono et al., 2013). Despite this progress, such FP-based monitoring of plant autophagy has been limited to Arabidopsis or cultured cells (Toyooka et al., 2006; Takatsuka et al., 2011; Kurusu et al., 2014).
In this study, we visualized autophagy of leaf chloroplasts and root plastids in living cells of rice plants using FP-OsATG8 fusions, chloroplast stroma-targeted (CT)-FP, FP-labeled Rubisco proteins, and an OsATG7 knockout mutant. These methods allowed us to establish that the importance of autophagic recycling of chloroplastic proteins is conserved in energy-limited leaves in rice and also, revealed unique features of autophagic degradation of plastids. This report shows the establishment of in planta monitoring methods for autophagy in rice and directly shows the progression of ATG-dependent autophagy and its involvement in chloroplast protein recycling.
RESULTS
In Planta Visualization of Autophagy in Rice
GFP-AtATG8a was previously successfully used to visualize the progression of starvation-induced autophagy in rice-cultured cells (Kurusu et al., 2014). Here, we set out to develop in planta autophagy visualization techniques for rice by producing transgenic rice expressing FP-OsATG8 fusion proteins. Previous searches of rice genomic DNA or complementary DNA (cDNA) databases identified several possible orthologs (OsATGs) of yeast and Arabidopsis ATG genes, among which were five or seven candidate OsATG8s (Chung et al., 2009; Xia et al., 2011). Because there was some uncertainty about these candidates, with some lacking corresponding cDNA information or having differences in cDNA data between the two previous reports, we began by data mining OsATG8 cDNA data, cloning, and direct sequencing. We confirmed the sequences of OsATG8a to OsATG8d (Supplemental Fig. S1A), named according to Chung et al., 2009. Compared with high identity of deduced amino acid sequences among OsATG8a to OsATG8c, the deduced amino acid sequence of OsATG8d was more similar to that of AtATG8i (Supplemental Fig. S1B). The C-terminal Gly residue in ATG8 is exposed through cleavage by ATG4 protease during the elongation of the autophagosomal membrane. OsATG8d does not possess an extra amino acid tail downstream of the Gly like that found in AtATG8h and AtATG8i (Supplemental Fig. S1B; Doelling et al., 2002; Hanaoka et al., 2002). To consider the consequences of these sequence differences, we chose OsATG8a and OsATG8d to produce the FP-OsATG8 fusion constructs.
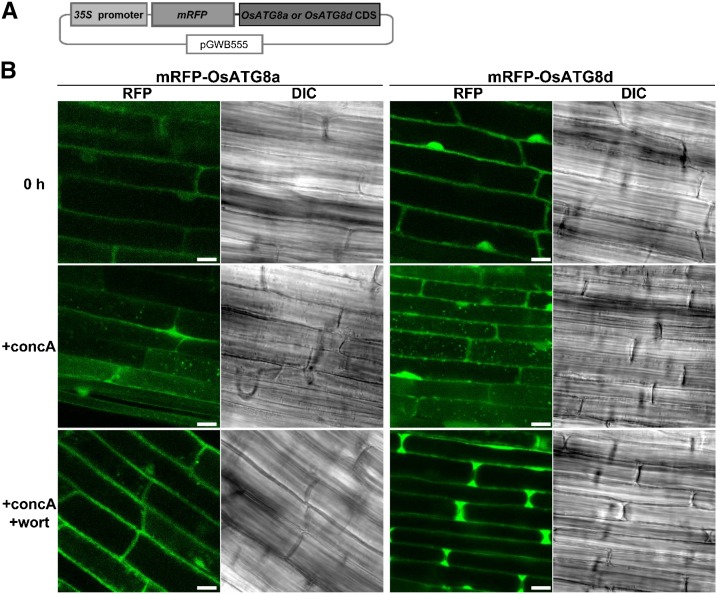
We generated transgenic rice expressing monomeric red fluorescent protein (mRFP)-OsATG8a or mRFP-OsATG8d fusions under the control of the Cauliflower mosaic virus (CaMV) 35S promoter (Fig. 1A). When roots of the transgenic plants were excised and immediately observed by laser-scanning confocal microscopy (LSCM), the RFP fluorescence was detected in the cytoplasm and nucleus (Fig. 1B). Treatment with concanamycin A, an inhibitor of vacuolar H+-ATPase, is used to facilitate observation of autophagic bodies by increasing the interior pH in the vacuolar lumen (Yoshimoto et al., 2004; Ishida et al., 2008). When excised roots of the transgenic plants were incubated with concanamycin A in nutrient-free solution for 6 h in darkness, we observed many vesicles exhibiting strong RFP signal and the spread of faint RFP signal in the central area of the root cells (Fig. 1B). In addition, vesicle structures were visible in differential interference contrast (DIC) images. Wortmannin is an inhibitor of phosphatidylinositol 3-kinases that can inhibit autophagic processes in Arabidopsis plants (Merkulova et al., 2014). The addition of wortmannin during concanamycin A treatment suppressed the mobilization of RFP signals into the vacuolar lumen in rice expressing mRFP-OsATG8a or mRFP-OsATG8d (Fig. 1B). These results indicate that FP-OsATG8 can be used to visualize the progression of autophagy in living cells of rice plants.
Figure 1.
In planta visualization of autophagy in rice. A, Schematic representation of the mRFP-OsATG8a or mRFP-OsATG8d fusion construct. Fusion proteins consisting of mRFP and OsATG8a or OsATG8d were expressed under the control of the CaMV 35S promoter. B, Visualization of autophagy in rice roots. Fresh roots of rice expressing mRFP-OsATG8a or mRFP-OsATG8d were excised and observed immediately (0 h) or after treatment for 6 h with 1 μm concanamycin A without (+concA) or with (+concA+wort) 5 μm wortmannin. The region approximately 5 mm from root tips was observed by LSCM. RFP fluorescence images and DIC images obtained simultaneously are shown. Bars = 10 μm.
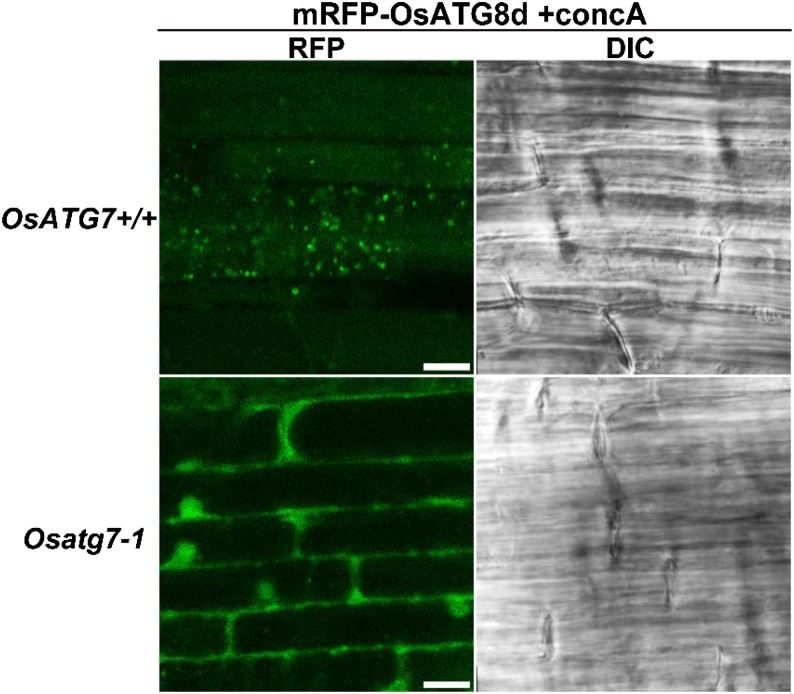
To examine whether the vesicles exhibiting mRFP-OsATG8 signals are autophagic bodies, we adopted a reverse genetic approach using the Tos17-insertional OsATG7 knockout mutant (Osatg7-1). We used mRFP-OsATG8d for additional analyses, because it tended to produce stronger and more stable signals than mRFP-OsATG8a in our rice lines (Fig. 1B). Because Osatg7-1 plants are male sterile because of defects in pollen germination (Kurusu et al., 2014), we first generated mRFP-OsATG8d-expressing rice in a background heterozygous for the Tos17 insertion in the OsATG7 locus. Homozygous Tos17-insertional plants (Osatg7-1), such as OsATG7 knockout mutant plants, and plants lacking the Tos17 insertion (OsATG7+/+), such as the corresponding wild-type plants, were characterized from the population of next generation by genomic PCR and used for imaging analyses. When roots of the resulting mRFP-OsATG8d-expressing OsATG7+/+ control and Osatg7-1 plants were excised and incubated with concanamycin A, the RFP-labeled vesicles and spread RFP signals in the vacuole were absent from the Osatg7-1 roots (Fig. 2). Therefore, we concluded that the vesicles visualized by mRFP-OsATG8 were, indeed, autophagic bodies and represent direct evidence of the progression of ATG-dependent autophagy in rice plants.
Figure 2.
Mobilization of mRFP-OsATG8 signal into the vacuole is not observed in OsATG7 knockout plants. Roots of OsATG7 knockout (Osatg7-1) and corresponding wild-type (OsATG7+/+) plants expressing mRFP-OsATG8d were excised and incubated for 6 h with 1 μm concanamycin A (+concA). RFP fluorescence images and DIC images obtained simultaneously are shown. Bars = 10 μm.
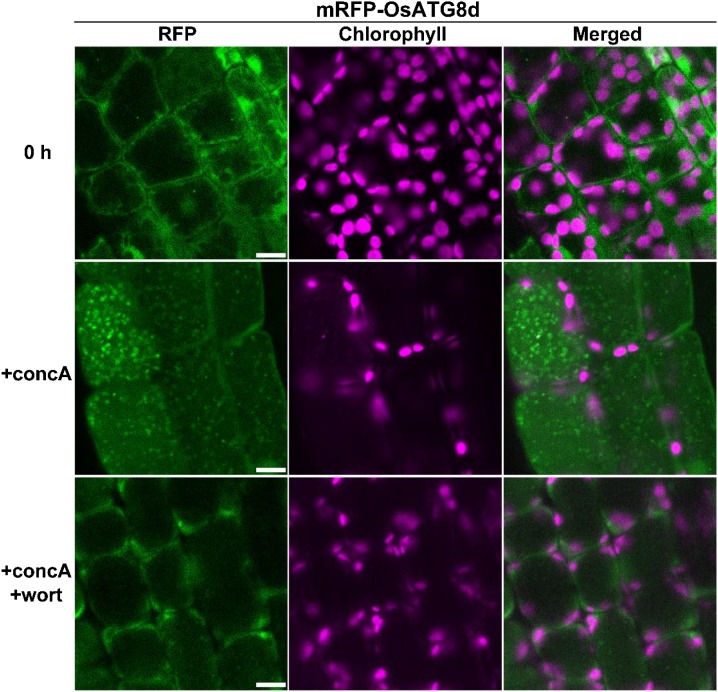
Although the mature leaf blade of rice is unsuitable for live-cell imaging by microscopy because of its solidness and small cell size, we recently established imaging methods using the first leaf of 5- to 7-d-old rice seedlings (Takahashi et al., 2014). In the mesophyll cells of the first leaf of mRFP-OsATG8d-expressing rice, RFP fluorescence was observed in the cytoplasm and nucleus (Fig. 3). After treatment with concanamycin A, many autophagic bodies and mobilization of mRFP-OsATG8d signals into vacuolar lumen were observed, and both were suppressed by the addition of wortmannin (Fig. 3). Collectively, these results confirm the existence of autophagy in rice mesophyll cells containing chloroplasts.
Figure 3.
Visualization of autophagy in mesophyll cells containing chloroplasts in rice. First leaves of mRFP-OsATG8d-expressing rice were excised and observed immediately (0 h) or after incubation for 20 h in 1 μm concanamycin A without (+concA) or with (+concA+wort) 5 μm wortmannin. RFP fluorescence appears green, and chlorophyll autofluorescence appears magenta. Bars = 10 μm.
Visualization of Autophagic Degradation of Chloroplasts by RCBs in Rice
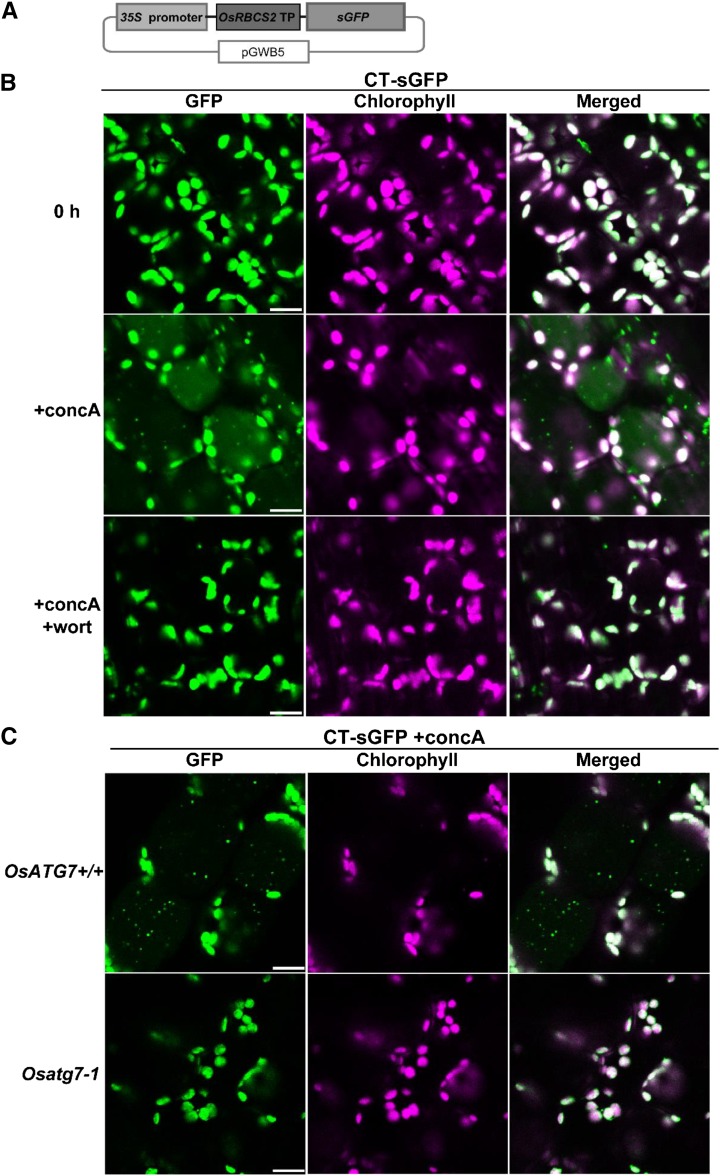
To visualize the behavior of stromal proteins in living cells of rice plants, we generated transgenic rice expressing CT synthetic GFP (sGFP) under the control of CaMV 35S promoter (Fig. 4A). When the first leaf was excised and immediately observed by LSCM, GFP fluorescence overlapped with chlorophyll autofluorescence in mesophyll cells (Fig. 4B), indicating that CT-sGFP was transported into leaf chloroplasts. The Mr of the CT-sGFP-corresponding band detected by immunoblotting was approximately 27 kD, which is the same size as recombinant GFP (rGFP; Supplemental Fig. S2A). This supports that almost all CT-sGFP fusion protein was present as the mature form after transport to chloroplasts, with the concomitant cleavage of the N-terminal transit peptide (CT). When leaf fragments were incubated with concanamycin A in nutrient-free solution for 20 h in darkness, we observed the spread of faint GFP signals and small vesicles exhibiting strong GFP signals in the central area of the mesophyll cells (Fig. 4B). The GFP-labeled vesicles were around 1 μm in diameter, did not exhibit chlorophyll signal, and appeared to move randomly (Fig. 4B; Supplemental Movie S1). These characteristics are identical to those of RCBs in Arabidopsis cells (Ishida et al., 2008). The addition of wortmannin during concanamycin A treatment suppressed the appearance of such vesicles (Fig. 4B) in the same manner as autophagic bodies visualized by mRFP-OsATG8d (Fig. 3). Furthermore, labeled vesicles failed to accumulate in leaves of Osatg7-1 stably expressing CT-sGFP upon incubation with concanamycin A (Fig. 4C). These findings indicate that the CT-sGFP-labeled vesicles are autophagic bodies that specifically contain chloroplast stroma (i.e. RCBs). Thus, we conclude that piecemeal degradation of chloroplasts by RCBs functions in rice in an ATG-dependent manner.
Figure 4.
Autophagy-dependent mobilization of stromal proteins into the vacuole by RCBs in rice. A, Schematic representation illustrating the construct for CT-sGFP expression. Fusion proteins consisting of the transit peptide (TP) of OsRBCS2 and sGFP were expressed under the control of the CaMV 35S promoter. B, Visualization of RCBs in rice leaves. The first leaf of CT-sGFP-expressing rice was excised and observed immediately (0 h) or after incubation for 20 h in the presence of 1 μm concanamycin A without (+concA) or with (+concA+wort) 5 μm wortmannin. C, RCB accumulation is not observed in OsATG7 knockout plants. First leaves of Osatg7-1 or the corresponding wild-type plant (OsATG7+/+) expressing CT-sGFP were excised and incubated for 20 h in the presence of 1 μm concanamycin A (+concA). GFP fluorescence appears green, and chlorophyll autofluorescence appears magenta. In merged images, overlapping signals appear white. Bars = 10 μm.
During the observation of RCBs in rice leaves, additional RCB-like dots exhibiting faint chlorophyll signal were frequently observed (Supplemental Fig. S3A). We compared the profiles of fluorescence among such dots, RCBs and chloroplasts (Supplemental Fig. S3B). Although chloroplasts exhibited the two fluorescent peaks corresponding GFP and chlorophyll, RCBs showed one fluorescent peak corresponding to GFP. The unknown dots exhibited broad fluorescence spectra without any clear peaks. Furthermore, such unknown vesicles were also observed in cells of rice ‘Nipponbare,’ in which no FPs were expressed (Supplemental Fig. S3, A and C). Thus, we concluded that the unknown dots were structures unrelated to RCBs or autophagy. Because such dots were frequently observed in both green and nongreen tissues of rice during fluorescence microscopy analysis, special attention should be paid when fluorescent signals are monitored in living cells of rice plants.
RCB Production in Excised Rice Leaves Is a Response to Energy Limitation Caused by Darkness
In Arabidopsis, RCB production in excised leaves responds to nutrient conditions: the presence of sugars or light energy strongly suppresses RCB production (Izumi et al., 2010). We checked such responses in rice leaves (Fig. 5). Around 27 RCBs were detected in the field of view (90 × 90 μm each) in leaves incubated with concanamycin A in nutrient-free solution for 20 h in darkness, whereas the addition of Suc led to a reduced number of RCBs (Fig. 5). The accumulation of RCBs was also suppressed by light irradiation, and this suppression was relieved by the addition of 3-(3,4-dichlorophenyl)-1,1-dimethylurea (DCMU), an inhibitor of photosynthetic electron transport (Fig. 5). These results indicate that RCB production during concanamycin A treatment of excised rice leaves is induced by energy limitation caused by the interruption of photosynthetic carbon fixation.
Figure 5.
Supplying external sugar or light energy suppresses RCB production. A, Visualization of RCBs under various conditions. First leaves of CT-sGFP-expressing rice were excised and incubated for 20 h in the presence of 1 μm concanamycin A (+concA) in darkness (Dark), with the addition of 2% (w/v) Suc in darkness (Dark +Suc), in light (Light), or with the addition of DCMU in the light (Light +DCMU). Images were obtained after incubation, and merged images are shown. GFP fluorescence appears green, chlorophyll autofluorescence appears magenta, and overlapping signals appear white. Bars = 10 μm. B, The number of accumulated RCBs in various incubation conditions. Excised leaves from four independent plants were incubated in the conditions described in A, and the number of accumulated RCBs in the field of view (90 × 90 μm) was counted. The data represent mean ± se (n = 4). Statistical analysis was performed by Tukey’s test; columns with the same letter are not significantly different (P ≤ 0.05).
Autophagic Recycling of Chloroplastic Proteins Is Accelerated by Energy Limitation in Individually Darkened Intact Leaves
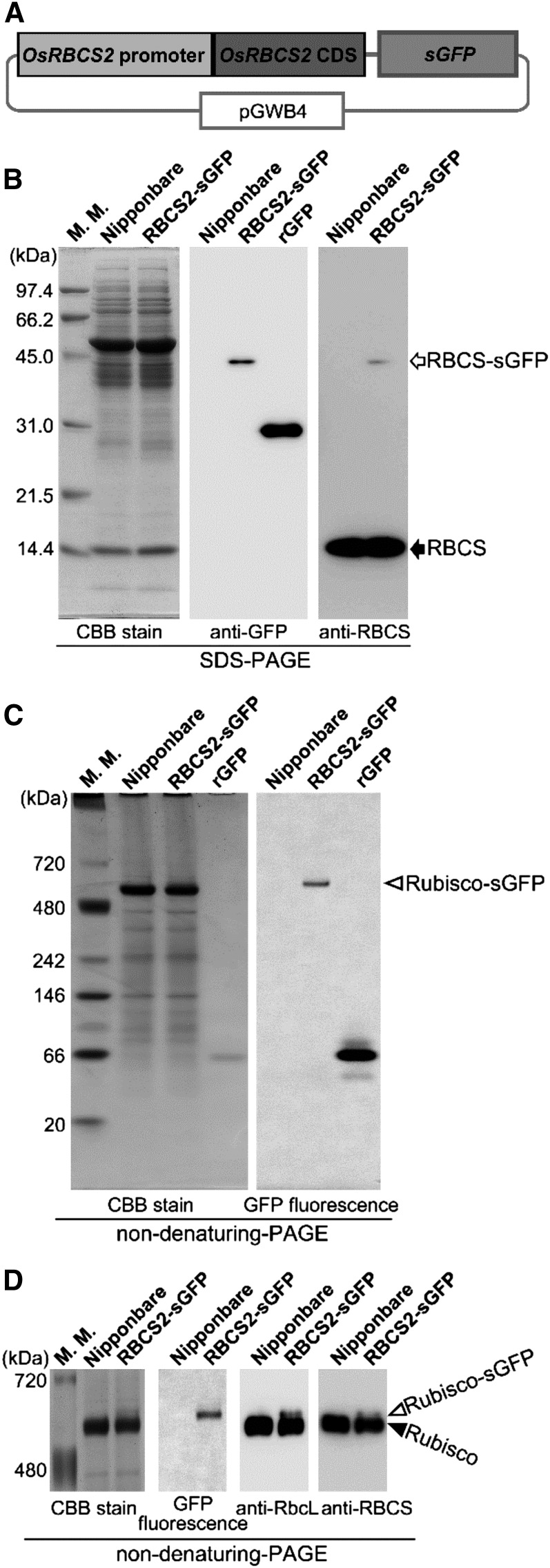
Direct visualization of autophagic bodies or RCBs requires inhibition of vacuolar lytic activities, such as by concanamycin A treatment (Figs. 1B, 3, and 4; Ishida et al., 2008). However, detection of autophagic flux without concanamycin A treatment is essential for investigating the activity of autophagy in intact tissues. One possible method for this is biochemical detection of autophagy-dependent processing of autophagy markers (Cheong and Klionsky, 2008; Mizushima et al., 2010; Suttangkakul et al., 2011). Released free FPs derived from vacuolar degradation of FP fusion markers, such as FP-ATG8 or FP-organelle marker proteins, can be detected by immunoblotting after SDS-PAGE or fluorescence detection after nondenaturing PAGE, because FPs are relatively stable in the vacuole (Cheong and Klionsky, 2008). Such biochemical detection is especially advantageous in leaf blades of rice, because the mature leaf blade is not amenable to observation by fluorescence microscopy. The release of free FP into the vacuole by autophagy-dependent degradation of Rubisco-FP fusion protein is a good indicator of chloroplast autophagy in intact leaves of Arabidopsis during natural or dark-induced senescence (Ono et al., 2013). To establish such a method for rice plants, we generated transgenic rice expressing rice Rubisco Small Subunit2 (OsRBCS2)-sGFP fusion under the OsRBCS2 promoter (Fig. 6A). OsRBCS2 is predominantly expressed in green tissues of rice plants (Suzuki et al., 2009). GFP fluorescence was detected in shoots but not roots in OsRBCS2-sGFP-expressing rice (Supplemental Fig. S4), unlike rice expressing CT-sGFP driven by the CaMV 35S promoter, in which GFP fluorescence was detected throughout the entire plant (Supplemental Fig. S4). These data indicate that the activity of the OsRBCS2 promoter in our constructs is consistent with its endogenous characteristics in rice.
Figure 6.
Transgenic rice expressing GFP-labeled Rubisco. A, Schematic representation illustrating the OsRBCS2-sGFP fusion construct. Fusion proteins consisting of OsRBCS2 and sGFP were expressed under the control of the OsRBCS2 promoter. B to D, Protein analysis in transgenic rice expressing OsRBCS2-sGFP fusion. Total soluble proteins (10 μg for Coomassie Brilliant Blue R250 stain, 1 or 2 μg for immunoblotting, and 10 μg for detection of GFP fluorescence) extracted from shoots of plants expressing the OsRBCS2-sGFP fusion (RBCS2-sGFP), shoots of wild-type rice ‘Nipponbare’ plants as a control, and rGFP were separated by SDS-PAGE (B), nondenaturing PAGE using 4% to 12% gradient gels (C), or nondenaturing PAGE using 5% gels (D) and either stained with Coomassie Brilliant Blue R250 and subjected to immunoblotting with anti-GFP, anti-RBCS, or anti-RbcL antibodies or observed with a fluorescent image analyzer to detect GFP fluorescence. The white arrow indicates the mature form of RBCS2-sGFP after cleavage of the OsRBCS2 transit peptide, and the black arrow indicates the mature form of native OsRBCS after cleavage of its transit peptide. White arrowheads indicate GFP-labeled Rubisco holoenzyme (Rubisco-sGFP), and the black arrowhead indicates native Rubisco holoenzyme. The sizes of molecular mass markers (in kilodaltons) are indicated on the left of the stained gels. CBB, Coomassie Brilliant Blue R250; M. M., mass marker.
The presence of RBCS-sGFP fusion protein of around 43 kD, which corresponds to the mature size after cleavage of the RBCS2 transit peptide, was confirmed by immunoblotting (Fig. 6B). We then investigated whether the RBCS-sGFP fusion proteins are incorporated to Rubisco holoenzyme, which comprises eight plastid-encoded Rubisco Large Subunit (RbcL) and eight nucleus-encoded RBCS. When soluble protein extracts from shoots of plants expressing OsRBCS2-sGFP were separated by nondenaturing PAGE using 4% to 12% gradient gels, GFP signal was detected as a single sharp band near the top of the native Rubisco holoenzyme (white arrowhead in Fig. 6C). The GFP fluorescence-exhibiting band and the native Rubisco band were more clearly separated by nondenaturing PAGE using 5% fixed concentration gels (Fig. 6D). The GFP fluorescence-exhibiting band was also detected by immunoblotting using anti-RbcL and anti-RBCS antibodies, confirming that this band corresponds to Rubisco holoenzyme (Fig. 6D). Therefore, we concluded that expressed OsRBCS2-sGFP fusion proteins were incorporated into Rubisco holoenzyme and present as GFP-labeled Rubisco (Rubisco-sGFP).
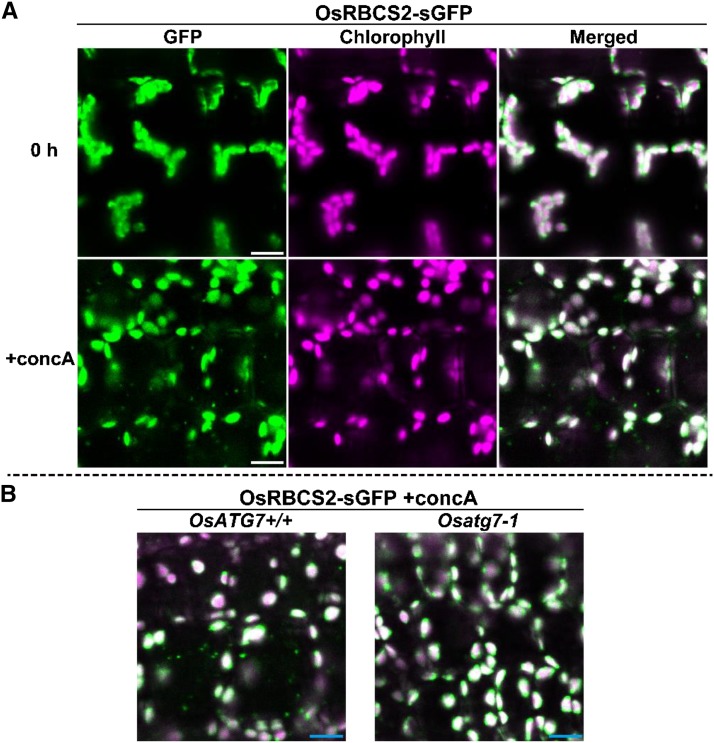
When first leaves of OsRBCS2-sGFP-expressing rice were excised and immediately observed by LSCM, GFP fluorescence overlapped with chlorophyll autofluorescence, indicating that Rubisco-sGFP was localized to chloroplasts (Fig. 7A). When leaf fragments were incubated with concanamycin A for 20 h in darkness, the accumulated RCBs appeared to move randomly in the vacuolar lumen, such as in the case of CT-sGFP-expressing plants (Fig. 7A; Supplemental Movie S2). When leaves of OsRBCS2-sGFP-expressing plants of Osatg7-1 or the corresponding wild-type (OsATG7+/+) background were subjected to concanamycin A treatment, RCBs accumulated in wild-type but not Osatg7-1 leaves (Fig. 7B). These results are direct evidence that Rubisco is mobilized into the vacuole through RCBs in rice leaves.
Figure 7.
Autophagy-dependent mobilization of Rubisco in the vacuole by RCBs in rice. A, Visualization of RCBs by GFP-labeled Rubisco in rice leaf. First leaves of OsRBCS2-sGFP-expressing rice were excised and observed immediately (0 h) or incubated for 20 h in the presence of 1 μm concanamycin A (+concA). B, Mobilization of GFP-labeled Rubisco is not observed in OsATG7 knockout plants. First leaves of OsATG7 knockout (Osatg7-1) or corresponding wild-type (OsATG7+/+) plants expressing OsRBCS-sGFP were excised and incubated for 20 h in the presence of 1 μm concanamycin A (+concA). GFP fluorescence appears green, and chlorophyll autofluorescence appears magenta. In merged images, overlapping signals appear white. In B, only merged images are shown. Bars = 10 μm.
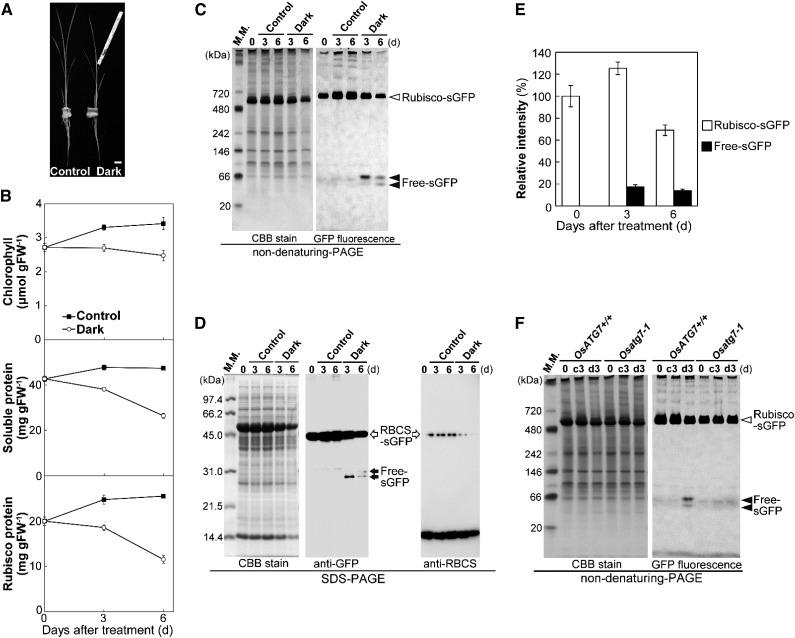
The responses of RCB production to incubation conditions in excised rice leaves indicated that energy limitation is an important factor inducing RCB production similar to the case in Arabidopsis leaves (Fig. 5; Izumi et al., 2010). The darkening of an individual leaf causes energy limitation in the darkened leaf, leading to acceleration of leaf senescence and increased accumulation of transcripts for several ATG genes in Arabidopsis and maize plants (Weaver and Amasino, 2001; Chung et al., 2009; Wada et al., 2009). We investigated whether chloroplast autophagy is induced in a darkened leaf of rice plants through detection of autophagy-dependent processing of Rubisco-sGFP. The fifth leaves, which are the mature leaves in 5-week-old plants, were individually darkened (Fig. 8A). The contents of chlorophyll, soluble protein, and Rubisco protein decreased earlier in darkened leaves than in control leaves of nontreated plants (Fig. 8B), indicative of accelerated senescence in darkened leaves of rice plants. When soluble protein extracts were separated by nondenaturing PAGE and GFP fluorescence was detected in the gel, two additional bands below the Rubisco-sGFP bands were detected only in darkened leaves, like that of rGFP (Figs. 6C and 8C). Rubisco-sGFP processing was further confirmed by immunoblotting after SDS-PAGE (Fig. 8D). The two bands representing free-sGFP, which reacted with anti-GFP antibody but not with anti-RBCS antibody, were detected only in darkened leaves. These findings indicate that the processing of Rubisco-sGFP and the release of peptides corresponding to free-sGFP occur in darkened rice leaves. Quantification of the fluorescence intensities of nondenaturing PAGE bands corresponding to Rubisco-sGFP and free-sGFP (values for the two bands were combined) from soluble protein extracts indicated that the free-sGFP bands increased until 3-d darkening and slightly decreased at 6 d (Fig. 8E). This decrease indicated that free-sGFP gradually declines in the vacuole, although GFP is relatively stable against vacuolar lytic activity. The relative amount of free-sGFPs at 3-d darkening was approximately 17% of the Rubisco-sGFP before dark treatment (Fig. 8E). Release of free-sGFPs in 3-d-darkened leaves was detected in OsRBCS2-sGFP-expressing rice of the wild-type background (OsATG7+/+) but not the Osatg7-1 background (Fig. 8F; Supplemental Fig. S5), indicating that the processing of Rubisco-sGFP is dependent on autophagy. These results indicate that chloroplast autophagy is induced during accelerated senescence in individually darkened leaves and that Rubisco-sGFP can be used as an indicator of the progression of chloroplast autophagy in intact rice leaves.
Figure 8.
Induction of autophagy-dependent processing of Rubisco-sGFP in individually darkened leaves. A, Image of individually darkened leaf treatment. The leaf blades of fifth leaves in 5-week-old plants were individually darkened using aluminum foil (Dark). The fifth leaves of nontreated plants were used as the control. Bar = 10 mm. B, Accelerated senescence in individually darkened leaves. Contents of chlorophyll, soluble protein, and Rubisco protein in control leaves (black squares) and individually darkened leaves (white circles) are shown. The data represent mean ± se (n = 4). C, Detection of Rubisco-sGFP processing by nondenaturing PAGE. Total soluble proteins from equal volumes of control leaves and darkened leaves of OsRBCS2-sGFP-expressing plants were separated by nondenaturing PAGE and either stained with Coomassie Brilliant Blue R250 or observed by a fluorescent image analyzer to detect the GFP fluorescence. D, Detection of RBCS2-sGFP processing by immunoblotting after SDS-PAGE. Total soluble proteins from equal volumes of control leaves and darkened leaves of OsRBCS2-sGFP-expressing plants were separated by SDS-PAGE and either stained with Coomassie Brilliant Blue R250 or subjected to immunoblotting with anti-GFP or anti-RBCS antibodies. E, Changes in relative intensities of Rubisco-sGFP (white columns) and free-sGFP (black columns) in darkened leaves. The intensity of the band corresponding to Rubisco-sGFP and the two bands corresponding to free-sGFP were measured, and relative intensity is shown, with Rubisco-sGFP at 0 d set to 100%. The data represent mean ± se (n = 4). F, Processing of Rubisco-sGFP is not detected in OsATG7 knockout plants. Total soluble proteins from equal volumes of leaves before treatment or 3-d control (c3) and 3-d-darkened leaves (d3) of OsATG7 knockout plants (Osatg7-1) or corresponding wild-type plants (OsATG7+/+) expressing OsRBCS2-sGFP were separated by nondenaturing PAGE and either stained with Coomassie Brilliant Blue R250 or observed by a fluorescent image analyzer to detect the GFP fluorescence. White arrowheads indicate GFP-labeled Rubisco holoeyzyme (Rubisco-sGFP), and black arrowheads indicate free-sGFP released from Rubisco-sGFP processing. White arrows indicate the mature form of OsRBCS2-sGFP after cleavage of the OsRBCS2 transit peptide, and black arrows indicate free-sGFP released from OsRBCS2-sGFP processing. The sizes of molecular mass markers (in kilodaltons) are indicated at the left of the stained gels. CBB, Coomassie Brilliant Blue R250; FW, fresh weight; M. M., mass marker.
We further compared the contents of chlorophyll, soluble protein, and Rubisco proteins in individually darkened leaves between Osatg7-1 and corresponding wild-type (OsATG7+/+) plants. The contents of soluble protein and Rubisco protein rapidly decreased in darkened leaves of Osatg7-1, similar to those of wild-type plants (Supplemental Fig. S6). The decline of chlorophyll contents was accelerated in darkened leaves of Osatg7-1 compared with the wild type (Supplemental Fig. S6). These phenomena are similar to those in individually darkened leaves of Arabidopsis atg mutants, in which the declines of chlorophyll and Rubisco protein content are accelerated, whereas chloroplast autophagy clearly contributes to Rubisco degradation (Wada et al., 2009; Ono et al., 2013).
Autophagic Degradation of Root Plastids in Rice
In rice expressing CT-sGFP, GFP fluorescence was detected in not only green tissues but also, nongreen tissues (Supplemental Fig. S4). When soluble protein extract from roots of CT-sGFP-expressing rice was subjected to immunoblotting using anti-GFP antibody, a CT-sGFP-derived band was detected, and its Mr corresponded to that of rGFP (Supplemental Fig. S2B), indicating that CT-sGFP is present as its mature form after transport into nongreen plastids of rice roots and concomitant cleavage of the N-terminal transit peptide. When roots were excised and immediately observed by LSCM, GFP fluorescence was detected in large organelles corresponding to nongreen plastids (Fig. 9A), similar to in CaMV 35S promoter-driven CT-GFP-expressing tobacco (Nicotiana tabacum; Hanson and Sattarzadeh, 2008). The induction of autophagy in rice roots was clear upon analysis of mRFP-OsATG8-expressing rice (Figs. 1 and 2). RCB-like autophagy in nongreen plastids has also been observed in dark-grown hypocotyls in Arabidopsis (Ishida et al., 2014). Therefore, we investigated whether root plastids are degraded by autophagy in rice plants.
Figure 9.
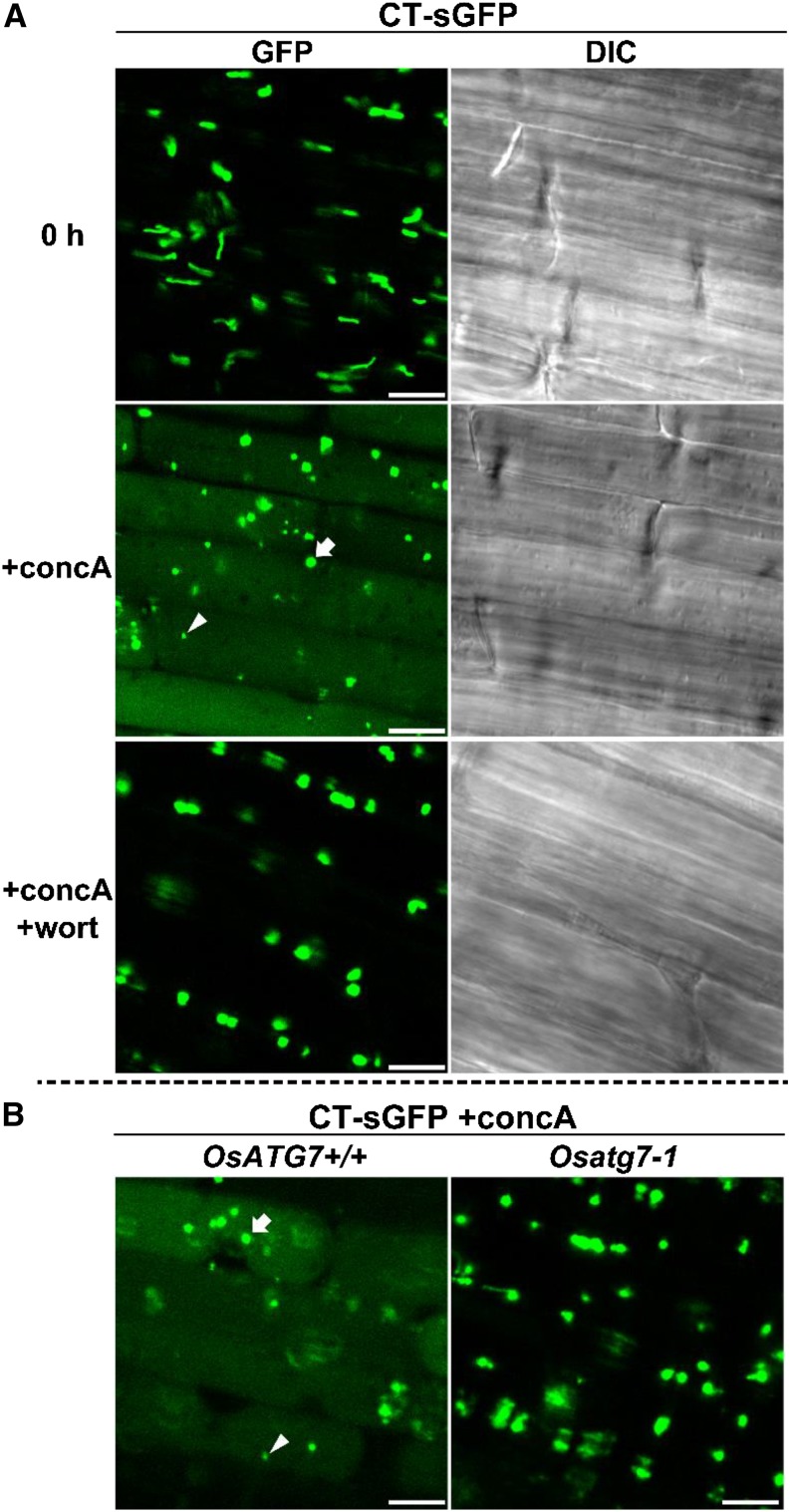
Autophagy-dependent mobilization of nongreen plastids into the vacuole in rice roots. A, Visualization of autophagic degradation of root plastids in rice. Roots of CT-sGFP-expressing rice were excised and observed immediately (0 h) or incubated for 20 h in the presence of 1 μm concanamycin A without (+concA) or with (+concA+wort) 5 μm wortmannin. GFP fluorescence images and DIC images obtained simultaneously are shown. B, Mobilization of root plastids into the vacuole is not observed in OsATG7 knockout plants. Roots of OsATG7 knockout (Osatg7-1) or corresponding wild-type (OsATG7+/+) plants expressing CT-sGFP were excised and incubated for 20 h in the presence of 1 μm concanamycin A (+concA). GFP fluorescence images are shown. White arrowheads indicate typical GFP vesicles of similar size as the RCBs derived from chloroplasts in Figure 4. White arrows indicate typical GFP vesicles of similar size as root plastids. Bars = 10 μm.
When excised roots of CT-sGFP-expressing rice were incubated with concanamycin A for 20 h in darkness, the spread of faint GFP signals and randomly moving vesicles exhibiting strong GFP signals were observed (Fig. 9A). These phenomena were suppressed by the addition of wortmannin (Fig. 9A) and observed in the wild-type background (OsATG7+/+) but not in Osatg7-1 (Fig. 9B). Thus, root plastids also become the targets of autophagic degradation. Interestingly, the randomly moving bodies exhibiting GFP signals were of various sizes: some of them were around 1 μm in diameter, similar to the size of RCBs in leaves (white arrowheads in Fig. 9), whereas others were around 2 to 4 μm in diameter, similar to the size of root plastids (white arrows in Fig. 9). Both types of GFP bodies appeared to move randomly (Supplemental Movie S3), indicating that both bodies were already delivered into the vacuole. When roots of a CT-GFP-expressing line of Arabidopsis, which was previously used for visualization of RCBs in leaves (Ishida et al., 2008), were incubated with concanamycin A, only small vesicles of around 1 μm in diameter were observed (Supplemental Fig. S7). The presence of these small vesicles is consistent with piecemeal degradation of nongreen plastids by RCB structures in Arabidopsis. It is likely that RCB-like piecemeal autophagy and chlorophagy-like whole incorporation simultaneously occurred for nongreen plastids in rice roots under the conditions of this study.
DISCUSSION
Numerous studies of plant autophagy, mainly using Arabidopsis plants, have suggested that autophagy could be of importance in agriculturally important processes, such as grain filling, fruit development, and defense responses to pathogen infection (Yoshimoto et al., 2009; Guiboileau et al., 2012; Ono et al., 2013). Conservation of ATG orthologs and autophagic machinery in crop plants was identified in maize and rice (Chung et al., 2009; Xia et al., 2011). Reverse genetic approaches to investigate its role have only recently commenced (Kurusu et al., 2014; Zhou et al., 2014). To develop such approaches further, proper methods to monitor autophagy in the respective plant species are essential, such as the FP-based monitoring that has been widely used in studies of autophagy in yeast, mammals, and Arabidopsis (Yoshimoto et al., 2004; Thompson et al., 2005; Cheong and Klionsky, 2008; Ishida et al., 2008; Mizushima et al., 2010). Here, we established such methods for rice, an important cereal, and directly showed the progression of autophagic recycling of leaf chloroplasts and root plastids in rice. Furthermore, we report that energy limitation is an important factor inducing chloroplast autophagy in rice leaves. This work is a significant step enabling direct investigation of autophagic activity in crop plants.
We previously established FP-based monitoring methods for chloroplast autophagy and investigated its role in Arabidopsis plants (Ishida et al., 2008; Wada et al., 2009; Izumi et al., 2010, 2013; Ono et al., 2013). A clearly common feature between rice and Arabidopsis autophagy found in this report was its induction by energy limitation caused by leaf darkening. During incubation of excised leaves of Arabidopsis, RCB production was suppressed by the presence of metabolic sugars whether added externally or produced during photosynthesis under light irradiation (Izumi et al., 2010). In rice leaves, RCB production was also suppressed by the addition of external Suc or under light irradiation (Fig. 5). Chloroplast autophagy is accelerated in individually darkened leaves of Arabidopsis (Wada et al., 2009; Ono et al., 2013), and the same phenomenon was observed in individually darkened leaves of rice by the detection of Rubisco-sGFP processing (Fig. 8). These results suggest the conserved importance of autophagy during induced senescence caused by energy limitation in a leaf.
The relative amount of free-sGFP in 3-d-darkened leaves was approximately 17% that of the Rubisco-sGFP before the treatment (Fig. 8E). Because free-sGFP is gradually degraded by the vacuolar lytic activity, even in darkened leaves (Fig. 8; Ono et al., 2013), the net amount of Rubisco-sGFP degraded by autophagy might be larger. However, the amount of Rubisco-sGFP did not decrease during 3-d darkening (Fig. 8E), which could be caused by de novo biosynthesis of Rubisco and RBCS2-sGFP proteins after the dark treatment. Therefore, we cannot calculate the autophagic contribution to Rubisco degradation at the induction stage of dark-induced senescence. Considering that the decrease of Rubisco-sGFP protein during 6-d darkening was 32%, the 17% autophagic Rubisco-sGFP degradation during first 3 d of darkening seems to account for the majority of Rubisco degradation during that period. These estimates at least provide convincing support for a substantial contribution of rice autophagy to Rubisco degradation during dark-induced senescence in mature leaf blades.
The contents of Rubisco protein rapidly decreased in darkened leaves of Osatg7-1 mutants, similar to that of corresponding wild-type plants (Supplemental Fig. S6), which is analogous to the case in Arabidopsis atg mutants (Wada et al., 2009; Ono et al., 2013). In Arabidopsis atg mutants, it is suggested that uncontrolled senescence phenomena, such as hyperaccumulation of ubiquitinated proteins, salicylic acid, and oxidized peroxisomes, lead to early cell death, with rapid loss of chlorophyll (Yoshimoto et al., 2009, 2014; Kim et al., 2013; Shibata et al., 2013; Munch et al., 2014). Similar phenomena could also take place during induced senescence of rice. The experimentally darkened leaves partly mimic the conditions of leaves naturally shaded by other leaves of the same plants or neighboring plants in canopies (Brouwer et al., 2012; Ono et al., 2013). Such conditions often occur in the canopy of rice paddy fields. The induction of chloroplast autophagy in individually darkened leaves of rice in this study is suggestive of its importance in nutrient recycling from naturally shaded leaves for grain filling in paddy fields. However, such effects on yield cannot be tested easily at this point, because the Osatg mutants isolated to date show male sterility caused by defective pollen development (Kurusu et al., 2014).
We found some differences in chloroplast autophagy between rice and Arabidopsis. In Arabidopsis, even if leaves were excised and incubated with concanamycin A for 20 h in darkness, RCBs were rarely seen in young leaves (Ishida et al., 2008). By contrast, many RCBs were observed in rice leaves, although we used young leaves (the first leaves of 5- to 7-d-old seedlings). This difference may be related to the different types of carbohydrate storage used in the two species. Although photoassimilates are stored as starch in Arabidopsis leaves, they are mainly stored as Suc in rice leaf blades and starch in the rice leaf sheath (Nakano et al., 1995). In Arabidopsis, low starch contents in leaves lead to active production of RCBs during diurnal changes of starch contents or in several starch-related mutants (Izumi et al., 2010). It is a possibility that the low starch in leaf blades also leads to active production of RCBs in rice, although detailed analysis of the relationships between leaf carbohydrate contents and RCB production in rice leaves is required.
Another difference between rice and Arabidopsis autophagy was observed for root plastids. When roots of CT-sGFP-expressing rice were incubated with concanamycin A, large GFP bodies were frequently transported into the vacuole (Fig. 9), which possibly corresponds to transport of entire root plastids by autophagy. We have observed only piecemeal autophagy through RCB-like structures in nongreen plastids of Arabidopsis to date (Supplemental Fig. S7; Ishida et al., 2014). However, it has been suggested that amyloplasts in columella cells of Arabidopsis roots are transported in their entirety by autophagy into the vacuole and rapidly degraded during water stress (Nakayama et al., 2012). Although the biological significance of autophagy for root plastids is still unclear, rice may become a useful tool for understanding the intracellular dynamics of autophagy for plastid degradation, because whole-plastid transport into the vacuole by autophagy is easily observed in rice roots.
In conclusion, we established FP-based monitoring methods for autophagy in rice and revealed conserved involvement of autophagy in chloroplast recycling, especially in energy-limited leaves. We also found that some features of rice autophagy are different from the previously characterized Arabidopsis autophagy. These methods pave the way for additional investigation of the activity and roles of autophagy in physiological processes in rice and other biologically and/or agriculturally important species.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
The vectors for generating transgenic rice (Oryza sativa) used in this study were created as follows. The protein coding regions of OsATG8a or OsATG8d were amplified from rice cDNA by reverse transcription (RT)-PCR, cloned into pENTR/D/TOPO vector (Invitrogen), and transferred to the pGWB555 vector (Nakagawa et al., 2007b) by an LR recombination reaction for creation of mRFP-OsATG8a and mRFP-OsATG8d expression constructs. For the CT-sGFP or the OsRBCS2-sGFP expression construct, the coding region of the OsRBCS2 transit peptide or a DNA fragment containing the promoter region and the protein coding region of OsRBCS2 was amplified from the pBIRS vector (Suzuki et al., 2007) and ultimately transferred to the pGWB5 or pGWB4 vector (Nakagawa et al., 2007a), respectively. All vectors were introduced into rice ‘Nipponbare’ by Agrobacterium tumefaciens-mediated methods (Toki et al., 2006). The Tos17-insertional OsATG7 knockout mutant Osatg7-1, in which Tos17 is inserted into first exon of OsATG7, was described previously (Kurusu et al., 2014). To obtain transgenic rice expressing above constructs in the Osatg7-1 background, vectors were introduced into heterozygous Osatg7-1 plants. From the progeny of those plants, homozygous Osatg7-1 plants and those lacking the Tos17 insertion (OsATG7+/+; which were used as the corresponding wild-type plants) were identified by genomic PCR. The primer sequences used for vector construction and genotyping are shown in Supplemental Table S1.
For the imaging analysis in rice, plants were grown on water for 5 to 14 d at 28°C under continuous light conditions with fluorescent lamps (40 μmol quanta m−2 s−1). For the analysis of Rubisco-sGFP processing in darkened leaves, plants were first grown on water for 21 d and then hydroponically grown on nutrient-rich solution in an environmentally controlled growth chamber at 24°C under a 14-h photoperiod with fluorescent lamps (250 μmol quanta m−2 s−1). The nutrient composition was described previously (Mae and Ohira, 1981). Fifth leaves of 5-week-old plants were covered with aluminum foil for dark treatment. For the imaging analysis in Arabidopsis (Arabidopsis thaliana), plants were grown on solid Murashige and Skoog medium (Ono et al., 2013) containing 2% (w/v) Suc and 0.4% (w/v) gelrite at 23°C under continuous light conditions with fluorescent lamps (100 μmol quanta m−2 s−1).
Sequence Analysis of OsATG8 Genes
Rice orthologs of yeast (Saccharomyces cerevisiae) ScATG8 and AtATG8 genes were searched for in the nucleotide databases using the BLAST program. Total RNA was extracted from rice seedlings using the RNeasy Kit (Qiagen), and first strand cDNA was synthesized with SuperScript III reverse transcriptase (Invitrogen). RT-PCR was performed to amplify the predicted coding region of OsATG8 candidates using gene-specific primers (Supplemental Fig. S1; Supplemental Table S1). RT-PCR products were inserted into pCR4/TOPO vector (Invitrogen) and sequenced.
Live-Cell Imaging with LSCM
LSCM was performed with a Nikon C1si System equipped with a CFI Plan apo VC 60× water-immersion objective (numerical aperture = 1.20; Nikon) or a Carl Zeiss LSM710 System equipped with a Plan-apochromat 63× oil-immersion objective (numerical aperture = 1.40; Carl Zeiss). GFP was excited with the 488-nm line of a multiargon ion laser, and RFP was excited with the 561-nm line of a diode-pumped solid-state laser. The fluorescence spectra excited with 488-nm lines of a multiargon laser between 450 and 700 nm were obtained at 10-nm resolution by a spectral detector of Nikon C1si System.
For the imaging in rice roots, roots of 7- to 14-d-old plants were used, and regions around 5 mm from root tips were observed. When roots were treated with concanamycin A or wortmannin, 10 to 15 mm from root tips was excised and incubated in a solution consisting of MES-NaOH (pH 5.5) and 1 μm concanamycin A with or without 5 μm wortmannin for 6 or 20 h at 28°C in darkness.
For the imaging in rice leaves, first leaves of 5- to 7-d-old seedlings were used as previously described (Takahashi et al., 2014). Thin layers of around 0.5 mm were peeled from first leaves and observed by LSCM. For inhibitor treatments, thin leaf layers were vacuum infiltrated using a 50-mL syringe containing MES-NaOH (pH 5.5) and 1 μm concanamycin A with or without 5 μm wortmannin and incubated in the same solution for 20 h at 28°C in darkness. When the responses of RCB production to Suc energy and light energy were examined, 2% (w/v) Suc was added to concanamycin A solution, or the samples were irradiated with light (40 μmol quanta m−2 s−1) with or without 10 μm DCMU. The number of accumulated RCBs was examined as previously described (Izumi et al., 2010) with slight modifications. After incubation of first leaves from four independent plants, three square regions (90 × 90 μm each) per plant were monitored by LSCM, and images were obtained. The number of accumulated RCBs in the images was counted.
Protein Analysis
SDS-PAGE and nondenaturing PAGE analyses were performed as described previously (Ishida et al., 2008; Ono et al., 2013). Rice leaves or roots were homogenized in HEPES-NaOH (pH 7.5) containing 14 mm 2-mercaptoethanol, 10% (v/v) glycerol, and protease inhibitor cocktail (Complete Mini; Roche). Total soluble proteins in extracts were quantified by Bradford assay (Bio-Rad) and subjected to nondenaturing PAGE or mixed with an equal volume of SDS sample buffer consisting of 200 mm Tris-HCl (pH 8.5), 2% (w/v) SDS, 0.7 m 2-mercaptoethanol, and 20% (v/v) glycerol, boiled for 3 min, and then subjected to SDS-PAGE. For nondenaturing PAGE, 4% to 12% gradient gels (Invitrogen) or 5% gels were used. GFP fluorescence on nondenaturing gels was detected by LAS-4000mini (Fujifilm) equipped with a blue light-emitting diode (460 nm Epi) and band-pass filter 510DF10. The intensity of the band corresponding to Rubisco-sGFP and the two bands corresponding to free-sGFP were measured by Multi Gauge software (Fujifilm). Immunoblotting was performed with anti-GFP antibody (1:5,000; Molecular Cloning), anti-RBCS antibody (1:1,000; Ishida et al., 1997), and anti-RBCL antibody (1:10,000; Ishida et al., 1997). Signals were developed with horseradish peroxidase-conjugated secondary antibodies (Pierce) and chemiluminescent reagent (Pierce) and detected using an LAS-4000mini.
Quantifications of chlorophyll, soluble proteins, and Rubisco protein were performed as previously described with slight modification (Izumi et al., 2010). Rice leaves were homogenized in HEPES-NaOH (pH 7.5) containing 14 mm 2-mercaptoethanol, 10% (v/v) glycerol, and protease inhibitor cocktail. Chlorophyll was determined by the method in Arnon (1949). The soluble protein content was measured in the supernatant of a portion of the homogenate by Bradford assay, with bovine serum albumin as the standard. Triton X-100 (0.1% [v/v]; final concentration) was added to the remaining homogenate. After centrifugation, the supernatant was mixed with an equal volume of SDS sample buffer, boiled for 3 min, and then subjected to SDS-PAGE. The Rubisco content was determined spectrophotometrically by formamide extraction of the Coomassie Brilliant Blue R-250-stained bands corresponding to RBCS and RbcL of Rubisco. Calibration curves were made with bovine serum albumin (Thermo Fisher Scientific).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers OsATG8a (AK059939, NM001066302), OsATG8b (AK121268, NM001060464), OsATG8c (AK121169, NM001067706), OsATG8d (NM001071754), and OsRBCS2 (AK061611).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. OsATG8 genes characterized in this study.
Supplemental Figure S2. Protein analysis in transgenic rice expressing CT-sGFP.
Supplemental Figure S3. Fluorescence spectra of chloroplasts, RCBs, and unknown dots observed in rice by LSCM.
Supplemental Figure S4. Detection of GFP fluorescence in whole rice plants expressing CT-sGFP or OsRBCS2-sGFP.
Supplemental Figure S5. Processing of OsRBCS-sGFP is not detected in OsATG7 knockout plants.
Supplemental Figure S6. Changes in chlorophyll, soluble protein, and Rubisco protein in individually darkened leaves of Osatg7 knockout rice.
Supplemental Figure S7. Visualization of autophagic degradation of nongreen plastids in Arabidopsis roots.
Supplemental Table S1. The list of primers used in this study.
Supplemental Movie S1. Movement of RCBs in the vacuolar lumen in a leaf of CT-sGFP-expressing rice.
Supplemental Movie S2. Movement of RCBs in the vacuolar lumen in a leaf of OsRBCS2-sGFP-expressing rice.
Supplemental Movie S3. Movement of mobilized plastids into the vacuolar lumen in a root of CT-sGFP-expressing rice.
Supplementary Material
Acknowledgments
We thank Dr. Tsuyushi Nakagawa for the gift of pGWB vectors, Dr. Maureen R. Hanson for the use of stroma-targeted GFP-expressing Arabidopsis, and the Katahira Technical Support Center, Technology Center for Research and Education Activities (Tohoku University) for providing the analytical instrument LSM710.
Glossary
- CaMV
Cauliflower mosaic virus
- cDNA
complementary DNA
- DCMU
3-(3,4-dichlorophenyl)-1,1-dimethylurea
- DIC
differential interference contrast
- LSCM
laser-scanning confocal microscopy
- RCB
Rubisco-containing body
- RT
reverse transcription
Footnotes
This work was supported, in part, by KAKENHI (grant nos. 26506001 to M.I., 25120702 to J.H., 24380037 to H.I., 25119703 to H.I., and 26660286 to H.I.); the Frontier Research Institute for Interdisciplinary Sciences, Tohoku University (Program for Creation of Interdisciplinary Research to M.I.); and the Ministry of Education, Culture, Sports, Science and Technology, Japan (GRENE/NC-CARP Project to A.M.).
Articles can be viewed without a subscription.
References
- Arnon DI. (1949) Copper enzymes in isolated chloroplasts: polyphenoloxidase in Beta vulgaris. Plant Physiol 24: 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bampton ET, Goemans CG, Niranjan D, Mizushima N, Tolkovsky AM (2005) The dynamics of autophagy visualized in live cells: from autophagosome formation to fusion with endo/lysosomes. Autophagy 1: 23–36 [DOI] [PubMed] [Google Scholar]
- Brouwer B, Ziolkowska A, Bagard M, Keech O, Gardeström P (2012) The impact of light intensity on shade-induced leaf senescence. Plant Cell Environ 35: 1084–1098 [DOI] [PubMed] [Google Scholar]
- Cheong H, Klionsky DJ (2008) Biochemical methods to monitor autophagy-related processes in yeast. Methods Enzymol 451: 1–26 [DOI] [PubMed] [Google Scholar]
- Chung T, Phillips AR, Vierstra RD (2010) ATG8 lipidation and ATG8-mediated autophagy in Arabidopsis require ATG12 expressed from the differentially controlled ATG12A AND ATG12B loci. Plant J 62: 483–493 [DOI] [PubMed] [Google Scholar]
- Chung T, Suttangkakul A, Vierstra RD (2009) The ATG autophagic conjugation system in maize: ATG transcripts and abundance of the ATG8-lipid adduct are regulated by development and nutrient availability. Plant Physiol 149: 220–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doelling JH, Walker JM, Friedman EM, Thompson AR, Vierstra RD (2002) The APG8/12-activating enzyme APG7 is required for proper nutrient recycling and senescence in Arabidopsis thaliana. J Biol Chem 277: 33105–33114 [DOI] [PubMed] [Google Scholar]
- Friedrich JW, Huffaker RC (1980) Photosynthesis, leaf resistances, and ribulose-1,5-bisphosphate carboxylase degradation in senescing barley leaves. Plant Physiol 65: 1103–1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiki Y, Yoshimoto K, Ohsumi Y (2007) An Arabidopsis homolog of yeast ATG6/VPS30 is essential for pollen germination. Plant Physiol 143: 1132–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghiglione HO, Gonzalez FG, Serrago R, Maldonado SB, Chilcott C, Curá JA, Miralles DJ, Zhu T, Casal JJ (2008) Autophagy regulated by day length determines the number of fertile florets in wheat. Plant J 55: 1010–1024 [DOI] [PubMed] [Google Scholar]
- Guiboileau A, Yoshimoto K, Soulay F, Bataillé MP, Avice JC, Masclaux-Daubresse C (2012) Autophagy machinery controls nitrogen remobilization at the whole-plant level under both limiting and ample nitrate conditions in Arabidopsis. New Phytol 194: 732–740 [DOI] [PubMed] [Google Scholar]
- Hanaoka H, Noda T, Shirano Y, Kato T, Hayashi H, Shibata D, Tabata S, Ohsumi Y (2002) Leaf senescence and starvation-induced chlorosis are accelerated by the disruption of an Arabidopsis autophagy gene. Plant Physiol 129: 1181–1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson MR, Sattarzadeh A (2008) Dynamic morphology of plastids and stromules in angiosperm plants. Plant Cell Environ 31: 646–657 [DOI] [PubMed] [Google Scholar]
- Ishida H, Izumi M, Wada S, Makino A (2014) Roles of autophagy in chloroplast recycling. Biochim Biophys Acta 1837: 512–521 [DOI] [PubMed] [Google Scholar]
- Ishida H, Nishimori Y, Sugisawa M, Makino A, Mae T (1997) The large subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase is fragmented into 37-kDa and 16-kDa polypeptides by active oxygen in the lysates of chloroplasts from primary leaves of wheat. Plant Cell Physiol 38: 471–479 [DOI] [PubMed] [Google Scholar]
- Ishida H, Yoshimoto K, Izumi M, Reisen D, Yano Y, Makino A, Ohsumi Y, Hanson MR, Mae T (2008) Mobilization of rubisco and stroma-localized fluorescent proteins of chloroplasts to the vacuole by an ATG gene-dependent autophagic process. Plant Physiol 148: 142–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi M, Hidema J, Makino A, Ishida H (2013) Autophagy contributes to nighttime energy availability for growth in Arabidopsis. Plant Physiol 161: 1682–1693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi M, Wada S, Makino A, Ishida H (2010) The autophagic degradation of chloroplasts via Rubisco-containing bodies is specifically linked to leaf carbon status but not nitrogen status in Arabidopsis. Plant Physiol 154: 1196–1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Lee H, Lee HN, Kim SH, Shin KD, Chung T (2013) Autophagy-related proteins are required for degradation of peroxisomes in Arabidopsis hypocotyls during seedling growth. Plant Cell 25: 4956–4966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurusu T, Koyano T, Hanamata S, Kubo T, Noguchi Y, Yagi C, Nagata N, Yamamoto T, Ohnishi T, Okazaki Y, et al. (2014) OsATG7 is required for autophagy-dependent lipid metabolism in rice postmeiotic anther development. Autophagy 10: 878–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B, Klionsky DJ (2004) Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell 6: 463–477 [DOI] [PubMed] [Google Scholar]
- Li F, Chung T, Vierstra RD (2014) AUTOPHAGY-RELATED11 plays a critical role in general autophagy- and senescence-induced mitophagy in Arabidopsis. Plant Cell 26: 788–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Vierstra RD (2012) Autophagy: a multifaceted intracellular system for bulk and selective recycling. Trends Plant Sci 17: 526–537 [DOI] [PubMed] [Google Scholar]
- Liu Y, Bassham DC (2012) Autophagy: pathways for self-eating in plant cells. Annu Rev Plant Biol 63: 215–237 [DOI] [PubMed] [Google Scholar]
- Mae T, Makino A, Ohira K (1983) Changes in the amounts of ribulose bisphosphate carbozylase synthesized and degraded during the life-span of rice leaf (Oryza sativa L.). Plant Cell Physiol 24: 1079–1086 [Google Scholar]
- Mae T, Ohira K (1981) The remobilization of nitrogen related to leaf growth and senescence in rice plants (Oryza sativa L.). Plant Cell Physiol 22: 1067–1074 [Google Scholar]
- Makino A, Sakuma H, Sudo E, Mae T (2003) Differences between maize and rice in N-use efficiency for photosynthesis and protein allocation. Plant Cell Physiol 44: 952–956 [DOI] [PubMed] [Google Scholar]
- Masclaux C, Quillere I, Gallais A, Hirel B (2001) The challenge of remobilisation in plant nitrogen economy. A survey of physio-agronomic and molecular approaches. Ann Appl Biol 138: 69–81 [Google Scholar]
- Merkulova EA, Guiboileau A, Naya L, Masclaux-Daubresse C, Yoshimoto K (2014) Assessment and optimization of autophagy monitoring methods in Arabidopsis roots indicate direct fusion of autophagosomes with vacuoles. Plant Cell Physiol 55: 715–726 [DOI] [PubMed] [Google Scholar]
- Mizushima N, Ohsumi Y, Yoshimori T (2002) Autophagosome formation in mammalian cells. Cell Struct Funct 27: 421–429 [DOI] [PubMed] [Google Scholar]
- Mizushima N, Yoshimori T, Levine B (2010) Methods in mammalian autophagy research. Cell 140: 313–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munch D, Rodriguez E, Bressendorff S, Park OK, Hofius D, Petersen M (2014) Autophagy deficiency leads to accumulation of ubiquitinated proteins, ER stress, and cell death in Arabidopsis. Autophagy 10: 1579–1587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Kurose T, Hino T, Tanaka K, Kawamukai M, Niwa Y, Toyooka K, Matsuoka K, Jinbo T, Kimura T (2007a) Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J Biosci Bioeng 104: 34–41 [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Suzuki T, Murata S, Nakamura S, Hino T, Maeo K, Tabata R, Kawai T, Tanaka K, Niwa Y, et al. (2007b) Improved Gateway binary vectors: high-performance vectors for creation of fusion constructs in transgenic analysis of plants. Biosci Biotechnol Biochem 71: 2095–2100 [DOI] [PubMed] [Google Scholar]
- Nakano H, Makino A, Mae T (1995) Effects of panicle removal on the photosynthetic characteristics of the flag leaf of rice plants during the ripening stage. Plant Cell Physiol 36: 653–659 [Google Scholar]
- Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y (2009) Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat Rev Mol Cell Biol 10: 458–467 [DOI] [PubMed] [Google Scholar]
- Nakayama M, Kaneko Y, Miyazawa Y, Fujii N, Higashitani N, Wada S, Ishida H, Yoshimoto K, Shirasu K, Yamada K, et al. (2012) A possible involvement of autophagy in amyloplast degradation in columella cells during hydrotropic response of Arabidopsis roots. Planta 236: 999–1012 [DOI] [PubMed] [Google Scholar]
- Ohsumi Y. (2001) Molecular dissection of autophagy: two ubiquitin-like systems. Nat Rev Mol Cell Biol 2: 211–216 [DOI] [PubMed] [Google Scholar]
- Ono Y, Wada S, Izumi M, Makino A, Ishida H (2013) Evidence for contribution of autophagy to rubisco degradation during leaf senescence in Arabidopsis thaliana. Plant Cell Environ 36: 1147–1159 [DOI] [PubMed] [Google Scholar]
- Phillips AR, Suttangkakul A, Vierstra RD (2008) The ATG12-conjugating enzyme ATG10 is essential for autophagic vesicle formation in Arabidopsis thaliana. Genetics 178: 1339–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata M, Oikawa K, Yoshimoto K, Kondo M, Mano S, Yamada K, Hayashi M, Sakamoto W, Ohsumi Y, Nishimura M (2013) Highly oxidized peroxisomes are selectively degraded via autophagy in Arabidopsis. Plant Cell 25: 4967–4983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suttangkakul A, Li F, Chung T, Vierstra RD (2011) The ATG1/ATG13 protein kinase complex is both a regulator and a target of autophagic recycling in Arabidopsis. Plant Cell 23: 3761–3779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y, Nakabayashi K, Yoshizawa R, Mae T, Makino A (2009) Differences in expression of the RBCS multigene family and rubisco protein content in various rice plant tissues at different growth stages. Plant Cell Physiol 50: 1851–1855 [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Ohkubo M, Hatakeyama H, Ohashi K, Yoshizawa R, Kojima S, Hayakawa T, Yamaya T, Mae T, Makino A (2007) Increased Rubisco content in transgenic rice transformed with the ‘sense’ rbcS gene. Plant Cell Physiol 48: 626–637 [DOI] [PubMed] [Google Scholar]
- Takahashi S, Teranishi M, Izumi M, Takahashi M, Takahashi F, Hidema J (2014) Transport of rice cyclobutane pyrimidine dimer photolyase into mitochondria relies on a targeting sequence located in its C-terminal internal region. Plant J 79: 951–963 [DOI] [PubMed] [Google Scholar]
- Takatsuka C, Inoue Y, Higuchi T, Hillmer S, Robinson DG, Moriyasu Y (2011) Autophagy in tobacco BY-2 cells cultured under sucrose starvation conditions: isolation of the autolysosome and its characterization. Plant Cell Physiol 52: 2074–2087 [DOI] [PubMed] [Google Scholar]
- Thompson AR, Doelling JH, Suttangkakul A, Vierstra RD (2005) Autophagic nutrient recycling in Arabidopsis directed by the ATG8 and ATG12 conjugation pathways. Plant Physiol 138: 2097–2110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toki S, Hara N, Ono K, Onodera H, Tagiri A, Oka S, Tanaka H (2006) Early infection of scutellum tissue with Agrobacterium allows high-speed transformation of rice. Plant J 47: 969–976 [DOI] [PubMed] [Google Scholar]
- Toyooka K, Moriyasu Y, Goto Y, Takeuchi M, Fukuda H, Matsuoka K (2006) Protein aggregates are transported to vacuoles by a macroautophagic mechanism in nutrient-starved plant cells. Autophagy 2: 96–106 [DOI] [PubMed] [Google Scholar]
- Wada S, Ishida H, Izumi M, Yoshimoto K, Ohsumi Y, Mae T, Makino A (2009) Autophagy plays a role in chloroplast degradation during senescence in individually darkened leaves. Plant Physiol 149: 885–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver LM, Amasino RM (2001) Senescence is induced in individually darkened Arabidopsis leaves, but inhibited in whole darkened plants. Plant Physiol 127: 876–886 [PMC free article] [PubMed] [Google Scholar]
- Xia K, Liu T, Ouyang J, Wang R, Fan T, Zhang M (2011) Genome-wide identification, classification, and expression analysis of autophagy-associated gene homologues in rice (Oryza sativa L.). DNA Res 18: 363–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Klionsky DJ (2007) Autophagosome formation: core machinery and adaptations. Nat Cell Biol 9: 1102–1109 [DOI] [PubMed] [Google Scholar]
- Xiong Y, Contento AL, Bassham DC (2005) AtATG18a is required for the formation of autophagosomes during nutrient stress and senescence in Arabidopsis thaliana. Plant J 42: 535–546 [DOI] [PubMed] [Google Scholar]
- Yoshimoto K. (2012) Beginning to understand autophagy, an intracellular self-degradation system in plants. Plant Cell Physiol 53: 1355–1365 [DOI] [PubMed] [Google Scholar]
- Yoshimoto K, Hanaoka H, Sato S, Kato T, Tabata S, Noda T, Ohsumi Y (2004) Processing of ATG8s, ubiquitin-like proteins, and their deconjugation by ATG4s are essential for plant autophagy. Plant Cell 16: 2967–2983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto K, Jikumaru Y, Kamiya Y, Kusano M, Consonni C, Panstruga R, Ohsumi Y, Shirasu K (2009) Autophagy negatively regulates cell death by controlling NPR1-dependent salicylic acid signaling during senescence and the innate immune response in Arabidopsis. Plant Cell 21: 2914–2927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto K, Shibata M, Kondo M, Oikawa K, Sato M, Toyooka K, Shirasu K, Nishimura M, Ohsumi Y (2014) Organ-specific quality control of plant peroxisomes is mediated by autophagy. J Cell Sci 127: 1161–1168 [DOI] [PubMed] [Google Scholar]
- Zhou J, Wang J, Yu JQ, Chen Z (2014) Role and regulation of autophagy in heat stress responses of tomato plants. Front Plant Sci 5: 174. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.